
Edited by Richard W. Aldrich, The University of Texas at Austin, Austin, TX, and approved December 1, 2020 (received for review April 19, 2020)
Author contributions: G.J.I., H.W., W.Z., and G.K.P. designed research; G.J.I., H.W., M.H., and B.L. performed research; G.J.I., H.W., and B.L. analyzed data; and G.J.I., H.W., W.Z., and G.K.P. wrote the paper.
1G.J.I. and H.W. contributed equally to this work.
2W.Z. and G.K.P. contributed equally to this work.
- Altmetric
NMDA receptors are glutamatergic channels whose activation controls the strength of excitatory synapses in the central nervous system. Agonist binding initiates a complex activation reaction that consists of a stepwise sequence of reversible isomerizations. In addition to previously identified steps in this series, which include agonist-induced closure of the ligand-binding lobes and the subsequent mechanical pulling by the ligand-binding domain on the gate-forming transmembrane helix, we identify a cross-subunit interaction that stabilizes open receptors and slows the rate of the current decay. Naturally occurring NMDA receptor variants lacking this interaction are pathogenic.
NMDA receptors are excitatory channels with critical functions in the physiology of central synapses. Their activation reaction proceeds as a series of kinetically distinguishable, reversible steps, whose structural bases are currently under investigation. Very likely, the earliest steps include glutamate binding to glycine-bound receptors and subsequent constriction of the ligand-binding domain. Later, three short linkers transduce this movement to open the gate by mechanical pulling on transmembrane helices. Here, we used molecular and kinetic simulations and double-mutant cycle analyses to show that a direct chemical interaction between GluN1-I642 (on M3 helix) and GluN2A-L550 (on L1-M1 linker) stabilizes receptors after they have opened and thus represents one of the structural changes that occur late in the activation reaction. This native interaction extends the current decay, and its absence causes deficits in charge transfer by GluN1-I642L, a pathogenic human variant.
Ionotropic glutamate receptors (iGluRs) are ligand-gated ion channels responsible for the majority of fast excitatory currents in the central nervous system (1). Among iGluRs, mammalian NMDA receptors are remarkable for their prolonged activations and Ca2+-rich currents. These biophysical properties make them directly responsible for synaptic processes that underlie complex brain functions such as learning, memory, and cognition (2). Their biophysical properties vary across life span and brain regions and between physiological and pathological states because of differential expression of subunits and complex regulatory mechanisms (3). Recently identified NMDA receptor mutations in patient cohorts provide important clues about how the physicochemical properties of this critical protein support physiological and behavioral phenomena (4, 5).
NMDA receptors are large heterotetrameric transmembrane proteins that assemble from two glycine-binding GluN1 subunits and two glutamate-binding GluN2 subunits. The obligatory GluN1 subunit is practically omnipresent across excitatory synapses, whereas GluN2 subunits, which exist as four distinct subtypes (A through D), have discrete expression patterns. Studies with recombinant receptors have demonstrated unique functional features and biological significance for NMDA receptors containing GluN2A or GluN2B subunits (6). Notably, a rapid replacement of GluN2B with GluN2A subunits marks a critical stage in synaptic development (7). Functionally, GluN2A- and GluN2B-containing receptors have similar pore conductance and calcium permeability properties (8); however, they differ markedly in the time course of their synaptic response, which is fully explained by the distinct kinetics of their activation mechanisms (9). Typically, GluN1/GluN2A receptors produce more robust electrophysiological data, whereas GluN1/GluN2B receptors are more amenable to structural studies.
All iGluR subunits have modular architectures with large extracellular and cytoplasmic domains and a short, membrane-embedded domain (TMD), which consists of three transmembrane helices (M1, M3, and M4) and an internally facing P-loop (M2). The external portion of each subunit consists of two stacked globular domains, the N-terminal (NTD) and ligand-binding (LBD) domains, for which several atomic-resolution structures exist (10, 11). In contrast, the cytoplasmic C-terminal domain (CTD), which is least conserved across subunits, appears largely disordered. Although critically important for the receptor’s cellular functions, the NTD and the CTD are dispensable for glutamate-dependent electric activity (12, 13). Therefore, the LBD and TMD layers and the short linkers that connect them, L1-M1, M3-L2, and L2-M4, perform the essential activation reaction, while the attached NTD and CTD layers provide critical modulation. Consistent with this view, the existing evidence suggests that the activation reaction starts with the binding of agonists to the LBDs of resting receptors, which facilitate subsequent local interactions and stabilize LBDs in closed, more compact, and less flexible conformations (2). This whole-domain conformational change repositions the protruding LBD-TMD linkers. Notably, the length and mobility of the LBD-TMD linkers correlates with channel opening probability, suggesting that they serve as mechanical transducers between LBDs and the TMD-situated gate (1415–16). Given their high degree of conservation across species, low toleration to mutations in humans, and structural proximity to the gate, linker residues very likely have additional and more specific roles in receptor activation aside from length and mobility. Notably, alanine-scanning mutagenesis showed distinct contributions by the side chains of several L1-M1 residues in gating (17, 18). However, because the available structures have limited resolution for these flexible linkers (10, 11, 192021–22), the mechanisms by which they control the gating equilibrium remain unknown.
In keeping with the multiplicity of intramolecular motions that transform resting receptors into open, current-passing proteins, NMDA receptors have similarly intricate kinetic signatures. Statistically derived models from both single-molecule patch-clamp recordings (2324–25) and single-molecule FRET studies (26, 27) showed that these proteins explore complex free-energy landscapes by transitioning stochastically between several closed and open states, which are distinguishable by their lifetimes. These functionally defined states likely represent entire families of closely related, rapidly interconverting structural conformers. Importantly, during activation, receptors occupy these kinetically defined microscopic states in a predictable sequence. A present objective is to describe the series of structural changes that underlie the activation reaction and to delineate their functional significance by integrating structural and functional information from multiple experimental approaches.
Here, we used mutagenesis guided by molecular dynamics simulations, kinetic modeling of single molecule currents, and double-mutant thermodynamic analyses to show that during gating, L550 on GluN2A L1-M1 linker couples energetically with I642 on GluN1 M3 helix through a direct cross-subunit chemical interaction. This additional contact forms late during gating and serves to stabilize an open, ion-conductive conformation. These results identify a cross-subunit state-specific interaction that mediates NMDA receptor gating and provide insight into how spontaneous naturally occurring mutations at this site may produce neuropsychiatric pathologies.
