
Edited by Richard M. Harland, University of California, Berkeley, CA, and approved February 24, 2021 (received for review August 26, 2020)
Author contributions: B.T., J.A.D., S.C.L., and M.C.M. designed research; B.T., J.A.D., and S.C.L. performed research; B.T. and J.A.D. contributed new reagents/analytic tools; B.T., J.A.D., S.C.L., and M.C.M. analyzed data; and B.T., J.A.D., and M.C.M. wrote the paper.
1B.T. and J.A.D. contributed equally to this work.
- Altmetric
TGF-β family heterodimeric ligands show increased or exclusive signaling compared to homodimeric ligands in both vertebrate and insect development as well as in therapeutically relevant processes, like osteogenesis. However, the mechanisms that differentiate heterodimer and homodimer signaling remain uncharacterized. We show that BMP antagonists do not account for the exclusive signaling of Bmp2/7 heterodimers in zebrafish development. We found that overexpressed homodimers can signal but surprisingly require two distinct type I receptors, like heterodimers, indicating a required activity of the heteromeric type I receptor complex. We further demonstrate that a canonical type I receptor function has been delegated to only one of these receptors, Acvr1. Our findings should inform both basic and translational research in multiple TGF-β family signaling contexts.
Heterodimeric TGF-β ligands outperform homodimers in a variety of developmental, cell culture, and therapeutic contexts; however, the mechanisms underlying this increased potency remain uncharacterized. Here, we use dorsal–ventral axial patterning of the zebrafish embryo to interrogate the BMP2/7 heterodimer signaling mechanism. We demonstrate that differential interactions with BMP antagonists do not account for the reduced signaling ability of homodimers. Instead, we find that while overexpressed BMP2 homodimers can signal, they require two nonredundant type I receptors, one from the Acvr1 subfamily and one from the Bmpr1 subfamily. This implies that all BMP signaling within the zebrafish gastrula, even BMP2 homodimer signaling, requires Acvr1. This is particularly surprising as BMP2 homodimers do not bind Acvr1 in vitro. Furthermore, we find that the roles of the two type I receptors are subfunctionalized within the heterodimer signaling complex, with the kinase activity of Acvr1 being essential, while that of Bmpr1 is not. These results suggest that the potency of the Bmp2/7 heterodimer arises from the ability to recruit both Acvr1 and Bmpr1 into the same signaling complex.
TGF-β family heterodimers participate in a broad variety of developmental and physiological contexts. BMP7/GDF7 heterodimers drive axon repulsion in the roof plate of the mouse neural tube (1), while GDF9/BMP15 heterodimers drive cumulus expansion in the ovary (23–4). BMP9/10 heterodimers circulate freely in the blood plasma where they regulate angiogenesis (5), and Nodal/Gdf3 heterodimers act in both mesendoderm induction (678910–11) and the specification of the left–right axis (6, 8, 10). In particular, BMP2/7 and related BMP4/7 heterodimers are required for the development of organisms as diverse as mice (12), zebrafish (13), and Drosophila (141516–17) and have been shown to outperform homodimers in a wide variety of cell culture and in vivo contexts (18192021222324252627282930313233–34). The large number and diverse roles of TGF-β family heterodimers strongly suggest that heterodimers are a general feature of TGF-β signaling, yet the mechanistic distinctions between heterodimer and homodimer signaling remain relatively unexplored.
The basic mechanism of TGF-β family signaling is well established (35). At the surface of the receiving cell, the ligand binds two type I receptors and two type II receptors, assembling a tetrameric complex (35) (Fig. 1A). The type II receptors then phosphorylate serines and threonines in the type I receptor GS domains (35) (Fig. 1B). This phosphorylation activates the type I receptors, which in turn phosphorylate R-Smads (35). Upon phosphorylation, R-Smads complex with the co-Smad Smad4, and together they accumulate in the nucleus where they regulate gene transcription (35). The biological activity of these signaling components is highly conserved throughout the animal kingdom: Receptors and ligands from mammals can effectively rescue loss of function in zebrafish and even Drosophila embryos (13, 36373839–40).

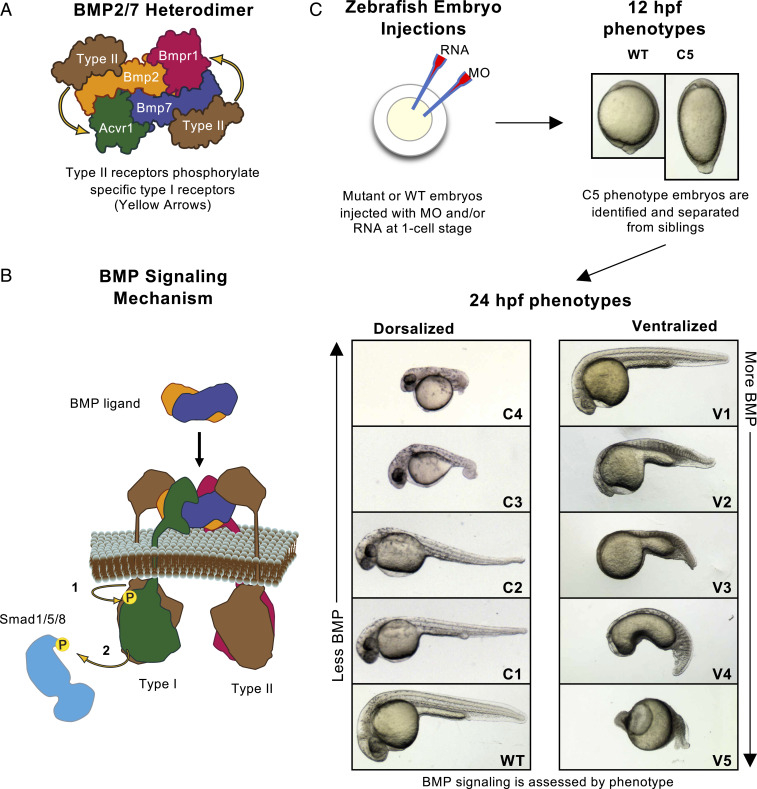
BMP heterodimer signaling and assays in the zebrafish embryo. (A) A BMP heterodimer contains four distinct receptor binding sites for two type I and two type II receptors. One type I receptor site resembles the BMP2 homodimer type I receptor binding site, predicted to bind Bmpr1. The other type I receptor binding site resembles the BMP7 homodimer type I receptor binding site, predicted to bind Acvr1. Yellow arrows indicate the type II-type I receptor interactions. (B) The BMP signaling mechanism: Type II receptors phosphorylate Type I receptors (1), which in turn phosphorylate and activate the transcription factor Smad1/5/8 (2). (C) Experimental schematic. Eggs are injected at the one-cell stage. Embryos are screened, photographed, and C5 phenotype embryos separated at 12 hpf. After 24 hpf, strongly dorsalized C5 embryos have died, and all other phenotypes are photographed and classified.
In the zebrafish, both Bmp2/7 and Nodal/Gdf3 function exclusively as heterodimers in early embryonic patterning (678–9, 13). Our laboratory previously found that Bmp2 and Bmp7 function nonredundantly in dorsal–ventral (DV) axial patterning (41, 42) and that only recombinant Bmp2/7 heterodimers can signal, whereas a combination of BMP2 and BMP7 homodimers cannot (13). Similarly, Nodal and Gdf3 are nonredundantly required for mesendodermal specification and left–right patterning (6, 7). In these contexts, Gdf3 homodimers are not secreted, while Nodal/Gdf3 heterodimers are (678–9). This differential secretion, however, does not explain the requirement of heterodimers, as Nodal homodimers are secreted but do not signal at physiological levels and show diminished activity in cell culture (7, 43). While BMP4/7 heterodimers are preferentially secreted in Xenopus (19), we do not find a preference for heterodimer versus homodimer secretion in the zebrafish embryo (13). Thus, the exclusive heterodimer signaling of these ligands likely lies downstream of secretion.
