
Edited by Jacqueline K. Barton, California Institute of Technology, Pasadena, CA, and approved October 7, 2020 (received for review May 8, 2020)
Author contributions: A.A.G. designed research; M.J.U., P.P., A.C., J.A.C.S., H.S.K., G.I., M.K., C.M., B.M., B.S., A.W.B., E.M.L., Z.F., K.L.N., B.N.-B., L.P., J.L., A.A., J.E.L.-C., Q.L., P.B., S.B., J.P., and Y.G.Y. performed research; P.B., S.B., J.P., and Y.G.Y. contributed new methods/analytic tools; M.J.U., P.P., A.C., J.A.C.S., H.S.K., G.I., M.K., C.M., B.M., B.S., A.L.K., H.O., and A.A.G. analyzed data; and M.J.U., P.P., A.C., and A.A.G. wrote the paper.
1M.J.U. and P.P. contributed equally to this work.
- Altmetric
- Bioinformatics-Guided Selection of a Prototypical Reflectin Variant
- Computational Prediction of the Structure of the Reflectin Variant
- Production and Characterization of the Reflectin Variant
- NMR Spectroscopy of the Reflectin Variant
- Hierarchical Assembly and Optical Properties of the Reflectin Variant
- Discussion and Significance
- Methods
- Supplementary Material
- Data Availability.
The investigation of protein-based materials has provided a better understanding of living systems and has led to the development of ubiquitous modern technologies. Within this context, unique cephalopod proteins called reflectins have exhibited promise for biophotonics and bioelectronics applications, but the exploration of reflectins as materials has been hindered by an incomplete understanding of their structures and properties. Here, we resolve the molecular-level structure of a model reflectin variant, establish a straightforward approach to controlling the assembly of this protein, and describe a correlation between its structural characteristics and light-manipulating properties. Taken together, our findings advance current understanding of reflectin-based materials, provide insight into the color-changing capabilities of cephalopods, and afford new opportunities in biochemistry, cellular biology, bioengineering, and optics.
Naturally occurring and recombinant protein-based materials are frequently employed for the study of fundamental biological processes and are often leveraged for applications in areas as diverse as electronics, optics, bioengineering, medicine, and even fashion. Within this context, unique structural proteins known as reflectins have recently attracted substantial attention due to their key roles in the fascinating color-changing capabilities of cephalopods and their technological potential as biophotonic and bioelectronic materials. However, progress toward understanding reflectins has been hindered by their atypical aromatic and charged residue-enriched sequences, extreme sensitivities to subtle changes in environmental conditions, and well-known propensities for aggregation. Herein, we elucidate the structure of a reflectin variant at the molecular level, demonstrate a straightforward mechanical agitation-based methodology for controlling this variant’s hierarchical assembly, and establish a direct correlation between the protein’s structural characteristics and intrinsic optical properties. Altogether, our findings address multiple challenges associated with the development of reflectins as materials, furnish molecular-level insight into the mechanistic underpinnings of cephalopod skin cells’ color-changing functionalities, and may inform new research directions across biochemistry, cellular biology, bioengineering, and optics.
Materials from naturally occurring and recombinant proteins are frequently employed for the study of fundamental biological processes and leveraged for applications in fields as diverse as electronics, optics, bioengineering, medicine, and fashion (123456789101112–13). Such broad utility is enabled by the numerous advantageous characteristics of protein-based materials, which include sequence modularity, controllable self-assembly, stimuli-responsiveness, straightforward processability, inherent biological compatibility, and customizable functionality (123456789101112–13). Within this context, unique structural proteins known as reflectins have recently attracted substantial attention because of their key roles in the fascinating color-changing capabilities of cephalopods, such as the squid shown in Fig. 1A, and have furthermore demonstrated their utility for unconventional biophotonic and bioelectronic technologies (1112131415161718192021222324252627282930313233343536373839–40). For example, in vivo, Bragg stack-like ultrastructures from reflectin-based high refractive index lamellae (membrane-enclosed platelets) are responsible for the angle-dependent narrowband reflectance (iridescence) of squid iridophores, as shown in Fig. 1B (1516171819–20). Analogously, folded membranes containing distributed reflectin-based particle arrangements within sheath cells lead to the mechanically actuated iridescence of squid chromatophore organs, as shown in Fig. 1C (15, 16, 21, 22). Moreover, in vitro, films processed from squid reflectins not only exhibit proton conductivities on par with some state-of-the-art artificial materials (23242526–27) but also support the growth of murine and human neural stem cells (28, 29). Additionally, morphologically variable coatings assembled from different reflectin isoforms can enable the functionality of chemically and electrically actuated color-changing devices, dynamic near-infrared camouflage platforms, and stimuli-responsive photonic architectures (27, 30313233–34). When considered together, these discoveries and demonstrations constitute compelling motivation for the continued exploration of reflectins as model biomaterials.

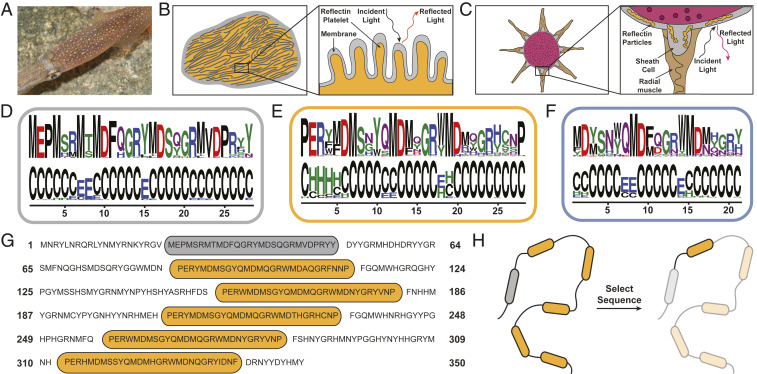
(A) A camera image of a D. pealeii squid for which the skin contains light-reflecting cells called iridophores (bright spots) and pigmented organs called chromatophores (colored spots). Image credit: Roger T. Hanlon (photographer). (B) An illustration of an iridophore (Left), which shows internal Bragg stack-like ultrastructures from reflectin-based lamellae (i.e., membrane-enclosed platelets) (Inset). (C) An illustration of a chromatophore organ (Left), which shows arrangements of reflectin-based particles within the sheath cells (Inset). (D) The logo of the 28-residue-long N-terminal motif (RMN), which depicts the constituent amino acids (Upper) and their predicted secondary structures (Lower). (E) The logo of the 28-residue-long internal motif (RMI), which depicts the constituent amino acids (Upper) and their predicted secondary structures (Lower). (F) The logo of the 21-residue-long C-terminal motif (RMC), which depicts the constituent amino acids (Upper) and their predicted secondary structures (Lower). (G) The amino acid sequence of full-length D. pealeii reflectin A1, which contains a single RMN motif (gray oval) and five RMI motifs (orange ovals). (H) An illustration of the selection of the prototypical truncated reflectin variant (denoted as RfA1TV) from full-length D. pealeii reflectin A1.
