
- Altmetric
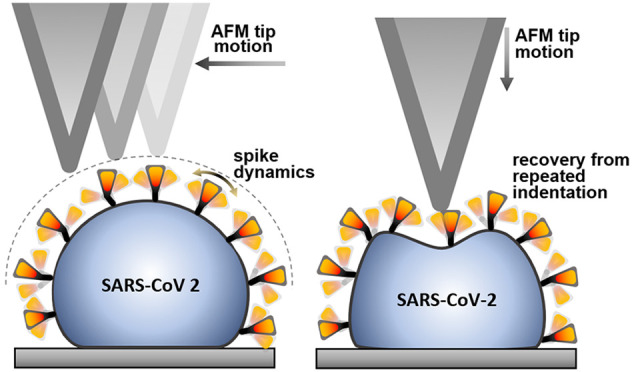
SARS-CoV-2, the virus responsible for the current COVID-19 pandemic, displays a corona-shaped layer of spikes which play a fundamental role in the infection process. Recent structural data suggest that the spikes possess orientational freedom and the ribonucleoproteins segregate into basketlike structures. How these structural features regulate the dynamic and mechanical behavior of the native virion are yet unknown. By imaging and mechanically manipulating individual, native SARS-CoV-2 virions with atomic force microscopy, here, we show that their surface displays a dynamic brush owing to the flexibility and rapid motion of the spikes. The virions are highly compliant and able to recover from drastic mechanical perturbations. Their global structure is remarkably temperature resistant, but the virion surface becomes progressively denuded of spikes upon thermal exposure. The dynamics and the mechanics of SARS-CoV-2 are likely to affect its stability and interactions.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the infective agent behind the current coronavirus disease (COVID-19) pandemic,1,2 is an enveloped ssRNA virus with a corona-shaped surface layer of spikes that are thought to play an important role in the infection mechanism.3−7 Structural information about the spike protein has been acquired either on crystals of purified protein5,8−10 or on fixed and frozen virus particles.11−13 It has been suggested that the spike hinges provide structural flexibility.11,12 High-resolution cryoelectron tomography observations indicate that the ribonucleoprotein (RNP) of SARS-CoV-2 is partitioned into spherical, basketlike structures.13 However, the surface dynamics and mechanical properties of native virions remain to be understood. Here, we employed atomic force microscopy (AFM) and molecular force spectroscopy14−16 to investigate the topographical and nanomechanical properties of native SARS-CoV-2 virions immobilized on an anti-spike-protein-functionalized substrate surface. The unique single-particle approach revealed that the surface layer of spikes on SARS-CoV-2 is highly dynamic; the virion is unusually compliant and resilient, and it displays an unexpected global thermal stability.
The topographical structure of individual SARS-CoV-2 virus particles bound to the substrate surface was imaged using AFM (Figure 1a). To increase the efficiency and specificity of binding, we employed a monoclonal anti-spike-protein antibody, which resulted in a nearly 100-fold enhancement in the density of substrate-bound virions (Figure S1). AFM images of glutaraldehyde-fixed SARS-CoV-2 revealed virions (Figure 1b) with somewhat variable dimensions (Table S1) and a rugged surface (Figure 1c). The mean central height of the virions (Figure S2), the structural parameter least sensitive to AFM tip convolution, was 62 nm (±8 nm, standard deviation (SD)) The height was smaller than the virion diameter measured in cryo-electron microscopic images,11−13 suggesting that the virus particles were partially flattened on the substrate. The 3D-rendered AFM image (Figure 1d) supported this interpretation and revealed that the rugged surface is due to the presence of protrusions which we identify as the spikes (S protein trimers) based on earlier topographical results on SARS-CoV17,18 and more recent cryo-electron microscopic11,12 data on the whole SARS-CoV-2 virion. In high-resolution (pixel size 5 Å) AFM images (Figure 1e and Figure S4a–d), individual S trimers could sometimes be resolved based on the characteristic triangular appearance. Visual inspection of the S trimers pointed at their positional (Figure S3c), flexural (Figure S3d), and rotational (Figure S4e) disorder in the viral envelope. The mean nearest-neighbor distance between the S trimers and their topographical height were 21 nm (±6 nm, SD) and 13 nm (±5 nm, SD), respectively (Figure S3 and Table S2). From the mean nearest-neighbor distance and the virion dimensions, and presuming that the spikes remain evenly distributed over the virus surface during sample preparation, we calculated that an average of 61 spikes cover the SARS-CoV-2 virus particle surface. This number exceeds those reported recently (24,11 26,13 and 4012), suggesting that the spike number is highly variable and may be regulated during virus assembly and maturation in the host cell.19 The flexural disorder observed here supports the interpretation of cryo-electron microscopic data,11−13 revealing a high degree of spike flexibility. We propose that the positional and rotational disorder of S trimers is due to their mobility in the virus envelope.

