
Edited by Dominique C. Bergmann, Stanford University, Stanford, CA, and approved November 5, 2020 (received for review September 6, 2020)
Author contributions: J.W.S. and M.J.S. designed research; J.W.S. and J.S. performed research; J.W.S. and J.S. analyzed data; and J.W.S. and M.J.S. wrote the paper.
- Altmetric
Plants possess the remarkable ability to grow and produce new organs throughout their lifespan, owing to the activities of persistent populations of pluripotent stem cells within their meristematic tips. Here we isolated individual cells from the microscopic shoot apical meristem (SAM) of maize and provide single-cell transcriptomic analysis of a plant shoot meristem. This study enabled an unbiased analysis of the developmental genetic organization of the maize shoot apex and uncovered evolutionarily divergent and conserved signatures of SAM homeostasis. The fine-scale resolution of single-cell analysis was used to reconstruct the process of shoot cell differentiation, whereby stem cells acquire diverse and distinct cell fates over developmental time in wild-type and mutant maize seedlings.
Plants maintain populations of pluripotent stem cells in shoot apical meristems (SAMs), which continuously produce new aboveground organs. We used single-cell RNA sequencing (scRNA-seq) to achieve an unbiased characterization of the transcriptional landscape of the maize shoot stem-cell niche and its differentiating cellular descendants. Stem cells housed in the SAM tip are engaged in genome integrity maintenance and exhibit a low rate of cell division, consistent with their contributions to germline and somatic cell fates. Surprisingly, we find no evidence for a canonical stem-cell organizing center subtending these cells. In addition, trajectory inference was used to trace the gene expression changes that accompany cell differentiation, revealing that ectopic expression of KNOTTED1 (KN1) accelerates cell differentiation and promotes development of the sheathing maize leaf base. These single-cell transcriptomic analyses of the shoot apex yield insight into the processes of stem-cell function and cell-fate acquisition in the maize seedling and provide a valuable scaffold on which to better dissect the genetic control of plant shoot morphogenesis at the cellular level.
Unlike animals where organogenesis is typically completed in juvenile stages, plants initiate new organs throughout the lifespan via the persistence of pluripotent stem-cell populations long after embryogenesis. In the plant shoot, these stem cells are housed within the shoot apical meristem (SAM), which gives rise to all of the aboveground organs of the plant (1). Canonical descriptions of SAM organization in flowering plants include a stem-cell niche within the central zone at the SAM tip, subtended by the stem-cell organizing center, and a peripheral zone surrounding the SAM flank that provides initial cells for organogenesis. The genetic maintenance and organization of the stem-cell niche, and how cells attain differentiated fates remain fundamental areas of interest in plant development.
Class I KNOTTED-LIKE HOMEOBOX (KNOX) genes broadly promote indeterminate cell identity in vascular plants, that is, a state in which cell proliferation and growth can continue indefinitely (2). This is in contrast to determinate cell identity, in which cell proliferation and growth cease in order to produce a tissue or organ with a predetermined size. KNOX down-regulation is a marker of cell differentiation and comprises an initial step in lateral organ identity acquisition at the SAM periphery. In Arabidopsis, stem-cell homeostasis is achieved via a negative feedback loop wherein the activity of the stem-cell-organizing transcription factor WUSCHEL (WUS) is repressed by binding of the small secreted peptide CLAVATA3 (CLV3) to the CLAVATA1 (CLV1) receptor (3). The canonical CLV–WUS signaling pathway and other receptor–ligand signaling complexes regulating WUS-mediated control of shoot meristem size are identified across the flowering plants (3).
In order to better understand the spatial organization of the maize SAM and the process of cell differentiation during plant development, we took a single-cell transcriptomic approach to achieve an unbiased sampling of cell types from the maize (Zea mays spp. mays) SAM and seedling shoot apex unimpeded by prior histological assumptions. Improved protocols for the isolation of living plant stem cells enabled this single-cell transcriptomic analysis of a plant vegetative shoot meristem. Two zones of cell identity are identified within the maize SAM: 1) a slowly dividing stem-cell domain at the SAM tip expressing genes with functions in genome integrity, and 2) a subtending population of cells undergoing transit-amplifying divisions. Although the CLV–WUS stem-cell homeostatic pathway is well described in a diverse array of angiosperm SAMs and in the inflorescence and floral meristems of maize (3), we find no evidence for a stem-cell organizing center expressing WUS in the maize SAM (4). Trajectory inference was used to identify dynamic gene expression patterns correlated with cell differentiation and ultimate cell fate in the seedling shoot. We find that ectopic expression of KNOTTED1 (KN1) in differentiating leaf primordia drives cellular transcriptomes to more differentiated states, which we ascribe to a role for KN1 in promoting sheath cell fate in leaves alongside its role in maintaining shoot indeterminacy in the SAM and stem. Taken together, this study reveals the landscape of cell states and the dynamics of cell-fate acquisition in the developing maize seedling shoot apex, as a first step toward further analyses of maize development at single-cell resolution.
Results
Single-Cell Transcriptomic Approach for the Analysis of Maize Vegetative SAM Cells.
Single-cell transcriptomic analyses of plant cells require the preparation of protoplasts, viable cells whose rigid, cellulosic cell walls are enzymatically removed. Previously, limitations in the recovery of viable protoplasts from SAM-enriched plant tissues have presented an obstacle to single-cell RNA sequencing (scRNA-seq) analyses of shoot meristems (5). To achieve a higher rate of viable cell recovery, we supplemented the protoplasting solutions with l-arginine (l-Arg), which modestly enhanced cell viability (SI Appendix, Fig. S1A). This finding was consistent with previous reports of enhanced cell viability of oat (Avena sativa) coleoptile protoplasts cultured in media supplemented with l-Arg (6). Increasing the pH of the media further enhanced protoplast viability (SI Appendix, Fig. S1B), in keeping with prior studies showing that the in vivo pH of SAM tissue in the herbaceous plant Chenopodium rubrum is two orders of magnitude more alkaline than typical plant protoplasting solutions (7). Together, these modifications to our protoplasting protocol improved cell viability between 10- and 30-fold, depending on the tissue.