There are two points downstream of secretion at which heterodimers could outperform homodimers: heterodimers may be resistant to BMP antagonists or heterodimers may assemble a more active receptor complex than homodimers. After secretion, dimeric BMP ligands can interact in the extracellular space with a variety of extracellular antagonists (44, 45). There is evidence in other systems that BMP antagonists differentially bind homodimers and heterodimers (17, 46). In Drosophila, Dpp/Scw heterodimers (homologous to Bmp2/7 heterodimers) preferentially bind the BMP antagonist Sog (homolog to Chordin in vertebrates) during DV patterning and within the wing disk (17, 47). In human cell culture, the BMP antagonist Noggin preferentially binds BMP homodimers to heterodimers (46). It remains unclear, however, whether extracellular BMP antagonists discriminate between heterodimer and homodimer signaling during zebrafish DV patterning.
A second hypothesis is that Bmp2/7 heterodimers assemble a distinct signaling complex with increased activity (13). BMP7 has been shown to coimmunoprecipitate with and signal through ACVR1 in cell culture (4849–50). Furthermore, cell culture studies have demonstrated that BMP2 and related BMP4 homodimers signal through BMPR1 receptors (35, 49). Biochemical affinity data show that BMPR1 binds the BMP2 homodimer strongly (Kd < 1 nM), while ACVR1 binds BMP7 homodimers only weakly (Kd > 500 nM) (29, 5051525354–55). As Bmp2/7 heterodimers have both BMP2-like and BMP7-like type I receptor binding sites, it is likely that they bind both Acvr1 and Bmpr1 (Fig. 1A). Supporting this, Acvr1l (the functional ortholog of ACVR1 in the zebrafish) and Bmpr1 only coimmunoprecipitate in zebrafish embryos that produce heterodimers (13) and are nonredundantly required in zebrafish DV patterning (13, 56, 57). Interestingly, these two receptors also function nonredundantly in other BMP heterodimer signaling contexts, including mouse gastrulation (10, 12, 5859606162–63), Xenopus DV patterning (646566–67), and Drosophila DV patterning (16, 68).
Here, we use zebrafish DV patterning to examine the mechanism of Bmp2/7 heterodimer signaling. We show that homodimers do not signal efficiently, even in the absence of all antagonists, countering the hypothesis that BMP antagonists preferentially block homodimers and thus allow heterodimers to signal. We also report that, while overexpressed Bmp2 homodimers can signal, they unexpectedly require both type I receptors Bmpr1 and Acvr1l. This is very surprising because BMP2 has no measurable affinity for ACVR1 (29, 51). We further demonstrate a differential kinase function between the two type I receptors. We find that Acvr1l kinase activity is required for signaling in DV patterning, whereas Bmpr1 kinase activity remarkably is not. These findings suggest that within the Bmp2/7 heterodimer signaling complex, Acvr1 exclusively phosphorylates Smad1/5/8, while Bmpr1 performs a required, kinase-independent signaling function.
Results
Physiological Levels of Homodimers Do Not Signal Even in the Absence of Antagonists.
Zebrafish DV patterning provides an excellent in vivo system to investigate the BMP signaling mechanism. Through a combination of BMP pathway mutants, antisense morpholino (MO), and RNA injection, we can readily eliminate and replace BMP signaling components and assess their effect on endogenous signaling (Fig. 1C). By 24 h postfertilization (hpf), zebrafish embryos manifest a well-characterized, dose-dependent spectrum of BMP phenotypes (69). BMP partial and complete loss-of-function mutants display a range of dorsalized phenotypes from the weakest class 1 (C1) phenotype displaying expanded dorsally derived gastrula tissues at the expense of some ventral tissues to the strongest class 5 (C5) phenotype, exhibiting radially expanded dorsal tissues at the expense of all ventral tissue and dying by 16 hpf (Fig. 1C) (69). BMP gain of function leads to an alternative range of ventralized phenotypes, V1 to V5, lacking progressively more dorsal tissue with concomitantly expanded ventral tissue, with V5 displaying the most severe ventralized phenotype lacking all dorsal tissue and radially expanded ventral tissue (Fig. 1C) (70).
Zebrafish embryos express three BMP antagonists: chordin, noggin, and follistatin (CNF) and the combined knockdown of these antagonists strongly ventralizes zebrafish embryos (SI Appendix, Figs. S1 and S2) (71, 72). To test whether these antagonists specifically interfere with homodimer signaling, we knocked down these three antagonists (CNF) in either bmp7a mutant embryos that only express Bmp2 homodimers or bmp2b mutant embryos that only express Bmp7 homodimers (Fig. 2 A and B). We found that CNF knockdown did not restore BMP signaling in bmp7a mutant embryos (Fig. 2C, columns 3 and 5). Since bmp7a mutant embryos have less total BMP ligand than wild-type (WT) embryos, we tested if providing additional Bmp2b, an amount that rescues bmp2b mutants (Fig. 2C, columns 1 and 2), could allow Bmp2b homodimers to signal. However, we found that this additional Bmp2b did not significantly restore ventral tissues, even in the absence of CNF (Fig. 2C, columns 4 and 6). We also assessed DV marker gene expression during gastrulation (Fig. 2 D–H). In the absence of Bmp2/7 heterodimers, the anterior neural marker otx2b, normally dorsally confined by BMP signaling (Fig. 2D) expands around bmp7a mutant embryos, reflecting the loss of BMP-specified ventral tissues (Fig. 2E). This expansion of otx2b in bmp7a mutant embryos is not reduced by CNF knockdown, the addition of bmp2b RNA, nor by the combination thereof (Fig. 2 F–H), consistent with a lack of Bmp2b homodimer signaling even in the absence of BMP antagonists.

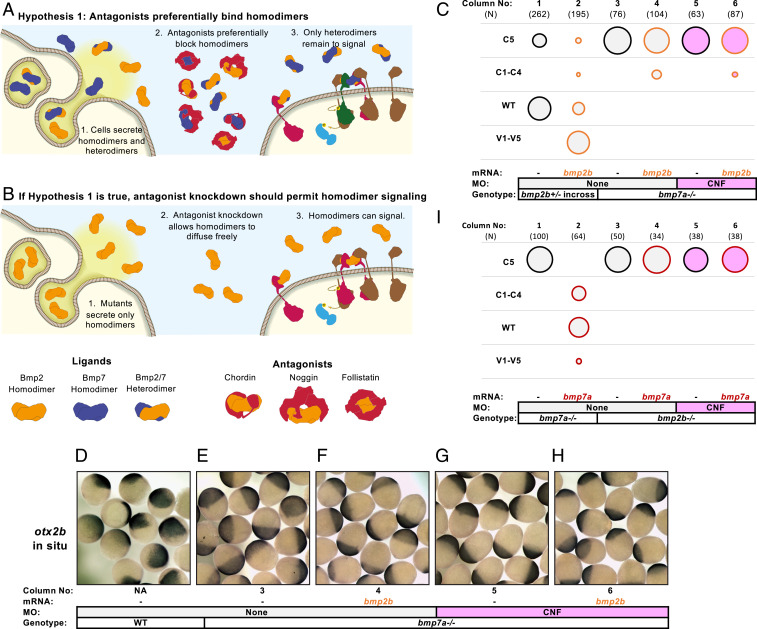
BMP homodimers fail to rescue embryos lacking BMP antagonists and heterodimers: In bubble plots, circle area reflects the percent embryos of a given phenotype (Left), fill color reflects the MO condition, and line color reflects the RNA injection condition. Ns are in brackets above each column. Injection conditions and genotypes are labeled below each column. Raw phenotype scores are shown in Dataset S1. MO concentrations and combinations are listed in SI Appendix, Table S4. Hypothesis 1: BMP heterodimers prevail because antagonists preferentially bind homodimers (A) and, if true, then BMP homodimers should signal in the absence of antagonists and heterodimers (B). (C) Bmp2 homodimers cannot signal at endogenous expression levels, even without CNF. Column 1: 1/4 of the offspring of a bmp2b+/− incross are C5 bmp2b−/−. Column 2: 15 pg FLAG-bmp2b RNA injection rescues most C5 mutant embryos and ventralizes nonmutants. Other columns: bmp2b homodimers fail to rescue bmp7a mutant embryos with or without CNF and additional bmp2b RNA. (D–H) Column numbers below images refer to the experimental condition of columns in C. The otx2b expression can be seen in the following: WT embryos (D), bmp7a mutant embryos (E), bmp7a mutants with additional bmp2b RNA (F), CNF MO-injected bmp7a mutants (G), and bmp7a mutants injected with CNF MO and bmp2b RNA (H). (I) Bmp7 homodimers cannot signal at endogenous concentrations, even without CNF. Column 1: bmp7a−/− embryos are C5 dorsalized. Column 2: 40 pg bmp7a RNA rescues DV patterning in bmp7a mutants. Other columns: Bmp7 homodimers fail to rescue bmp2b mutants with or without CNF and additional bmp7a RNA.
