
Edited by Scott V. Edwards, Harvard University, Cambridge, MA, and approved December 23, 2020 (received for review October 16, 2020)
Author contributions: H.Z. designed research; H.J., H.-W.X., L.Z., and N.Z. performed research; H.Z. contributed new reagents/analytic tools; H.J., H.-W.X., L.Z., P.J., and H.Z. analyzed data; and H.J. and H.Z. wrote the paper.
1H.J., H.-W.X., and L.Z. contributed equally to this work.
- Altmetric
The sense of taste provides key information on diet, but evolution of taste receptor genes in vertebrates is sometimes unable to predict their feeding ecology. Here we use behavioral experiments and functional assays to demonstrate the loss of sweet taste despite the conservation of sweet receptor genes in insectivorous bats. Although sweet taste receptor genes were highly conserved between frugivorous and insectivorous bats at the sequence level, our behavioral experiments revealed dramatic divergence in two bat species with distinct diets: the insectivorous bat showed no preference for natural sugars, whereas the frugivorous bat showed strong preferences for sucrose and fructose. Our cell-based assays from multiple representative bat species across the phylogeny further supported the behavioral preference tests.
The evolution of taste perception is usually associated with the ecology and dietary changes of organisms. However, the association between feeding ecology and taste receptor evolution is unclear in some lineages of vertebrate animals. One example is the sweet taste receptor gene Tas1r2. Previous analysis of partial sequences has revealed that Tas1r2 has undergone equally strong purifying selection between insectivorous and frugivorous bats. To test whether the sweet taste function is also important in bats with contrasting diets, we examined the complete coding sequences of both sweet taste receptor genes (Tas1r2 and Tas1r3) in 34 representative bat species. Although these two genes are highly conserved between frugivorous and insectivorous bats at the sequence level, our behavioral experiments revealed that an insectivorous bat (Myotis ricketti) showed no preference for natural sugars, whereas the frugivorous species (Rousettus leschenaultii) showed strong preferences for sucrose and fructose. Furthermore, while both sweet taste receptor genes are expressed in the taste tissue of insectivorous and frugivorous bats, our cell-based assays revealed striking functional divergence: the sweet taste receptors of frugivorous bats are able to respond to natural sugars whereas those of insectivorous bats are not, which is consistent with the behavioral preference tests, suggesting that functional evolution of sweet taste receptors is closely related to diet. This comprehensive study suggests that using sequence conservation alone could be misleading in inferring protein and physiological function and highlights the power of combining behavioral experiments, expression analysis, and functional assays in molecular evolutionary studies.
The sense of taste plays a crucial role in selecting and ingesting food items, and is therefore of fundamental importance for the survival and growth of animals (1, 2). Among the five basic tastes (sweet, umami, bitter, salty, and sour) in mammals (3, 4), the sweet/umami and bitter tastes are mediated by Tas1rs and Tas2rs, respectively, both of which are G protein-coupled receptors (1). The receptors of the sweet and umami tastes encoded by Tas1r genes are heterodimers. Specifically, Tas1r3 can bind to Tas1r2 to form the sweet taste receptor, which responds to dietary carbohydrates; Tas1r3 can also bind to Tas1r1 to form the umami taste receptor, which recognizes dietary proteins (56–7). The bitter taste receptors, encoded by Tas2r genes, mainly detect dietary toxins (89–10).
The evolution of taste perception is usually associated with the ecology and dietary changes of organisms (1112–13). A well-known case is the Tas2r gene family, which shows great variation in family size among species (11). In general, a higher number of Tas2r genes are found in herbivorous and insectivorous vertebrates compared to carnivorous species, indicating that the proportion of potential dietary toxins largely shapes the Tas2r repertoire size (11, 14). Another interesting example is the convergent pseudogenization of the umami-specific taste receptor gene (Tas1r1) in the giant panda and red panda (15, 16), two species that have also evolved a specialized bamboo diet that rendered the umami taste redundant (16). Similarly, pseudogenization of the sweet-specific taste receptor gene (Tas1r2) was detected in many carnivorous mammals that swallow food whole, which may have made the sweet taste useless in food choice (12, 17).
Despite that such close links were observed between diet and taste receptor evolution, mismatches between feeding ecology and taste receptor evolution have also been identified in a number of vertebrates (18, 19). Such mismatches are exemplified by the sweet taste receptor gene Tas1r2 in bats. Among all mammalian orders, Chiroptera (bats) shows the highest diversity in terms of diet, which includes insects, small vertebrates, fruits, nectar, pollen, flowers, foliage, and even blood (20); bats thus offer an excellent model system for studying molecular signatures of dietary changes (21). Despite the clear dietary differences among bats, especially in the proportion of carbohydrates, our previous study of 42 insectivorous and frugivorous bat species found no inactivating mutations in exon 6 of Tas1r2 (22). Moreover, no significant variation in selective pressure on Tas1r2 was detected between these two groups with distinct diets, leading to the suggestion that diet may not play a major role in the evolution of Tas1r2 in bats (22). However, while a pseudogene is a good indicator of loss of original function, a complete and intact gene does not necessarily reflect the full function of a protein because functional loss could also result from mutations in regulatory regions that abolish gene expression, or from missense mutations in coding regions that lead to a change in protein structure (23). In other words, one cannot directly infer functional conservation from sequence conservation alone. As a result, the correlation between diet and sweet taste receptor evolution in bats remains inconclusive.