Given reflectins’ demonstrated significance from both fundamental biology and applications perspectives, some research effort has been devoted to resolving their three-dimensional (3D) structures (30, 31, 35363738–39). For example, fibers drawn from full-length Euprymna scolopes reflectin 1a and films processed from truncated E. scolopes reflectin 1a were shown to possess secondary structural elements (i.e., α-helices or β-sheets) (30, 31). In addition, precipitated nanoparticles and drop-cast films from full-length Doryteuthis pealeii reflectin A1 have exhibited β-character, which was seemingly associated with their conserved motifs (35, 36). Moreover, nanoparticles assembled from both full-length and truncated Sepia officinalis reflectin 2 variants have demonstrated signatures consistent with β-sheet or α-helical secondary structure, albeit in the presence of surfactants (38). However, such studies were made exceedingly challenging by reflectins’ atypical primary sequences enriched in aromatic and charged residues, documented extreme sensitivities to subtle changes in environmental conditions, and well-known propensities for poorly controlled aggregation (12, 14, 15, 3031–32, 3435363738–39). Consequently, the reported efforts have all suffered from multiple drawbacks, including the need for organic solvents or denaturants, the evaluation of only polydisperse or aggregated (rather than monomeric) proteins, a lack of consensus among different experimental techniques, inadequate resolution that precluded molecular-level insight, imperfect agreement between computational predictions and experimental observations, and/or the absence of conclusive correlations between structure and optical functionality. As such, there has emerged an exciting opportunity for investigating reflectins’ molecular structures, which remain poorly understood and the subject of some debate.
Herein, we elucidate the structure of a reflectin variant at the molecular level, demonstrate a robust methodology for controlling this variant’s hierarchical assembly, and establish a direct correlation between its structural characteristics and optical properties. We first rationally select a prototypical reflectin variant expected to recapitulate the behavior of its parent protein by using a bioinformatics-guided approach. We next map the conformational and energetic landscape accessible to our selected protein by means of all-atom molecular dynamics (MD) simulations. We in turn produce our truncated reflectin variant with and without isotopic labeling, develop solution conditions that maintain the protein in a monomeric state, and characterize the variant’s size and shape with small-angle X-ray scattering (SAXS). We subsequently resolve our protein’s dynamic secondary and tertiary structures and evaluate its backbone conformational fluctuations with NMR spectroscopy. Finally, we demonstrate a straightforward mechanical agitation-based approach to controlling our truncated reflectin variant’s secondary structure, hierarchical self-assembly, and bulk refractive index distribution. Overall, our findings address multiple challenges associated with the development of reflectins as materials, furnish molecular-level insight into the mechanistic underpinnings of cephalopod skin cells’ color-changing functionalities, and appear poised to inform new directions across biochemistry, cellular biology, bioengineering, and optics.
Bioinformatics-Guided Selection of a Prototypical Reflectin Variant
We began our experiments by evaluating the primary sequences reported for various reflectin isoforms via a bioinformatics approach. For this purpose, we expanded upon prior work that analyzed four reflectins from one cephalopod species (37) and performed a broader evaluation of 51 reflectins from six cephalopod species: that is, quantified their sequence homology, defined conserved motifs, and predicted the motifs’ secondary structures (see SI Appendix, Supplementary Text for details). Based on our analysis, we subclassified the 238 discovered motifs into 38 N-terminal ones present in ∼74.5% of the reflectin sequences (denoted as RMN), 159 internal ones present in ∼92% of the reflectin sequences (denoted as RMI), and 41 C-terminal ones present in ∼74.5% of reflectin sequences (denoted as RMC) (Fig. 1 D–F). The 28-residue-long RMN motifs featured relatively little sequence diversity, started with an MEPM segment, contained multiple methionine and aspartic acid dyads (MD-dyads), and were composed almost entirely of basic, acidic, hydroxylic, and sulfur-containing amino acids (Fig. 1D). These motifs’ predicted secondary structures consisted primarily of random coils, with possible β-sheets or β-bridges in proximity to the MD-dyads (Fig. 1D). The 28-residue-long RMI motifs featured a somewhat higher sequence diversity, generally started with a PER segment, ended with the amino acid P, contained MD-dyads, and were enriched in various polar and charged amino acids (Fig. 1E). These motifs’ predicted secondary structures again consisted primarily of random coils, with α-helical character near the PER segment and possible β-sheets or β-bridges near the MD-dyads (Fig. 1E). The 21-residue-long RMC motifs featured more substantial sequence diversity, contained multiple MD-dyads, and were generally enriched in basic, acidic, hydroxylic, and amidic amino acids (Fig. 1F). These motifs’ predicted secondary structures yet again consisted primarily of random coils, with α-helical or β-sheet/β-bridge character in proximity to the MD-dyads (Fig. 1F). Our detailed analysis yielded a categorization of most of the reported full-length reflectins’ conserved motifs according to their frequencies, amino acid compositions, and predicted secondary structures.
We next leveraged our bioinformatics analysis to select a prototypical truncated reflectin variant likely to recapitulate the essential properties of full-length reflectins and to prove tractable during molecular-level structural characterization. As the parent protein for our truncated variant, we specifically chose D. pealeii reflectin A1 (RfA1), for which the sequence is shown in Fig. 1G, because this isoform contains prototypical RMN and RMI motifs separated by variable linker regions, features a >60% homology with most of the reported reflectins, exhibits reasonable aqueous solubility, possesses well-studied self-assembly characteristics, demonstrates valuable optical functionality, and is found in multiple types of squid skin cells (15, 18, 22, 27, 32, 33, 3536–37, 39, 40). From full-length RfA1’s amino acid sequence, we selected a prototypical truncated reflectin variant (RfA1TV) consisting of the first RMI motif and its preceding linker region, as shown in Fig. 1H, for several key reasons. First, our bioinformatics analysis indicated that analogs of RfA1TV’s RMI motifs are found in 92% of the identified reflectin isoforms (see above and Dataset S1). Additionally, multiple reports have suggested that RfA1’s internal motifs possess a propensity for forming β-sheets and α-helices (36, 37), and our own analysis has likewise predicted some secondary structure for RfA1TV’s RMI motif and linker region, as shown in Fig. 2A. Finally, a previous study found that an analogous recombinant reflectin from E. scolopes reflectin 1a, although not monomeric in aqueous solution or characterized at the single-molecular level, reasonably captured several of its less tractable parent protein’s self-assembly characteristics (31). Given such considerations, we postulated that the systematic study of RfA1TV’s structure and properties would enhance understanding of full-length RfA1, and by extension, of the entire reflectin protein family.

