
Edited by John D. Lipscomb, University of Minnesota, Minneapolis, MN, and accepted by Editorial Board Member Marcetta Y. Darensbourg November 28, 2020 (received for review July 29, 2020)
Author contributions: M.J.M., S.R.P., K.H., C.D.O., E.P.B., J.M.B., and A.K.B. designed research; M.J.M., S.R.P., K.H., C.D.O., and J.M.B. performed research; M.J.M., S.R.P., K.H., C.D.O., J.M.B., and A.K.B. analyzed data; and M.J.M., S.R.P., K.H., C.D.O., E.P.B., J.M.B., and A.K.B. wrote the paper.
1Present address: Department of Molecular, Cell, and Cancer Biology, University of Massachusetts Medical School, Worcester, MA 01605.
- Altmetric
The enzyme SznF assembles the N-nitrosourea pharmacophore of the drug streptozotocin. Its N-oxygenase domain resembles heme-oxygenase (HO) and belongs to an emerging superfamily of HO-like diiron enzymes (HDOs) with unstable metallocofactors that have resisted structural characterization. Insight into cofactor dynamics from our prior investigation of SznF’s N-oxygenation reactions suggested an approach that has yielded a structure of a functionally assigned HDO with its diiron cofactor intact. Conformational changes accompanying cofactor (dis)assembly explain its instability, and the observation of an unanticipated glutamate ligand that is conserved in only a subset of HDO sequences provides a potential basis for top-level assignment of enzymatic function. Our results thus provide a roadmap for structural and functional characterization of novel HDOs.
In biosynthesis of the pancreatic cancer drug streptozotocin, the tridomain nonheme-iron oxygenase SznF hydroxylates Nδ and Nω′ of Nω-methyl-l-arginine before oxidatively rearranging the triply modified guanidine to the N-methyl-N-nitrosourea pharmacophore. A previously published structure visualized the monoiron cofactor in the enzyme’s C-terminal cupin domain, which promotes the final rearrangement, but exhibited disorder and minimal metal occupancy in the site of the proposed diiron cofactor in the N-hydroxylating heme-oxygenase–like (HO-like) central domain. We leveraged our recent observation that the N-oxygenating µ-peroxodiiron(III/III) intermediate can form in the HO-like domain after the apo protein self-assembles its diiron(II/II) cofactor to solve structures of SznF with both of its iron cofactors bound. These structures of a biochemically validated member of the emerging heme-oxygenase–like diiron oxidase and oxygenase (HDO) superfamily with intact diiron cofactor reveal both the large-scale conformational change required to assemble the O2-reactive Fe2(II/II) complex and the structural basis for cofactor instability—a trait shared by the other validated HDOs. During cofactor (dis)assembly, a ligand-harboring core helix dynamically (un)folds. The diiron cofactor also coordinates an unanticipated Glu ligand contributed by an auxiliary helix implicated in substrate binding by docking and molecular dynamics simulations. The additional carboxylate ligand is conserved in another N-oxygenating HDO but not in two HDOs that cleave carbon–hydrogen and carbon–carbon bonds to install olefins. Among ∼9,600 sequences identified bioinformatically as members of the emerging HDO superfamily, ∼25% conserve this additional carboxylate residue and are thus tentatively assigned as N-oxygenases.
Enzymes with histidine- and carboxylate-coordinated dinuclear iron cofactors catalyze biologically, biomedically, and environmentally important reactions in organisms from bacteria to humans (1, 2). Most of these enzymes use dioxygen as a cosubstrate and cleave strong C–H, O–H, or N–H bonds in their reactions. Extensively studied members of this functional class share the conserved four-helix–bundle architecture initially recognized in the iron-storage protein ferritin (SI Appendix, Fig. S1) (3). Examples of ferritin-like diiron oxidases and oxygenases (FDOs) include the β-subunit of class Ia ribonucleotide reductase (4, 5), soluble methane monooxygenase (6789–10), stearoyl acyl carrier Δ9 desaturase (11, 12), toluene/o-xylene monooxygenase (131415–16), and arylamine N-oxygenases from the biosynthetic pathways that build the antibiotics aureothin (AurF) (171819–20) and chloramphenicol (CmlI) (21222324–25). Each of these proteins reduces O2 at its Fe2(II/II) cofactor to form a µ-peroxo-Fe2(III/III) intermediate (1). In some cases, binding of an additional protein (8) or the substrate (11) triggers this step. The initial peroxide-level intermediates can convert to more reactive high-valent [Fe(IV)-containing] complexes by reductive or redox-neutral O–O bond cleavage (4, 26272829–30). Some FDO substrates, such as those of the N-oxygenases, likely react directly with the initial midvalent peroxide adducts (181920–21, 23). Regardless of the pathway, the reactions of FDO proteins most commonly generate µ-(hydr)oxo-Fe2(III/III) product clusters, which must be reduced in situ for subsequent turnovers (1, 2). Thus, the diiron clusters of the FDOs act as stable cofactors, cycling through multiple redox states in discrete oxidative and reductive half-reactions without dissociation from the protein scaffold.