In this study, we analyzed the full-length coding sequences of both sweet taste receptor genes (Tas1r2 and Tas1r3) in 34 bat species to test whether these two genes are highly conserved at the sequence level, as previously reported (22). We performed behavioral experiments to test whether behavioral taste preference for natural sugars is similar between insectivorous and frugivorous bat species. We next conducted gene expression experiments to test whether both sweet taste receptor genes are expressed in the taste tissue. Finally, we examined the responsiveness of the sweet taste receptors of insectivorous and frugivorous bats to three natural sugars using a cell-based functional assay with a heterologous expression system.
Results
Collection of Complete Coding Sequences of Tas1r2 and Tas1r3 in Bats.
We collected the sequences of bat sweet taste receptor genes from three independent sources. First, TblastN searches were conducted to identify Tas1r2 and Tas1r3 from 33 publicly available bat genome assemblies, and from the three additional bat genome assemblies generated in the present study (see Materials and Methods; SI Appendix, Table S1). We noted that the assembly of Eonycteris spelaea was generated solely using PacBio long-read sequencing data, which may contain many sequencing errors (24). Given that there are no short-read sequencing data to confirm the accuracy of the annotated Tas1r2 and Tas1r3 in E. spelaea, both of these sequences were eliminated from further analysis. Both genes with full-length coding sequences were successfully identified in most of the bat genomes (Fig. 1 and SI Appendix, Table S1), with some exceptions. Specifically, in one bat species (Myotis lucifugus), we detected no BLAST hits of Tas1r3; in two bat species (Eidolon helvum and Myotis brandtii), we found abundant “N”s in the coding region of Tas1r2 and detected only four exons in Tas1r3; and, in one bat species (Murina aurata), numerous “N”s were detected in the coding region of Tas1r3. All of these sequences were excluded from subsequent analyses (SI Appendix, Table S1). We also found that both genes are pseudogenized in the common vampire bat (Desmodus rotundus) due to frame-shifting indels (Fig. 1 and SI Appendix, Table S1). Second, we amplified and sequenced the full-length coding sequences of Tas1r2 for two bat species (Glossophaga soricina and Myotis ricketti) and Tas1r3 for four bat species (Rhinolophus ferrumequinum, G. soricina, Myotis davidii, and M. ricketti; SI Appendix, Tables S1 and S2). Third, we retrieved two bat Tas1r2 sequences from a previous study (22). In total, our collection includes 34 Tas1r2 sequences (33 complete genes and 1 pseudogene) and 32 Tas1r3 sequences (31 complete genes and 1 pseudogene) from the representative bat lineages (34 bat species) with diverse diets (Fig. 1). For simplicity, these bats are classified into three dietary groups: 1) insectivorous bats, which primarily feed on insects and small vertebrates; 2) frugivorous bats, which primarily ingest fruits or nectar; and 3) sanguivorous bats, which exclusively feed on blood (Fig. 1). We did not differentiate between frugivorous and nectarivorous species because we assume that they similarly consume abundant natural sugars in their diets.

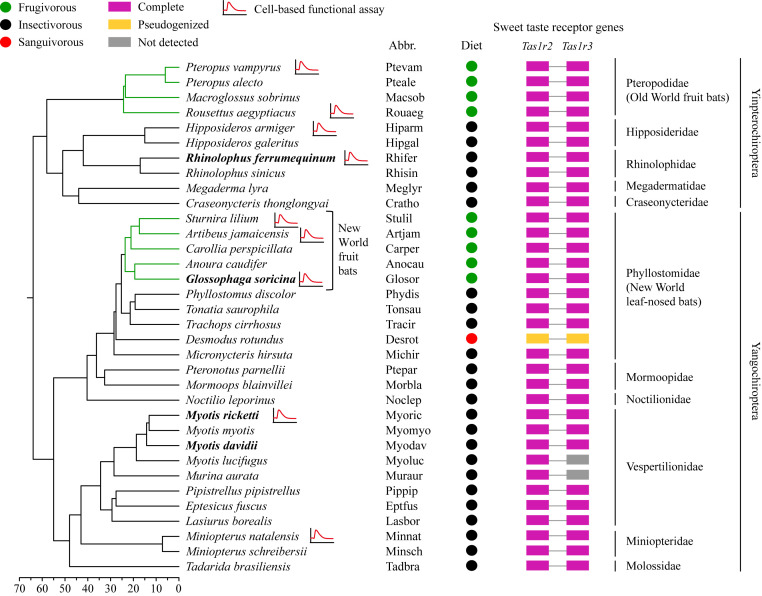
The species tree depicting the evolution of Tas1r2 and Tas1r3 in 34 bat species. The bat phylogeny with divergence time was obtained from previous studies (5253545556–57). Four bat species with newly acquired sequences that were generated via PCRs are shown in bold. Dietary niches are indicated by colored circles, and frugivorous lineages are denoted by green branches. The response line indicates that the sweet taste receptors of these species were selected for cell-based functional assays. “Not detected” indicates that the full-length coding sequence was not detected.
Sequence Conservation of Sweet Taste Receptor Genes across Bats.