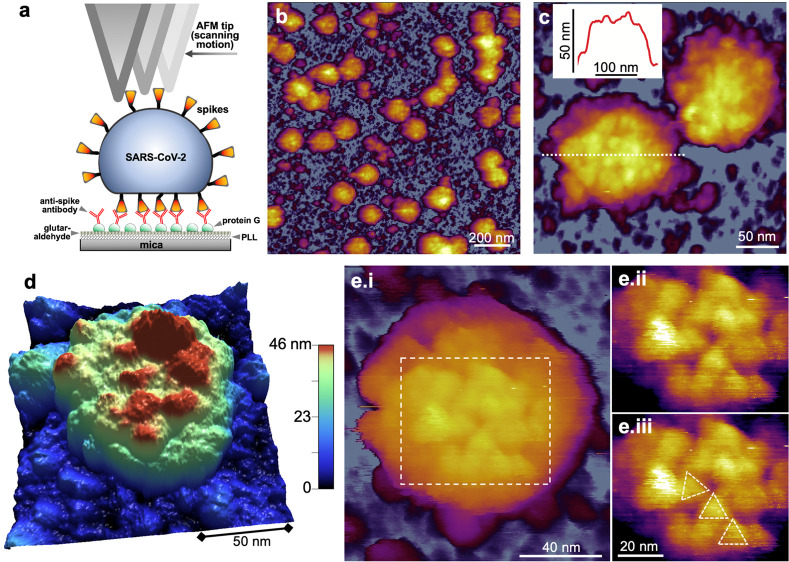
Topographical structure of SARS-CoV-2 virions treated with 5% glutaraldehyde. (a) Schematics of scanning substrate-surface-bound virions with the AFM tip. PLL: poly-l-lysine. (b) AFM image of an overview (1.5 × 1.5 μm) sample area. (c) Zoomed-in AFM image of SARS-CoV-2 virions. The virion surfaces are covered with protrusions that correspond to spikes (S protein trimers). Inset: Topographical profile plot measured along the center of one of the virions (dotted line). The profile plot reveals a rugged surface. (d) 3D-rendered image of a SARS-CoV-2 virion. A partially flattened virus particle is observed, pointing at a global flexibility of the virion. (e) High-resolution AFM image of a SARS-CoV-2 virion displaying axial view of S trimers. (i) AFM image of the entire virion. (ii) Enlarged and contrast-enhanced image of the rectangular area. (iii) Same AFM image with overlaid triangles indicating S trimer orientation. The spikes apparently display translational, rotational, and flexural disorder owing to their flexibility.
To circumvent the effects caused by chemical fixation and to uncover the spike dynamics in situ, we investigated the topography of unfixed, native SARS-CoV-2 virions (Figure 2). Unexpectedly, we were unable to resolve S trimers on the virion surface at any of the investigated scanning strengths (Figure S5); rather, the virus particles displayed a blurred, smooth surface (Figure 2a). The mean central height of the native virions was 83 nm (±7 nm, SD), which is significantly greater than that observed for the fixed one (Figure 2b). We interpret the blurring of virion topography as the result of aperture error caused by time averaging of spike movement within the sampling region of each image pixel, hence the increase in virion height is caused by the AFM tip scanning an apparent dynamic surface (Figure 2c). Most plausibly, spike motion is dictated by the Brownian dynamics of the receptor-binding-domain (RBD) trimer, which may then be thought of as a tethered particle. Spike mobility in the virus envelope may contribute further to the observed dynamics. An alternative explanation for the observed blurred virion surface is that the spikes evade the moving AFM cantilever tip, which then scans the envelope surface. However, while it relies on a similarly dynamic spike behavior, a reduced virion height should have been observed. We speculate that the rapid spike motion revealed by these experiments may contribute to an efficient dynamic search by the virion on the surface of the targeted host cell, which explains why SARS-CoV-2 is at least as infective as the influenza virus20 in spite of its fewer spikes (up to ∼60 in SARS-CoV-2 versus up to ∼350 in influenza A21).