To capture cells from the microscopic seedling SAM, we manually harvested protoplasts from dissected apices comprising the SAM plus the two most recently initiated leaf primordia (SAM + P2). After filtering, six biological replicates captured a total of 327 cells for scRNA-seq analyses (median transcripts detected = 8,955, median genes detected = 2,221) (Fig. 1A and SI Appendix, Fig. S2). We first used k-means clustering to classify transcriptionally similar cells. Next, we performed dimensionality reduction using uniform manifold approximation and projection (UMAP), which plotted the seven resulting clusters in two-dimensional space. Owing to the abundance of cycling cells in the SAM + P2 tissues (49% in S/G2/M phase), we regressed out variation contributed by the cell cycle on cell clustering (Dataset S1). We assigned cell-type identities to clusters based on their expression of known shoot apex marker genes (Dataset S2) and identified clusters corresponding to major cell types derived from the epidermis, primordia, and vasculature, along with indeterminate cell types from the SAM (Fig. 1B and SI Appendix, Fig. S3). Intriguingly, instead of forming discrete, isolated cell clusters, the majority of SAM-enriched cells exhibited a continuum of intermediate identity states (Fig. 1B), suggesting that differentiation is highly dynamic and continuous in the maize SAM and young leaf primordia.

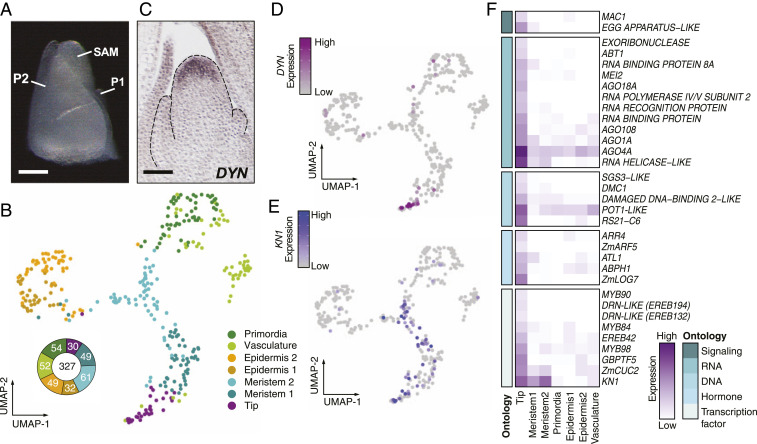
Transcriptomic signatures of stem-cell identity and maintenance in the maize SAM. (A) Cells were isolated from the SAM plus two most recently initiated leaf primordia (SAM + P2). (B) Dimensionality reduction and cell classification for cells in the SAM + P2 dataset. Donut graph shows number of cells within each classification. (C) RNA in situ hybridization with antisense probe to DYNAMIN (DYN) in medial longitudinal section of the SAM. (D) DYN, a marker for the SAM tip and putative stem-cell domain, in the SAM + P2 dataset correlates in expression with the putative tip cell population. (E) Expression of KN1 marks a broader indeterminate cell population that includes the tip cell population. (F) Heatmap of select differentially expressed genes in the tip domain grouped based on functional ontology.
Control of Genomic Integrity Among Maize Stem Cells.
We first sought to identify signatures of the stem-cell population of the maize SAM. The tip of the maize SAM is thought to house the stem cells (4, 8), which are essential for generating the aboveground somatic tissue of the maize plant as well as cells that give rise to the germline. Cell clustering analysis independently identified a transcriptionally distinct cell population in which DYNAMIN (DYN), a previously identified marker of tip cells in the maize SAM, was up-regulated within a broader population of meristematic cells expressing KN1 (Fig. 1 C–E) (4). We therefore designated cells belonging to this cluster as the putative stem cells residing within the SAM tip and used differential expression (DE) analysis to identify 89 genes preferentially expressed within this population (Fig. 1F and Dataset S2) (4). Among these were genes with confirmed or predicted roles in intercellular signaling, small RNA biogenesis, DNA maintenance, response to the plant hormones auxin and cytokinin, and transcriptional regulation (Fig. 1F). Closer inspection of the numerous genes (false discovery rate [FDR] for gene ontology [GO] term enrichment = 0.05; Dataset S3) involved in RNA biogenesis suggested that tip cells are engaged in RNA-dependent gene silencing activities. For example, the stem-cell-enriched SUPRESSOR OF GENE SILENCING3-LIKE (SGS3-LIKE), RNA POLYMERASE IV/V SUBUNIT2, and ARGONAUTE4a (AGO4a) all encode members of the RNA-directed DNA methylation pathway that maintains heterochromatin at repetitive, retrotransposon-enriched, maize genomic regions (9). Indeed, maintenance of heterochromatin is likewise essential for the genomic stability and homeostasis of stem-cell populations in animals (10). However, unlike animals where germline cells are specified and sequestered during early embryonic stages, plants lack a segregated germ cell lineage during vegetative stages of development (11). Up-regulation of genes involved in DNA repair-related processes, such as a PROTECTION OF TELOMERES1-LIKE (POT1-LIKE) and a DNA-DAMAGE BINDING2-LIKE gene likely reflect the advantage of maintaining high genomic fidelity among cells that have the potential for both somatic and germ cell fate (12). Collectively, these data suggest that cells in the maize SAM tip are engaged in genome protective functions consistent with their plant stem-cell identity.
Rates of Cell Division Are Kept Low in the SAM Stem-Cell Population.