In the last decades, O2-reactive, His/carboxylate-coordinated diiron cofactors have been identified in other protein architectures, including HD domains (31), metallo-β-lactamase folds (32), TIM barrels (33, 34), and HEAT-repeat domains (35, 36). The seven-helix architecture first recognized in heme-oxygenase (HO) (SI Appendix, Fig. S1) (37) has emerged recently as a second privileged diiron scaffold that may rival the ferritin-like fold in the versatility of transformations it supports (2). At least four HO-like diiron enzymes (HDOs)—a pair of N-oxygenases (383940–41) and a pair of olefin-forming lyase/oxidase (desaturase) enzymes (424344–45)—have been described to date, and several others have been tentatively identified within biosynthetic gene clusters (SI Appendix, Fig. S2) (464748–49). The presumptive enzymatic activity of the first structurally characterized protein in this new superfamily, a Chlamydia trachomatis protein associated with death domains (CADD), is still unknown (50, 51). The first HDO to be assigned an enzymatic function, UndA, oxidatively decarboxylates dodecanoic (lauric) acid to undecene (42). An X-ray crystal structure of UndA (Protein Data Bank [PDB] ID code 4WWJ) revealed only a single iron ion with His2/carboxylate protein coordination within the relatively open HDO core, and it was initially suggested that UndA uses the observed monoiron(II) center for O2 activation (42). A pair of later studies showed, by contrast, that the iron cofactor is dinuclear (43, 44). One of these studies also characterized a µ-peroxo-Fe2(III/III) complex on the reaction pathway (44). In that work, concerted efforts to solve a structure (PDB ID code 6P5Q) with both cofactor subsites fully occupied were unsuccessful, and regions of the protein implicated in providing ligands for the second iron site (core α3) gave weak electron density, indicative of structural disorder. In roughly concurrent studies, the aforementioned pair of HDO N-oxygenases, involved in biosynthesis of streptozotocin (SznF) (38, 39) and azomycin (RohS) (41), also failed to retain their predicted diiron cofactors. For the case of SznF, multiple attempts at cofactor reconstitution, either prior or subsequent to crystallization, did not yield a high-quality structure of the holoenzyme, although partially metallated forms were successfully characterized (SI Appendix, Fig. S3) (38). The apparent conservation of diiron cofactor instability among the first several functionally assigned proteins in the emerging HDO superfamily (38, 39, 41, 42, 44) was taken as evidence that they might operate without a stably bound, in situ-recycled cofactor (40). Regardless of the reason for it, no structure of a catalytically relevant (diferrous or diferric) holoenzyme HDO complex for a functionally assigned member of this emerging superfamily has yet been reported, owing to this shared feature. The structures that do exist either lack full occupancy of the metal ions (SznF and UndA) (38, 42, 44), have persistent disorder in one or more of the ligands (SznF and UndA) (38, 42, 44), or lack corroborating evidence of the metal-ion identity and oxidation state (CADD) (50, 51). These limitations diminish the value of the existing structural data for understanding HDO reactivities and mechanisms.
SznF has, in addition to its HDO central domain, an N-terminal dimerization domain and a C-terminal monoiron cupin domain (SI Appendix, Fig. S4). The HDO domain sequentially hydroxylates each of the two unmethylated guanidino nitrogen atoms of Nω-methyl-l-arginine (l-NMA), with Nδ being targeted first, before the cupin domain rearranges the triply modified l-Arg to Nδ-hydroxy-Nω-methyl-Nω′-nitroso-l-citrulline (SI Appendix, Fig. S2), the proposed source of the N-nitrosourea pharmacophore of streptozotocin (38). The first reported structures of SznF had a single iron ion in the cupin domain but did not reveal the structure of the HDO diiron cofactor (SI Appendix, Fig. S3). In a later study, we showed that, far from being incompetent for iron binding, the HDO domain of the protein can readily acquire Fe(II) from solution and use it to capture O2 in an N-oxygenating µ-peroxo-Fe2(III/III) complex (40). Decay of this complex in the absence or presence of substrate yields an Fe2(III/III) cluster that disintegrates to uncoupled Fe(III) species in minutes to hours. Here we have used this understanding of cofactor (dis)assembly dynamics to solve the structure of SznF with its diiron cofactor intact, thus providing a view of a mechanistically relevant reactant complex for a member of this enzyme superfamily. Comparison of an improved structure of SznF with its HDO domain in the metal-free (apo) form with that of the Fe2(II/II) complex affords a detailed structural description of the process of Fe(II) acquisition and the major protein conformational changes accompanying it. Together, the results provide a structural rationale for the diiron cofactor instability seen in each of the initially described HDO systems.