Results
All-Atom Targeted MD Simulation Reveals a Putative Cross-Subunit Interaction in the Open State.
The functional properties of recombinant and native GluN1/GluN2A receptors have been characterized extensively using single-molecule current recordings. Both statistically derived kinetic models from one-channel patch-clamp recordings (2324–25) and single-molecule FRET measurements (26, 27) have demonstrated that these proteins explore a complex free energy landscape by transitioning stochastically between several closed and open states. These statistically defined states likely represent entire families of closely related rapidly interconverting structural conformers. Separately, cryo-EM studies showed that in any given experimental condition, receptors can exist as arrays of structural configurations (20, 21). A current challenge is to integrate the information obtained with these separate approaches to assign functional identity onto atomic structures and to delineate the pathways of sequential conformational changes that define activation (28). Previously, we used computational approaches to envision the structural movements that may occur during opening for CTD-lacking GluN1/GluN2A receptors and identified a small number of sites where pairs of residues changed their relative proximity during the process (29). Notably, these pairs clustered spatially, forming hotspots of movement. Many patient-derived missense mutations map to these hotspots, suggesting that subtle structural perturbations at these sites alter receptor function in biologically significant ways. One possibility is that the change in interresidue distance between some of these pairs correlates with the making or breaking of state-dependent interactions and alters the receptor’s opening equilibrium. The low resolution of the coarse-grained approach we used in this previous work prevented us from observing state-dependent side-chain motions that we could select for experimental testing. Therefore, we aimed to simulate the open or active state with atomic-resolution models.
To reduce simulation cost, we focused on the smallest receptor assembly that preserves ligand-dependent gating. Such a minimal receptor, GluN1ΔNΔC/GluN2AΔNΔC, consists of linked LBD and TMD layers, and lacks the larger NTD and CTD layers, which have a purely regulatory role (Fig. 1 A, Top). To reproduce the activation trajectory explored previously with our coarse-grained flexible fitting approach, we performed all-atom targeted molecular dynamics (MD) simulation of the minimal receptor using as target a putative “active” conformation (see Materials and Methods) (20). Next, we subjected the system to three independent 700-ns MD simulations in which we restrained LBD to relax the TMD further (see Materials and Methods). While these trajectories may not reflect a fully open channel (SI Appendix, Fig. S1 and Video S1), comparing initial and final conformations, we observed substantial pore dilation, consistent with partial channel opening (Fig. 1 A, Bottom). Next, we searched for putative atomic interactions formed during the simulation. Due to the dominance of nonpolar residues in the TMD, we focused on van der Waals (vdW) energies and selected intersubunit residue pairs which satisfied the following criteria: 1) average pairwise vdW energy was <−1 kcal/mol (SI Appendix, Table S1) and 2) at least one of the residues in the pair has been found mutated in patients (ClinVar, accessed March 2020). We further focused this study on pairs of residues located proximal to the gate and more specifically at the intersubunit M1-M3 interface, which has been highlighted in recent studies as critical for gating (17, 30, 31). Only two residue pairs met these criteria: GluN1 I642-GluN2A F549 and GluN1 I642-GluN2A L550. To evaluate computationally whether the change in interresidue distance may be coupled with pore opening, we calculated the Pearson correlation (PC) coefficient between their intercentroid distances and the intercentroid distances of pairs of residues situated at the GluN2A upper gate (across A650 residues) and lower gate (across N614 residues). We found significant negative correlation for the I642-L550 pair (PC = −0.46 and −0.46, with P < 0.0001), but not for the I642-F549 pair (PC = 0.07 and 0.13, with P = 0.14). Given that the predicted change in distance within the I642 and L550 pair correlates strongly with pore dilation at both upper and lower gates, we were motivated to test the hypothesis that they may form a state-dependent interaction that contributes to gating.

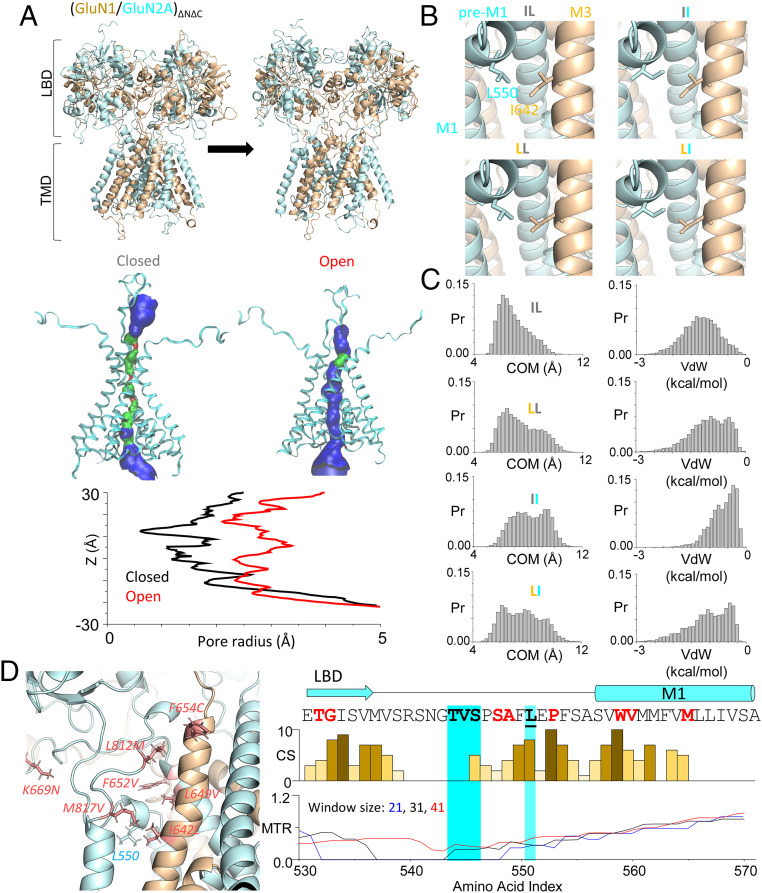
All-atom MD simulation predicts new cross-subunit interaction in the open/active state. (A, Top) All-atom structural model of (GluN1/GluN2A)ΔNΔC in closed and open conformations. (A, Bottom) HOLE-calculated pore images and radii in closed (Left, black) and open (Right, red) conformations. (B) Close-ups illustrate the predicted structural proximity between GluN1-I642 (on LBD/M1-linker, tan) and GluN2A-L550 (on M3-helix, cyan) in the four constructs examined. (C) Histograms of inter–side-chain center-of-mass (COM) distance (Left) and vdW contact energy (Right) between GluN1-I642 and GluN2A-L550 in the WT (IL) and three mutants (LL, II, and LI). (D, Left) GluN2A-L550 maps to a region of high disease-associated mutation density. (D, Right) Residues on the GluN2A pre-M1 region are evolutionarily conserved (conservation score, CS) and have low mutation tolerance ratios (shown with three sliding scales) in human populations. Bold indicates previously identified hotspot residues; red indicates disease-associated mutations (reported and predicted).