Increased rates of cell division increase the chances for spontaneous mutations during genome replication. We therefore asked if stem cells in the maize SAM tip exhibit differences in cell division rate. Estimates of cell-cycle stage generated during cell-cycle regression (Materials and Methods) indicated that the fraction of SAM + P2 cells in G1 phase decreased as differentiation progressed, indicative of higher rates of cell division (Fig. 2A). For example, the SAM tip population contains a larger fraction of cells in G1 phase than the remainder of the meristem, leaf primordia, or vasculature cell populations, suggesting a lower cell division rate among the stem cells. In order to test this, we performed RNA in situ hybridization on HISTONE H3 and CYCLIN1 transcripts that accumulate in cells at S phase and G2/M phase, respectively. We next subdivided medial SAM sections into five bins of equal height along the proximodistal axis and inferred the spatial distribution of cell division stages by image thresholding on HISTONE H3 and CYCLIN1 staining (Fig. 2 B–E). Cells in bin 1 that comprises the tipmost region of the SAM consistently had the lowest number of dividing cells. When considered together with our detection of transcripts encoding factors that promote genomic and epigenomic stability in the SAM tip (Fig. 1F), a low cell division rate among the stem-cell population can explain how plants avoid unfavorable increases in genetic load over successive generations in the absence of a segregated germline early in ontogeny. A reduced rate of cell division at the SAM tip is likewise consistent with the low number of cell divisions that are predicted to occur between formation of the maize zygote and the gametophytes contributing to the next generation (13). Cells in the remainder of the SAM, below the tip, showed higher rates of cell division, similar to transit-amplifying cell divisions found in animal stem-cell niches (14). These proliferative cell divisions in transit-amplifying cells generate the anlagen for determinate lateral organs, obviating the requirement for high rates of stem-cell divisions. Finally, we observed that in axillary meristems (AMs), the highest concentration of dividing cells typically occurs closer to the AM tip as compared to the SAM (Fig. 2D), suggesting that cell division patterns are dynamically regulated in different shoot meristem types, possibly due to relative differences in meristem activity.

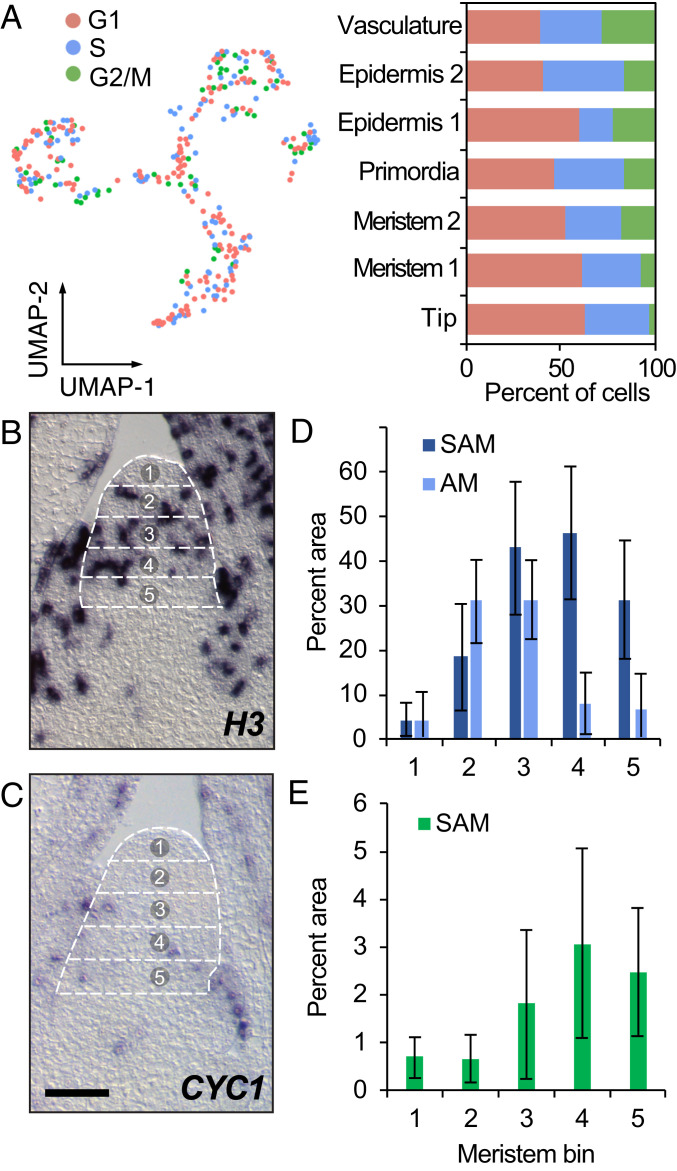
Cell division dynamics throughout the maize SAM. (A) Estimated cell division stage of cells in the SAM + P2 datasets. Bar charts show fraction of cells in each stage among cell clusters. (B and C) RNA in situ hybridization with antisense probe to H3 (B; SAM, n = 9; AM, n = 4) and CYC1 (C; SAM, n = 7) in medial longitudinal sections of the SAM showing bins (outlined in a dotted line grid and numbered) used for quantification of the proportion of cells in (D) S phase and (E) G2/M phase. (Scale bars, 100 µm.)
Divergence in SAM Stem-Cell Regulation.
We next sought to analyze the cell-specific expression patterns of regulators of stem-cell maintenance within the SAM + P2 tissues. FON2-LIKE CLE PROTEIN1 (FCP1) was the only CLV3-like ligand-encoding transcript detected in meristem tip cells (Fig. 3A). RNA in situ hybridization identified weak expression of FCP1 just below the SAM tip, as well as the originally reported expression in the SAM periphery and leaf primordia (15). The FCP1 peptide–ligand is perceived by the FEA3 receptor to repress stem-cell identity (15). FEA3 transcripts show low and diffuse accumulation in the SAM periphery and heightened expression in leaf primordia (Fig. 3B). Other maize transcripts encoding predicted leucine-rich repeat (LRR) receptors exhibited similar accumulation patterns (Dataset S4), with higher expression in the SAM periphery and primordia but a lack of a strong SAM-specific expression profile as is seen for CLV1 in Arabidopsis (16). This could reflect a role for LRR receptors in inhibiting stem-cell identity outside of the SAM tip domain, as described previously for the FCP1–FEA3 ligand–receptor system (15). Notably, mutations in the CLV1 homolog of maize causes enlarged inflorescence meristems, but do not affect vegetative SAM size (3).