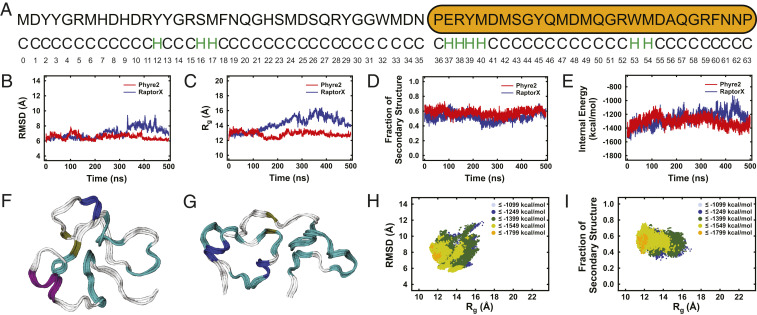
(A) The amino acid sequence (Upper) and predicted secondary structure (Lower) of the prototypical truncated reflectin variant (RfA1TV). Note that the amino acids are sequentially numbered, the conserved RMI motif is indicated by an orange oval, and the predicted helices are colored green. (B) A plot of RfA1TV’s RMSD as a function of time for representative Phyre2-initiated (red trace) and RaptorX-initiated (blue trace) simulations. (C) A plot of RfA1TV’s Rg as a function of time for representative Phyre2-initiated (red trace) and RaptorX-initiated (blue trace) simulations. (D) A plot of RfA1TV’s fraction of secondary structure as a function of time for representative Phyre2-initiated (red trace) and RaptorX-initiated (blue trace) simulations. (E) A plot of RfA1TV’s internal potential energy as a function of time for representative Phyre2-initiated (red trace) and RaptorX-initiated (blue trace) simulations. (F) A snapshot of RfA1TV from the end of a representative Phyre2-initiated simulation, which shows random coils (white), α-helices (purple), 3–10 helices (dark blue), β-strands (yellow), and β-turns (cyan). (G) A snapshot of RfA1TV from the end of a representative RaptorX-initiated simulation, which shows random coils (white), α-helices (purple), 3–10 helices (dark blue), β-strands (yellow), and β-turns (cyan). (H) A plot of RfA1TV’s internal potential energy as a function of RMSD and Rg for an ensemble of conformational snapshots extracted from 10 independent simulations. (I) A plot of RfA1TV’s internal potential energy as a function of the fraction of secondary structure and Rg for an ensemble of conformational snapshots extracted from 10 independent simulations.
Computational Prediction of the Structure of the Reflectin Variant
Having selected our prototypical truncated reflectin variant, we initially obtained computational insight into this protein’s likely 3D structure. For this purpose, we performed MD simulations for RfA1TV modified with an N-terminal methionine necessary for bacterial expression (Fig. 2A) and then analyzed the resulting computationally predicted structures (see SI Appendix, Supplementary Text for details). Here, we note that the starting atomic coordinates were obtained from both Phyre2 (41) and RaptorX (42), which are two common protein structure prediction webservers, thus avoiding possible initial bias. For representative Phyre2-initiated and RaptorX-initiated simulations, RfA1TV’s root-mean-square displacement (RMSD), radius of gyration (Rg), fraction of secondary structure, and internal potential energy (defined as the sum of the intramolecular bonded and nonbonded contributions) are all shown as a function of time in Fig. 2 B–E, and RfA1TV snapshots from the end of these two simulations are shown in Fig. 2 F and G (see also Datasets S2 and S3). During the Phyre2-initiated simulation, RfA1TV’s conformation was dynamic and fluctuated between an RMSD of ∼6 Å and ∼8 Å, an Rg of ∼12 Å and ∼14 Å, a fraction of secondary structure of ∼0.45 and ∼0.73, and an internal energy of approximately −1,100 and −1,500 kcal/mol (Fig. 2 B–E, red traces). At the end of the simulation, RfA1TV possessed an Rg of ∼12.9 Å, a partially ordered conformation with a fraction of secondary structure of ∼0.60 (corresponding to ∼0.55 β-structures and ∼0.05 helices), and an internal energy of approximately −1,275 kcal/mol (Fig. 2F). During the RaptorX-initiated simulation, RfA1TV’s conformation was especially dynamic and fluctuated between an RMSD of ∼6 Å and ∼9 Å, an Rg of ∼12 Å and ∼16 Å, a fraction of secondary structure of ∼0.35 and ∼0.7, and an internal energy of approximately −900 and −1,600 kcal/mol (Fig. 2 B–E, blue traces). At the end of the simulation, RfA1TV possessed a relatively large Rg of ∼13.9 Å, an analogous partially ordered conformation with a fraction of secondary structure of ∼0.58 (corresponding to ∼0.52 β-structures and ∼0.06 helices), and a slightly different internal energy of approximately −1,323 kcal/mol (Fig. 2G). Here, the two representative snapshots’ z-scores (defined as the relative deviation of the total energy of a protein from the energy distribution expected for random conformations) were −1.35 and −2.25 and therefore within the range reported for the X-ray and NMR structures of similarly sized proteins, implying that our models were physically reasonable (see SI Appendix, Supplementary Text and Fig. S1). The representative simulations predicted that RfA1TV would likely feature significant conformational flexibility but would consistently maintain some secondary structural order.
Because our initial simulations indicated a highly dynamic protein, we evaluated the conformational landscape accessible to RfA1TV. Toward this end, we performed an additional four Phyre2-initiated and four RaptorX-initiated simulations (for a total of 10 simulations) and then formulated corresponding internal potential energy surfaces from an ensemble of 40,000 extracted snapshots (see SI Appendix, Supplementary Text for details). The internal potential energy of RfA1TV as a function of the RMSD and Rg for the structural ensemble is shown in Fig. 2H. The landscape revealed that RfA1TV’s RMSD values varied from ∼5.5 Å to ∼12 Å, while its Rg values ranged from ∼11 Å to ∼17 Å, reinforcing the notion that RfA1TV was dynamic and interconverted between many possible conformations. Here, the protein’s lowest-energy conformations were clustered together near RMSD values of ∼8 Å and Rg values of ∼12 Å. The internal potential energy of RfA1TV as a function of the fraction of secondary structure and Rg for the structural ensemble is shown in Fig. 2I. The landscape revealed that RfA1TV’s fraction of secondary structure varied from ∼0.3 to ∼0.75, while its Rg values ranged from ∼11 Å to ∼17 Å, corroborating the idea that RfA1TV maintained some partial secondary structural order. Interestingly, the corresponding internal potential energy of RfA1TV as a function of the different types of secondary structure and Rg indicated that our protein’s lowest-energy predicted structures featured comparatively smaller sizes and consisted almost entirely of β-sheets, β-turns, and β-bends (SI Appendix, Fig. S2). The multiple simulations again predicted that RfA1TV would sample a broad conformational landscape but remain at least partially folded, portending favorably for the experimental investigation of the protein’s secondary and tertiary structures.
Production and Characterization of the Reflectin Variant
After gaining computational insight into RfA1TV’s likely conformation and structure, we produced the unlabeled and isotopically labeled forms of the protein. For this purpose, we expressed, purified, and characterized RfA1TV (which was modified with an N-terminal methionine) by drawing upon protocols reported for D. pealeii RfA1 and RfA2 (23, 25, 32) and standard isotopic labeling strategies for various proteins (see SI Appendix, Supplementary Text for details) (43, 44). Here, we routinely monitored key steps in the process via sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), thus facilitating optimization of the procedure (see SI Appendix, Fig. S3 for a typical gel). For isolated unlabeled RfA1TV, a high-performance liquid chromatography (HPLC) trace, which revealed excellent purity, is shown in SI Appendix, Fig. S4A; a matrix-assisted laser desorption ionization (MALDI) spectrum, which confirmed the expected mass of ∼7,862 Da, is shown in SI Appendix, Fig. S4B; and a sequence map obtained from tryptic digestion in tandem with mass spectrometry, which indicated a coverage of ∼100%, is shown in SI Appendix, Fig. S4C. For isolated 13C- and 15N-labeled RfA1TV, an HPLC trace, which again revealed excellent purity, is shown in SI Appendix, Fig. S5A; a MALDI spectrum, which showed a mass of ∼8,286 Da and a labeling efficiency of >98%, is shown in SI Appendix, Fig. S5B; and a sequence map obtained from tryptic digestion in tandem with mass spectrometry, which indicated a coverage of ∼100% is shown in SI Appendix, Fig. S5C. In general, our protocols reliably furnished the native unlabeled and isotopically labeled RfA1TV in reasonable yields of at least tens of milligrams per liter.