Bmp2b homodimers are expected to signal through a complex containing two Bmpr1 receptors (49), and the Bmp2/7 heterodimer is expected to signal through a complex containing only one (Fig. 1A) (13). It is therefore possible that zebrafish embryos do not express the sufficient Bmpr1 receptor to enable Bmp2b homodimer signaling. To test this possibility, we simultaneously provided additional bmp2b and bmpr1aa, amounts that rescue their respective loss-of-function phenotypes, in bmp7a mutants. However, we again found no sign of homodimer signaling, even in the absence of CNF (SI Appendix, Fig. S3).
We found that Bmp7 homodimers behave in a similar manner to Bmp2 homodimers. Bmp7 homodimers did not signal in patterning bmp2b mutants, even in the absence of CNF (Fig. 2I, columns 3 and 5). As with bmp2b, the addition of a rescuing amount of bmp7a mRNA (Fig. 2I columns 1 and 2) did not restore signaling (Fig. 2I column 4), even when antagonists were also knocked down (Fig. 2I column 6). Together, these results indicate that preferential binding of the BMP antagonists CNF to homodimers does not account for the exclusive signaling by Bmp2/7 heterodimers.
Overexpressed Bmp2 Homodimers Can Signal, Requiring Both Type I Receptors Bmpr1 and Acvr1.
While it is clear that endogenous levels of Bmp2b homodimer are insufficient to restore signaling in bmp7a mutants (Fig. 2C column 3), additional bmp2b messenger RNA (mRNA) did rescue a small number of these embryos to the less-dorsalized C4 phenotype (Fig. 2C, columns 4 and 6). Though our previous experiments clearly demonstrate that endogenous levels of Bmp2 homodimer cannot signal in DV patterning (13), we found that 5- to 40-fold bmp2b overexpression could signal and ventralized bmp7a mutants (Fig. 3A columns 1 and 2 and SI Appendix, Fig. S4), consistent with previous results (41, 70, 73).

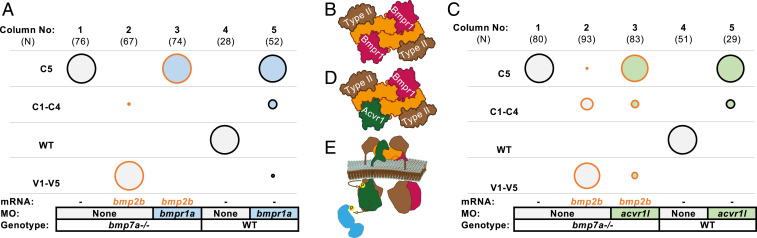
Overexpressed Bmp2 homodimers require both Bmpr1 and Acvr1 to signal: The HA-bmp2b RNA injected is 5- to 40-fold (5 to 10 pg) above the rescuing concentrations (0.25 to 1 pg) shown in SI Appendix, Fig. S4. Raw phenotype scores are shown in Dataset S2. MO concentrations and combinations are listed in SI Appendix, Table S4. (A) Overexpressed Bmp2 homodimers require Bmpr1 to signal. The bmp2b overexpression ventralizes bmp7−/− embryos (columns 1 and 2). The bmpr1a MO knockdown abates signaling caused by bmp2b overexpression (column 3). Efficacy of the bmpr1a MO mix is affirmed in WT embryos (columns 4 and 5). (B) In one model, the BMP2 homodimer forms a Bmpr1–Bmpr1 complex. (C) Overexpressed Bmp2 homodimers require Acvr1 to signal. Overexpressed bmp2b RNA ventralizes bmp7−/− embryos (columns 1 and 2). The acvr1l MO knockdown abates signaling caused by bmp2b overexpression (column 3). Efficacy of the acvr1l MO is affirmed in WT embryos too (columns 4 and 5). (D) In another model, the BMP2 homodimer binds to Acvr1 and Bmpr1. (E) Bmpr1–Acvr1 heteromeric complex is necessary for BMP2 homodimer signaling in the zebrafish.
Bmp2 homodimer signaling is expected to require Bmpr1 and to be independent of Acvr1. Previous studies show that BMP2 homodimers bind with high affinity to and signal through BMPR1 (49, 55) (Fig. 3B), and BMP2 homodimers have no detectable affinity for ACVR1 (29, 51). Depleting Bmpr1 in WT zebrafish embryos blocks endogenous BMP signaling (Fig. 3A, columns 4 and 5), as previously reported (13). To test the requirement for Bmpr1 in Bmp2 homodimer signaling, we induced Bmp2 homodimer signaling by overexpressing Bmp2b in bmp7a mutant embryos and then depleted Bmpr1. Bmpr1 deficiency completely reversed the ventralization induced by Bmp2 homodimer signaling (Fig. 3A, columns 1 to 3). Thus, Bmpr1 is required for Bmp2 homodimer signaling, consistent with previous results (49, 55).
We further tested the requirement of Acvr1 under the same Bmp2b homodimer overexpression conditions (Fig. 3C, columns 1 and 2). If Bmp2 homodimers signal exclusively through Bmpr1 receptors (Fig. 3B), Bmp2 homodimer signaling should be unaffected by Acvr1l knockdown. In vitro ACVR1 has no detectable affinity for BMP2 and binds weakly to BMP7 (29, 51). We demonstrated the efficacy of our acvr1l MO knockdown by replicating the MZ-acvr1l mutant C5 phenotype in WT embryos (Fig. 3C, columns 4 and 5) (56). Interestingly, the ventralizing activity of overexpressed Bmp2 homodimers was blocked in embryos lacking Acvr1 receptors (Fig. 3C, column 3). These results demonstrate that, surprisingly, Acvr1 is required for Bmp2b homodimer signaling, implying that Acvr1 performs an essential signaling function distinct from Bmpr1 (Fig. 3 D and E).
Acvr1 Kinase Function Is Essential for Gastrula Bmp2/7 Heterodimer Signaling.