Next, we examined whether changing the side chain at I642 and/or L550 affected the predicted interaction. We ran separate open-state MD simulation for minimal receptors carrying the conservative single-residue substitutions GluN1I642L (LL) or GluN2AL550I (II) or the double substitution GluN1I642L/GluN2AL550I (LI; see Materials and Methods). We found that relative to receptors containing the wild-type (WT) pair (IL), conservative mutations weakened the I642-L550 interaction by significantly right-shifting the distributions of the center-of-mass distance (Kolmogorov–Smirnov test statistics compared to IL: LL, 0.16; II, 0.39; LI, 0.24, P < 0.0001 for all) and the vdW energy (Kolmogorov–Smirnov test statistics compared to IL: LL, 0.22; II, 0.54; LI, 0.23, P < 0.0001 for all) (Fig. 1C and SI Appendix, Fig. S2). Together, these measurements support a plausible role for GluN1-I642/GluN2A-L550 side chains, and therefore their putative interaction, in stabilizing the open state. Notably, whole genome sequencing in select patient populations identified the GluN1I642L variation as likely pathogenic (ClinVar ID: 421935). This observation further supports the hypothesis that subtle structural changes at this site have profound consequences for the gating properties of the receptor and for the wellbeing of individuals carrying mutations at this site.
Consistent with this hypothesis, both GluN1-I642 and GluN2A-L550 reside in regions that are highly conserved in iGluR subunits across species and are mutation intolerant in humans. GluN1-I642 maps to the highly conserved M3 transmembrane helix, just one turn below the invariant SYTANLAAF sequence, which forms the ligand-controlled pore gate at the level of alanine S + 6 (32). Likewise, GluN2A-L550 maps to the highly conserved S2-M1 linker (Fig. 1 D, Right), whose residues have been implicated by structural and functional studies as important for NMDA receptor activity (17, 30, 33). The observation that this linker hosts many recently identified pathogenic missense mutations further supports the hypothesis that its residues influence receptor activation (Fig. 1 D, Left) (17, 29).
Both GluN1-I642 and GluN2A-L550 Are Necessary for Normal Gating in Full-Length Receptors.
To explore experimentally whether the subtle structural alterations produced by conservative substitutions at GluN1-I642 and GluN2A-L550 have any functional consequences on receptor activation, we examined the gating mechanism of substituted receptors. We recorded currents from cell-attached patches containing one copy of full-length WT GluN1/GluN2A (IL), GluN1I642L/GluN2A (LL), GluN1/GluN2AL550I (II), or GluN1I642L/GluN2AL550I (LI) in saturating agonist (glutamate and glycine) and divalent-free solutions. In these conditions, all detected events reflect the intrinsic gating mechanism of the observed channel in real time, and the recorded signal is amenable to modeling and kinetic analyses (34). With this approach, we found that relative to the WT, GluN1I642L/GluN2A, GluN1/GluN2AL550I, and GluN1I642L/GluN2AL550I receptors had profoundly changed activity patterns (Fig. 2A and Tables 1 and 2). The mutations caused substantial (∼8-fold) decrease in burst open probability (Po, burst): from 0.7 ± 0.1, for WT to 0.1 ± 0.01 for GluN1I642L/GluN2A (P = 7 × 10−4) and GluN1/GluN2AL550I (P = 5 × 10−3) and to 0.003 ± 0.001 for GluN1I642L/GluN2AL550I (P = 4 × 10−6). Mean open durations (MOT) were ∼4-fold shorter, decreasing from 4.5 ± 0.7 ms for the WT to 1.3 ± 0.3 ms (P = 8 × 10−4) for GluN1I642L/GluN2A, 1.9 ± 0.4 ms (P = 2 × 10−3) for GluN1/GluN2AL550I, and 0.7 ± 0.1 (P = 5 × 10−4) for GluN1I642L/GluN2AL550I. Mean closed durations within bursts were >10-fold longer, increasing from 1.3 ± 0.3 ms for the WT to 23.0 ± 3.9 ms for I642L (P = 1 × 10−5), 39.9 ± 10.3 ms for L550I (P = 3 × 10−5), and 244.8 ± 55.4 (P = 5 × 10−3) for GluN1I642L/GluN2AL550I (Fig. 2B and Tables 1 and 2).

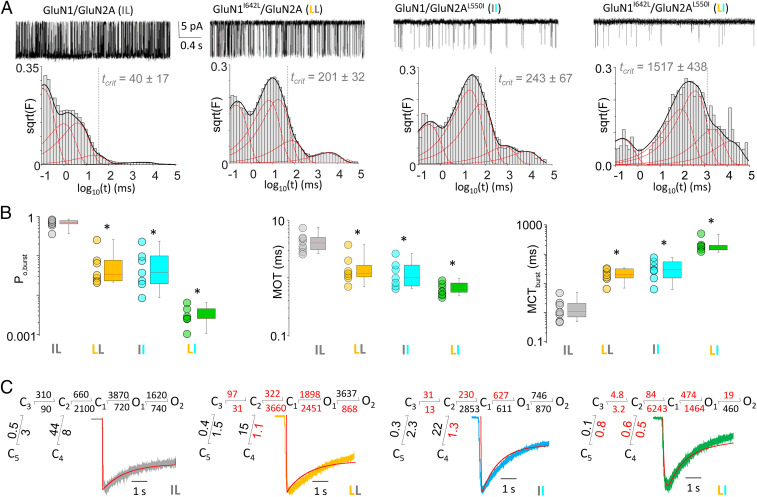
Side chains of both GluN1-I642 and GluN2A-L550 contribute to gating in full-length receptors. (A, Top) Unitary currents (3 s) for WT (IL; n = 8), GluN1I642L/GluN2A (LL; n = 7), GluN1/GluN2AL550I (II; n = 8), and GluN1I642L/GluN2AL550I (LI; n = 7) receptors. (A, Bottom) Corresponding histograms of closed event durations with the fitted exponential functions (red curves) and the composite probability density function (black curve). Bursts were defined using thresholds (tcrit) calculated for each recording. (B) Plots illustrate open probability (Po), MOT, and mean closed (MCT) times for bursts of activity in each recording. *P < 0.05, independent t test relative to WT. (C) Reaction mechanism for each receptor with rates estimated by fitting the indicated models to all event durations across all files in each data set. Red indicates statistical significance compared to WT; P < 0.05, independent t test. Lines represent the predicted macroscopic response of each model (red) superimposed with experimentally recorded whole-cell current responses to glutamate (5 s, 1 mM) and normalized to peak.