We next developed solution conditions that would mitigate the difficult to control aggregation reported for many reflectin variants (3031–32, 3435363738–39) and thus maintain RfA1TV in a primarily monomeric state. Toward this end, we solubilized our protein in minimal ionic strength buffer (i.e., 20 mM ammonium formate) at different pH values and protein concentrations, initially interrogated such solutions with dynamic light scattering (DLS), and then further probed some of them with circular dichroism (CD) spectroscopy, UV-visible absorption spectroscopy (UV-Vis), and fluorescence spectroscopy (see SI Appendix, Supplementary Text for details). The DLS volume distributions and average hydrodynamic radii (RH) obtained for low-concentration RfA1TV solutions with different pH values are shown in SI Appendix, Fig. S6. For pH values of ≤∼4, the measurements revealed small radii of <∼2 nm but for pH values of ≥∼5, the measurements revealed large radii of >∼300 nm. The complementary DLS volume distributions and average RHs obtained for low-pH solutions with different RfA1TV concentrations are shown in SI Appendix, Fig. S7. For concentrations of ≤∼5 mg/mL, the measurements again revealed small radii of <∼1.5 nm, but for concentrations of ≥∼7.5 mg/mL, the measurements revealed large radii of >∼14 nm. Based on the DLS experiments, RfA1TV achieved its smallest RH at pH values of ≤∼4 and for concentrations of ≤∼5 mg/mL, in agreement with our and others’ findings for RfA1 (32, 3536–37, 40). As such, we studied presumably monomeric RfA1TV solubilized in pH = 3.2, 20 mM ammonium formate buffer at concentrations of ≤∼5 mg/mL, as shown in Fig. 3A in more detail. For such solutions, the DLS number, volume, and intensity distributions indicated a dominant particle population with an RH of ∼1.4 nm, which was consistent with MD predictions for RfA1TV’s size (note also the small subpopulations of larger particles that account for <2% of the total) (Fig. 3B). The UV-Vis absorption spectra revealed a main peak at ∼280 nm, corresponding to absorption by the aromatic amino acids, as well as little tailing at higher wavelengths, confirming negligible aggregation into larger particles (Fig. 3C) (32, 40). The fluorescence emission spectra exhibited a peak at ∼350 nm associated with RfA1TV’s tryptophan residues, as expected from prior observations for RfA1 (Fig. 3C) (32, 35). The CD spectra featured a distinct negative peak at ∼200 nm and multiple smaller negative shoulders between ∼210 nm to ∼240 nm, in agreement with our findings for RfA1 (32), thus suggesting that RfA1TV was similarly weakly ordered or interconverted between multiple conformations (Fig. 3D) (45, 46). Importantly, we discovered that the RfA1TV solutions were relatively stable, with little change in their characteristics over periods of weeks. The combined biophysical characterization experiments indicated that RfA1TV’s behavior was comparable to that of the parent reflectin under analogous conditions and established the foundation (i.e., a stable monomeric state) necessary for studying the protein’s overall geometry.

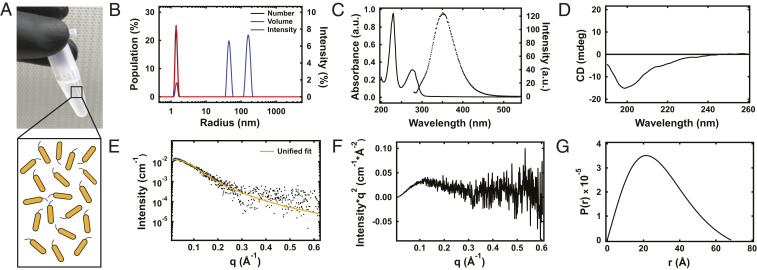
(A) A digital camera image of a solution of primarily monomeric RfA1TV in a low pH ammonium formate buffer (Upper) and a corresponding cartoon of monomeric RfA1TV (Lower Inset). (B) Representative number (black trace), volume (red trace), and intensity (blue trace) distributions obtained for RfA1TV in pH = 3.2, 20 mM ammonium formate buffer. (C) Representative UV-Vis absorption (black solid trace) and fluorescence (black dotted trace) spectra obtained for RfA1TV in pH = 3.2, 20 mM ammonium formate buffer. (D) A representative CD spectrum obtained for RfA1TV in pH = 3.2, 20 mM ammonium formate buffer. (E) A representative plot of the scattering intensity I(q) as a function of the scattering vector q obtained for RfA1TV in pH = 3.2, 20 mM ammonium formate buffer (black squares), and the corresponding unified fit of the experimental data (orange trace). (F) A Kratky plot of the scattering intensity times the square of the scattering vector I(q) × q2 as a function of the scattering vector q for RfA1TV. The plot corresponds to the scattering intensity profile in E. (G) A representative plot of the pair-wise distance distribution function P(r) as a function of the distance r for RfA1TV. The plot corresponds to the scattering intensity profile in E.
We subsequently pursued an improved understanding of monomeric RfA1TV’s global shape and folding state. Toward this end, we probed well-dispersed unlabeled RfA1TV solubilized under the optimized low pH, minimal ionic strength buffer conditions with SAXS (see SI Appendix, Supplementary Text for details). The background-subtracted experimental scattering intensity I(q) as a function of the scattering vector q, along with the corresponding fit obtained via a unified approach, are shown in Fig. 3E. The unified fit’s low-q (<0.07 Å−1) Guinier region revealed an Rg value of 16.6 ± 0.3 Å, which correlated with an experimental Rg/RH ratio of 1.18 that was indicative of deviation from a globular shape for RfA1TV (36, 47), and the fit’s high-q (>0.07 Å−1) Porod region revealed a Porod exponent of ∼3, which was likewise indicative of a complex shape for the protein (4849–50). The Kratky plot of I(q) × q2 as a function of q for our scattering intensity profile is shown in Fig. 3F. This characteristic plot featured a peak at q ≈ 0.12 Å−1 that was followed by a decay and a plateau at intermediate to high q values, which suggested that RfA1TV was at least partially folded (51, 52). The pair-wise distance distribution function P(r) generated from our scattering intensity profile is shown in Fig. 3G. The distribution exhibited a single main peak at a distance of r ≈ 21 Å and tailed to a maximum dimension of Dmax ≈ 68 Å, corroborating the notion of a nonspherical, partially folded protein (53, 54). Notably, the scattering intensity profiles simulated for our two representative MD snapshots also generally resembled the experimental scattering intensity (SI Appendix, Fig. S8). Altogether, the combined measurements were consistent with computational predictions for monomeric RfA1TV and paved the way for further exploration of the protein’s secondary and tertiary structures.