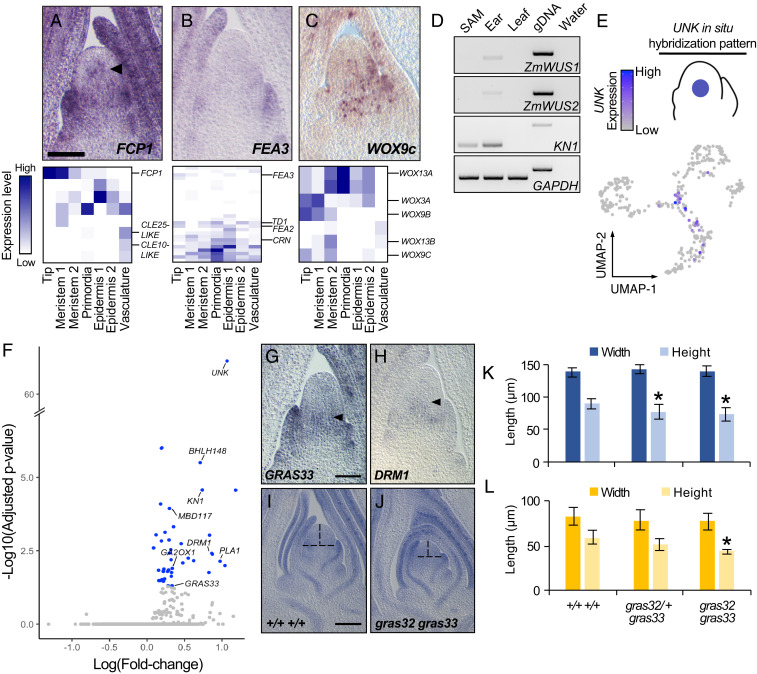
Characterization of the SAM core. (A–C) RNA in situ hybridization with antisense probes to FCP1 (A), FEA3 (B), and WOX9c (C) (Top) and DE analysis along with paralog gene expression in the SAM + P2 dataset (Bottom). (D) RT-PCR of ZmWUS1/2, GAPDH, and KN1 transcripts from the maize SAM and the three most recently initiated leaf primordia (SAM), 2-mm immature ears (ear), mature leaf 3 tissue (leaf), genomic DNA (gDNA), and a no template control (water). (E) Cells expressing the UNKNOWN marker gene in the SAM + P2 dataset. The illustration reflects the published UNKNOWN gene expression pattern (4). (F) Volcano plot showing the differential expression of genes (blue) in cells expressing the UNKNOWN SAM core marker gene. (G and H) RNA in situ hybridization of antisense probes to GRAS33 (G) and DRM1 (H) in the SAM. (I and J) Toluidine Blue-O stained medial longitudinal section of the SAM from normal siblings (I) and gras32 gras33 double mutants (J). Vertical and horizontal dashed lines indicate SAM height and width, respectively. (K and L) Quantification of SAM (n = 5 to 8) (K) and AM (n = 3) (L) height and width in normal siblings and gras32 gras33 double mutants; error bars represent SD about the mean, two-tailed Student’s t test, *P < 0.05. (Scale bars, 100 µm.)
In Arabidopsis, the stem-cell promoting transcription factor WUS is negatively regulated by CLV1–CLV3 function, to control the size of the meristematic stem-cell pool (3). WUS is mobile and is expressed in the organizing center (OC) below the stem-cell domain from where it is trafficked to promote stem-cell fate in the SAM tip. A similar, stem-cell organizing ZmWUS function is described in the maize inflorescence meristem (3). However, we did not detect ZmWUS-expressing cells in the SAM, although transcripts of several maize WUS homologs, including ZmWOX3a, ZmWOX9b, and ZmWOX9c were identified (Fig. 3C). The single Arabidopsis WOX9 homolog promotes cell proliferation in meristematic tissues upstream of WUS function, but belongs to a more ancient, functionally divergent WOX clade that lacks the repressive WUS box (17, 18). Overall, this suggests that the maize coorthologs WOX9b and WOX9c are unlikely to be functionally homologous to WUS. Moreover, although WOX3A does encode a WUS box and is detected in the SAM (Fig. 3C), its expression pattern is not cell-type specific, inconsistent with a well-defined OC. Thus, our data identify no candidate WOX gene expressed in the maize B73 SAM that is likely to function as a stem-cell organizing center, homologous to WUS in Arabidopsis and ZmWUS in the maize inflorescence meristem. Indeed, past bulk RNA-seq analyses of the SAM have failed to identify transcripts from the two paralogous maize WUS genes, ZmWUS1 and ZmWUS2 (4, 19). To extend these findings, we performed RT-PCR analyses in an attempt to detect the accumulation of ZmWUS1 and ZmWUS2 transcripts. While both transcripts were detectable in immature ears containing inflorescence meristems, we did not detect their expression in vegetative shoot apical tissue comprising the SAM and the three most recently initiated leaf primordia (Fig. 3D). Together, these results suggest that the canonical CLV1–CLV3–WUS pathway has been bypassed in the maize B73 SAM, with changes in the spatial expression patterns of corresponding maize paralogs as a defining feature.
WUS-Independent Functions for Maize HAM-LIKE Genes in the SAM Core.