To infer the evolutionary patterns of bat Tas1r2 and Tas1r3, we estimated the ω values along the established phylogeny of bats (Fig. 1) using a likelihood method (25). We conducted a suite of selection tests on dataset I (33 complete coding sequences of bat Tas1r2) and dataset II (31 complete coding sequences of bat Tas1r3; SI Appendix, Table S3). We did not include the pseudogenized sequences of the common vampire bat, which were analyzed in detail elsewhere (22, 26, 27). We first examined whether both Tas1r2 and Tas1r3 have undergone purifying selection in bats by comparing two models. Under the assumption of the same ω value for all branches, the ω value was estimated to be 0.189 for Tas1r2, which is significantly less than 1 (P < 0.001; SI Appendix, Table S3, shows comparison between models). A similar result was observed in the dataset of Tas1r3 (ω = 0.175, P < 0.001; SI Appendix, Table S3). These findings suggest that bat Tas1r2 and Tas1r3 are under strong purifying selection and exposed to similar functional constraints. We next asked whether the bat sweet taste receptor genes have evolved under different selective pressures in frugivorous and insectivorous bats (Fig. 1). Our results of Tas1r2 showed that model C (which allows two different ω values for frugivorous and insectivorous bats) did not fit the data significantly better than model B (which assumes the same ω value for all lineages; P = 0.075; SI Appendix, Table S3). Similarly, we did not find model F to have a significantly better fit over model E for the dataset of Tas1r3 (P = 0.059; SI Appendix, Table S3). Our results indicate that a similar level of purifying selection has acted on the sweet taste receptor genes of frugivorous and insectivorous bats, concordant with our previous tests on partial coding sequences of Tas1r2 (22).
Behavioral Indifference of an Insectivorous Bat toward Natural Sugar.
To test whether the bat sweet taste receptor genes are responsible for detecting natural sugars, and thus would directly dictate the behavioral taste responses, we performed “two-bottle” preference tests (Materials and Methods) for two bat species, including one insectivorous bat (M. ricketti) and one frugivorous bat (Rousettus leschenaultii; Fig. 2). As expected, the frugivorous bat displayed behavioral preferences for the three natural sugars over water (Fig. 2 A and B). In contrast, the insectivorous bat showed no preference for any sugar at any concentration tested (Fig. 2 C–F), indicative of the loss of an ability to perceive sugar. Notably, M. ricketti avoided fructose at the highest concentration (20% wt/vol; Fig. 2E). To verify our modified preference test (Materials and Methods), which works well for insectivorous bats, we used it to test behavioral taste preferences of M. ricketti to one bitterant, quinine hydrochloride, and found that the animals showed strong aversion to this compound (SI Appendix, Fig. S1 and Movie S1). This indicates that the bitter taste perception is retained in Myotis bats as previously suggested (19) and that our behavioral preference test worked properly.

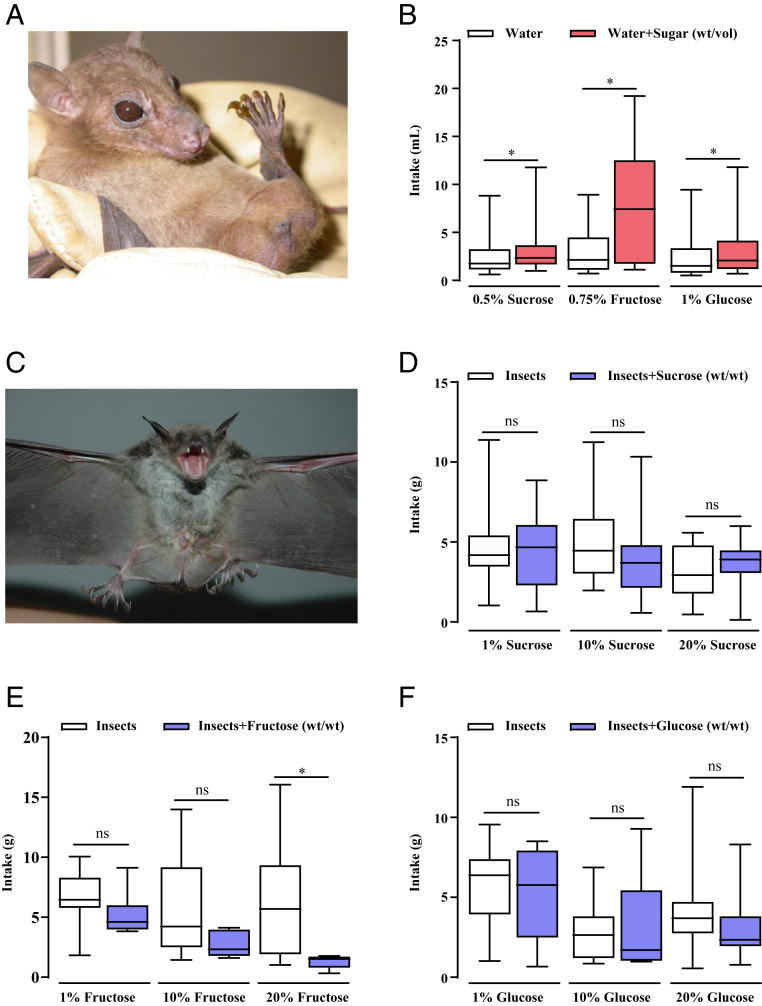
Behavioral preferences of two bat species toward natural sugars. (A) R. leschenaultii, a frugivorous bat species. (B) Results of preference tests to sucrose, fructose, and glucose (R. leschenaultii, n = 10 to 14). (C) M. ricketti, an insectivorous bat species. (D–F) Results of preference tests to sucrose, fructose, and glucose at three concentrations (M. ricketti, n = 9 to 18). Data are expressed as the mean ± SD (*P < 0.05, Student’s t test).
Tas1r2 and Tas1r3 Are Expressed in Both Frugivorous and Insectivorous Bats.