NMR Spectroscopy of the Reflectin Variant
Having obtained isotopically labeled RfA1TV and validated conditions for maintaining the protein in a monomeric state, we sought to gain detailed experimental insight into our variant’s conformation and secondary structure. For this purpose, we studied solutions containing well-dispersed 13C- and 15N-labeled RfA1TV with two-dimensional (2D) heteronuclear single quantum coherence (1H-15N HSQC) NMR spectroscopy, which furnished specific chemical shifts for the polypeptide backbone’s amide bonds, as well as with 3D HNCA, HNCO, HN(CO)CA, HNCACB, and CBCA(CO)NH experiments, which afforded a sequential backbone assignment (see SI Appendix, Supplementary Text for details). For isotopically labeled RfA1TV, a 1H-15N HSQC NMR spectrum exhibiting 61 different cross-peaks is shown in Fig. 4A. The spectrum featured eight well-separated peaks between 15N chemical shifts of ∼108 to ∼111 ppm and 1H chemical shifts of ∼7.8 to ∼8.4 ppm, which corresponded to glycines (i.e., G4, G14, G21, G30, G31, G44, G51, and G58); four independent peaks between 15N chemical shifts of ∼115 to ∼117 ppm and 1H chemical shifts of ∼8.2 to ∼8.4 ppm, which corresponded to serines (i.e., S16, S23, S26, and S43); and three easily identifiable cross-peaks between 15N chemical shifts of ∼117 to ∼119 ppm and 1H chemical shifts of ∼8.2 to ∼8.5 ppm, which corresponded to histidines (i.e., H7, H9, and H22). The spectrum’s remaining 46 cross-peaks were clustered between δ's of ∼117 ppm to ∼125 ppm in the 15N dimension and δ's of ∼7.8 to ∼8.7 ppm in the 1H dimension, and although they were closely spaced together and often overlapping, these peaks could nonetheless be assigned to the remaining amino acids. The 1H-15N HSQC spectrum’s combination of well-separated and overlapping peaks suggested that RfA1TV was partially folded, in agreement with our MD simulations and SAXS measurements (Figs. 2 B–I and 3 E–G). Furthermore, for RfA1TV’s amino acids, a corresponding Ramachandran plot of the torsion angles extracted from the backbone assignment is shown in SI Appendix, Fig. S9. This plot demonstrated that 18 residues were between ψ angles of approximately 120° and 150° and ϕ angles of appoximately −70° and −120° (i.e., the β-sheet region), 39 residues were between ψ angles of approximately −40° and 0° and ϕ angles of approximately −95° and −60° (i.e., the right-handed helix region), and four residues were between ψ angles of approximately 10° and 35° and ϕ angles of approximately 70° and 85° (i.e., the left-handed helix region). The experimental Ramachandran plots resembled those generated for our two representative MD snapshots, with the differences presumably arising from the protein’s conformational flexibility (SI Appendix, Fig. S9). This analysis hinted that RfA1TV possessed some secondary structure, in agreement with the bioinformatics and MD predictions (Fig. 2A and SI Appendix, Fig. S2). Altogether, the measurements elucidated the geometry of RfA1TV’s polypeptide backbone and confirmed the likely presence of specific associated secondary structural characteristics.

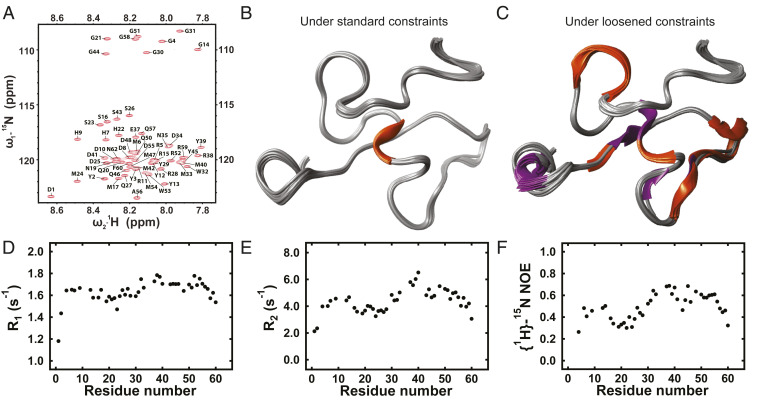
(A) A representative 1H-15N HSQC spectrum obtained for RfA1TV in solution, wherein the cross-peaks are labeled with the corresponding amino acids. (B) Overlaid representative models of the 3D structure of RfA1TV visualized under standard constraints, which show the presence of random coils (colored gray) and a helical domain (colored orange). (C) Overlaid representative models of the 3D structure of RfA1TV visualized under loosened constraints, which show the presence of random coils (colored gray), helical domains (colored orange), and β-strands (colored purple). (D) A plot of the longitudinal (R1 = 1/T1) relaxation rate as a function of the amino acid number for RfA1TV. (E) A plot of the transverse (R2 = 1/T2) relaxation rate as a function of the amino acid number for RfA1TV. (F) A plot of the steady-state {1H}-15N heteronuclear NOE profile as a function of the amino acid number for RfA1TV. Note that the amino acid numbering scheme in A, D, E, and F corresponds to the one in Fig. 2A.
We proceeded to determine the relative spatial arrangement of RfA1TV’s amino acids and consequently resolve the protein’s secondary and tertiary structures in three dimensions. Toward this end, we probed solutions of monomeric 13C- and 15N-labeled RfA1TV with HBHA(CO)NH, (H)CC(CO)NH, HCCH-TOCSY, (H)CCH-TOCSY, 15N-edited HSQC-NOESY, 13C-edited HSQC-NOESY, and 15N-/13C-edited HSQC-NOESY experiments, which together furnished both chemical shifts and distance restraints for the identifiable residues (see SI Appendix, Supplementary Text for details). From these data and our sequential backbone assignment, we calculated, refined, and visualized a plausible structural ensemble for RfA1TV (i.e., 20 low-energy models of the protein’s structure) (see SI Appendix, Supplementary Text for details). To enable a conservative evaluation of the structure, a representative ensemble model for RfA1TV under standard constraints of a minimum three residue helix or β-strand and a ≥−0.5 kcal/mol hydrogen bond energy is shown in Fig. 4B (see also Dataset S4). The conservative model indicated that RfA1TV primarily consisted of random coils and contained a single α-helix located within the RMI motif at P36-E37-R38. This α-helix was separated by an ∼0.4 Å van der Waals overlap distance from 17 polar, charged, and aromatic amino acids (e.g., aspartic acid, arginine, tyrosine, tryptophan, asparagine, glutamine, methionine, and serine) and appeared stabilized by hydrogen bonding interactions with the residues (e.g., between the backbone carbonyl of R38 and the hydroxyl of S16; the pendant guanidino functionality of R38 and the carboxyl of D34; the backbone carbonyl of E37 and the acetamide of Q50; and the backbone carbonyl of P36 and the amide proton of Y12). To assess the presence of subtle or transient structural characteristics, the same representative ensemble model for RfA1TV under loosened constraints of a minimum two residue helix or β-strand and a ≥−0.05 kcal/mol hydrogen bond energy is shown in Fig. 4C. The less conservative model indicated that RfA1TV contained four α-helices located at F18-N19-Q20, M24-D25-S26-Q27-R28, N35-P36, and Q46-M47-D48-M49-Q50 and two β-strands located at Y12-Y13 and E37-R38-Y39. Here, the consistent observation of helical or β-secondary structure at P36-E37-R38-Y39 matched the bioinformatics analysis for this amino acid tract in the conserved RMI motif and in RfA1TV (Figs. 1E and 2A). These findings were further corroborated by an assessment of the measured carbonyl and α-carbon chemical shifts with respect to those reported for disordered proteins, which showed a predisposition toward α-helical structure at P36-E37-R38-Y39-M40 (as well as at R5-M6-H7-D8-H9-D10-R11-Y12-Y13) (SI Appendix, Fig. S10). Moreover, the scattering intensity profiles generated for one NMR model resembled RfA1TV’s experimental scattering intensity (SI Appendix, Fig. S8), and the z-score of −0.48 calculated for the model was within the range expected for similarly sized proteins (SI Appendix, Fig. S1). Overall, the NMR-derived ensemble model, whose accuracy was reinforced by the good agreement across multiple techniques and methods, indicated that RfA1TV possessed a defined but seemingly dynamic 3D structure.