Results
X-Ray Crystal Structure of SznF with an Intact Diiron Cofactor Reveals a Seventh Cofactor Ligand in the HDO Domain.
In previously reported structures of SznF (PDB ID codes 6M9S and 6M9R), the HDO domain was deficient in iron (SI Appendix, Fig. S3), despite concerted efforts to reconstitute or isolate the protein with an oxidized cofactor (38). Our recent recognition that the Fe2(III/III) complex is labile (40) suggested that crystallization of the apo protein followed by a brief soak with Fe(II) might allow for structural characterization of the holoenzyme. We produced SznF in Escherichia coli cultures supplemented with Mn(II) to limit Fe(II) uptake during overexpression and purification. We removed the Mn(II) by chelation to produce the naive [not previously reacted with Fe(II) and O2] apo protein. We then crystallized the protein anoxically and soaked crystals in a mildly acidic solution of Fe(II) for 5 to 10 min prior to collecting diffraction data (SI Appendix, Table S1). This approach yielded a 1.66-Å-resolution structure with high occupancy (67 to 100%) of all four Fe(II) binding sites in the two HDO domains of the SznF dimer (PDB ID code 6VZY) (Fig. 1A). We verified the identity of the Fe2(II/II) cofactor by collecting anomalous diffraction datasets at the Fe and Mn X-ray K-edge absorption peaks (Fig. 1A and SI Appendix, Fig. S4). The structure confirms that the diiron cofactor is held within core helices 1 to 3 by the conserved 3-His/3-carboxylate ligand set (SI Appendix, Fig. S1) predicted from sequence alignments of known HDOs.

![X-ray crystal structures of Fe2(II/II)–SznF (A) and apo SznF (B) reveal a conformational change in core α3 (C and D) upon iron binding. 2FO − FC maps (purple or pink mesh) are shown contoured at 1.5σ [Fe2(II/II)–SznF] or 1.0σ (apo SznF). An iron anomalous difference map (orange mesh) is shown contoured at 3.0σ. Selected amino acid side chains are shown in stick format. Fe(II) ions and water molecules are shown as orange and red spheres, respectively.](/dataresources/secured/content-1765847508655-72160df9-0c14-42aa-a7e9-c1d2a5852604/assets/pnas.2015931118fig01.jpg)
X-ray crystal structures of Fe2(II/II)–SznF (A) and apo SznF (B) reveal a conformational change in core α3 (C and D) upon iron binding. 2FO − FC maps (purple or pink mesh) are shown contoured at 1.5σ [Fe2(II/II)–SznF] or 1.0σ (apo SznF). An iron anomalous difference map (orange mesh) is shown contoured at 3.0σ. Selected amino acid side chains are shown in stick format. Fe(II) ions and water molecules are shown as orange and red spheres, respectively.
In addition to verifying the six predicted iron ligands, the structure identifies an additional coordinating side chain, Glu189. CADD and UndA—two other structurally characterized HDOs—lack a coordinating residue at this position (SI Appendix, Fig. S5). The SznF Glu189 carboxylate binds in bidentate mode to Fe1 (Fig. 1A and SI Appendix, Fig. S6). Interestingly, FDO N-oxygenases, such as AurF and CmlI, have an analogous extra Fe1 ligand relative to their C/O–H–cleaving counterparts (SI Appendix, Figs. S1 and S7) (17, 22). In the FDOs, the extra ligand is a His provided by the same core helix (core α4) that harbors the Glu and His ligands in the second of the tandem pair of EXXH motifs that identifies the scaffold (SI Appendix, Fig. S1). In the HDO enzyme, SznF, Glu189 is instead contributed by one of the auxiliary helices (aux α2) (SI Appendix, Figs. S1 and S7). Despite these differences, the extra ligand serves to differentiate the cofactor subsites in both scaffolds, giving a higher coordination number for Fe1 than for Fe2. Comparison of the occupancies of the two cofactor subsites in SznF—90 to 100% for subsite 1 versus 60 to 80% for subsite 2—implies that subsite 1 has greater iron(II) affinity, as one might expect in light of the increased coordination number of Fe1. In the SznF Fe2(II/II) cofactor, the six-coordinate Fe1 has a distorted octahedral geometry, whereas the five-coordinate Fe2 has roughly square-pyramidal symmetry. Each site additionally coordinates a water molecule trans to a His ligand (Fig. 1A and SI Appendix, Fig. S6). Interestingly, these presumably labile waters are separated by a distance of only 2.8 Å, and they project toward one another into a solvent-lined cavity that could potentially harbor the l-NMA or Nδ-hydroxy-Nω-methyl-l-arginine (l-HMA) substrate (see below). Their locations might approximate the positions of the O atoms of the bridging peroxide in the detected intermediate. However, formation of a µ-peroxo-Fe2(III/III) complex would likely require significant contraction along the Fe–Fe vector: The ∼5.0-Å separation observed in the structure is much greater than those found in most structurally characterized µ-peroxo-Fe2(III/III) complexes with similar ligand sets (<3.4 Å) (1).