To further investigate the organization of the maize SAM, we examined gene expression patterns among cells derived from a recently reported domain situated in the center, or “core” region of the SAM, which is marked by the expression of GRMZM2G049151, a gene of unknown function (UNKNOWN [UNK]) (4). We identified cells within the core region by transcript accumulation of GRMZM2G049151 and used DE analysis to characterize their expression profiles (Fig. 3 E and F and Dataset S5) (4). SAM core cells show up-regulated expression of the PLASTOCHRON1 (PLA1) gene. PLA1 promotes cell division and growth in an auxin-dependent manner and is also expressed within multiple maize organs and tissues (20). Indeed, our analyses of cell division dynamics in the SAM indicated that the core region had higher cell proliferation activity relative to the SAM tip (Fig. 2). The auxin-promoted dormancy-associated gene DRM1 (21) and the maize HAIRY MERISTEM3 (HAM3) homolog GRAS33 were also up-regulated in the SAM core, as confirmed by RNA in situ hybridizations (Fig. 3 F–H and SI Appendix, Fig. S4 B and C). Arabidopsis HAM genes are expressed in the organizing center, SAM periphery, and in leaf primordia. AtHAM genes promote SAM maintenance through their physical interaction with WUS and also activate the formation and maintenance of AMs that give rise to lateral branches (22, 23). To determine if GRAS33 activity in the core domain has a conserved role in maintenance of the maize SAM, we generated double mutants between GRAS33 and its paralog GRAS32 and analyzed SAM size in seedlings (SI Appendix, Fig. S4A). Compared to the wild-type (WT) siblings, gras32/+ gras33 seedlings and gras32 gras33 double mutants displayed shorter SAMs (Fig. 3 I–K). Only gras32 gras33 SAMs possessed shorter AMs, likely owing to genetic redundancy between these factors in AMs (Fig. 3L). These data suggest that maize GRAS32 and GRAS33, like their Arabidopsis homologs, have roles in regulating SAM homeostasis from a stem-cell surrounding region that overlaps the maize SAM core domain. These results further suggest that unlike in Arabidopsis, the maize SAM tip is not subtended by a WUS-expressing, stem-cell organizing center. Rather, this SAM core region displays signatures of auxin-regulated growth processes, as suggested by the up-regulated expression of PLA1 and DRM1. Thus, the maize SAM core may be functionally akin to the tip-subtending SAM regions expressing HAM genes in Arabidopsis, with maize HAM genes having a potential WUS-independent SAM regulatory function.
Cell Differentiation Follows a Continuum of Transcriptional States.
Given that the individual transcriptomes of cells within the maize shoot apex fit a continuum of cell differentiation states (Fig. 1B), we aimed to determine the dynamic changes in gene expression that accompany this developmental progression. We applied a principal graph algorithm to identify a branching path among the embedded cellular coordinates in the UMAP projection, which we used to infer the differentiation trajectory of cells in the SAM + P2 dataset. Each cell was then assigned a pseudotime value based on its distance along the resulting path, relative to a specified pseudotime start position within the SAM tip cell population (Fig. 4A). As expected, we found that pseudotime progression is associated with the transition of cells from indeterminate to determinate cell fates; KN1 a key marker of indeterminate meristematic identity is highly expressed early in pseudotime and declines as pseudotime progresses (Fig. 4A) (24). To survey the transcriptional changes associated with cell differentiation, we performed DE analysis to identify transcript accumulation patterns that significantly correlate with pseudotime progression. In total, over 2,000 genes exhibited significant changes in expression over pseudotime (Dataset S6). Hierarchical clustering grouped each transcript according to expression pattern and identified several patterns of transcript accumulation that correspond to particular stages of cell differentiation (Fig. 4B).

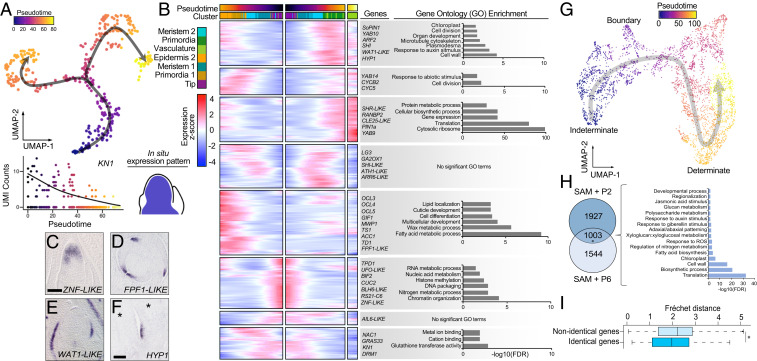
Tracing the gene expression patterns associated with cell differentiation. (A) (Top) Pseudotime values and trajectories for cells in the SAM + P2 dataset. (Bottom) KN1 gene expression declines over pseudotime consistent with its transcript accumulation patterns from RNA in situ hybridization studies. (B) Heatmap of ∼2,000 genes that show correlated changes in gene expression along the inferred trajectory clustered based on their expression patterns. Cells are mirrored along the central axis prior to the trajectory branch point. Representative genes and significant GO term enrichments for each cluster are shown. (C–F) Transcript accumulation patterns for trajectory-correlated genes showing high expression levels at early, intermediate, and late points in the trajectory. RNA in situ hybridization of antisense probes to ZNF-LIKE in a medial longitudinal section of the SAM shows early trajectory expression (C), FPF1-LIKE in transverse section above the SAM and WAT1-LIKE in medial longitudinal section of the SAM show late trajectory expression (D and E), and HYP1 in longitudinal section below the SAM (F). Asterisks in F indicate leaf primordia. (G) A subset of cells from the SAM + P6 dataset (see SI Appendix, Fig. S5B for inferred cluster identities) were reclustered and trajectory inference was used to assign cell pseudotime scores along a transition from indeterminate to determinate cell fates. (H) Overlap of trajectory-correlated genes from the SAM + P2 and SAM + P6 datasets and their GO term enrichment (*, hypergeometric test, P = 1.2E-320). (I) Box plot showing median Fréchet distance for identical and nonidentical genes in the SAM + P2 and SAM + P6 datasets, box edges represent the 75th and 25th percentiles, Wilcoxon rank-sum test, *P = 5.2E-9.