The nonredundant requirement of both Acvr1 and Bmpr1 in Bmp2/7 heterodimer (13) and Bmp2 homodimer signaling (Fig. 3) suggests that these two type I receptors have distinct and essential functions within these signaling complexes. Canonically, type I receptors phosphorylate the Smad signal transducer (35), but it is unclear whether both type I receptors within the signaling complex perform this function. In one hypothesis, the kinase function of the type I receptors is redundant, and heterodimer signaling can proceed with either kinase. In a second hypothesis, the cumulative kinase activity of both type I receptors is needed for signaling and both kinases are required. In a third hypothesis, the kinase function is specialized to one of the receptors. In this scenario, one receptor would require kinase activity, while the other receptor would not.
We distinguished between these hypotheses by using modified, kinase-dead versions of both Acvr1 and Bmpr1. As the intracellular domains of Acvr1 and Bmpr1 are highly conserved (55% identical, 67% similar; see Methods), we generated constructs bearing two equivalent mutations in both receptors. One well-characterized mutation directly eliminates the ATP binding site (acvr1l-K232R and bmpr1aa-K256R) (7475–76). The second mutation replaces the threonines and serines in the type I receptor GS domain with inert valines and alanines, preventing the type II receptor from phosphorylating and activating the type I receptor (acvr1l-VAAA and bmpr1aa-VVAAA) (74).
To test the requirement of Acvr1 kinase function, we knocked down endogenous Acvr1l and replaced it by injecting RNA either of WT acvr1l, kinase-dead acvr1l-K232R, or GS-domain mutant acvr1l-VAAA (Fig. 4A). As expected, WT RNA restored WT morphology to Acvr1l-deficient embryos (Fig. 4A, columns 1, 5, and 6 and Fig. 4 B–D). The expression of both Acvr1l-K232R and Acvr1l-VAAA in WT embryos was mildly dominant negative, while the overexpression of WT Acvr1l was not (Fig. 4A, columns 2, 3, and 4 and SI Appendix, Fig. S5A), consistent with previous results (57). Neither Acvr1l-K232R nor Acvr1l-VAAA could rescue Acvr1l-deficient embryos (Fig. 4A, columns 7 and 8 and Fig. 4 E and F), demonstrating that Acvr1l kinase function is essential for BMP signaling.

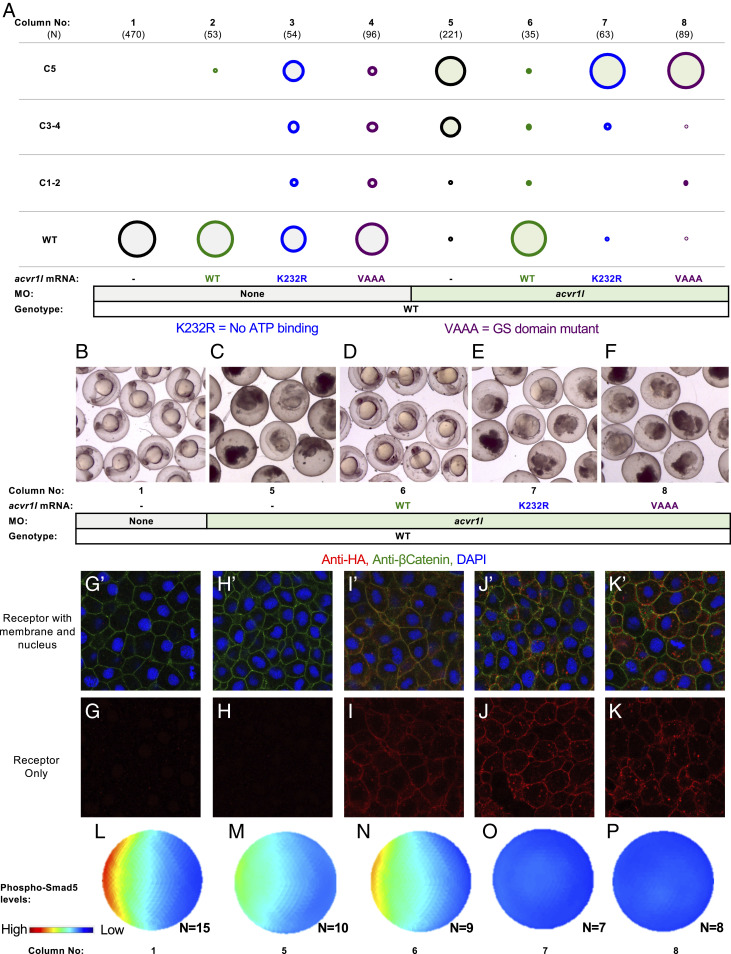
Acvr1l kinase function is essential for BMP signaling. (A) Kinase-dead Acvr1l cannot signal and restore DV patterning. WT embryos injected with acvr1l-HA RNA (100 pg), kinase-dead acvr1l-K232R-HA RNA (60 pg), and GS-domain mutant acvr1VAAA-HA RNA (100 pg), with or without acvr1l MO. MO concentrations and combinations are listed in SI Appendix, Table S4. Raw phenotype scores are in Dataset S3. (B–F) The acvr1l MO-injected embryos at ∼24 hpf. Injection conditions and corresponding columns in A are labeled below the images. Uninjected embryos are WT (B) Acvr1l-deficient C5 embryos lyse by 24 hpf (C). (D) WT acvr1l-HA RNA fully rescues Acvr1l-deficient embryos. (E and F) Acvr1l-deficient embryos injected with (E) acvr1l-K232R-HA RNA or (F) acvr1l-VAAA-HA RNA remain C5 dorsalized and lyse by 24 hpf. (G–K) Immunostaining against HA-tagged Acvr1l receptor with injection conditions labeled above the images. Embryos ≥3 were imaged for each condition. G’–K’ are the same images as G–K but showing DAPI-stained nuclei (blue) and membrane-localized β-catenin (green). (L–P) Quantified and averaged phospho-Smad5 immunostained embryos from various Acvr1l injection conditions. Corresponding columns in A are labeled on the Bottom. (L) Uninjected embryos display a WT phospho-Smad5 gradient. (M) Acvr1l-deficient embryos display reduced phospho-Smad5. (N) WT acvr1l RNA restores phospho-Smad5 signaling in Acvr1l-deficient embryos, while acvr1l-K232R RNA (O) and acvr1l-VAAA RNA (P) do not.
All acvr1l RNAs were tagged with a C-terminal HA epitope, allowing us to visualize receptor expression and localization by HA immunofluorescence. We costained early gastrula embryos with DAPI to label the nucleus and β-catenin to highlight the membrane (Fig. 4 G’–K’). From this analysis it is evident that while nonfunctional, both Acvr1l-K232R and Acvr1l-VAAA are expressed and localized to the membrane, like the WT receptor (Fig. 4 G–K).
Finally, we quantified the level of BMP signaling activity in the zebrafish embryo during gastrulation to test whether kinase-dead Acvr1l could signal at levels too low to rescue the embryonic phenotype. We immunostained embryos for phosphorylated Smad5 (P-Smad5) and then quantified the P-Smad5 gradient in control embryos (Fig. 4L), Acvr1l-deficient embryos (Fig. 4M), and Acvr1l-deficient embryos injected with either WT (Fig. 4N), acvr1l-K232R (Fig. 4O), or acvr1l-VAAA RNA (Fig. 4P) (77, 78). We found that kinase-dead and GS-domain mutant Acvr1l failed to restore P-Smad5 signaling in Acvr1l-depleted embryos. Altogether, these results demonstrate that the kinase function of Acvr1l is required for BMP signaling in DV patterning. Furthermore, it provides strong evidence against the hypothesis that the kinase functions of Acvr1l and Bmpr1 are redundant. Rather, these results are consistent either with the hypothesis that both kinases are required, or the hypothesis that kinase activity is specialized to one type I receptor.