The long intermetallic separation in reduced SznF is enforced by both local and global structural features. The cluster has a single bridging ligand, the carboxylate of Glu215 from core α2, which coordinates in a µ-1,3 mode. By contrast, reduced complexes of FDOs generally have shorter metal–metal distances (<4 Å) with additional bridging protein ligands (either two or three bridging Glu side chains) (1). In some cases, a μ-η1,η2 mode of one of these carboxylates is associated with contraction along the Fe–Fe axis (52). No such bridge is present in reduced SznF (SI Appendix, Fig. S7). The extended Fe–Fe vector in SznF may arise from irregularities within the four helices that provide the ligands. Aux α2 and core α1 contain significant loop/turn interruptions. Core α2 and core α3 exhibit more subtle deviations from canonical backbone H bonding that result in curvature of the helical axis. These irregularities would, in general, be expected to increase flexibility in the vicinity of the diiron cofactor (SI Appendix, Fig. S8). Interestingly, three of the four ligand-providing helices of the HDO scaffold face the solvent. Their surface exposure, coupled with the aforementioned structural distortions, likely underpins the unusual metal-binding properties of SznF. The large separation of the Fe(II) ions and single carboxylate bridge connecting them may also contribute to the observed cofactor instability (40).
Visualization of a Conformational Change in Core Helix 3 Accompanying Diiron Cofactor Assembly.
To understand the structural changes that accompany diiron cofactor assembly in SznF, we solved the structure of the naive apo protein (PDB ID code 6XCV), omitting the Fe(II) soak before harvesting the crystals for data collection. The resulting diffraction datasets, collected to a resolution of 1.8 Å, yielded more well-defined electron density for the protein than prior datasets with an apo HDO domain (Fig. 1B). The entire segment spanning core helix α3—both backbone and side chains—could be definitively modeled, whereas it had previously exhibited partial disorder (SI Appendix, Fig. S3). In this apo structure, the segment containing residues 311 to 318, with three cofactor ligands (His311, Asp315, and His318), adopts an ordered but nonhelical conformation. By contrast, in the iron-bound structure, the same stretch is essentially α-helical (with three canonical backbone interactions absent), and core α3 is uninterrupted. Remodeling of secondary structure in this region thus appears to be a key part of diiron cofactor assembly in SznF (Fig. 1 C and D).
In addition to core α3 backbone remodeling, substantial movements of cofactor-coordinating side chains are associated with iron binding. While the histidine and carboxylate ligands contributed by aux α2, core α1, and core α2 adopt new rotamers, the ligands in core α3 change position even more drastically. Asp315 undergoes the greatest shift, moving ∼7 Å from the surface of the protein to fold into the cavity that harbors the diiron cofactor. Interestingly, this side chain is a ligand only to Fe2, the subsite of lesser occupancy. Repositioning of Asp315 is likely a crucial part of configuring the second subsite. The structure suggests a multistep assembly pathway in which loading of subsite 1 initiates refolding of core α3 to bring subsite 2 ligand Asp315 into position to coordinate the second Fe(II).
Discussion
Role of Core α3 Dynamics in Diiron Cofactor Instability of HDOs.
Comparison of the structure of core α3 in the HDO domain of SznF with those of its counterparts in UndA and CADD (Fig. 2 A–H) reveals correlations that may be functionally significant. CADD appears to be an outlier in the emerging superfamily (at least at this early stage) in having a stable cluster that allows it to be crystallized with bound metal (50). As in Fe2(II/II)–SznF, core α3 of CADD, which we presume to have been characterized in its oxidized Fe2(III/III) state, is fully α-helical in the published structure. By contrast, UndA shares with SznF the property of diiron cofactor instability and still has not been crystallographically characterized with its diiron cluster fully intact (44). Core α3 in the UndA structure is partially disordered, as in the previously reported structures of SznF (38), and the Asp315 equivalent, Asp198, resides in or near this unmodeled stretch (SI Appendix, Fig. S5). Substitution of this residue abolished activity in UndA, implying that it is a diiron cofactor ligand, and its disorder suggests a conformational change during assembly of the O2-reactive complex (44), as proposed here for SznF. In UndA, an additional event, fatty acid (substrate) binding, is necessary to produce the reactive state. It is not known whether substrate binding precedes, interrupts, or follows binding of the two Fe(II) ions in UndA. It seems likely that Fe(II) binding in subsite 2 with coordination of the mobile core α3 carboxylate—brought into place by a conformational change driven by binding of Fe(II) (to subsite 1) and/or the substrate—could be an important step, as in SznF. Moreover, the structural features associated with conformationally gated Fe(II) binding in subsite 2 may be conserved across the larger superfamily. Core α3 is completely surface-exposed in all three structurally characterized HDOs. The mobile segment in SznF core α3 (HIHIDQHH) consists entirely of polar or β-branched side chains. Burial of such a sequence in the interior of an α-helix might be disfavored in the absence of compensating metal–ligand interactions. Alignment of the corresponding sequences in the functionally assigned HDOs (Fig. 2I) shows that all are rich in polar and/or branched amino acids.