| Receptor | Unitary current (pA) | Burst Po | MOT (ms) | Burst MCT (ms) | Events | n | Total burst time (min) |
| GluN1/GluN2A | 8.9 ± 0.6 | 0.73 ± 0.06 | 4.4 ± 0.6 | 1.2 ± 0.6 | 4,663,279 | 8 | 419 |
| GluN1I642L/GluN2A | 7.8 ± 0.9 | 0.08 ± 0.03* | 1.3 ± 0.3* | 23.0 ± 4.0* | 118,494 | 7 | 31 |
| GluN1/GluN2AL550I | 8.2 ± 0.3 | 0.08 ± 0.03* | 1.7 ± 0.4* | 39.9 ± 10.3* | 22,141 | 7 | 14 |
| GluN1I642L/GluN2AL550I | 5.4 ± 0.3* | 0.003 ± 0.001* | 0.7 ± 0.1* | 244.8 ± 55.4* | 5,682 | 7 | 21 |
| GluN1I642A/GluN2A | 4.4 ± 0.7* | 0.07 ± 0.02* | 1.3 ± 0.2* | 36.4 ± 4.5* | 47,732 | 7 | 17 |
| GluN1/GluN2AL550A | 7.0 ± 0.4 | 0.08 ± 0.01* | 1.9 ± 0.1* | 32.4 ± 5.5* | 24,170 | 12 | 10 |
| GluN1I642A/GluN2AL550A | 5.8 ± 0.3* | 0.033 ± 0.003* | 1.0 ± 0.1* | 1429.3 ± 6.4* | 30,097 | 7 | 12 |
Data are represented as mean ± SEM. *P < 0.05 independent t test compared to GluN1/GluN2A.

| Receptor | Closed components (ms) | Open components (ms) | |||||
| τ1 | τ2 | τ3 | τfast | τLow | τMedium | τHigh | |
| GluN1/GluN2A | 0.1 | 1.3 | 5.3 | 0.1 | 2.6 | 6.3 | 13.8 |
| GluN1I642L/GluN2A | 0.2 | 9.1 | 38.5 | 0.2 | 1.6 | 4.2 | – |
| GluN1/GluN2AL550I | 0.3 | 20.1 | 81.4 | 0.5 | 3.4 | – | – |
| GluN1I642L/GluN2AL550I | 0.2 | 138.1 | 525.3 | 0.8 | 6.8 | – | |
| GluN1I642A/GluN2A | 0.3 | 7.2 | 27.3 | 1.3 | 3.3 | – | – |
| GluN1/GluN2AL550A | 0.1 | 11.7 | 46.0 | 0.2 | 2.1 | – | – |
| GluN1I642A/GluN2AL550A | 0.2 | 7.4 | 31.8 | 0.1 | 0.9 | 2.1 | – |
The substantially shorter openings and longer closures observed in all mutants show that the side chains of both GluN1-I642 and GluN2A-L550 are required for the characteristically high Po of GluN1/GluN2A receptors. However, they do not offer clues as to how these residues increase channel activity. To ascertain a potential kinetic mechanism that would explain the role of the examined side chains, we used our previously validated kinetic model (2, 35) and fitted it to the four data sets described above (Fig. 2C). The reaction mechanisms deduced with this approach predicted macroscopic responses that were very similar in time course with the whole-cell currents we recorded experimentally in transfected HEK293 (Human Embryocarcinoma Kidney) cells (Fig. 2C and SI Appendix, Table S2). The kinetic mechanisms obtained can serve two important objectives. They can help to ascertain the functional effect of the pathologic mutant and therefore suggest potential courses of treatment. Additionally, they can help further inquire into the mechanism by which the two residues participate in gating.
Pathogenic Variant has Accelerated Deactivation and Reduced Two-Pulse Potentiation.
Presently, there are no reports of variations at GluN2A L550 in human populations. In contrast, GluN1 I642 appears more forgiving to substitutions; specifically, variants with the conservative GluN1 I642L replacement were recently identified and classified as pathogenic. We used the kinetic mechanism deduced above to investigate how this conservative substitution may affect the macroscopic responses produced by GluN1-1aI642L/GluN2A following physiologic stimulation patterns. We used the reaction mechanism derived from our kinetic modeling of one-channel recordings (Fig. 2C) to simulate responses to brief applications of glutamate (5 ms, 1 mM). As expected from the substantially shorter mean open times, responses simulated with the model derived for the pathogenic mutant (LL) had faster deactivation times relative to the WT (IL) (Fig. 3B). To test this prediction, we recorded currents from lifted whole-cells expressing the LL mutant, in conditions similar to our single channel recordings. As predicted, the measured deactivation time course for GluN1I642L/GluN2A receptors was ∼2-fold shorter (42 ± 10 ms; P = 0.001) compared to that of WT receptors (83 ± 12 ms). This close accord between simulation and experimental recording represents additional validation for the model proposed above and motivated us to explore additional possible behavioral changes for the mutant.