Early in pseudotime, genes enriched for stem-cell functions in RNA metabolism and chromatin organization give way to genes enriched for glutathione transferase and cation-binding activities as well as expression of DRM1 and GRAS33, which are expressed among cells in the SAM core. Next, cells progress through a putative boundary domain identity, marked by up-regulation of an ARABIDOPSIS THALIANA HOMEOBOX GENE1-LIKE (ATH1-LIKE) gene, LIGULELESS3 (LG3), and GA2OX1. ATH1 promotes organ boundary formation in Arabidopsis and antagonizes activity of the growth phytohormone gibberellic acid (GA) alongside GA2OX1-promoted GA catabolism (25, 26). In addition, LG3 is expressed in specific boundaries during leaf development (27). After progressing to this putative shoot boundary domain, the cellular transcriptomes of SAM + P2 cells resolve into either epidermal or ground and vascular cell identities. The lack of a transcriptionally distinct lineage of undifferentiated epidermal cells (protoderm) early in the trajectory is notable, given that the cell lineage of the outer protodermal layer is separate from that of underlying cells, even within the SAM tip (1). Together, this suggests that despite their cell lineage differences, the distinctive transcriptional profiles among cell types in the SAM tip become detectable only after exiting the boundary regions of the SAM. Indeed, transcript accumulation of epidermal marker genes such as LTP1, LTP3, and OCL4 has been shown to first occur in the protodermal layer in the SAM periphery, outside the SAM tip (4, 19). Increased sampling of the maize SAM may be necessary to uncover the more subtle expression differences that distinguish tip protodermal cells from underlying cell populations.
As expected, we found that epidermal cell differentiation correlates with up-regulation of the OUTER CELL LAYER (OCL) homeodomain leucine zipper IV transcription factor-encoding genes that promote epidermal cell identity (28). On the other hand, cells fated to become leaf primordia and/or vasculature exhibit up-regulation of auxin response genes and transcripts associated with cell wall, chloroplast, organ development, and cell proliferation. In addition, primordia and vascular cells are significantly enriched for transcripts related to translation, suggesting a large burst in protein synthesis accompanies leaf initiation and expansion. Selected genes significantly associated with pseudotime progression were examined by RNA in situ hybridization to validate their expression patterns along the developmental trajectory (Fig. 4 C–F). A gene expressed early in the pseudotime trajectory, ZINC FINGER DOMAIN-LIKE (ZNF-LIKE), showed expression in the SAM tip (Fig. 4C). Meanwhile, FLOWERING PROMOTING FACTOR1-LIKE (FPF1-LIKE) and WALLS ARE THIN1-LIKE (WAT1-LIKE) transcripts are up-regulated later in pseudotime and accumulate in differentiating vasculature and leaf margin cells (Fig. 4 D and E). Lastly, HYBRID PROLINE-RICH PROTEIN1 (HYP1) transcripts are also up-regulated later in the pseudotime, albeit detected at later stages of ontogeny (Fig. 4F). Together, these results support the notion that the transcriptomes of differentiating cells are highly dynamic across a continuum defined by pseudotime progression.
Genes Patterning Indeterminate and Determinate Cell Fates Are Expressed Across Leaf Ontogeny.
After leaf initiation at the SAM periphery, cells continue to proliferate in the leaf proximal region well beyond the P6 stage (i.e., the sixth leaf from the SAM), necessitating continued maintenance of indeterminate and determinate zones at the junction of the leaf and stem across ontogeny (29). To analyze this later patterning and differentiation process we prepared tissues comprising ∼3 mm of the maize shoot apex from 2-wk-old seedlings, dissected to include the six most recently initiated leaf primordia plus the SAM (SAM + P6; SI Appendix, Fig. S5A) for scRNA-seq analysis. In total, we captured the transcriptomes of 12,967 protoplasts (in two biological replicates) using microfluidic droplet capture (median transcripts detected = 14,992, median genes detected = 4,965). We performed dimensionality reduction and found cell clusters corresponding to epidermal, vascular, leaf primordial, indeterminate, and cell cycle states, which were remarkably similar to the cell types we detected at earlier stages of ontogeny in the SAM + P2 dataset (SI Appendix, Figs. S5B and S6–S8 and Datasets S7–S9). However, cells within our SAM + P6 dataset are overwhelmingly derived from later stages of shoot ontogeny (i.e., P4 to P6), owing to the markedly increased size of these older leaf primordia and associated stems. For example, although we identified indeterminate cells based on their expression of the transcription factor gene KN1 (SI Appendix, Fig. S7C), we did not identify cells with transcriptomic signatures of the seedling SAM stem-cell niche in our SAM + P6 dataset (Fig. 1 C–F). Nonetheless, we identified many of the signatures of the indeterminate-to-determinate cell-fate transition (SI Appendix, Fig. S9).
We hypothesized that the transcriptomic signatures of cell differentiation would be similar at both early and late stages of leaf development, reflecting the iterative and modular patterning of the plant shoot system. To test this, we again used trajectory inference to assign cells a pseudotime value reflective of their position in the indeterminate-to-determinate transition and utilized DE analysis to identify genes with pseudotime-correlated expression patterns (Fig. 4G). Overall, we identified ∼3,000 genes that met a stringent significance cutoff (adjusted [adj.] P <1E-100) and compared them to pseudotime-correlated genes in the SAM + P2 dataset. We found that approximately one-third of the transcripts showed a significant correlation with pseudotime in both datasets (hypergeometric test, P <1E-100), suggesting a core module of genes controls the indeterminate-to-determinate cell transition across ontogeny, which includes KN1 (Fig. 4H and Dataset S10). We used RNA in situ hybridization to validate these patterns and confirmed shared genes significantly associated with cell differentiation in both the SAM + P2 and SAM + P6 datasets (SI Appendix, Fig. S10). In addition, expression curves for identical genes in both datasets have a lower median Fréchet distance than nonidentical genes, indicating similar expression behavior across ontogeny (Fig. 4I). Among the 1,003 shared genes, GO functions related to translation, cell wall, organ polarity, auxin, and gibberellin-related processes were enriched, likely reflecting the roles of auxin and gibberellic acid hormones in promoting differentiation in opposition to KN1, which imposes indeterminacy (2, 26).