We investigated the expression patterns of Tas1r2 and Tas1r3 in two bat species (M. ricketti and R. leschenaultii; three individuals per species). Eight tissues that are known to express these two genes in other mammals (2829–30) were examined in our study (SI Appendix, Fig. S2). As a positive control, we also examined the expression of PKD2L1, a candidate sour taste receptor gene (31), in the taste tissue. The taste tissue we chose is at the posterior of the tongue, where circumvallate papillae are located, because circumvallate papillae are large and round and therefore easy to identify, and typically house over 100 taste buds (32). Both Tas1r2 and Tas1r3 genes were found to be expressed in all eight tissues in both species, and PKD2L1 was also detected to express in the taste tissue (SI Appendix, Fig. S2). These findings suggest that sweet taste receptor genes (Tas1r2 and Tas1r3) are expressed in both insectivorous and frugivorous bats.
Functional Loss of Sweet Taste Receptor Genes in Insectivorous Bats.
Cell-based functional assays were performed to test whether sweet taste receptors of frugivorous and insectivorous bats show similar responses to natural sugars. We first examined the responsiveness of sweet taste receptors of seven bat species in vitro, including two species of the Old World fruit bats (Rousettus aegyptiacus and Pteropus vampyrus), one species of the New World fruit bat (Sturnira lilium), and four species of insectivorous bats (Rh. ferrumequinum, Hipposideros armiger, Miniopterus natalensis, and M. ricketti; Fig. 3). Three natural sugars (sucrose, fructose, and glucose), which are the most abundant sugars in fruits and nectars (33), were used as potential ligands in functional assays. We found that the sweet taste receptors of the two Old World fruit bats responded to sucrose and fructose, but not to glucose (Fig. 3 and SI Appendix, Fig. S3). Unexpectedly, the sweet taste receptors of the other five bat species (including one New Word fruit bat and four insectivorous bats) showed no response to any of the three natural sugars (Fig. 3 and SI Appendix, Fig. S3). We further obtained the dose-dependent curves for all bat sweet taste receptors examined (Fig. 3). The sweet taste receptors of the two Old World fruit bats showed dose-dependent responses to sucrose and fructose with different levels of sensitivity, but no detectable responses to glucose even at the maximum concentration used in this assay (100 mM; Fig. 3). The sweet taste receptor of R. aegyptiacus appears to be more sensitive than that of P. vampyrus to sucrose and fructose (Fig. 3). The sweet taste receptors of the other five bat species could not be activated by any of the three sugars at any concentration tested (Fig. 3). To further explore sweet taste sensitivity in the New World fruit bats, we next examined two additional species of New World fruit bats (Artibeus jamaicensis and Glossophaga soricina) using the same functional assays. While A. jamaicensis showed no response to any of the three natural sugars, G. soricina clearly showed a response to sucrose, although the sensitivity is relatively low (Fig. 3). We also confirmed that the absence of detectable response of some bat Tas1r2/Tas1r3 to sugars was not due to absent or low expression of Tas1rs in heterologous cells (SI Appendix, Fig. S4). The expression levels of bat Tas1r2 and Tas1r3 in HEK293 cells after transduction were assessed by immunocytochemistry using an anti-herpes simplex virus (HSV) antibody and an anti-Flag antibody. We observed similar coexpression levels (∼10%) for Tas1r2/Tas1r3 of different bat species (SI Appendix, Fig. S4). Additionally, we observed that all nine bat sweet receptors examined were able to respond to an artificial sweetener, neohesperidin dihydrochalcone (NHDC; SI Appendix, Fig. S5), which is known to bind to the transmembrane domain of the human sweet taste receptor (34), demonstrating that these receptors are all functional in our heterologous expression system. Our findings revealed that the sweet taste perception mediated by the Tas1r2–Tas1r3 heterodimer is intact in the two Old World fruit bats and one New World fruit bat, but lost in all four insectivorous bats examined and two additional New World fruit bats. These results also suggest an evolutionary convergence of sweet taste perception that was independently evolved in two distantly related lineages: Old World and New World fruit bats (Fig. 1).


Responses of bat sweet taste receptor to natural sugars. HEK293 cells transiently expressing bat sweet taste receptor and Gα16-gust44 were assayed by calcium mobilization for responses to three natural sugars. (A, C, and E) Quantitative analysis of responses of bat sweet taste receptor to sucrose (100 mM), fructose (100 mM), and glucose (100 mM; mean ± SEM; ***P < 0.001, one-way ANOVA). (B, D, and F) Dose-dependent responses of bat sweet taste receptors to sucrose, fructose, and glucose. Mock-transfected cells were used as negative controls.