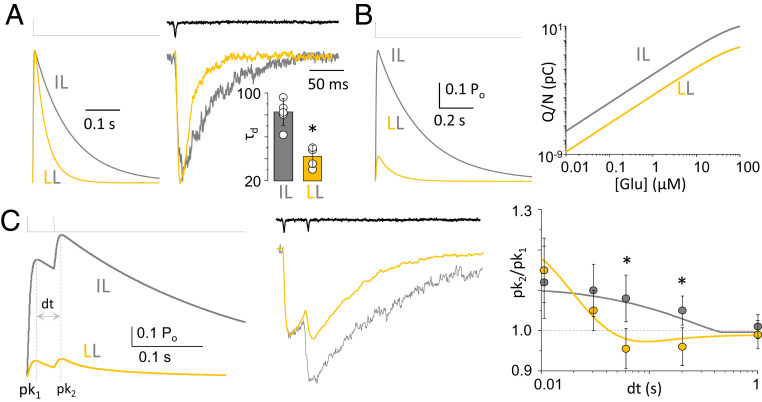
Pathogenic variant deactivates faster. (A, Left) Predicted macroscopic responses from GluN1/GluN2A (IL; gray) and GluN1I642L/GluN2A (LL; gold) to glutamate exposure (5 ms, 1 mM) normalized by peak amplitude. (A, Right) Experimentally recorded currents from lifted cells expressing the indicated receptors. Open-tip potential shown above (black). (Inset) Summary of measured deactivation times. (B, Left) Predicted macroscopic responses as in A, shown with absolute amplitudes. (B, Right) Predicted charge transfer (Q/N) per channel in response to one 1-ms pulse. (C, Left) Predicted current response to a two-pulse stimulus. Fractional current potentiation (pk2/pk1) is shown in response to a two-pulse protocol relative to interpulse duration (dt). (C, Middle) Empirically recorded currents using a two-pulse protocol, normalized to first peak. Open-tip potential is shown above (black). (C, Right) Summary of fold-change of second-to-first peak for predicted (solid lines) and empirically recorded currents. *P < 0.05, independent t test relative to WT.
Given the large change in open probability we observed for the mutant in single-channel records, we anticipated large changes not only in deactivation time course but also in current amplitude and therefore in the total charge transferred during activation. We used the model to simulate a macroscopic response to a brief pulse from a defined number of channels and determined that in addition to being faster to decay, currents from GluN1I642L/GluN2A receptors were also drastically smaller in actual amplitude (Fig. 3B). Together, the kinetic changes summarized by the kinetic model we propose here predict that the pathogenic receptors will transfer ∼100-fold less charge relative to WT receptors over a wide range of glutamate concentrations (Fig. 3B).
Lastly, we asked whether the pathogenic mutation affected the receptor’s frequency-dependent potentiation function. Macroscopic NMDA receptor currents exhibit submaximal peak current when stimulated with one short (<10 ms) pulse of glutamate. Depending on the interval at which it is administered, a second pulse can engage additional receptors to produce a larger response (2, 24). The model predicted that relative to the WT, the mutant will have reduced potentiation to a second pulse (Fig. 3 C, Left). Recordings with a double-pulse protocol allowed us to measure the change in amplitude of the second to the first pulse for WT and mutant receptors. As predicted by the model, we found significant reduction in the potentiation by a second pulse. Additional simulations predicted that this change will be significant for interpulse intervals in the 60 to 200 ms range (Fig. 3 C, Right).
Together, these simulations and measurements show that the GluN1-I642 side chain plays an important role in gating such that even a conservative substitution at this position (I/L) reduces considerably channel activity and affects several biophysical parameters important for synaptic function.
Interactions between GluN1-I642 and GluN2A-L550 Contribute to Receptor Gating.
The results and analyses described above for GluN1-I642 and GluN2A-L550 show that the side chains of both these residues are critically involved in setting the receptor’s gating kinetics, specifically by extending openings and shortening closures. They are also consistent with predictions from the MD simulation that by moving closer together during channel activation, they may form additional contacts and stabilize open states. However, these results do not prove direct interaction between the two.
To determine whether GluN1-I642 and GluN2A-L550 interact energetically during gating, we set up to perform double-mutant cycle analysis on this pair (36). Briefly, we compared the change in the free energy of the activation upon introducing single-residue substitutions at the positions investigated and then the corresponding double substitution. In this classic approach, if the change in free energy caused by the double mutation is simply the additive effect of single mutations, the residues likely contribute to the gating reaction independently; in contrast, if the double mutation causes changes in free energy that are distinct from the sum of changes due to single mutations, the two residues likely interact during the observed process, and the magnitude of the difference correlates with the strength of the interaction.
For these thermodynamic analyses, we used initially the records described above for WT GluN1/GluN2A (IL), GluN1I642L/GluN2A (LL), and GluN1/GluN2AL550I (II). In addition, we recorded from GluN1I642L/GluN2AL550I (LI). Here, we selected bursts of activity, which represent repeated stochastic excursions between resting and open states, to more closely mimic the closed-to-open trajectory simulated by the MD simulation. This was done by identifying in each record and excluding from analyses periods when the receptors were closed for long time periods, indicative of sojourns into desensitized states (34, 37, 38) (Fig. 4 and Tables 1 and 2). Kinetic modeling of bursts revealed that relative to single mutations, the double mutation produced a larger, synergistic increase in the free energy of gating (1.66 kJ mol−1) indicative of strong functional coupling between these residues (Fig. 4A). This value is within reported energies for vdW contacts (0.4 to 4 kJ/mol), and considering our predicted center-of-mass distance between these residues (6.9 Å), it is consistent with an attractive force that would stabilize the open state.