Because our combined experimental and computational analyses showed that RfA1TV was only partially ordered, we proceeded to further explore the protein’s anticipated conformational flexibility. For this purpose, we investigated solutions of 13C- and 15N-labeled RfA1TV with a combination of backbone amide longitudinal 15N relaxation (R1), transverse 15N relaxation (R2), and steady-state {1H}-15N heteronuclear nuclear Overhauser effect (NOE) measurements (see SI Appendix, Supplementary Text for details). Here, we note that cross-peak overlap in the 1H-15N HSQC spectra afforded accurate analyses of the relaxation rate and NOE enhancement measurements for only 44 amino acids. The longitudinal R1 and transverse R2 relaxation rate profiles obtained for RfA1TV are shown in Fig. 4D and Fig. 4E, respectively. The two profiles, which were characteristic of a predominantly disordered protein, revealed the lowest relaxation rates for amino acids at the N terminus, suggesting that this region was relatively flexible, and higher relaxation rates for amino acids within the conserved RMI motif, suggesting that it was more structured than the rest of the protein (note that the largest values of R1 > 1.7 s−1 and R2 > 5.5 s−1 were found for E37, R38 and Y39) (5556–57). The steady-state {1H}-15N heteronuclear NOE profile obtained for RfA1TV is shown in Fig. 4F. This profile, which corroborated the notion of a predominantly disordered protein, revealed low NOE values of ∼0.3 for amino acids near the N terminus, indicating substantial backbone flexibility for this region, and higher NOE values of ≥0.5 for many of the amino acids within the RMI motif, suggesting enhanced backbone rigidity relative to the rest of the protein for the motif (note that the largest values of ∼0.7 were observed for E37, R38, and Y45) (5556–57). Interestingly, the random coil index order parameter (RCI-S2) calculated from various experimentally determined secondary chemical shifts was >∼0.6 for M33-D34-N35-P36-E37-R38 (as well as for H7-D8-H9-D10 and D25-S26), implying reduced backbone flexibility and the presence of secondary structure within these tracts (SI Appendix, Fig. S11) (58, 59). The measurements and calculations were fully consistent with our NMR-derived ensemble model of RfA1TV’s 3D structure and provided a more nuanced understanding of this partially ordered protein’s dynamic nature.
Hierarchical Assembly and Optical Properties of the Reflectin Variant
After elucidating the secondary and tertiary structures of RfA1TV, we developed a method for directly controlling the protein’s degree of structural order. To achieve this goal, we drew inspiration from literature precedent for the mechanical force-induced folding of unstructured proteins (6061–62) and characterized solution-borne RfA1TV’s secondary structure as a function of gentle mechanical agitation in water with CD spectroscopy (see SI Appendix, Supplementary Text for details). Prior to mechanical agitation, the CD spectra revealed a negative peak near ∼200 nm and weak negative shoulders between ∼210 nm and ∼240 nm, suggesting that the protein was not strongly ordered or sampled multiple conformations (Fig. 5 A, Left) (45, 46). After 6 to 12 h of agitation, the CD spectra revealed the emergence of a small positive band near ∼190 nm, a slight red shift for the negative peak near ∼200 nm, and increased prominence for the negative shoulders between ∼210 nm and ∼240 nm, hinting that the protein now possessed some β-character (Fig. 5 A, Center) (45, 63). After 12 to 36 h of agitation, the CD spectra revealed a distinct combination of a positive peak near ∼200 nm and a negative peak at ∼220 nm, which was a fingerprint of β-sheet formation (Fig. 5 A, Right) (45, 63). Interestingly, we found that RfA1TV exhibited analogous secondary structural changes when mechanically agitated in either water or 20 mM ammonium formate buffer (albeit on slightly different time scales), implying that the process featured a weak ionic strength dependence at low pH (Fig. 5A and SI Appendix, Fig. S12A). Here, to better understand the observed structural transitions, we examined the internal potential energy as a function of different types of secondary structure and Rg obtained for RfA1TV from our MD simulations (SI Appendix, Fig. S2). These landscapes indicated that the lowest energy conformational snapshots generally possessed smaller sizes consistent with folding (i.e., Rg values of ∼12 Å) and featured relatively high proportions of β-bends, β-turns, and β-sheets (i.e., total fractions of ∼0.3 to ∼0.65), thus providing an energetic rationale for our findings. Overall, the experiments and simulations implied that mechanical agitation drove the conversion of RfA1TV from a partially ordered higher-energy conformation containing random coils and α-helices into a folded lower-energy one containing primarily β-sheets.

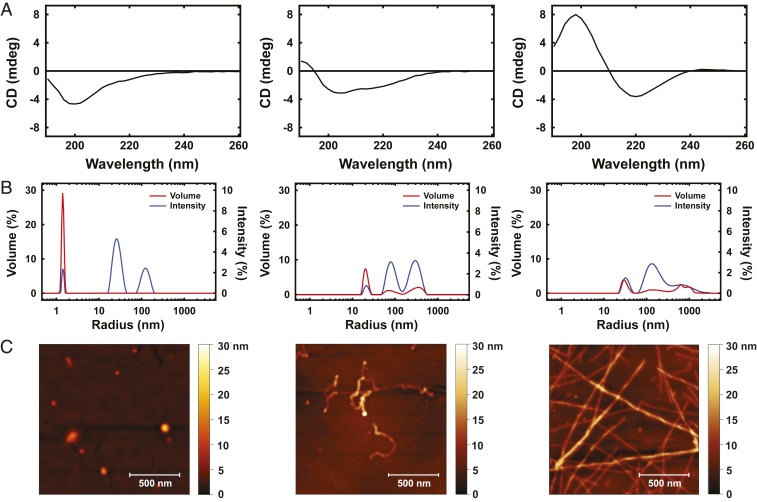
(A) Representative CD spectra for aqueous solutions of RfA1TV obtained after 0 h (Left), 6 to 12 h (Center), or 12 to 36 h (Right) of mechanical agitation. (B) Representative volume (red trace) and intensity (blue trace) distributions for aqueous solutions of RfA1TV obtained after 0 h (Left), 6 to 12 h (Center), and 12 to 36 h (Right) of mechanical agitation. (C) Representative AFM images of RfA1TV architectures obtained after 0 h (Left), 6 to 12 h (Center), and 12 to 36 h (Right) of mechanical agitation. (Scale bars, 500 nm.)