Ectopically Expressed KN1 Accelerates Leaf Cell Differentiation by Promoting Sheath Cell Fate.
Our single-cell transcriptomic analyses consistently distinguished between indeterminate cell types expressing KN1 and determinate cell types with low KN1 expression. Given the role of KN1 in promoting indeterminacy (2, 30), we sought to determine the effects of ectopic KN1 expression on cell-fate progression. The maize leaf comprises a distal, photosynthetic leaf blade and a proximal sheath that inserts into the leaf node and surrounds the stem. A hinge-like auricle and a fringe of epidermally derived tissue called the ligule develop at the boundary demarcating the blade and sheath. This proximodistal patterning begins in young leaf primordia (31). Dominant, gain-of-function mutations in KN1 confer its misexpression in developing leaf primordia, (reviewed in ref. 32), resulting in the formation of ectopic sheath identity in the midst of blade tissue (24, 31, 33). In support of this model, we found the Kn1-O/+ seedlings had significantly increased sheath length compared to their WT siblings (Fig. 5A). At the boundary between sheath and blade, LIGULELESS1 (LG1) gene function is required for formation of the ligule and auricle (27, 34). In Kn1-O/+ plants, ectopic knots of sheath tissue occur in the blade, coupled with adjacent ligule formation at this ectopic blade–sheath boundary (Fig. 5B and SI Appendix, Fig. S11). In Kn1-O lg1 double mutants, however, no ligules are formed at these ectopic blade–sheath boundaries, providing further evidence that the knots in KnO-1/+ mutant leaf blades comprise patches of misplaced sheath-like identity (Fig. 5B and SI Appendix, Fig. S11).

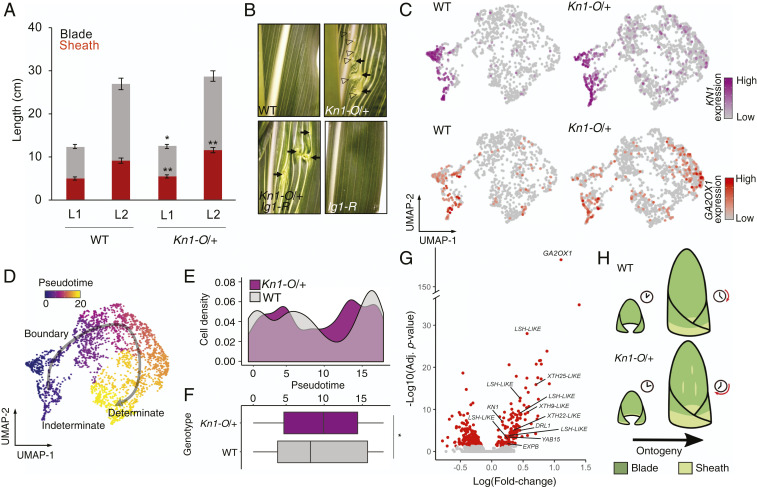
Ectopic expression of KN1 accelerates leaf cell differentiation toward sheath fate. (A) Blade and sheath lengths in seedling leaf 1 (L1) and leaf 2 (L2) in WT and Kn1-O/+ siblings. Error bars indicate SD about the mean, Student’s two-tailed t test, n = 17, *P < 0.05, **P < 0.001. (B) Genetic interaction between Kn1-O and lg1-R in mature leaf blade. Arrowheads indicate ectopic ligule and arrows indicate knots with sheath-like identity. (C) Expression of KN1 and GA2OX1 in cells from normal (WT) and Kn1-O/+ siblings and (D) the resulting indeterminate-to-determinate cell differentiation trajectory reconstruction. (E) Density plot showing cell distribution over pseudotime from WT and Kn1-O/+ siblings. (F) Box plot displaying median pseudotime value for cells from WT and Kn1-O/+ siblings, box edges represent the 75th and 25th percentiles, Wilcoxon rank sum test, P = 0.003. (G) Volcano plot showing differentially expressed genes (red) in Kn1-O/+ determinate cells ectopically expressing GA2OX1 versus WT determinate cells. (H) Model for the effect of overexpression of KN1 in leaves. Increased sheath growth and ectopic sheath development in Kn1-O/+ promotes more rapid progression of leaf ontogeny.