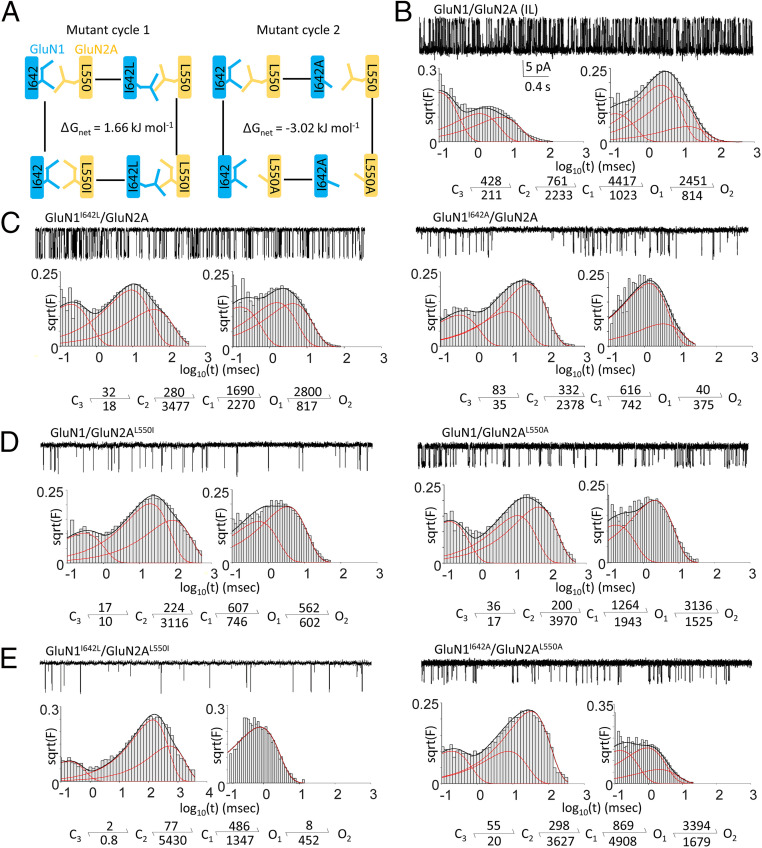
Double-mutant cycle analyses for GluN1I642 and GluN2AL550. (A) Diagrams for the two cycles examined, with calculated free energy changes (ΔΔGnet) due to mutual interactions during gating (B–E). Representative 3 s of continuous single-channel currents, with event duration histograms and kinetic activation mechanism for the indicated receptors. All rate constants (s−1) were derived from global fits to burst data pooled across all the recordings for each construct.
Classic double mutant cycle analyses, rather than introducing complementary mutations as we have done above, typically use alanine as the substitution of choice. These side chain truncations ablate both the specific interactions between the two residues and any additional putative interactions by each side chain with neighboring residues (nonspecific). Therefore, we repeated our measurements and analyses with receptors containing GluNI642A, GluN2AL550A, or the double mutation GluN1I642A/GluN2AL550A (Fig. 4 and Tables 1 and 2). Using single-molecule recordings, we observed that GluN1I642A/GluN2A shows smaller unitary current amplitude and MOT than GluN1/GluN2AL550A (Table 1). Previously, a study of GluN2D found the L575A mutation (corresponding to L550 in GluN2A) resulted in low Po and slow time course of MK-801 inhibition (31). Another study of GluN2A found the L550A mutation resulted in lower Po and MOT (17). These observations are consistent with our MD simulation results in which I642 formed several vdW interactions with F549, L550, Y645, M817, and V820, whereas L550 only interacted with I642 (SI Appendix, Table S1). Thus, we expected the truncation of the entire I642 side chain to destabilize the open state more severely than a mutation of L550. For this more drastic structural change, we calculated that interactions between the side chains of GluN1-I642 and GluN2A-L550 contributed a total of 3.02 kJ mol−1 to the gating reaction. Given our results from MD simulation that simple side-chain isomerization (I/L and L/I) mutations shifted the overall center-of-mass distance and vdW energy toward less stable open configurations (Fig. 1C), we can conclude that the large coupling energy we measured from single-molecule recordings reflects the contribution of contacts between GluN1-I642 and GluNA2A-L550. In addition, the larger change observed with side-chain truncations (I/A and L/A) suggests that in addition to the specific interactions probed above, additional steric effects with neighboring residues are involved as well. Based on these results, we propose that GluN1-I642 and GluN2A-L550 side chains facilitate the NMDA receptor gating reaction considerably through both mutual vdW contacts and additional interactions with surrounding residues.
NMDA receptors have complex activation reactions whose adequate kinetic description must include at a minimum three closed states and two open states (34). Although the structural correlates of these statistically defined states are unknown, a multistate kinetic model accounts well for all observed microscopic and macroscopic receptor behaviors (24). Importantly, it also shows how the channel’s biophysical properties support its physiological function at synapses (2). Therefore, in addition to considering a global closed-open gating reaction for each arm of the thermodynamic cycle (as in Fig. 4A), we aimed to resolve individual kinetic steps along the activation pathway for each mutant (Figs. 4 B–E and 5A and Table 1). With these values, we calculated the change in free energy contributed by single and double mutations (ΔΔGint) and therefore the coupling constants (θ) between the two residues at each step along the gating sequence (Fig. 5A). With this more detailed analysis, we found, unsurprisingly, that perturbing interactions between GluN1-I642 and GluN2A-L550 affected to some degree all the transitions considered. However, the final step (O1 to O2) was most drastically changed. During this step, the native interactions stabilized open states, as predicted by MD simulation, and lowered the energy barrier for the O1 to O2 transition.

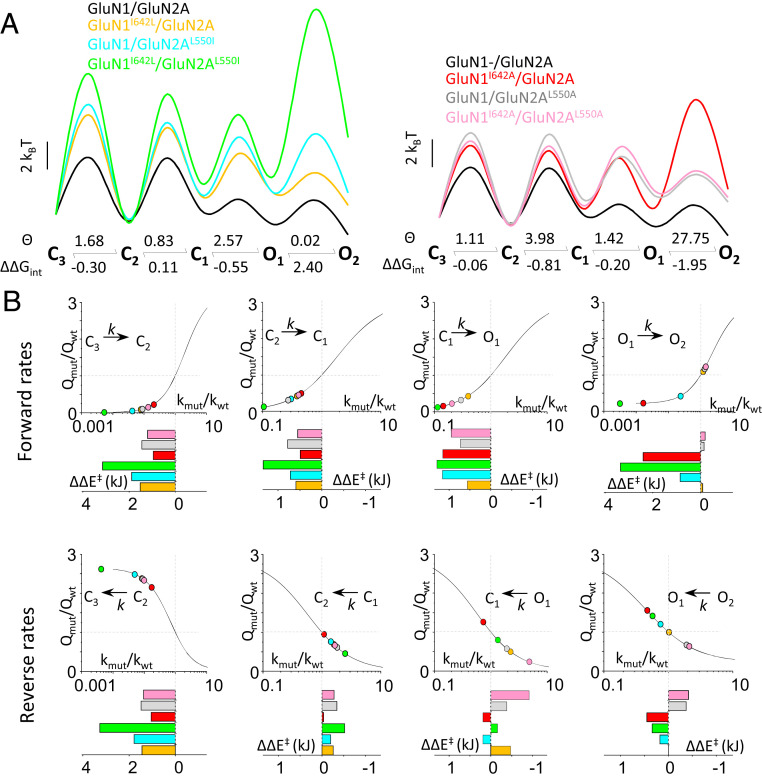
Interactions between GluN1-I642 and GluN2A-L550 stabilize active states and decrease barriers to opening. (A) Effects of side-chain isomerization (Left) and side-chain truncation (Right) on free energy landscapes (Top) calculated from measured rate constants (given in Fig. 3) and breakdown of coupling constants (Θ) and free energy contributions (ΔΔGint in kilocalorie per mole) per each transition (below). (B) In each panel, the indicated WT rate (kwt) was scaled iteratively by an order of magnitude (kmut), and the resulting kinetic model was used to calculate the change in charge transfer (Qmut/Qwt) following synaptic-like stimulation. Lines represent exponential fits to the simulated data points, and experimentally measured rate constants are mapped as colored circles. Bar graphs illustrate the change in the height of the transition state energy ΔΔE‡ (kilojoules) for each mutant.