We next sought to ascertain whether the mechanical agitation-induced structural transitions discovered for RfA1TV were accompanied by assembly of the protein into larger hierarchical architectures. Toward this end, we characterized RfA1TV’s aggregation state as a function of gentle mechanical agitation both when dispersed in solution with DLS and when cast onto substrates with atomic force microscopy (AFM) and transmission electron microscopy (TEM) (see SI Appendix, Supplementary Text for details). Before mechanical agitation, the DLS volume and intensity distributions indicated that RfA1TV was primarily monomeric with an RH of ∼1.5 nm but had also assembled into larger nanostructures with apparent RH values of ∼25 nm and ∼120 nm (note that such aggregates accounted for <2% of the total particle population) (Fig. 5 B, Left). The corresponding AFM images revealed that the nanostructures resembled rounded ellipsoidal nanoparticles and typically featured sizes of tens to hundreds of nanometers (note that monomeric protein could not be identified) (Fig. 5 C, Left). After 6 to 12 h of agitation, the DLS volume and intensity distributions indicated that RfA1TV was no longer monomeric and had assembled into a trimodal distribution of nanostructures with apparent RH values of ∼20 nm, ∼80 nm, and ∼300 nm (Fig. 5 B, Center). The corresponding AFM images revealed that some of the nanostructures resembled nanoparticles but that many of them possessed contorted beads-on-a-noodle-type morphologies, were frequently interconnected, and featured lengths of several hundred nanometers to several microns (Fig. 5 C, Center). After 12 to 36 h of agitation, the DLS volume and intensity distributions indicated that RfA1TV had now assembled into smaller nanostructures with RH values of ∼30 nm and larger polydisperse nanostructures with apparent RH values of ∼80 nm to >∼1,200 nm (Fig. 5 B, Right). The corresponding AFM images revealed that a few scattered nanostructures still resembled larger nanoparticles but that most of them possessed twisted plate-like extended morphologies characteristic of amyloid-type fibrils containing β-sheets and featured variable lengths of up to several microns (Fig. 5 C, Right) (6465–66). Notably, TEM images obtained for the larger aggregates likewise revealed contorted beads-on-a-noodle–type and elongated fibrillar morphologies indicative of amyloidogenesis, further supporting the interpretation of the AFM observations (SI Appendix, Fig. S13) (66). Here, we found that RfA1TV assembled into nanostructures with comparable size distributions when mechanically agitated in either water or 20 mM ammonium formate buffer (albeit on slightly different time scales), again intimating a weak ionic strength dependence for the assembly process (Fig. 5B and SI Appendix, Fig. S12B). Taken together, these experiments not only provided further confirmation of the conversion of RfA1TV’s secondary structure into β-sheets upon mechanical agitation, but also showed that this protein could be coaxed into forming a diverse set of architectures under straightforward conditions.
We last investigated whether the mechanically induced changes in RfA1TV’s secondary structure and overall assembly state were correlated with an evolution of the protein’s optical properties. For this purpose, we characterized the phase of our RfA1TV-based architectures in solution with holotomography (67, 68) and extracted the associated 3D refractive indices via standard methods (see SI Appendix, Supplementary Text for details). For the nanoparticles formed without mechanical agitation, the representative phase images confirmed the ellipsoidal geometry previously observed for RfA1 (36) and indicated a comparatively high contrast relative to the solution environment (Fig. 6A). The corresponding quantitative 3D refractive index maps revealed symmetric core-shell–type distributions, wherein the refractive index progressively increased upon moving from the surfaces to centers of the nanoparticles, with maximum refractive indices of 1.48 ± 0.01 (Fig. 6B and SI Appendix, Fig. S14 A and B). For the contorted nanostructures formed upon mechanical agitation, the representative phase images confirmed the expected beads-on-a-noodle–type morphologies and generally indicated a greater contrast for the beads (i.e., residual internal nanoparticles) than for the connecting noodles (i.e., newly formed fibrils) relative to the solution environment (Fig. 6C). The corresponding quantitative 3D refractive index maps revealed nonuniform distributions, wherein the nanoparticles again featured core-shell type gradients but the fibrils featured comparatively small differences between their exteriors and interiors, with maximum refractive indices of 1.45 ± 0.03 (Fig. 6D and SI Appendix, Fig. S14 C and D). For the extended fibrils formed upon mechanical agitation, the representative phase images confirmed the anticipated plate-like fibrillar morphologies and indicated a comparatively low contrast relative to the solution environment (Fig. 6E). The corresponding quantitative 3D refractive index maps revealed far more uniform distributions, wherein the plate-like fibrils featured subtle changes between their exteriors and interiors, with maximum refractive indices of 1.40 ± 0.02 (Fig. 6F and SI Appendix, Fig. S14 E and F). Notably, our hierarchical architectures’ appearances and sizes in solution were all in good agreement with those found by AFM and TEM on solid substrates (Fig. 5C and SI Appendix, Fig. S13). These measurements together demonstrated a direct relationship between RfA1TV’s optical properties (i.e., refractive index) and degree of structural order (i.e., β-sheet content) within the context of hierarchical architectures assembled under the influence of mechanical force.

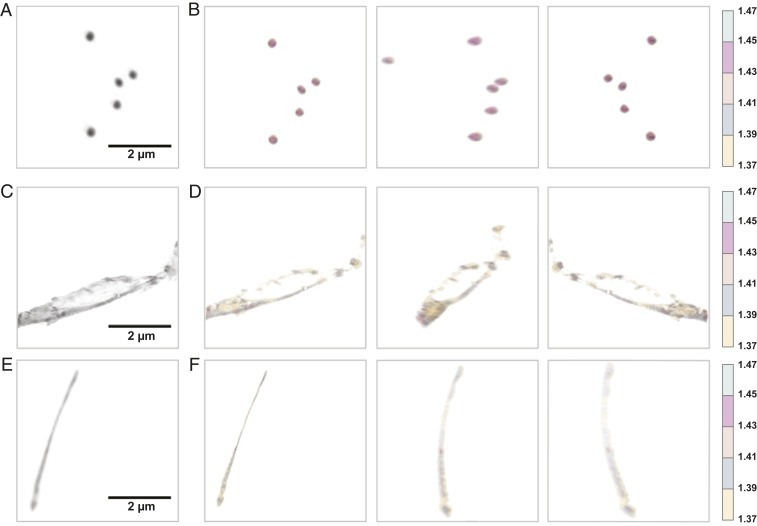
(A) A representative phase image for a grouping of ellipsoidal nanoparticles formed without mechanical agitation. (Scale bar, 2 µm.) (B) The corresponding 3D refractive index maps of these ellipsoidal nanoparticles from three different perspectives. Note that the colored legend (Right) indicates the refractive index range. (C) A representative phase image for a section of a contorted beads-on-a-noodle–type nanostructure formed upon mechanical agitation. (Scale bar, 2 µm.) (D) The corresponding 3D refractive index maps of this beads-on-a-noodle–type nanostructure from three different perspectives. Note that the colored legend (Right) indicates the refractive index range. (E) A representative phase image for an isolated extended plate-like fibril formed upon mechanical agitation. (Scale bar, 2 µm.) (F) The corresponding 3D refractive index maps of this plate-like fibril from three different perspectives. Note that the colored legend (Right) indicates the refractive index range.