To identify molecular mechanisms underlying functional loss of sweet taste receptors of insectivorous bats and New World fruit bats, we examined the responsiveness of cross-species receptor pairs to sucrose (100 mM; Fig. 4). Specifically, we selected one Old World fruit bat (R. aegyptiacus), one New World fruit bat (S. lilium), and two insectivorous bats (H. armiger and M. ricketti) to generate six cross-species receptor pairs (Fig. 4A). For example, RouT2/HipT3 is the pair of Tas1r2 of R. aegyptiacus and Tas1r3 of H. armiger (Fig. 4A). Our results showed that RouT2/HipT3 and HipT2/RouT3 pairs showed no response to sucrose (Fig. 4B), suggesting that both Tas1r2 and Tas1r3 of H. armiger have lost their function involved in sugar detection. In contrast, MyoT2/RouT3 and StuT2/RouT3 pairs showed clear responses to sucrose, whereas RouT2/MyoT3 and RouT2/StuT3 pairs cannot be activated by sucrose (Fig. 4B), indicating that Tas1r3, not Tas1r2, is responsible for the loss of sugar-binding function in S. lilium and M. ricketti. Furthermore, we constructed chimeric receptors by replacing the Tas1r venus flytrap domain (VFD) of nonsweet tasters with that of R. aegyptiacus, which can respond to sugars (Fig. 4C). For example, Rou-HipT2 was constructed by replacing the Tas1r2 VFD of H. armiger with that of R. aegyptiacus (Fig. 4C). We then examined the responses of three mismatched receptor pairs (Rou-HipT2/Rou-HipT3, MyoT2/Rou-MyoT3, and StuT2/Rou-StuT3) to sucrose and found that all these three receptor pairs can respond to sucrose (Fig. 4D). These results indicate that mutations in VFD of Tas1r2/Tas1r3 should, at least partially, account for the loss of sugar sensitivity in insectivorous bats and New World fruit bats, and highlight that VFD of Tas1r2/Tas1r3 is crucial for sugar detection. These findings also showed that the loss of sugar sensitivity in one species can be fully rescued by replacement with a critical functional domain from another species, which in turn demonstrated that our cell-based functional assays must have been working properly in the heterologous expression system.

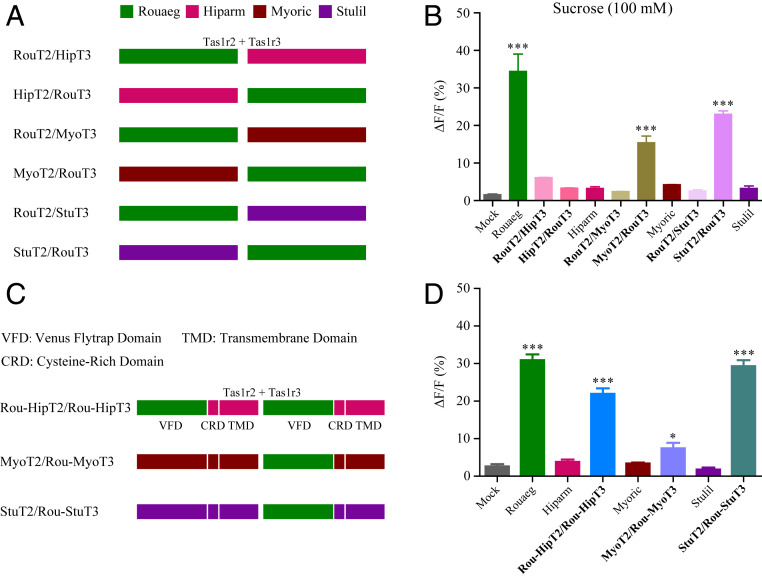
Responses of mismatched receptor pairs to natural sugars. (A) The schematic diagrams of cross-species receptor pairs. (B) Quantitative analysis of responses of four bat sweet taste receptors (Rouaeg, Hiparm, Myoric, and Stulil) and six cross-species receptor pairs (RouT2/HipT3, HipT2/RouT3, RouT2/MyoT3, MyoT2/RouT3, RouT2/StuT3, and StuT2/RouT3) to sucrose (100 mM; mean ± SEM; ***P < 0.001, one-way ANOVA). (C) The schematic diagrams of chimeric receptors. (D) Quantitative analysis of responses of four bat sweet taste receptors and three chimeric receptor pairs (Rou-HipT2/Rou-HipT3, MyoT2/Rou-MyoT3, StuT2/Rou-StuT3) to sucrose (100 mM; mean ± SEM; ***P < 0.001, one-way ANOVA). Mock-transfected cells were used as negative controls.
Discussion
We conducted comprehensive comparative analyses of coding-sequence molecular evolution, behavioral preference tests, gene-expression experiments, and cell-based functional assays to test whether diet plays a role in sweet taste perception in bats. We found that the sweet taste receptor genes are intact in all bats examined except the common vampire bat, as both Tas1r2 and Tas1r3 were conserved at the sequence level. In contrast, our behavioral experiments showed that the insectivorous bat (M. ricketti) exhibited no preference for natural sugars, while the frugivorous bat (R. leschenaultii) preferred sugars over water. In accordance with these results, our cell-based assays revealed an obvious functional divergence among the sweet taste receptors: while those of two Old World frugivorous bats and one New World fruit bat were activated by natural sugars, the receptors of four insectivorous bats and two New World frugivorous bats were insensitive to any natural sugars tested. Our findings thus suggest that diet must have played a role in the evolution of the sweet taste receptor in bats.
After identifying bat sweet taste receptor genes, we confirmed that both Tas1r2 and Tas1r3 are lost in the common vampire bat (D. rotundus), as reported earlier in previous studies of partial coding sequences (22, 27). This finding is also supported by taste behavioral preference tests in this species (35). Such pseudogenization events have also been identified in the sweet taste receptor genes of some carnivorous mammals, leading to the inability of these animals to detect natural sugars (12). We did not detect any inactivating mutations in the coding sequences of Tas1r2 and Tas1r3 in frugivorous (or nectarivorous) and insectivorous bats, indicative of strong functional constraints. We did not differentiate between frugivorous and nectarivorous species under the assumption that they similarly consume abundant natural sugars in their diets. Selection pressure tests detected signals of strong purifying selection on the two genes, and no signals of differential selection on Tas1r2 and Tas1r3 were identified between frugivorous and insectivorous bats, supporting the findings of our previous study of Tas1r2 (22).