Kinase-Dead Bmpr1 Can Restore BMP Signaling in Bmpr1-Deficient Embryos.
We next tested the kinase function of Bmpr1. While zebrafish only have one representative of the Acvr1 subfamily, acvr1l, zebrafish have four bmpr1 genes: bmpr1aa, bmpr1ab, bmpr1ba, and bmpr1bb, all of which contribute to DV patterning (13). Zebrafish bmpr1aa and bmpr1ab are orthologous to mammalian bmpr1a (also called alk3), and bmpr1ba and bmpr1bb are orthologous to mammalian bmpr1b (also called alk6). Since bmpr1a gene function contributes significantly more to DV patterning than the bmpr1b genes (13), we generated bmpr1aa; bmpr1ab double mutants (79).
Since bmpr1aa and bmpr1ab function redundantly to each other and bmpr1ab−/− adults are viable and fertile, we generated bmpr1aa+/−; bmpr1ab−/− adult fish. Incrossing bmpr1aa+/−; bmpr1ab−/− fish produced three-quarters of embryos that displayed a WT phenotype with the genotypes, bmpr1aa+/+; bmpr1ab−/− or bmpr1aa+/−; ab−/− (Fig. 5A, column 1 and Fig. 5B). The remaining one-quarter of embryos were bmpr1aa−/−; bmpr1ab−/− double mutants, which were strongly dorsalized to a C4 phenotype (Fig. 5A, column 5 and Fig. 5C). The injection of WT bmpr1aa RNA rescued these bmpr1aa; bmpr1ab double mutants to a much less dorsalized phenotype (mostly to C2, with some fully rescued to WT) (Fig. 5A, column 6 and Fig. 5D).

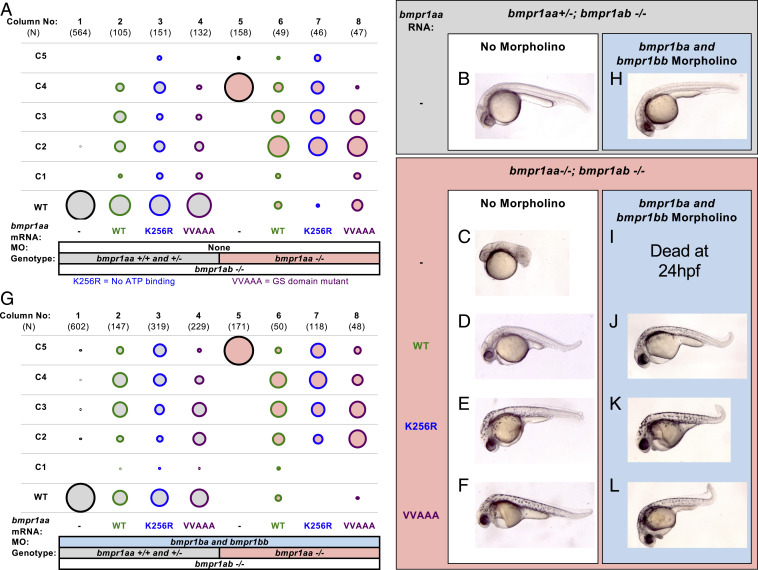
Kinase-dead Bmpr1 can restore Bmp signaling in Bmpr1-deficient embryos: (A and B) The bubble-plot fill color reflects genotype condition. Raw phenotype scores are in Dataset S4. (A) Kinase-dead Bmpr1 can rescue bmpr1aa−/−; bmpr1ab−/− double mutant embryos. Column 1: uninjected bmpr1aa+/+; bmpr1ab−/− and bmpr1aa+/−; bmpr1ab−/− embryos are phenotypically WT. Columns 2-4: bmpr1aa+/−; bmpr1ab−/− embryos injected with WT bmpr1aa RNA (40 to 80 pg) (column 2), kinase-dead bmpr1aa-K256R RNA (20 to 80 pg) (column 3), or GS-domain mutant bmpr1aa-VVAAA RNA (80 pg) (column 4). Column 5: bmpr1aa−/−; bmpr1ab−/− double mutants are C4 dorsalized. Columns 6 to 8: bmpr1aa−/−; bmpr1ab−/− double mutants injected with WT bmpr1aa RNA (column 6), bmpr1aa-K256R RNA (column 7), or bmpr1aa-VAAA RNA (column 8). (B–F) Representative-rescued embryos from bmpr1 genotypes and injection conditions in the following: a bmpr1aa+/−; bmpr1ab−/− embryo (A and B), bmpr1aa−/−; bmpr1ab−/− double mutants (C–F), uninjected (C), injected with WT bmpr1aa RNA (D), injected with bmpr1aa-K256R RNA (E), and injected with bmpr1aa-VAAA RNA (F). (G) Kinase-dead Bmpr1 rescues the simultaneous depletion of all four zebrafish Bmpr1 receptors. Experimental conditions in all columns are as in A, except that all embryos are additionally injected with a combination of bmpr1ba and bmpr1bb MO. MO concentrations and combinations are listed in SI Appendix, Table S4. H–L is the same as B–F but with additional bmpr1ba and bmpr1bb MO injection.
Unlike Acvr1l, overexpression of WT Bmpr1aa in bmpr1aa+/+; bmpr1ab−/− and bmpr1aa+/−; bmpr1ab−/− embryos was mildly dorsalizing (Fig. 5A, column 2). This may be due to the ability of Bmpr1 to bind BMP ligand with high affinity (51, 52, 54), and thus its overexpression may sequester ligand and prevent the formation of functional signaling complexes. The dominant-negative activity of WT Bmpr1aa necessitated that we titrate several concentrations of the WT RNA to identify an optimal concentration that minimized the dominant-negative effect and maximally rescued the mutant phenotype to a less-dorsalized phenotype (SI Appendix, Figs. S5B and S6). We were able to efficiently rescue these embryos to the much less dorsalized C2 phenotype (Fig. 5A, column 6 and Fig. 5D). The strongly dorsalized C4 bmpr1aa; bmpr1ab double mutants recovered a complete body axis and only lacked some ventral tail tissue (Fig. 5D). Because BMP signaling patterns tail tissues much later in development than more anterior tissues (44, 80, 81), it is likely that the optimized level of mRNA injected does not persist long enough to rescue DV patterning at these later stages of development.
To test the kinase function of Bmpr1a, we injected mRNA encoding kinase-dead or GS-domain mutant bmpr1aa and assayed their ability to rescue the C4 phenotype of bmpr1aa; ab double mutants. We found that these RNAs exhibited a similar dominant-negative activity to the WT RNA when overexpressed in bmpr1aa+/+; bmpr1ab−/− and bmpr1aa+/−; bmpr1ab−/− embryos (Fig. 5A, columns 3 and 4 and SI Appendix, Fig. S6). Surprisingly, unlike Acvr1l, both kinase-dead bmpr1aa-K256R RNA and GS-domain mutant bmpr1aa-VVAAA RNA could rescue the double mutant embryos with a similar efficiency to the WT RNA (Fig. 5A, columns 7 and 8 and Fig. 5 E and F). These results suggest that the essential function of Bmpr1 receptors during DV patterning in the zebrafish, unlike Acvr1l, is not the kinase function.