Discussion and Significance
From the perspective of protein-based materials, we believe that our study surmounts multiple critical challenges hindering reflectins’ technological development to date, including the lack of an experimental molecular-level structure, imperfect control over self-assembly during processing, and an incomplete understanding of structure–function relationships. First, we have reported the 3D structure of a truncated reflectin variant, which reveals that the protein is partially ordered, possesses substantial conformational flexibility, and features α-helical character within its conserved RMI motif. Given the selected variant’s homology with most known reflectins’ highly repetitive sequences, our computationally and experimentally validated structure provides additional context for these proteins’ classification as intrinsically disordered and establishes a valuable conceptual framework for their further rational engineering. Second, we have shown controlled assembly of our truncated reflectin variant into diverse hierarchical architectures (i.e., nanoparticles, contorted beads-on-a-noodle–type nanostructures, and extended plate-like fibrils) under straightforward conditions (i.e., in water without organic solvents, denaturants, or surfactants) via variation of a single experimental parameter (i.e., mechanical agitation). Assuming further optimization for reduced polydispersity and translation to various full-length reflectin isoforms, our processing strategy holds promise for custom-tailoring the nano- and microscale morphologies of reflectin-based films for particular biophotonic and bioelectronic applications. Third, we have found that the architectures formed by our truncated reflectin variant exhibit a specific correlation between their degree of structural order (i.e., fraction of β-character) and intrinsic optical properties (i.e., refractive indices) via 3D holotomography. Based on reflectins’ validated utility for chemically, mechanically, and electrically actuated appearance-changing platforms, the demonstrated programmability of reflectin-based architectures’ refractive indices may furnish protein-based devices with tunable light-manipulating capabilities thus far only encountered in nature. Altogether, the powerful presented methodology and associated discoveries appear to advance the study of reflectins on multiple distinct fronts and could help unlock their full potential as materials.
From the perspective of cephalopod biology, we postulate that our study provides fundamental insight into cephalopod skin cells’ tunable color-changing functionalities, for which the underlying molecular mechanisms are not yet completely understood. For example, in early work on Pterygioteuthis microlampas squid iridophores, the reflection (and diffuse scattering) of light was determined by the spacing (and nanostructure) of the lamellae comprising these cells’ Bragg stack-like ultrastructures, which was hypothesized to be influenced by tensional forces from the attached musculature (69). In subsequent work on Lollingucula brevis squid iridophores, switching from noniridescent to iridescent states was accompanied by morphological changes inside the cells’ ultrastructures, wherein the lamella transitioned from consisting of nanoparticle/filament networks to containing solid plates (70). In more recent work for Doryteuthis opalescens and D. pealeii squid iridophores, the intensity of the reflected light was governed by the refractive index contrast within the cells’ constituent ultrastructures, for which changes were proposed to originate from reversible condensation of reflectin-based nanoparticles into a dense gel (17, 35). Here, we have found 1) that our truncated reflectin variant forms hierarchical architectures (i.e., nanoparticles, beads-on-a-noodle-type nanostructures, and extended plate-like fibers) with morphologies superficially resembling those inside noniridescent and iridescent iridophores’ Bragg stack-like ultrastructures (35, 69, 70); and 2) that such hierarchical architectures’ controllable refractive index distributions (∼1.37 to ∼1.48) closely match the physiologically relevant ones measured for iridophores’ reflectin-based structures (average of ∼1.44) and isolated reflectin-containing platelets (range of ∼1.38 to ∼1.48) (19, 20). Moreover, we have established that the evolution of our RfA1TV-based architectures’ refractive index distributions under applied force (i.e., mechanical agitation) can be directly correlated with coupled transformations of RfA1TV’s secondary structure from primarily disordered to consisting of β-sheets and of the architectures’ morphologies from nanoparticle-like to fibrillar. The combined observations for our reflectin variant-based architectures in vitro therefore imply that the reversible tandem changes in reflectance intensity, refractive index, and internal morphology demonstrated for iridophores’ ultrastructures in vivo (17, 19, 20, 69, 70) might likewise be driven by the reversible transition of full-length reflectins from disordered to ordered states (potentially under the influence of tension). As such, our study forges plausible mechanistic connections across inquiries and hypotheses spanning nearly 50 years of cephalopod cellular biology research, and given that mechanical tension-induced iridescence was recently reported for reflectin particle-containing chromatophore sheath cells (22), may hold even broader relevance for better understanding the tunable optical functionality of various other cephalopod skin components.
When considered outside the context of cephalopods, our work could ultimately be extended to help address outstanding fundamental questions and even inspire methodological advances across multiple other fields and disciplines. As one example, there exists a philosophical debate in biochemistry about how to most meaningfully draw parallels between the behavior of disordered proteins in artificial dilute environments (e.g., buffers in cuvettes) and in natural crowded environments (e.g., cellular organelles) (71). Together with our recent seminal demonstration of reflectin expression in mammalian cells (40), the findings herein represent a foundation for the comparative evaluation of the structures and properties of isotopically labeled reflectin-derived model proteins in vivo and in vitro (with both holotomography and NMR techniques). Additionally, amyloid fibril formation has been explored for numerous unstructured or misfolded proteins and conclusively linked to the progression of several human neurological disorders, but despite much progress, the transient and less stable intermediate aggregates involved in amyloidogenesis remain difficult to characterize (72). The synergistic combination of our reflectin variants’ high refractive index contrast and holotomography’s 3D profiling capabilities represents an enticing framework for the label-free, high-resolution, real-time, interrogation of fibrillization mechanisms in varied environments. Furthermore, one of the most significant modern research thrusts in biotechnology involves the realization of new methodologies for making cells and tissues transparent and thus amenable to 3D visualization (73). If reflectin variants can be stably expressed at sufficiently high-volume fractions in mammalian cells, our recent and current findings may together enable the engineering of living systems for which the light-scattering properties and transparencies are reversibly modulated via mechanically actuated subcellular structural transformations. Finally, gradient refractive index materials have historically underpinned the performance of numerous optical technologies, including flat lenses that reduce aberration in imaging applications and core-shell fibers that decrease modal dispersion in communications applications, but the manufacturing of polymeric materials with sophisticated 3D refractive index profiles still remains fraught with technical difficulties (74). In principle, the synthesis and exploration of hybrid reflectin-derived polymers could lead to the development of soft materials with 3D refractive index distributions that are not readily accessible via conventional fabrication techniques. Given the above considerations, our work holds the potential to inform new research directions and furnish exciting scientific opportunities across biochemistry, cellular biology, bioengineering, and optics.
Methods
See the SI Appendix, Supplementary Text for detailed experimental procedures that describe the 1) bioinformatics analysis of the reported reflectins; 2) MD simulations of the truncated reflectin variant; 3) expression, purification, and characterization of the unlabeled and 13C- and 15N-labeled truncated reflectin variants; 4) DLS of the truncated reflectin variant; 5) UV-Vis absorption spectroscopy of the truncated reflectin variant; 6) fluorescence spectroscopy of the truncated reflectin variant; 7) CD spectroscopy of the truncated reflectin variant; 8) SAXS measurements and analyses of the truncated reflectin variant; 9) NMR spectroscopy of the truncated reflectin variant; 10) mechanically induced self-assembly of the truncated reflectin variant; 11) AFM of the truncated reflectin variant; 12) TEM of the truncated reflectin variant; and 13) holotomographic microscopy of the truncated reflectin variant.
Acknowledgements
We thank the Laser Spectroscopy Labs and the Mass Spectrometry Facility at the University of California, Irvine for access to experimental facilities; North Carolina State University’s Office of Information Technology for access to its Henry2 High-performance Computing Cluster; the Central European Research Infrastructure Consortium-European Research Infrastructure Consortium for access to experimental facilities and financial support; and Timothy Meerloo for performing the transmission electron microscopy measurements. This work was supported by Air Force Office of Scientific Research Grants FA9550-17-1-0024, FA9550-16-1-0296, and FA9550-14-1-0144 (to A.A.G.) and the Slovenian Research Agency Grant P1-0242 (to J.P.).
Data Availability.
All study data are included in the article and supporting information.
References
1
2
3
4
5
6
7
8
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
 Structure, self-assembly, and properties of a truncated reflectin variant
Structure, self-assembly, and properties of a truncated reflectin variant