Importantly, we cannot directly infer that sweet taste perception mediated by the Tas1r2–Tas1r3 heterodimer is conserved in frugivorous and insectivorous bats because an intact coding sequence does not indicate an intact gene function and/or animal behavior (36, 37). Therefore, we performed behavioral preference tests on R. leschenaultii (a congeneric species of R. aegyptiacus) and found strong behavioral preference toward natural sugars (Fig. 2). Similar behavioral preference was also recorded in two other Old World frugivorous bats (38). Cell-based assays consistently demonstrated that the sweet taste receptors of two Old World frugivorous bats (R. aegyptiacus and P. vampyrus) showed clear dose-dependent responses to sucrose and fructose, although not to glucose (Fig. 3). These results are similar to a previous cell-based functional assay in which the giant panda was shown to detect sucrose and fructose but not glucose (39). The disparity between the cell-based assays and behavior experiments (Figs. 2 and 3) for glucose sensing may have resulted from the technical limitation of our heterologous system, in which we were unable to use glucose at higher concentrations due to nonspecific cellular responses (39). An alternative explanation is the possible existence of a second type of glucose sensor (40). In addition, the sweet taste receptor of one New World fruit bat (G. soricina) also showed clear response to sucrose, although not to fructose and glucose, which is consistent with behavioral evidence of sugar preference in two New World fruit bats, including G. soricina (41). Clearly, such evidence suggests that all Old World fruit bats and some New World fruit bats may have the normal sweet taste perception. Similarly, nectar-feeding hummingbirds have been found to evolve the perception of sweet taste by transformation of the canonical umami receptor (42), despite the lack of sweet taste receptor gene Tas1r2 in the common ancestor of birds (43). In contrast, we did not observe behavioral preference for natural sugars in the insectivorous bat species (M. ricketti; Fig. 2). Consistent with this result, our cell-based assays showed that the sweet taste receptors of the four insectivorous bats were not activated by any of the sugars tested in this study (Fig. 3), possibly due to mutations on critical functional domains of their receptors (37). Our subsequent assays on chimeric receptors supported this view and confirmed the functional importance of VFD domain of Tas1r2/Tas1r3 on sugar detection. One plausible explanation is that the natural food of those bats, insects, contains little sugar; hence, the sweet taste of insectivorous bats appears to have been useless, relaxing the functional constraint on the sweet taste. A similar case is the low sensitivity of sweet taste in colobine monkeys, which mainly consume leaves (37). This specialized feeding habit may have resulted in the low opportunity to consume natural sugars (44). Notably, in these two cases, both insect-eating and leaf-eating animals possess complete coding sequences of Tas1r2 and Tas1r3. On the contrary, we noticed that, despite a frugivorous diet, the sweet taste receptor of two New World fruit bats (S. lilium and A. jamaicensis) did not respond to any natural sugars tested (Fig. 3). This is noteworthy since, to the best of our knowledge, sweet taste preference has not been demonstrated in S. lilium and A. jamaicensis by behavioral experiments. Alternatively, the common loss of sensitivity to natural sugars in these two New World frugivorous species could have been shaped by phylogenetic constraints, which may have resulted from the inability of sugar detection in their common ancestor. Thus, more investigations should be carried out to test whether the sweet taste preference is truly lost in some New World fruit bats in the future.
We also examined the expression patterns of Tas1r2 and Tas1r3 in the same two bat species used in our behavioral experiments. In line with previous studies (2829–30), our results showed that both genes in the two species are expressed throughout the body, including many extraoral tissues (SI Appendix, Fig. S2). This finding suggests that these two proteins may have other important biological functions (28, 30), which explains the sequence conservation of the two genes across the bat phylogeny. The expressions of Tas1r2 and Tas1r3 are detectable in the taste tissue of insectivorous bat M. ricketti and frugivorous bat R. leschenaultii. This finding rejects the possibility of that the loss of sugar-tasting ability in insectivorous bats could have resulted from mutations in regulatory regions of their sweet taste receptors that abolish gene expression.
Two alternative scenarios can be used to explain the evolution of sweet taste receptor gene function in bats. Either the responses to sugars was lost once in the common ancestor of all extant bats, but subsequently regained in the Old World fruit bats (Pteropodidae) and some New World fruit bats (Glossophaginae), or the sense of sweet taste was lost frequently—multiple times across the Yangochiroptera lineages and once in the common ancestor of insectivorous bats in the Yinpterochiroptera—but retained in Glossophaginae and Pteropodidae (Fig. 1). Which scenario reflects the true evolutionary history? The best way to solve this might be reconstruction of the ancestral protein, followed by gene synthesis, expression, and functional characterization using cell-based assays (45). If the resurrected receptor of the ancestor of all extant bats shows clear responses to natural sugars, the results would support the second scenario. Otherwise, the first scenario may be accurate. Unfortunately, the present study was unable to accurately reconstruct the ancestral sequences of Tas1r2 and Tas1r3 for the common ancestor of all extant bats because many variable amino acid sites were reconstructed with low posterior probabilities, even though we tried different models and methods (25, 46).
Our comprehensive comparative analysis may help solve some mismatches between feeding ecology and taste receptor evolution (18). For those mammals such as obligated carnivores, folivores, and insectivores, who possess intact sweet taste receptor genes even though their diets include little sugar, their abilities of sweet taste perception need to be reevaluated in future studies using cell-based functional assays and/or behavioral experiments. More generally, our findings support the suggestion that phenotypic diversity cannot always be predicted from genotypes alone, as previously proposed based on an analysis of parallel opsin loss in neotropical bats (36). Together, our study detects loss of sweet taste despite the conservation of sweet receptor genes in insectivorous bats, suggests that the loss of sweet taste may be more common in vertebrates than previously thought, and highlights the power of functional assays and behavioral experiments in molecular evolutionary studies.