State-dependent Interactions Differentially Stabilize Activation and Deactivation Transition States.
In addition to the expected destabilization of open conformations, we found that disrupting the native interactions between GluN1-I642 and GluN2A-L550 also increased the transition barriers along the entire gating reaction and affected asymmetrically the forward and reverse transitions. This observation has implications for understanding the subtle internal dynamics that make this receptor’s gating sequence and therefore the mechanistic basis of their macroscopic output. A theoretical study examining the effect of state-dependent perturbations at NMDA receptors found that kinetic changes of similar magnitude affect differentially the receptor’s electrical output according to the specific kinetic transition affected (39). Therefore, we set up to estimate how the observed changes in each rate constant and the corresponding change in activation energy impacted the magnitude of the macroscopic response.
First, we used the rate constants measured for WT receptors (Fig. 4B) to simulate macroscopic currents in response to brief glutamate pulses and to calculate the resulting charge transfer (Qwt) as one measure of potential impact on biological function (2). Next, we scaled each rate constant 10-fold in both directions to estimate the sensitivity of charge transfer (Qn/Qwt) to changes in specific rate constants (Fig. 5B, black lines). Onto the estimated charge versus rate relationship, we mapped the empirical rate constants measured for the six mutants investigated in this study. This analysis shows that absent the native interactions afforded by GluN1-I642 and GluN2A-L550, all forward rates were substantially slower, indicative of larger activation barriers, and resulted in substantial decrease in charge transfer. Both side-chain isomerizations (I/L) and side-chain truncations (I/A and L/A) were influential, suggesting that both specific and nonspecific interactions at this sensitive site contribute to channel activation in WT receptors. In contrast, changes in the reverse rates were generally smaller and had only mild influence on charge transfer, except for the last step in the deactivation sequence (C3 ← C2), for which the rate was substantially slower. This last step represents functionally a transition from receptors with very high glutamate affinity into receptors from which glutamate can dissociate with measurable rates (60 s−1/site) (23, 24); it sets the kinetics of macroscopic decay, and for this reason, it has a steep influence on charge transfer. Together, these simulations point to the possibility that the interactions examined here make asymmetric contributions to the opening and closing reactions. This possibility merits further investigation. The overall pattern of influence on specific rate constants is consistent with the hypothesis suggested by our initial all-atom MD simulation that GluN1-I642 and GluN2A-L550 form native-state–dependent interactions with each other that stabilize open states and shift the closed-open gating equilibrium to support longer responses.
Discussion
In this study, we identified a state-dependent, cross-subunit, cross-dimer interaction in NMDA receptors and estimated its energetic contribution to the stability of the open state in WT GluN1/GluN2A receptors. We used targeted molecular dynamics simulation to model the opening trajectory of minimal NMDA receptor channels, consisting of linked LBD and TMD layers. This analysis identified atomic contacts between GluN1-I642 and GluN2A-L550 that form during opening. Both GluN1-I642 and GluN2A-L550 map to regions of primary sequence that are highly conserved in iGluR subunits across species and have low mutation tolerance ratios in human populations. Specifically, GluN1-I642 resides deep within the M3 helix, below the strictly invariant SYTANLAAF sequence, which hosts the ligand-controlled gate. Notably, GluN1-I642L variant was identified as likely pathogenic based on exome sequencing analyses in large patient populations (40). Conversely, GluN2A-L550 resides on a short membrane-proximal helix that links the lower lobe of the LBD with the M1 transmembrane helix. Previous work has implicated residues on this short helix as critical for the gating of all iGluRs (17, 33). Our results support the hypothesis that direct interactions between GluN1-I642 and GluN2A-L550 form during pore opening as an intrinsic step in the physiological activation of full-length NMDA receptors (Fig. 6).

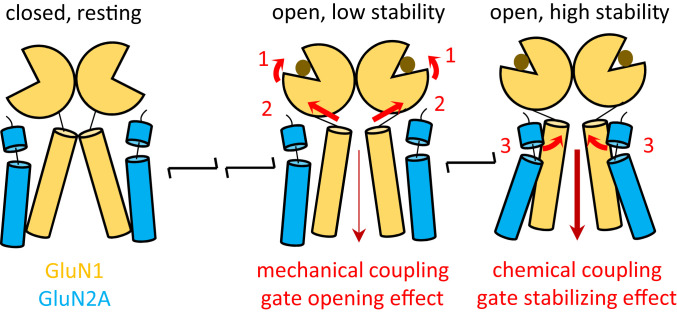
Proposed roles of linkers during gating. As receptors transition to open states upon agonist binding and cleft closure (1), mechanical tension on M3 linkers mediates coupling between the ligand-binding domain and the M3 helices to open the gate (2). As the gate opens, specific interactions between residues in GluN1 M3 of one GluN1/GluN2 dimer and the pre-M1 helix on the other GluN1/GluN2 dimer serve to stabilize the open gate (3).