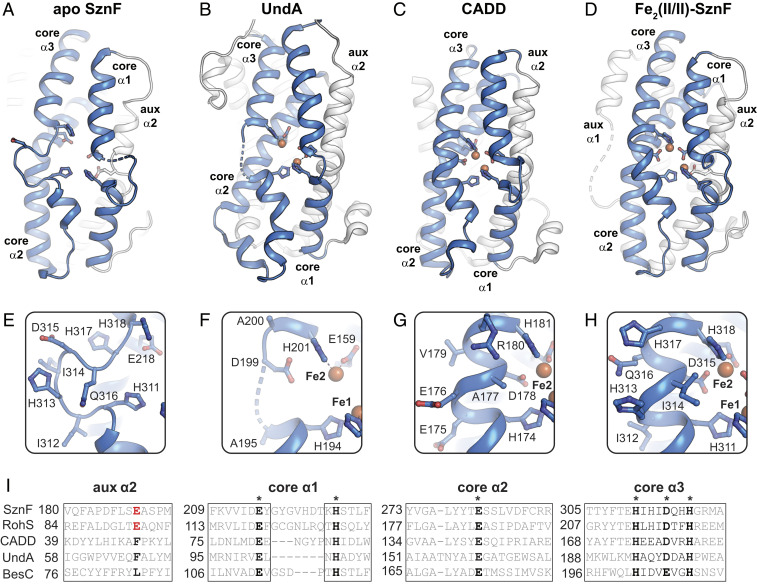
Structural comparison and sequence alignment of functionally diverse HDO proteins. Apo SznF (A and E) (PDB ID code 6XCV) (38) and UndA (B and F) (PDB ID code 6P5Q) (44) X-ray structures lack full-occupancy cofactors and exhibit disrupted or disordered regions in core α3. CADD (C and G) (PDB ID code 1RCW) (50) and the iron-bound form of SznF (D and H) (PDB ID code 6VZY) contain full-occupancy diiron cofactors and maintain a canonical α-helical structure throughout core α3. A sequence alignment (I) of characterized N-oxygenases (SznF, RohS), oxidative desaturases (BesC, UndA), and HDO protein of unknown function (CADD) shows that the Glu ligand in aux α2 of SznF is conserved in the N-oxygenases but absent in CADD and the desaturases. However, all enzymes conserve branched and polar amino acids in the metal-binding region of core α3, suggesting that all HDOs characterized to date are equipped to employ the conformationally gated iron-binding mechanism described here for SznF.
Identification of the Substrate Binding Site in the SznF HDO Domain by Computational Modeling Guided by Structural Comparisons.
Cavity-mapping analysis (53) of the Fe2(II/II)–SznF structure revealed a pocket near the cofactor with sufficient volume (>1,000 Å3) to accommodate the substrate (SI Appendix, Fig. S9). The pocket resides on the same face of the metal cluster as the solvent ligands postulated to approximate the positions of the peroxide oxygen atoms in the intermediate. Substrate bound within this pocket would be ideally poised for direct nucleophilic attack on the peroxide, as proposed for the FDO N-oxygenases (18, 21). Despite the ionic character of both SznF HDO substrates, this hypothetical binding site is largely hydrophobic. Only a single charged side chain, that of Asp185, projects into the pocket. Asp185 resides on aux α2, which also contains the Glu189 ligand. Aux α2 is interrupted near these side chains by a type II β-turn insertion (Fig. 3A). Glu189 is located at the tip of this turn. The motif both enables a closer approach of the coordinating carboxylates and exposes additional backbone polar functional groups that could interact with the substrate. This interruption could at least partly complement the charges of the substrates via exposed helical dipoles. Specifically, the bisection of aux α2 by the β-turn exposes negative and positive dipoles that could orient near the α-ammonium and α-carboxylate groups, respectively. The turn is flanked at its C terminus by Met193. Along with other hydrophobic groups in the substrate pocket, the Met193 side chain could contribute to selectivity for the N-methylated l-Arg over the more prevalent, unmethylated form. Thus, aux α2 not only delivers a ligand that was not anticipated from sequence comparisons with other HDOs but may provide multiple substrate-binding determinants.