Materials and Methods
Genomic Data.
The genome assemblies of the 33 bat species investigated here were retrieved from the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/, last accessed February 18, 2019; SI Appendix, Table S1). In addition, we generated draft assemblies for three species of New World bats (Pteronotus parnellii, Trachops cirrhosus, and S. lilium) at 50 to 100× coverage, with two Illumina paired-end libraries (SI Appendix, Table S1). These assemblies are deposited in Dryad Digital Repository (47).
Genome Mining.
Following a previous study (48), we performed TblastN searches (49) against each of the 36 bat genome sequences (SI Appendix, Table S1) to identify two sweet taste receptor genes, Tas1r2 and Tas1r3, using the protein sequences of human sweet taste receptor genes as queries. The coding sequences of these two genes were determined using GeneWise (50) and checked manually. We classified the annotated genes into three categories: complete genes, partial genes, and pseudogenes. Complete genes refer to those that have full-length coding sequences and a proper start and stop codon; partial genes refer to those with a truncated open reading frame as a result of incomplete genome sequencing; pseudogenes refer to those with inactivated mutations. All identified sequences are provided in Dataset S1.
Gene Sequencing.
Genomic DNA of three bat species was isolated from liver or wing punch samples (SI Appendix, Table S1) using Qiagen DNeasy Kits, following the manufacturer’s instructions. A suite of primers (SI Appendix, Table S2) were designed based on the alignments of known sequences and used to amplify the full-length coding sequences of bat Tas1r2/Tas1r3. PCRs were conducted as previously described (19). PCR products were purified and sequenced in both directions. The generated sequences were deposited into GenBank under accession numbers MK955939 to MK955942 and MT940227 to MT940228.
Evolutionary Analysis.
Using the MUSCLE program (51), we aligned the deduced amino acid sequences of bat Tas1r2 and Tas1r3. The pseudogenized sequences of sweet taste receptor genes in D. rotundus were not included because they are too divergent to allow a reliable alignment. The corresponding nucleotide sequence alignments were generated based on the amino acid alignments, with all gaps removed. We then performed evolutionary analyses for each gene by estimating the ratio (ω) of the rate of nonsynonymous substitutions to the rate of synonymous substitutions using the codeml program implemented in PAML v. 4 (25). Likelihood ratio tests were applied to compare nested models. The input tree was the species tree taken from previous studies (5253545556–57).
Sweet Compounds.
We selected three natural sugars (sucrose, fructose, and glucose; Sigma-Aldrich) and one artificial sweetener (NHDC; TCI Chemicals) to examine the responsiveness of bat sweet taste receptors in vitro. These compounds were dissolved in Dulbecco’s phosphate-buffered saline (DPBS) or in a mixture of dimethyl sulfoxide and DPBS. The highest concentrations of natural sugars and the artificial sweetener were 100 mM and 4 mM, respectively.
Construction of Bat Tas1r2 and Tas1r3 Expression Plasmids.
Full-length coding sequences of bat Tas1r2 and Tas1r3 were chemically synthesized. Given the codon usage preference in different organisms, we performed codon optimization for expression in HEK293 cells. The optimized coding sequences were then inserted into the expression vector pcDNA3.1, with the Kozak sequence introduced before the start codon. Bat Tas1r2 and Tas1r3 were fused with the HSV glycoprotein D epitope and 3×Flag tags at the C terminal, respectively. All constructs were verified by Sanger sequencing.
Functional Assay of the Sweet Taste Receptor in Bats.
We performed the functional assays as previously described (39). Briefly, HEK293 cells (PEAKrapid) were cultured in Opti-MEM medium supplemented with 5% fetal bovine serum. Healthy cells were seeded onto 96-well plates at a density of 50,000 cells per well. After 14 to 16 h, the cells were transiently transfected with plasmids that express the bat sweet taste receptor and a chimeric G protein Gα16-gust44 (0.067 μg per well for each plasmid) using Lipofectamine 2000 (0.5 μL per well). Cells transfected with Gα16-gust44 alone were used as negative controls. After 48 h of transfection, the cells were washed once with assay buffer DPBS and loaded with 50 μL Fluo-4 AM (2.5 μM; Invitrogen), a Ca2+ indicator, for 1 h in the dark at room temperature. The dyed cells were washed three times with DPBS, maintained in 50 μL DPBS, and assayed for their response to sweet compounds using a FlexStation 3 Microplate Reader (Molecular Devices). Relative fluorescence units (excitation at 494 nm, emission at 516 nm, cutoff at 515 nm) were recorded every 2 s over a period of 200 s. DPBS supplemented with 2× ligands were added after 30 s. All experiments were performed three times. The percentage of change in fluorescence (ΔF) from the baseline fluorescence (F) was used to quantify the calcium mobilization in response to ligands.
Immunocytochemistry Assay.