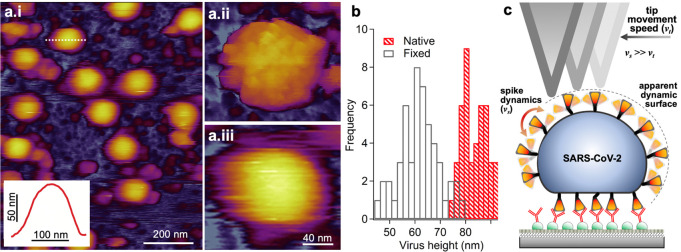
Topographical analysis of native, unfixed SARS-CoV-2 virions. (a.i) AFM image of an overview (0.8 × 1 μm) sample area. Inset: Topographical profile plot measured along the horizontal diameter of one of the virions (dotted line), revealing a smooth surface. (a.ii,iii) Magnified image of a fixed and native virion, respectively, for better comparison. Surface protrusions are not resolved in the native virus particle, but a blurred, smooth topography is observed. (b) Distribution of the topographical maximal central height of fixed and unfixed SARS-CoV-2 virions obtained from particle analysis (see Figure S2). The bimodal distribution seen in both histograms may be associated with spike position. Mean particle height (±SD) of fixed and unfixed virions are 62 ± 8 and 83 ± 7 nm, respectively. Unfixed virions have a particle height significantly larger than that of the fixed ones. (c) Schematics explaining the dynamically enhanced height of the unfixed virion.
We investigated the mechanical properties of SARS-CoV-2 by lowering the cantilever tip on the vertex of individual virions selected on the AFM image (Figure 3a). The virion was indented by pressing the tip downward (Figure 3b) with constant velocity (typically 0.5 μm/s) until a preset maximum force, measured by the cantilever deflection, was reached (typically 2–3 nN). Such a nanomechanical manipulation did not result in permanent topographical changes (Figure 3c and Figure S6) in spite of completely compressing the virion so that the tip reached all the way to the substrate, resulting in a wall-to-wall deformation (Figure 3d). In the initial stage of indentation, immediately following the landing of the tip on the virion, we observed a linear force response devoid of discrete mechanical events (Figure S7), which allowed us to measure virion stiffness (Table S3). Mean stiffness was 13 pN/nm (±5 pN/nm, SD), which makes SARS-CoV-2 the most compliant virus investigated so far.22,23 Virion stiffness is somewhat lower than that measured for the influenza virus lipid envelope,24 suggesting that the elasticity of SARS-CoV-2 is dominated by its envelope, and the RNP contributes little to the overall viral mechanics. The elastic regime was followed by a yield point marking the deviation from the linear force response and the onset of force-induced structural transitions which continued to take place until total compression. Unlike in other viruses,14,25 force did not drop to near zero values following mechanical yield, indicating that virion collapse or breakage was not evoked in spite of the drastic mechanical perturbation. Conceivably, the mechanical yield is made possible by the force-induced rearrangement of the basketlike structures of the RNPs.13 Subsequent to indentation, we retracted the cantilever. Remarkably, the virion generated forces up to several hundred pN during retraction, suggesting that structural recovery was taking place, possibly driven by the restitution of the initial RNP arrangement. The process continued until the initial virion height was re-established. The mean force–spectroscopic height was 94 nm (±10 nm, SD), which is comparable to that obtained from topographical data. The differences between the indentation and retraction force traces revealed a force hysteresis indicating that part of the mechanical energy invested in distorting the virion was dissipated as heat (Figure S8). We were able to continue the indentation–retraction cycles up to 100 times, but the virions never broke or collapsed (Figure 3e,f). Rather, both the indentation and retraction force traces relaxed after about 10 mechanical cycles, resulting in a minimized force hysteresis corresponding to a dissipated heat of ∼25 aJ. The force response may potentially be explained by two other mechanisms alternative to virion compression. The first one is the force-induced virion rolling on the substrate. We exclude this possibility because, due to the presence of anti-spike-protein antibodies on the surrounding surface, the process is expected to be completely irreversible. The second is the sideways slippage of the cantilever tip off the virion surface. We exclude this possibility based on a calibration of the cantilever’s lateral torsion (Figure S9) and because this process is expected to be completely reversible. We can only speculate about the mechanisms behind the persistent structural self-healing of SARS-CoV-2. Conceivably, the process involves the dynamic interaction between the RNA, protein, and lipid components. Notably, in some (∼1%) of the retraction traces, we observed sawtooth-shaped force responses, the peak forces of which fall between ∼210 and 330 pN (Figure 3f). The most plausible explanation is that the force peaks correspond to the mechanically driven unfolding of S protein domains, which is supported by the similar unfolding forces calculated with molecular dynamics simulation.26 Altogether, the SARS-CoV-2 virion is a mechanically stable, remarkably compliant and surprisingly resilient nanoparticle.