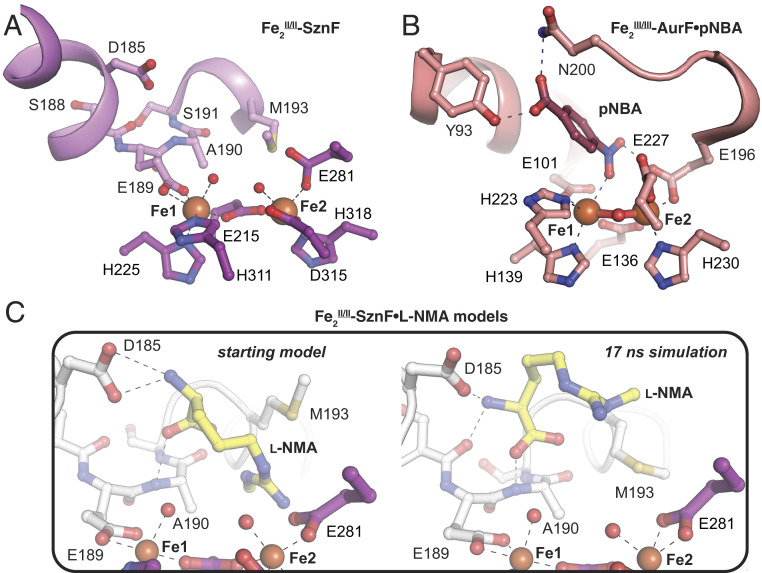
In aux α2 (violet in A; white in C), E189 contributes an unanticipated seventh ligand. Other predicted substrate-binding determinants are also present in this motif. A comparison of the metal coordination environment and putative substrate- or product-binding pockets in SznF (PDB ID code 6VZY) (A) and AurF (PDB ID code 3CHT) (B) reveals similarities between the HDO and FDO N-oxygenases. An energy-minimized docking model of the SznF substrate, l-NMA, before (Left) and after (Right) molecular dynamics simulation (C) shows that substrate H-bonding interactions with aux α2 can be accommodated. Selected amino acids and substrates are shown in stick format. Fe(II) ions and water molecules are shown as orange and red spheres, respectively.
Initial efforts to obtain an experimental structure of SznF in complex with one of its substrates (l-NMA or l-HMA) have been unsuccessful. In an attempt to gain further insight into the enzyme–substrate interactions, we used computational docking and molecular dynamics to assess whether and how the aux α2 functional groups contribute to binding and proper orientation of l-NMA. We reasoned that the substrate α-amine and α-carboxylate groups might interact with the side chain of Asp185 and the backbone amide of Ala190, respectively. We subjected an energy-minimized docking model of this Fe2(II/II)–SznF•l-NMA complex to molecular dynamics simulations (Fig. 3C and SI Appendix, Fig. S10), showing that the substrate can be accommodated in the open cavity above the diiron cofactor while maintaining the predicted contacts with the Asp185 carboxylate and Ala190 amide nitrogen within the β-turn. To test this model, we prepared the D185L variant of SznF. The variant protein supports accumulation of the µ-peroxo-Fe2(III/III) complex upon exposure of its Fe2(II/II) complex to O2, as seen in stopped-flow absorption experiments analogous to those in the published work on the wild-type (WT) enzyme (SI Appendix, Fig. S11A) (40). The kinetics of formation and decay of the intermediate in the D185L SznF reaction are very similar to those seen with the WT protein (SI Appendix, Fig. S11 A, Inset). In sequential-mixing experiments, the preformed intermediate was found to react with l-NMA extremely sluggishly (SI Appendix, Fig. S11B), with an observed first-order rate constant (kobs) linearly dependent on the substrate concentration [i.e., showing no sign of saturation, by contrast to the behavior of the WT protein (40)] (SI Appendix, Fig. S11C). The second-order rate constant for the reaction in the D185L variant is less than that for the reaction in the WT protein by a factor of almost 4,000. This deficit corresponds to a loss of ∼4 to 5 kcal/mol of binding energy caused by the substitution, consistent with loss of the carboxylate–ammonium charge pair in the enzyme–substrate complex predicted by the docking model.