HEK293 (PEAKrapid) cells were seeded onto poly-lysine–coated coverslips in 12-well plates and transfected with a bat Tas1r2 and a Tas1r3 construct (0.3 µg per well for each construct), along with Gα16-gust44 (0.3 µg per well), by Lipofectamine (1.5 µL per well). After 2 d of transfection, cells were washed twice with phosphate-buffered saline (PBS) buffer and fixed by 4% paraformaldehyde in PBS for 20 min. Cells were then washed once with PBS and incubated with 0.1% Triton X-100 in PBS for 15 min. After they were washed with PBS, cells were incubated for 1 h in 10% fetal bovine serum in PBS to block unspecific binding. Next, an anti-HSV antibody (ab19355; Abcam; 1:500) and an anti-Flag (66008-3-lg; Proteintech; 1:500) in PBS supplemented with 10% fetal bovine serum were applied overnight at 4 °C. A donkey anti-rabbit secondary antibody conjugated with Alexa Fluor 488 (ANT024; AntGene Biotechnology; 1:800) and a goat anti-mouse secondary antibody conjugated with Alexa Fluor 594 (R37121; Thermo Fisher Scientific; two drops in 1 mL PBS supplemented with 10% fetal bovine serum) were then used for detection of the HSV and 3×Flag tags. The nucleus was stained with DAPI. Images were captured with confocal laser scanning microscopy (Leica TCS SP8). To evaluate the coexpressions of bat Tas1r2 and Tas1r3, at least six areas were counted.
Behavioral Experiments.
Wild-caught bats (M. ricketti and R. leschenaultii) were individually maintained in captivity at the Institute of Zoology at Guangdong Academy of Sciences and were trained to access food prior to behavioral trials. The use and care of the bats in this study were reviewed and approved by the ethics committees of the Institute of Zoology at Guangdong Academy of Sciences and Wuhan University. All samples were lawfully acquired, and their use conformed to the national and local laws and regulations.
The Rickett’s big-footed bat (M. ricketti) usually eats insects and seldom drinks water in captivity. We thus modified the two-bottle preference test for this insectivorous species to include bowls instead of bottles. We prepared two plastic bowls, one containing mashed insects (mealworms) and the other containing a homogeneous mixture of mashed insects and one of the test compounds [sucrose (1%, 10%, 20%, wt/wt), fructose (1%, 10%, 20%, wt/wt), glucose (1%, 10%, 20%, wt/wt), and a bitterant, quinine hydrochloride (0.4%, wt/wt)]. The bowls were placed on opposite sides of each cage. The animals (n = 9 to 18) had access to food from both bowls for 12 h. The experiment was repeated after the bowls were switched to opposite sides to control for the position effect.
The Leschenault’s rousette fruit bat (R. leschenaultii) is able to drink water in captivity, so we used the two-bottle preference test. Two bottles were placed on opposite sides of the cage; one contained plain water and the other contained the test compound dissolved in water. The animals (n = 10 to 14) could drink from either bottle for 12 h. The bottles were switched to opposite sides of the cage, and the experiment was repeated. Test solutions include sucrose (0.5%, wt/vol), fructose (0.75%, wt/vol), and glucose (1%, wt/vol), which are similar to those used in nectar-eating bats (41). Food intake was measured and compared between the experimental and control groups using a Student’s t test. The data are expressed as the mean ± SD.
Real-Time qPCR.
To study the expression patterns of bat Tas1r2 and Tas1r3, we conducted real-time qPCR experiments for one insectivorous bat species (M. ricketti, n = 3 individuals) and one frugivorous bat species (R. leschenaultii, n = 3 individuals). All bat specimens were captured in the field and euthanized in the lab. A total of eight tissues (brain, taste tissue, liver, spleen, lung, kidney, stomach, and intestine) were sampled and stored at −80 °C after freezing in liquid nitrogen. Total RNA was extracted using RNAiso Plus (Takara) from all tissues and reverse-transcribed to cDNA using the PrimeScript RT reagent Kit with gDNA Eraser. The gene expression levels of bat Tas1r2 and Tas1r3 were determined with SYBR Premix Ex Taq II (Takara) on an Applied Biosystems 7500 Real-Time PCR System. The qPCR primers spanning two exons are listed in SI Appendix, Table S4. The housekeeping gene SNRPD3 was used as a control to normalize the expression levels of bat Tas1r2 and Tas1r3. PKD2L1 was used as a positive control to confirm the specificity of dissected taste tissue.
Acknowledgements
We thank Dr. Nancy Simmons from the American Museum of Natural History for providing the tissues of four bat species and Bing-Jun Wang, Qian Wang, and Zhen Wang from Wuhan University for technical assistance. This work was supported by the National Natural Science Foundation of China (31722051, 31672272), Natural Science Foundation of Hubei Province (2019CFA075), and the Ten-thousand Talents Program (to H.Z.). Additionally, H.J. was supported by the National Natural Science Foundation of China (32000385), China National Postdoctoral Program for Innovative Talents (BX20200255), China Postdoctoral Science Foundation (2020M672407), and Hubei Provincial Postdoctoral Foundation; L.Z. by the GDAS Special Project of Science and Technology Development (2017GDASCX-0107, 2018GDASCX-0107); and P.J. by the National Institute on Deafness and Other Communication Disorders at the NIH (R01DC010842).
Data and Materials Availability.
All data that support the conclusions are present in the paper and/or the supplementary materials. The generated sequences were deposited to GenBank under accession numbers MK955939 to MK955942 and MT940227 to MT940228. Draft assemblies newly sequenced for three species of New World bats (P. parnellii, T. cirrhosus, and S. lilium) are deposited in the Dryad Digital Repository (https://doi.org/10.5061/dryad.0k6djh9z1) (47).
References
2
4
7
10
11
12
13
14
15
16
17
18
19
20
21
22
24
25
26
27
28
29
30
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
51
52
53
54
55
56
57
 Loss of sweet taste despite the conservation of sweet receptor genes in insectivorous bats
Loss of sweet taste despite the conservation of sweet receptor genes in insectivorous bats