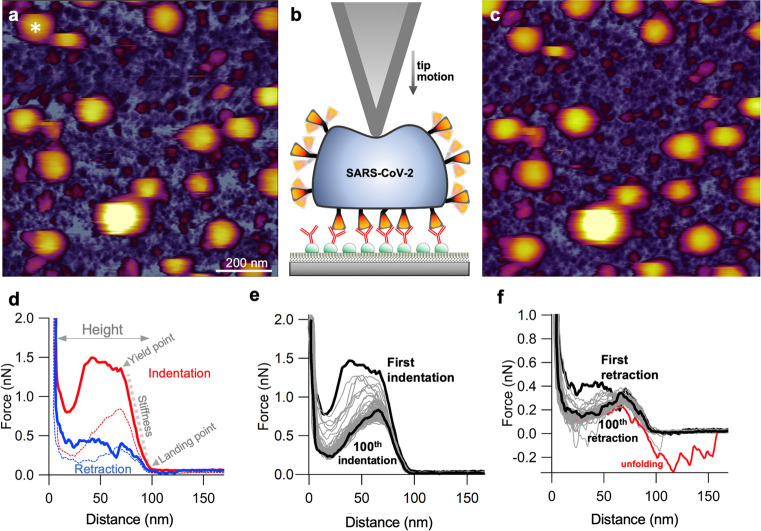
Single-particle force spectroscopy of SARS-CoV-2 virions. (a) AFM image of an overview (1 × 1 μm) sample area prior to nanomechanical manipulation. Asterisk shows the virion selected for nanomechanical manipulation. (b) Schematics of the force spectroscopy experiment. The virion is indented with the AFM tip until a preset force (typically 2–3 nN) is reached. (c) AFM image of the same overview sample area following nanomechanical manipulation. We could not detect any topographical sign of permanent structural change. (d) Example of a force versus distance curve obtained during a single indentation–retraction cycle. From the slope of the indentation curve (gray dotted line) and the distance between the landing point and substrate limit of the trace, we obtained the stiffness and the force–spectroscopic height of the virions, respectively. Red and blue dotted lines indicate indentation and retraction data, respectively, obtained in the 100th nanomechanical cycle. (e) Force versus distance curves obtained during repeated indentation of a single SARS-CoV-2 virion. (f) Matching force versus distance curves obtained during retraction. In some traces, force sawteeth (red trace) correspond to the unfolding of domains in a surface protein, plausibly within the S trimer.
To assess the thermal stability of SARS-CoV-2, we explored the topographical changes of virions exposed to high-temperature treatment (Figure 4). The sample was exposed to temperatures of 60 (Figure 4a), 80 (Figure 4b), and 90 °C (Figure 4c) for 10 min then cooled to 20 °C for AFM imaging. Remarkably, although virion density dropped from 22 (at 60 °C) to 18 virions/μm2 (at 90 °C), the virus particles remained on the substrate surface and their global appearance was only slightly altered. The mean peak height of the viral particles increased slightly (83 ± 12, 88 ± 11, and 90 ± 9 nm at 60, 80, and 90 °C, respectively), and virion topography became somewhat faceted, but the particles retained their blurred, smooth surface. To test whether spikes were still present following thermal exposure, we fixed the sample with 5% glutaraldehyde (Figure 4d). Although the rugged topography, seen in chemically fixed SARS-CoV-2 virions (Figure 1c), was partially restored, the spikes were much fewer and less distinct, and their trigonal shape (Figure 1e) could not be resolved (Figure 4e), suggesting they became thermally denatured. Furthermore, the smooth areas interspersed between rugged regions indicate that thermal treatment resulted in a progressive dissociation of the S trimers from the virion surface. Thus, the SARS-CoV-2 virion displays an unexpected global thermal stability, which is likely related to their aerosol and surface stabilities.27 However, the conformational response of the spike proteins observed here eventually leads to the heat-induced inactivation of SARS-CoV-2.