The model of the l-NMA complex provides a basis to compare the substrate-binding pocket in SznF with those in other HDOs and FDOs. Some published structures of UndA include bound lauric acid (SI Appendix, Figs. S5 and S12) (42, 44) and, although none of these have high occupancies of iron subsite 2, they do reveal direct coordination of the substrate carboxylate by Fe1, in approximately the same location as the Glu189 ligand in SznF. Correspondingly, aux α2 of UndA lacks the β-turn insertion that harbors the extra carboxylate ligand and is instead uninterrupted in all published structures (42, 44). The carboxylate of Glu189 in SznF and the substrate carboxylate in UndA might thus have, in a sense, analogous functions. If this carboxylate–Fe1 interaction is necessary to set the geometry and tune the electronic structure of the cofactor for O2 capture, the difference between the UndA and SznF structures would explain why the O2-addition step is markedly accelerated by substrate binding in the former HDO (44) but not in the latter (40). Despite this difference, aux α2 of UndA, like the corresponding helix in SznF, contributes groups that interact with the substrate: The hydrophobic side chains of Val65 and Phe68 both contact the alkyl chain of the bound lauric acid (42, 44). The comparison suggests that analysis of aux α2 sequences in HDOs might aid in identifying their substrates and perhaps engineering their specificities and outcomes.
Although the FDO and HDO scaffolds have different topologies, the N-oxygenases in the two superfamilies appear to use a conserved coordination sphere (3-His/4-carboxylate) that, in both cases, includes an increased coordination number of their Fe1 site in comparison with their C/O–H–activating counterparts (SI Appendix, Figs. S1, S7, and S12). However, in the FDOs, the extra ligand resides on a core helix that, across the entire superfamily, contributes two other cofactor ligands (17), whereas, in SznF, it extends from an auxiliary helix that contributes only this ligand, and only in a subset of the superfamily (SI Appendix, Fig. S1). AurF is the only FDO N-oxygenase crystallographically characterized to date with its product bound (17). The two systems share a similar relative disposition of the substrate/product and cofactor (Fig. 3 A and B). In AurF, the 4-nitrobenzoic acid (pNBA) product is held over Fe1 by polar contacts between the carboxylate of the product and side chains of Tyr93 and Asn200 from core helices α1 and α3, respectively. In the Fe2(II/II)–SznF•l-NMA model, the approach and number/type of contacts are similar. Our model predicts that the substrate is held over Fe1 by the aforementioned interactions with its α-ammonium and α-carboxylate groups. The FDO N-oxygenases have also resisted characterization in their substrate-bound forms, perhaps due to relatively low substrate affinities. In both the FDO and HDO systems, the failure of the substrate to impact the kinetics of O2 addition may indicate that this step precedes substrate binding in the preferred pathway (18, 21, 40). In this scenario, the Fe2(II/II)–SznF•l-NMA complex would be out of sequence and challenging to capture in the crystal.
Bioinformatic Analysis of the HDO Superfamily.
Comparison of the known structural and functional characteristics of SznF and UndA highlights potentially predictive correlations. The presence of the carboxylate ligand from aux α2 may 1) obviate direct substrate coordination to the diiron cofactor and thus its triggering of O2 addition and 2) stabilize the diiron–O2 adduct with respect to O–O bond cleavage, favoring oxidation only of substrates that can attack as nucleophiles. Conversely, the absence of the extra carboxylate should leave this site vacant for substrate coordination, triggering of O2 addition, and potential progression to a more reactive complex capable of H• abstraction from an aliphatic carbon (e.g., C3 of lauric acid, as proposed in recent studies of UndA) (44). We searched sequence databases to segregate potential HDOs into groups that conserve the aux α2 β-turn insertion harboring the extra Glu from those that do not. In a sequence similarity network (SSN) generated from the ∼9,600 predicted HDO proteins that we identified in sequence databases (Fig. 4), the reference sequences (SznF and RohS [validated N-oxygenases], UndA and BesC [validated desaturases], and CADD [unknown function]) are located in different groups, consistent with their distinct activities and targets. Within the top 80 clusters in the network (ranked by number of sequences), we searched for conservation of an aux α2 β-turn and coordinating carboxylate by manual alignment to SznF and other reference sequences. Predicted protein sequences in eight of these clusters (clusters 1, 7, 9, 11, 17, 22, 70, and 76) conserve features of the insertion/extra-ligand motif (SI Appendix, Fig. S13 and Table S2). Clusters 9 and 11 contain the known N-oxygenases RohS and SznF, respectively. We propose that the other six clusters might also contain heteroatom oxygenases with properties similar to those of SznF. One of these groups, cluster 1, is the largest of the SSN, containing 1,780 sequences, roughly 20% of the HDO sequence pool evaluated here. Cluster 1 is enriched in sequences from nonpathogenic Streptomyces and Mycobacterium organisms (SI Appendix, Fig. S13). Each of the remaining four clusters contains 50 to 200 sequences, and these are similarly dominated by predicted proteins from environmental aquatic and soil microbes, as opposed to pathogens or commensal bacteria. Apart from cluster 11, in which all sequences are sufficiently long (SI Appendix, Fig. S14) to suggest fusion to a separate (e.g., cupin) domain, the remaining predicted N-oxygenases are uniformly shorter, indicating that they likely act as simple N-oxygenases. Testing of these top-level functional predictions will almost certainly expand the known repertoire of the emerging HDO superfamily.