Having demonstrated that kinase-dead Bmpr1 can rescue embryos lacking Bmpr1a, we next tested whether kinase-dead Bmpr1 could also rescue embryos lacking both Bmpr1a and Bmpr1b. To simultaneously eliminate Bmpr1a and Bmpr1b, we incrossed bmpr1aa+/−; bmpr1ab−/− fish and injected the embryos with a combination of MOs targeting bmpr1ba and bmpr1bb (13). Knockdown of bmpr1ba and bmpr1bb caused no phenotype in bmpr1aa+/+; bmpr1ab−/− or bmpr1aa+/−; bmpr1ab−/− embryos (Fig. 5G, column 1 and Fig. 5H). However, it increased the dorsalization of double mutants from a C4 to a C5 phenotype (Fig. 5G, column 5 and Fig. 5I), indicating a complete loss of BMP signaling, consistent with previous results (13, 79). We found that both bmpr1aa-K256R and bmpr1aa-VVAAA RNA rescued bmpr1a double mutants, also deficient for Bmpr1b, from the most severe C5 dorsalization to a weakly dorsalized C2 phenotype, similar to WT bmpr1aa RNA (Fig. 5G, columns 6 to 8 and Fig. 5 J–L). These results show that even when all Bmpr1 receptors are deficient, kinase-dead and GS-domain mutant Bmpr1 are still capable of restoring BMP signaling in DV patterning.
These results are inconsistent with the hypothesis that the cumulative kinase activity of Bmpr1 and Acvr1 is necessary for BMP signaling. The requirement of Acvr1 kinase activity is additionally inconsistent with the hypothesis that Acvr1 and Bmpr1 kinase activity are redundant. Both observations, however, are consistent with a model in which the kinase activity is specialized to one receptor within the heterodimer signaling complex, which in this case is Acvr1.
Discussion
Here, we provide evidence that the function of the Bmp2/7 heterodimer is to recruit two distinct classes of type I BMP receptor, Acvr1, and Bmpr1, together in the same signaling complex. We find that Bmp2/7 heterodimers and, surprisingly, overexpressed Bmp2 homodimers both require Acvr1l and Bmpr1 to signal. Bmp2 homodimers at physiological levels are ineffective at signaling, even in the absence of BMP antagonists, dispelling a role for BMP antagonists in the preferential inhibition of homodimers in signaling. We also found that providing additional Bmpr1, the known Bmp2 receptor, did not facilitate Bmp2 homodimer signaling. This suggests that during DV patterning, all BMP signaling, including overexpressed Bmp2 homodimer signaling, must go through a complex containing Acvr1 and Bmpr1. Moreover, we have demonstrated that Bmpr1 and Acvr1 have distinct functions within the heterodimer signaling complex, as Acvr1l kinase activity is required while Bmpr1 kinase activity is not.
Our results demonstrate that Bmp2 homodimers can signal in the zebrafish embryo when present at much higher concentrations than heterodimers, indicating a relative lack of potency of Bmp2 homodimers. As both endogenous Bmp2/7 heterodimers and overexpressed Bmp2 homodimers require Acvr1 for signaling, this diminished potency may arise from the inability of Bmp2 to interact with Acvr1 (29, 51). Hence, Bmp2 homodimers may only signal once their concentration is high enough to force an interaction with Acvr1 (Fig. 3 D and E). While Bmp2 does not bind ACVR1 in vitro (29, 51), our results are consistent with an earlier observation in cell culture (49), in which ACVR1 knockdown reduced BMP2 homodimer signaling by 60%, suggesting that Acvr1–Bmpr1 heteromeric complexes account for a significant portion of BMP2 homodimer signaling (49). Similarly, Bmp7 overexpression can rescue bmp2 mutants in the zebrafish (37), and BMP7 homodimers can bind both BMPR1 and ACVR1 in vitro (5051–52). Future studies will be needed to determine whether overexpressed Bmp7 homodimer signaling depends on both classes of type I receptor. It is possible that all BMP signaling in the zebrafish gastrula, both by endogenous heterodimers and by overexpressed homodimers, requires the formation of an Acvr1–Bmpr1 heteromeric signaling complex.
Interestingly, within the Bmp2/7 signaling complex, the low-affinity ligand-binding receptor Acvr1 provides the necessary kinase activity, while the kinase activity of the high-affinity ligand-binding receptor Bmpr1 is dispensable (5051–52). This particular association of a low-affinity receptor with the essential signal-transducing kinase activity may enhance the responsiveness of the BMP receptor complex. Two high-affinity type I receptors could result in an overly stable, potentially hyperactive receptor complex, unable to respond to the rapidly changing signaling environment during the massive cell movements of gastrulation (82, 83). A low-affinity receptor could destabilize the complex, rendering it more sensitive to rapid fluctuations in BMP concentration (82, 83). Coupling the kinase function to the lower affinity receptor would also ensure that only fully assembled complexes signal (82, 83). Moreover, too many high-affinity receptors at the cell surface could potentially prevent BMP from diffusing throughout the embryo (83). Indeed, even slight overexpression of the high-affinity Bmpr1 receptor has a dominant-negative effect on BMP signaling (SI Appendix, Fig. S5) (13).
It is also surprising that the kinase activity of Bmpr1 is neither necessary nor sufficient to potentiate BMP signaling. In other contexts, Bmpr1 is a capable kinase, as others have observed that BMP2 homodimers can signal through a Bmpr1–Bmpr1 homomeric complex (49), and constitutively active Bmpr1 can activate BMP signaling in cell culture and within the zebrafish embryo (37, 8485–86). These results make it all the more remarkable that such Bmp2 homodimer–Bmpr1 homomeric complexes seem unable to signal in the highly conserved process of axial DV patterning.
It is possible that Bmpr1 kinase activity is blocked by the inhibitory Smad, Smad6, thus restricting the kinase function to Acvr1 in the signaling complex. Smad6 preferentially binds residues unique to BMPR1 near the ATP binding site and down-regulates BMPR1-specific signaling (85). Up-regulation of Smad6 could specifically inhibit Bmpr1 by inhibiting signaling from Bmpr1 homomeric complexes but permitting heteromeric signaling through Acvr1. Both smad6 genes, smad6a and smad6b, as well as smad7, another inhibitory Smad that can equally inhibit Bmpr1 and Acvr1, are expressed during DV patterning in zebrafish (87), but their roles in early vertebrate development remain relatively unexplored (8889909192–93).
Another mechanism that could explain the differential kinase functions may rely on the relative position of Acvr1 and Bmpr1 to the type II receptors within the signaling complex. Just as the heterodimer contains two unique type I receptor sites, each monomer contains a unique type II receptor binding site (Fig. 1A) (54). Similar to BMP type I receptors, there are two distinct classes of BMP type II receptor, BMPR2 and ACVR2 (87, 94). ACVR2 has been shown to have a higher affinity for BMP7 than BMP2, while BMPR2 has no binding preference for BMP2 or BMP7 and has a lower affinity for both ligands than ACVR2 (51, 52). Functional experiments suggest that BMPR2 is specifically required for BMP2 homodimer signaling and that ACVR2 preferentially mediates BMP7 homodimer signaling (49, 55). Crystal structures suggest that in a BMP2/7 heterodimer–receptor complex, the BMPR1 kinase domain is closest to the type II receptor bound to BMP7, and the ACVR1 kinase domain is closest to the type II receptor bound to BMP2 (Fig. 1A) (53). If type II receptors preferentially interact with the most proximal type I receptor within the complex (95), differences in the requirements of Acvr1 and Bmpr1 kinase activity may arise from their relative positions within the complex and their association with specific type II receptors.