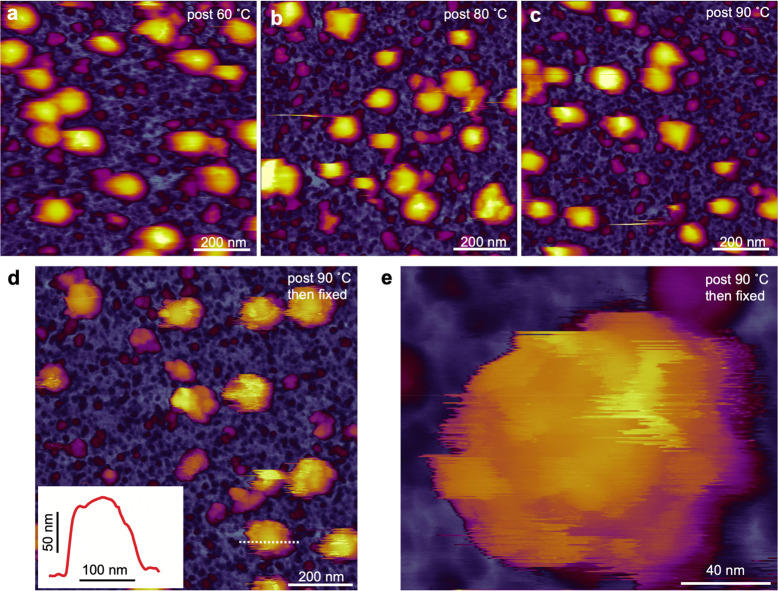
Effect of temperature change on the topographical structure of SARS-CoV-2. The sample was heated for 10 min at 60 (a), 80 (b), and 90 °C (c) then cooled back to 20 °C prior to AFM imaging. The virions persist on the substrate surface with their global structure nearly intact, but the topography becomes progressively more rugged, pointing at the gradual disappearance of the dynamic surface smoothing hence reduction of spike dynamics. (d) AFM image of an overview (1 × 1 μm) sample area following thermal treatment (90 °C for 10 min) and glutaraldehyde (5%) fixation. Inset: Topographical profile plot measured along the center of one of the virions (dotted line). The rugged surface topography is partially restored, but large areas on the virions are devoid of spikes. (e) High-resolution AFM image of a heat-exposed (90 °C for 10 min) and fixed (5% glutaraldehyde) SARS-CoV-2 virion. Shallow surface protrusions are present.
In conclusion, the atomic force microscopic imaging and nanomechanical measurements revealed that the SARS-CoV-2 virion is highly dynamic, compliant, and resilient, and it displays remarkable mechanical and global thermal stabilities. While the dynamics of the surface spikes may play an important role in the unusually high infectivity of the virus, its mechanical and self-healing properties may also ensure adaptation to a wide range of environmental circumstances. Considering its capability of exploring viruses under native conditions, the single-particle approaches16 employed here may be important in uncovering not only the mechanistic details behind viral infection but also the viral response to potential therapies.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.0c04465.
Supplementary figures, tables, and experimental details (PDF)
Author Contributions
B.K., Z.K., B.P., and M.S.Z.K. conceived the experiments; Z.K. and B.P. purified the SARS-CoV-2 samples; B.K. and M.S.Z.K. performed AFM imaging and force spectroscopy measurements and analyzed the data; B.K. and M.S.Z.K. wrote paper; M.S.Z.K. secured financial support for the project. All authors critically read and revised the manuscript. B.K. and Z.K. contributed equally.
Notes
The authors declare no competing financial interest.
Acknowledgments
We gratefully acknowledge the help of Mónika Drabbant and Hedvig Tordai with experimental preparations and measurements. We thank Levente Herényi for theoretical discussions and reading the manuscript. This work was funded by grants from the Hungarian National Research, Development and Innovation Office (K124966 to M.K.; National Heart Program NVKP-16-1-2016-0017; Thematic Excellence Programme; National Bionics Programme ED_17-1-2017-0009). The research was financed by the Higher Education Institutional Excellence Programme of the Ministry for Innovation and Technology in Hungary, within the framework of the Therapeutic Development thematic programme of the Semmelweis University. HCEMM, a teaming grant associated with the European Molecular Biology Laboratories, has received funding from the European Union’s Horizon 2020 research and innovation programme under Grant Agreement No. 739593.
 Topography, Spike Dynamics, and Nanomechanics of Individual
Native SARS-CoV-2 Virions
Topography, Spike Dynamics, and Nanomechanics of Individual
Native SARS-CoV-2 Virions