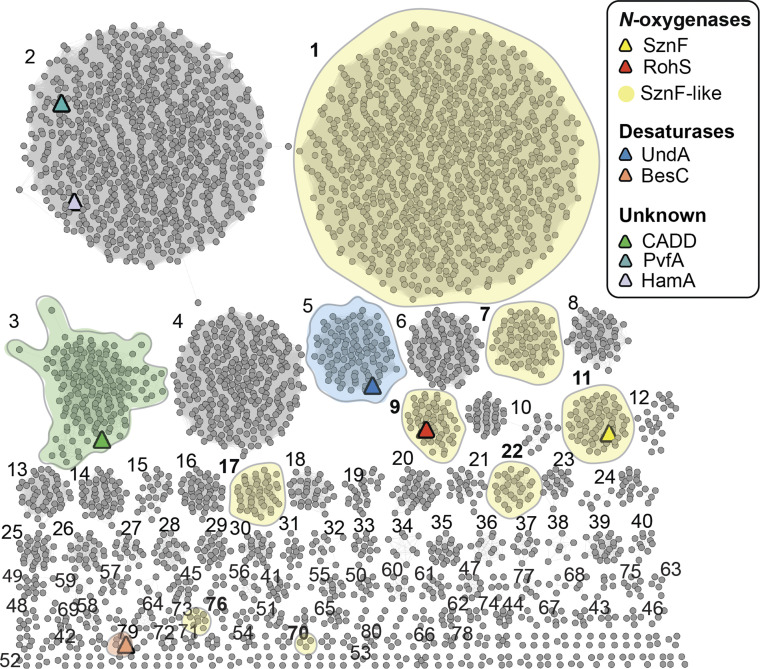
Bioinformatic analysis of the HDO superfamily. A sequence similarity network of 9,600 predicted HDO sequences (alignment score 51) shows that many sequence clusters remain uncharacterized at the structural and/or functional level. Homologs of known N-oxygenases (red, yellow), oxidative desaturases (blue, peach), or proteins of unknown function (green, lavender, teal) are indicated by colored triangles. Clusters designated SznF-like are shaded yellow and numbered in bold, indicating conservation of the extra Glu ligand in aux α2.
Materials and Methods
Detailed materials and methods are provided in SI Appendix.
X-Ray Crystallographic Characterization of Apo and Fe2(II/II)–SznF.
Apo N-terminally His6-tagged SznF was overexpressed and purified as detailed in SI Appendix. SznF was crystallized under anoxic conditions in a stringently apo form in 16% polyethylene glycol 4000, 0.1 M MgCl2, and 0.1 M Tris, pH 8.5. To obtain the Fe(II)-bound form of the enzyme, crystals were soaked for 5 to 10 min in precipitant solution supplemented with 10 mM ferrous ammonium sulfate. Data collection and refinement procedures are described in SI Appendix.
Bioinformatic Analysis of HDO Enzymes.
An initial pool of 42,581 unique sequences annotated as either heme-oxygenase or heme-oxygenase–like proteins was extracted from InterPro (November 2019 release). Using length filters and a custom Python script, this pool was analyzed for conservation of the HDO core helix diiron ligands (SI Appendix, Fig. S1) to obtain a final set of ∼9,600 candidate HDO enzymes as input for SSN generation with the Enzyme Function Initiative enzyme similarity tool (54). Using manual and automated sequence alignments and homology model generation, 8 of the top 80 (ranked by number of sequences) sequence clusters were annotated as likely N-oxygenases on the basis of conservation of the additional carboxylate ligand and β-turn motif in aux α2.
Acknowledgements
We gratefully acknowledge use of the resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by the Argonne National Laboratory under Contract DE-AC02-06CH11357. Use of Life Sciences Collaborative Access Team Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). The National Institute of General Medical Sciences and National Cancer Institute Structural Biology Facility at the Advanced Photon Source (GM/CA@APS) has been funded in whole or in part with federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). The Eiger 16M detector at GM/CA@APS was funded by NIH Grant S10 OD012289. This work was additionally supported by NIH Grants GM119707 (to A.K.B.) and GM138580 (to J.M.B.).
Data Availability.
The X-ray crystallography datasets reported in this article have been deposited in the Protein Data Bank (ID codes 6XCV and 6VZY).
References
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
34
35
36
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
 Structure and assembly of the diiron cofactor in the heme-oxygenase–like domain of the N-nitrosourea–producing enzyme SznF
Structure and assembly of the diiron cofactor in the heme-oxygenase–like domain of the N-nitrosourea–producing enzyme SznF