The Bmp2/7 heterodimer mechanism is likely to be shared by many other animals. Recently, BMP2/7 and BMP4/7 heterodimers were shown to be indispensable for BMP signaling during early mouse development through studies of a dominant-negative BMP7 ligand mutant (12). Importantly, this study also confirmed the presence of BMP heterodimers in vivo in the mouse embryo by Western blot (12). The role of BMP2/7 heterodimers in early development is not limited to vertebrates, as Drosophila DV patterning also requires BMP heterodimers (16). Moreover, as in the zebrafish, Drosophila DV patterning requires the nonredundant function of two type I receptors, one from the Acvr1 class (Saxophone) and one from the Bmpr1 class (Thickveins) (68). Indeed, Acvr1 and Bmpr1 are nonredundant in almost every characterized BMP heterodimer signaling context (10, 13, 56575859–60, 64, 66, 68). BMP2/7 heterodimer signaling may even precede the bilateral body plan, as distinct representatives of both the BMP2/4 and BMP5/6/7 ligand classes as well as the Acvr1 and Bmpr1 receptor classes are present in cnidarians (96). While the two ligand classes are often coexpressed and have nonredundant roles in patterning (97), the specific function of heterodimers remains unexplored in these animals.
BMP heterodimers also exhibit increased potency in nondevelopmental contexts. In particular, BMP2/7 and BMP4/7 heterodimers are superior at inducing new bone tissue or regenerating bone tissue in a variety of cell culture and in vivo contexts (18, 2021222324252627282930313233–34). Indeed, a chimeric BMP2-Activin ligand was generated, combining the high BMPR1 affinity of BMP2 with the high ACVR1 affinity of Activin, which is both much more potent and has fewer side effects than BMP2 homodimers in a primate bone regeneration model (98). These results suggest that the synergistic relationship between Bmpr1 and Acvr1 is likely to have clinical relevance in humans.
The subfunctionalization of Bmpr1 and Acvr1 that we identified appears to act similarly in other heteromeric signaling complexes of the TGF-β family (4, 99). In the mouse ovary, BMP15/GDF9 heterodimers, also known as cumulin (2), signal through SMAD2/3 but surprisingly require BMPR1B in addition to ACVR1B/C (activin/Nodal type I receptors, also known as ALK4 and ALK7) receptors (4). Interestingly, similar to zebrafish DV patterning, BMPR1 kinase activity is not required for this signaling (4). Another example is during an epithelial–mesenchymal transition, when ACVR1 signals in the same complex as TGFBR1 (99). This particular context is somewhat different from ours, as here TGFBR1 actually phosphorylates ACVR1, yet it shows that type I receptor subfunctionalization is more wide-spread within the TGF-β superfamily. The Nodal/Gdf3 heterodimer, which signals in mesendodermal induction and left–right patterning (678–9, 43), is only known to require one type I receptor, Acvr1b, but additionally requires an EGF-CFC coreceptor (100). The specific arrangement of these components within the Nodal signaling complex remains unresolved (10), raising the possibility that, like the BMP2/7 heterodimer, the Nodal/Gdf3 heterodimer assembles an asymmetric complex.
Our results decisively shift the focus of BMP2/7 heterodimer signaling from the ligand to the receptor complex. We have shown that within this complex, the Smad phosphorylation role has been delegated to Acvr1, but the mechanisms that enforce this remain to be determined. Further studies into the specific functions of receptors within the BMP heterodimer signaling complex will not only inform BMP signaling and early development but will likely enhance our understanding of other TGF-β family signaling pathways and reveal additional targets and strategies for BMP-based therapies.
Methods
Zebrafish.
All procedures involving zebrafish were approved by the University of Pennsylvania Institutional Animal Care and Use Committee. All adult zebrafish were housed in a 28 °C facility on a 13 h light, 11 h dark cycle, in accordance with institutional and national regulatory standards. We used the mutant strains snhsb1aub/sb1aub (a null allele of bmp7a) (41), swrtd24c/+ (a null allele of bmp2b) (101), and bmpr1absa28/sa28 (a null allele of bmpr1ab) (102). An additional null bmpr1aap3 mutant allele was generated using CRISPR Cas9 technology (79). The mutant strains swrtdc24, bmpr1absa28, and bmpr1aap3 were genotyped using the Kompetitive allele specific PCR amplification (KASPar) method (LGC Genomics) (103). Information about the assay for bmpr1absa28 can be obtained from the Sanger mutagenesis project (102). KASPar primer info for bmpr1aap3 is described in (79). Sequence provided to LGC Genomics for swrtdc24 genotyping primers is: ACTTCCTGAACGAGTTTGAGCTACGCTTGCTCAATATGTTCGGATTGAAG[C/T]GAAAACCCACCCCAAGCAAATCGGCAGTGGTCCCTCAGTACATGCTGGAC. DNA from adult fin tissue or zebrafish embryos was obtained either using the HotShot method (104) or lysis buffer (15 mM Tris pH 8.3, 75 mM KCl, 2.35 nM MgCl2, 0.3% Tween 20, 0.3% Nonidet P-40, 0.0015% Gelatin) (105). Homozygous snhsb1aub embryos were rescued to adulthood by injecting bmp7a mRNA into one-cell stage embryos. The homozygous mutant genotype was confirmed by incrossing these fish and identifying those that produced 100% mutant offspring, which display a C5 phenotype (41).
Constructs and mRNAs.
All constructs were cloned into the plasmid pCS2+. The plasmids FLAG-bmp2b, HA-bmp2b, and FLAG-bmp7a are previously described (13). All Bmpr1 receptor constructs contain a carboxyl-terminal V5 tag. Overlap extension PCR was used to generate two catalytically inactive bmpr1aa variants, Bmpr1aa-VVAAA and Bmpr1aa-K256R. As bmpr1aa contains an intrinsic SP6 termination signal, reducing the efficiency of in vitro RNA synthesis, we additionally utilized overlap extension to replace this sequence with a synonymous sequence lacking the termination signal. For Bmpr1a-VVAAA, the coding sequence was changed to substitute valine and alanine for serine and threonine in the GS domain. For Bmpr1a-K256R, the coding sequence was changed to substitute arginine for lysine at position 256. All constructs are listed in SI Appendix, Table S1. All Acvr1l receptor constructs contain a carboxyl-terminal HA tag. As with Bmpr1, overlap extension PCR was used to generate two catalytically inactive Acvr1l variants, Acvr1l-VAAA and Acvr1l-K232R. For Acvr1l-VAAA, the coding sequence was changed to substitute valine and alanine for serine and threonine in the GS domain, and for Acvr1l-K232R, the coding sequence was changed to substitute arginine for lysine at position 232. PCR amplicons were cloned into pCS2+ by In-Fusion cloning (Clontech). To assure they contained the expected sequence, all constructs were sequenced. Sequence modifications to the intrinsic bmpr1aa SP6 termination site, the GS domains, and catalytic sites are listed in SI Appendix, Table S2. All mRNAs were transcribed using the SP6 mMessage mMachine kit (Ambion).
MOs.
All MOs were described previously and were obtained from Gene Tools LLC. MOs were reconstituted in Danieau solution at 25 mg/mL MO sequences are listed in SI Appendix, Table S3. All MO concentrations and mixtures are listed in SI Appendix, Table S4. MOs were sourced from the publications (13, 71, 106).
MO and mRNA Injection.
Embryos were collected for injection by 15 min postfertilization. All injections were performed in E3 media at 22 °C during the one-cell stage. For multiple injections, MO mixtures and RNAs were always kept in separate needles. This allowed us to independently test the potency of each MO mix and each RNA for every single injection. The mRNAs were diluted from stocks to working concentrations into 0.1 M KCl and 0.05 or 0.1% phenol red solution (Sigma). MOs were diluted from stocks to working concentrations in 1× Danieau and 0.05 or 0.1% phenol red solution (Sigma) (107). Each injection needle was calibrated to deliver 1 or 1.5 nL mRNA or MO. Working concentrations of all RNAs from every RNA synthesis were titrated based on their embryonic phenotypic effect. A rescuing level for HA-bmp2b, FLAG-bmp7a, bmpr1aa-V5, and acvr1l-HA mRNA was determined by injecting the mRNA into corresponding mutant or morphant embryos. For catalytically inactive acvr1l and bmpr1a variants, we injected a range of mRNA concentrations with the maximum concentrations showing a dominant-negative effect when injected into WT embryos. After injection, embryos were incubated at 28 or 31.5 °C to accelerate development. Between sphere and shield stage (4 to 6 hpf), infertile and damaged embryos were removed from the experiment.