The GluN2A subunit is the most abundant GluN2 subunit in adult neurons. The shift from GluN2B to GluN2A subunit expression marks an important stage in the normal development of central synapses. This change in subunit expression is accompanied by kinetic and pharmacologic changes consistent with an increased expression of the faster GluN2A-containing receptors, at the expense of the slower gating GluN2B-containing receptors. All empirical structural models available so far for functional NMDA receptors, whether from X-ray crystallography or cryoEM, describe GluN2B-containing proteins, which generate more complex patterns of single-channel activity and are more difficult to track with kinetic analyses. To facilitate direct inferences between structural and functional changes, we report here an all-atom structural model for GluN1/GluN2A receptors lacking the NTD and the CTD. This model allowed us to simulate the receptor’s opening reaction and to identify residues that may form new atomic contacts in the open state. Among these, the interaction between GluN1-I642 and GluN2A-L550 is a cross-subunit interaction specific to open receptors that we have characterized in NMDA receptors. Notably, it occurs between subunits that in the LBD layer belong to separate GluN1/GluN2A dimers. To test whether this interaction occurs in WT receptors and whether it contributes to their activation mechanism, we introduced single and double substitutions at these two positions in full-length receptors and used single-molecule recordings, coupled with kinetic modeling, to examine how each perturbation affected gating rate constants. Results from two double-mutant cycle analyses provide strong evidence that energetic coupling between these two native residues contributes substantially to the large open probabilities and slow decay kinetics of full-length GluN1/GluN2A receptors. Given that these two residues are conserved in GluN1/GluN2B receptors, it is likely that they play a similar role in both these subtypes.
A conservative substitution at one of the residues in the pair we examined, GluN1-I642L, was previously identified as pathogenic. Our results provide a mechanistic hypothesis for the pathogenicity of this variant. We propose that the I-to-L isomerization disrupts native cross-subunit atomic interactions in GluN1/GluN2A receptors and that this subtle perturbation, by destabilizing open pore conformations, causes the dramatic change we report here in receptor function. At the microscopic level, receptors have severely reduced open probabilities because of both shorter openings and longer closures. At the macroscopic level, GluN1I642L/GluN2A receptors produce currents that are much smaller in amplitude and decay faster, and as a consequence, they mediate much-reduced charge transfer. Given the importance of these NMDA receptor biophysical properties for the receptor’s physiologic role and given the ubiquitous expression of GluN1 subunits in the central nervous system, it is plausible that the GluN1-I642L variant will cause notable changes in excitability, synaptic integration and plasticity, and information processing. Cellular consequences of these biophysical changes may include deficits in signal integration across dendritic arbors and altered short-term and long-term synaptic plasticity, with likely repercussions on overall excitability, information processing, and storage. However, given the simplified experimental conditions employed here, additional work will be necessary to evaluate the exact effects of GluN1-I642L on NMDA receptor-mediated synaptic response and which of the synaptic changes cause the clinical pathology.
In this study, we focused specifically on the contributions to NMDA receptor gating by the GluN1-I642 and GluN2A-L550 side chains and their mutual interactions. For this reason, we recorded currents in divalent-free external solutions, which allowed us to associate reliably and specifically the on/off behavior of the electrical signal with open/closed conformations of the pore and thus exclude other mechanisms of current interruption. By the same token, these controlled conditions also prevented us from observing possible effects on many other critical channel properties such as calcium permeability, calcium-dependent block and inactivation, or voltage-dependent block by magnesium, all of which have critical roles in the NMDA receptor synaptic response and its effects on brain function. We noted however, that doubly substituted receptors had lower unitary current amplitudes, suggesting possible contributions by these residues to the physicochemical properties of the permeation pathway (Table 1). Of the singly substituted constructs tested, GluN1-I642A was sufficient to produce the same reduction in current amplitude as the double alanine substitution, suggesting that GluN1 residues on M3 are dominant in setting channel permeability, consistent with previous observations from receptors with Lurcher mutations (41). This is also consistent with evidence that pharmacological modulators that target residues in the pre-M1 linker region can control channel permeation properties.
Taken together, the data and results presented here provide direct empirical support for a chemical coupling mechanism by which a residue on a TMD-LBD linker can access and interact with residues beyond the gate on M3 to contribute to NMDA receptor gating. Therefore, in addition to the direct mechanical coupling provided by the backbone of S2-M3 linkers within same-subunit LBD-TMD (14), which transduces LBD movement to the M3-located gate, we propose here that a direct and specific chemical interaction between residues on the S2-M1 linker with residues on M3, belonging to GluN1 and GluN2A subunits from separate dimers, serves to stabilize open pore conformations. Thus, LBD-TMD linkers play multiple roles in supporting the gating reaction: by providing both the necessary mechanical tension to initially open the gate and the subsequent chemical coupling with residues below the gate to stabilize open conformations (Fig. 6). Uniquely among iGluRs, NMDA receptors have strongly concerted activation mechanisms, which confine channel opening to conditions in which both agonists, glutamate and glycine, are present. Structurally, it is supported by multiple-cross subunit contacts within the NTD and LBD layers. Here, we demonstrate that the interaction between GluN1-I642 and GluN2A-L550 also provides a cross-subunit, cross-dimer interaction, which is intrinsic to the activation sequence of NMDA receptors.
Materials and Methods
Detailed methods can be found in the SI Appendix. In brief, a truncated GluN1/GluN2A homology model (GluN1 397–838; GluN2A 397–842) was generated with the SWISS-MODEL server using a GluN1/GluN2B crystal structure as template (11). Targeted molecular dynamics simulations were performed with NAMD V2.9b2 using a putative open-state structure as the target conformation (20). In all MD trajectories (700 ns for the WT and 200 ns for the mutants), only the last 150 ns was kept for energetic analysis, and the last 10 ns was used for HOLE calculation (42). To measure interresidue vdW energies, we used the NAMDEnergy module in the VMD program (43, 44). Rat GluN1-1a (U08261), GluN2A (M91561) engineered to harbor the indicated substitutions at GluN1-I642, and GluN2A-L550 were transiently expressed in HEK293 cells. Single-channel currents were recorded using cell-attached patch clamp electrophysiology in divalent-free, high-pH, saturating agonist conditions. A five-closed, two-open state kinetic model (23) was fitted to event duration histograms to derive rate constants, which were used in thermodynamic double-mutant cycle analysis as described previously (45). All simulations of macroscopic currents using empirically derived rate constants were performed in MATLAB 2017a (Mathworks) with the built-in matrix exponential function, expm.
Acknowledgements
We thank Dr. Jamie Abbott for helpful discussion and critiques of the manuscript. We thank Evan Synor and Michael Steward for contributions to data collection. The NIH supported this project through awards R01NS097016 and R01NS108750.
Data Availability.
All study data are included in the article and supporting information.
References
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
44
 Cross-subunit interactions that stabilize open states mediate gating in NMDA receptors
Cross-subunit interactions that stabilize open states mediate gating in NMDA receptors