Phenotypic Evaluation and Imaging.
Dorsalization and ventralization phenotypes were assessed between 24 and 36 hpf on an 11-point scale, with dorsalized phenotypes ranging from C5 (complete dorsalization) to WT and ventralized phenotypes ranging from WT to V5 (complete ventralization) (69, 70). As C5 embryos lyse at around 16 hpf and die before 24 hpf, all embryos were additionally observed at 10 to 14 hpf (starting at the end of gastrulation), when extreme dorsalization manifests as elongation of the body axis. Strongly dorsalized embryos were separated at this stage and kept at room temperature (24 °C) to postpone lysis. Visibly dorsalized embryos at 10 to 14 hpf that lysed by 24 hpf were counted as C5s, while other embryos that died before 24 hpf were excluded from the experiment. Incubating dorsalized embryos at room temperature also allowed for the genotyping of lysed C5 embryos, as C5s incubated at 28 °C were too decomposed to provide useful DNA.
All injected embryos were photographed twice (bright field) in E3 media with a Leica IC80HD camera, once at 12 hpf and again between 24 and 36 hpf. In experiments that required genotyping, mutant embryos were sorted by phenotype, dehydrated in methanol after the final time point, and then distributed into 96-well plates for lysis and genotyping. All phenotypic data presented were collected from at least three separate injection experiments. Results from equivalent injection conditions on different days were pooled for final presentation and analysis, provided all the controls worked.
To obtain embryo images in Fig. 4, embryos were anesthetized in Tricaine (108) mounted in 1% low melt agarose and then photographed at 4× magnification with a Leica IC80HD camera. We photographed each embryo at multiple focal planes to create depth stacks. Embryos were then removed from the agarose and genotyped. The time-consuming nature of this experiment meant that images were taken over an ∼16 h time period covering 24 to 40 hpf. After genotyping, good depth stack photos of embryos were translationally registered using the Image J plugin “Stack Reg” (109) and projected into high resolution images using the Image J plugin “Extended Depth of Field” (110). These images were minimally adjusted in photoshop to correct for white balance and to remove distracting debris from the background.
Whole-Mount In Situ Hybridization.
Embryos were collected between 80 and 90% epiboly (8 to 9 hpf) and fixed in 4% PFA phosphate-buffered saline (PBS). Whole-mount in situ hybridizations were performed as described previously (71, 81, 111). All probes, otx2b (112), dlx3b (113), tfap2a (114), and cyp26a (71), were used as previously published. Embryos were cleared and mounted in glycerol as described (111) and imaged with a Leica IC80HD camera.
Immunostaining and Imaging of Epitope-Tagged Receptors.
For the immunostaining of epitope-tagged receptors, injected embryos were collected at shield stage and fixed overnight in 4% formaldehyde in PBS. Washing, blocking, and staining was performed as described (77, 78). To visualize the HA-tagged receptors, we used rabbit anti-HA primary antibody (Invitrogen 71-5500) diluted to 1:100 and an anti-rabbit–alexa594 secondary antibody (Invitrogen A11037) diluted to 1:500. The primary mouse IgG1 anti-β-catenin (Sigma C7207) antibody diluted to 1:1,000 and the secondary anti-mouse–IgG1 alexa488 (Invitrogen A21121) diluted to 1:500 were used to visualize membrane β-catenin. The nucleus was visualized with a 1:500 dilution of 300 μM DAPI (Invitrogen D3571) in a 20 min PBS with Triton X-100 (PBST) wash immediately after the removal of secondary antibody, followed by four additional 30 min PBST washes. After staining, embryos were dehydrated first in 50% methanol in PBST, then dehydrated into 100% methanol, and stored in the dark at 4 °C before imaging.
Embryos were then cleared using BABB: a 1:2 ratio of benzyl alcohol (Sigma B-104) and benzyl benzoate (Sigma B-6630) as described (77, 78). Embryos were mounted animal pole down with silicon wafers as described (77, 78). Embryos were imaged using a Zeiss LSM880 confocal microscope with a C Plan-Apochromat 63×/1.4 Oil DIC M27 objective. DAPI was excited with the 405 nm laser at 0.2% power and detected with a gain of 618, Alexa 488 was excited with the 488 nm laser at 2.8% power and detected with a gain of 573, and Alexa 594 was excited with the 561 nm laser and a gain of 800. Images were acquired as z stacks of 20 to 40 slices, and the best slice was selected for use in Fig. 4. Image brightness was adjusted in Fiji and the 594 channel was equally brightened in all images by setting the maximum intensity to 100. Brightness in the DAPI and 488 channels was modulated to normalize differences in brightness.
Immunostaining, Imaging, and Analysis of Phospho-Smad5.
For the immunostaining and quantification of phospho-Smad5, shield-stage embryos were fixed in 4% formaldehyde PBS. Washing, blocking, and staining was performed as described (77, 78). Phospho-Smad was detected with the primary rabbit anti-PSmad1/5/9 (Cell Signaling Technology, 13820) and the secondary goat anti-rabbit alexa647 (Invitrogen A-21245) at 1:500. To visualize the nucleus, Sytox green (Fisher S7020) 1:2,000 was added simultaneously with the secondary antibody diluted in blocking solution. After staining, embryos were dehydrated and stored, as described above.
On the day of imaging, embryos were cleared with BABB as described (77, 78). Embryos were mounted, either the animal pole up or down, with silicon wafers. Imaging was performed using a Zeiss LSM880 confocal microscope with an LD LCI Plan-Achromat 25×/0.8 lmm Corr DIC M27 multi-immersion lens in the oil-immersion setting. A single bead from a calibration slide (Thermo Fisher Scientific Cat#F369009, Well A1) was imaged once each hour of imaging to account for fluctuations in laser power over time. Imaging was performed as described (78), except embryos were imaged in a single 225 × 225 mm frame, and pixel dwell time was reduced to 0.77 μs. Images of embryos were converted into tiffs and analyzed using the MATLAB analysis developed (78).
Identity and Similarity Scores.
The zebrafish Bmpr1aa and Acvr1l intracellular domains were scored for amino acid identity and similarity groups. The peptide sequences were obtained from the Ensembl database (115), with the intracellular domain defined as the first residue after the annotated transmembrane domain and extending to the C terminus. Peptide sequences were aligned using a free MAFT alignment web tool (116, 117). Identity and similarity were scored from this alignment using another webtool (118), with the default similarity groups: GAVLI, FYW, CM, ST, KRH, DENQ, and P.
Acknowledgements
This study was supported by NIH Grants R01-GM056326 and R35-GM131908, the NIH Training Program in Developmental Biology 5T32HD007516, and the American Cancer Society Postdoctoral Fellowship Grant #PF-11-134-01-DDC to J.A.D. We thank Andrea Stout for assistance with use of the University of Pennsylvania Department of Cell and Developmental Biology microscopy core confocal microscope, Derek Stemple and the Sanger Centre for sending the bmpr1ab allele, and Hannah Greenfield and William Jones for comments on the manuscript.
Data Availability
All study data are included in the article and/or supporting information.
References
1
2
3
4
5
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
107
108
109
110
111
112
113
114
116
117
 BMP heterodimers signal via distinct type I receptor class functions
BMP heterodimers signal via distinct type I receptor class functions