To examine the effects of ectopic KN1 expression on pseudotime progression in the maize seedling shoot, we isolated protoplasts from normal WT and Kn1-O/+ sibling SAM + P6 tissue and subjected them to scRNA-seq, capturing a total of 2,761 cells spanning an indeterminate-to-determinate cell identity transition (median transcripts detected = 7,235, median genes detected = 3,427 (SI Appendix, Fig. S12). While KN1 and its direct target GA2OX1 were primarily expressed only in indeterminate cells in WT, both genes were more broadly expressed in determinate cell types in the Kn1-O/+ background (KN1 adj. P < 1E-8, GA2OX1 adj. P < 0.01, Fig. 5C). Trajectory reconstruction revealed a shift in cell-fate progression in Kn1-O/+ relative to WT cell populations (Fig. 5 D and E). Surprisingly, cells from the Kn1-O/+ background were significantly more advanced in their pseudotime progression than cells from WT (Fig. 5F, Wilcoxon rank sum test, P = 0.003), suggesting that KN1 activity in the leaf promotes rather than delays cell differentiation. To evaluate the gene expression signatures associated with ectopic KN1 activity, we compared the differentially expressed genes in cells ectopically expressing GA2OX1 from the Kn1-O/+ background to determinate cells from the WT background. By targeting cells ectopically expressing the KN1 direct-target gene GA2OX1, we increased the likelihood of analyzing cells in which ectopic KN1 is actively modulating gene expression. As expected, ectopic expression of GA2OX1 is associated with increased KN1 expression (Fig. 5G). However, we also detected up-regulated expression of key determinacy-promoting genes such as DROOPING LEAF1 (DRL1) and YABBY15 (YAB15), suggesting that these cells have retained or partially enhanced determinacy (35). Alongside these genes we observed up-regulated expression of several genes encoding putative cell-wall-modifying enzymes as well as five genes belonging to the LIGHT SENSITIVE HYPOCOTYL (LSH) family (Dataset S11). Intriguingly, RNA in situ hybridization revealed that transcripts of one of the up-regulated LSH-LIKE genes (GRMZM2G816289) accumulates at leaf primordia bases, in a domain that overlaps that of the developing sheath (SI Appendix, Fig. S10H). Taken together, these results reveal that when misexpressed in leaves, KN1 promotes sheath cell differentiation and accelerates the ontogenetic progression of leaf development as reflected by changes in cell transcriptomes over pseudotime (Fig. 5H).
Discussion
Here we present a single-cell transcriptomic survey of a plant vegetative shoot apex including the stem cells housed within the SAM. This technique provides the key advantage of uncovering cell types in an unbiased fashion, without strict reliance on histological or genetic markers. Critically, we identify known and novel markers of the putative SAM stem-cell niche and show that it is characterized by increased expression of DNA methylation and DNA repair-related genes, as well as a low cell division rate. This observation supports the notion that only a subset of cells at the SAM tip has the specialized properties of stem cells and may underlie the ability of plants to maintain high intergenerational genetic fidelity despite the absence of embryonic segregation of the germline as is found in animals (8). In addition, a low cell division rate and the up-regulated expression of genes with genome protective functions within the same cells highlights a convergently evolved solution to maintaining stem cells in postembryonic plant and animal development (11, 36). Questions are raised regarding the lineage contributions of these stem cells to the developing plant. For example, do cells comprising the SAM tip make significant contributions to lateral organs or are they preferentially destined to contribute to germ fates as described in the “meristem d’attente” (“meristem in waiting”) model proposed by early histological studies (1)? In the latter view, plant lateral organs are mostly derived from the transit-amplifying cell population that subtends the stem-cell domain, and the SAM tip makes minimal contributions to organogenesis until reproductive stages. Emerging cell lineage tracing methodologies may shed light on these questions (37). Meanwhile, we find a lack of evidence for a WUS-expressing organizing center in the B73 maize inbred vegetative SAM, unlike what is modeled in other angiosperms, suggesting that genes with other, noncanonical, stem-cell organizing functions may be at play.
Furthermore, by leveraging the continuum of cells ranging from indeterminate to determinate cell identities, we reconstruct differentiation trajectories for cells of the seedling vegetative shoot apex, offering unprecedented resolution into shoot cell differentiation. Many of the genes that are dynamically expressed along the trajectories at the leaf initiation stage show similar accumulation patterns at postinitiation stages of leaf ontogeny, demonstrating the iterative process of plant shoot patterning.
Surprisingly, we found that perturbing the shoot differentiation process via ectopic expression of the homeobox transcription factor gene KN1 accelerates, rather than delays, cell differentiation in the seedling shoot. This is in contrast to previous models of KN1 action, in which KN1 promotes stem-cell indeterminacy in the SAM, and ectopic KN1 expression in leaf primordia delays or reverses the maturation schedule of maize leaves (38). Indeed, ectopic class I KNOX gene expression has been shown to drive the formation of ectopic meristems in eudicot leaves (2). However, we propose that ectopic KN1 expression accelerates pseudotime progression in the monocot maize leaf by promoting increased and/or premature formation of sheath tissue. Clonal analyses of cell-fate acquisition in the wild-type maize leaf demonstrate that cells fated to adopt blade identity emerge from the SAM first, while cells that will become sheath emerge later in ontogeny (29). Notably, sheath is the last part of the maize leaf to emerge from the SAM and stem and is the last part to elongate. In this way, the young maize leaf primordia within our wild-type SAM + P6 seedling samples are predominantly composed of cells that are fated to form blade. Multiple studies have noted that KN1 transcript and protein accumulate in the extreme proximal bases of maize leaf primordia, starting from the time of leaf inception in the P0, and concluded that KN1 specifies proximal sheath identity in these leaf bases (24, 39). At the same time, sheath tissue emerges from the SAM/stem much later in ontogeny than blade tissue; indeed, studies of polar auxin transport inhibitors have shown that the most proximal region of the sheath does not emerge from the intercalary meristem of the stem until as late as P4 (40). Intriguingly, transcripts of the maize class I KNOX genes KN1 and RS1 accumulate in the the intercalary meristem of maize (41). Thus, while these proximal sheath cells are more “meristematic” in the sense of their proximity to and recent emergence from an indeterminate growth zone, acquisition of sheath cell fate occurs late in the developmental ontogeny of the leaf. We therefore propose a model in which misexpression of KN1 advances leaf ontogeny by promoting ectopic patches of sheath in young leaf primordia that are mostly blade tissue (Fig. 5H), thereby accelerating the progression of cell-fate acquisition in the maize leaf.
Materials and Methods
Plant Materials and Growth Conditions.
Plants for scRNA-seq, in situ hybridization, and gras allele phenotyping were grown in 72-well trays in a Percival A100 growth chamber under 16-h days, a day temperature of 29.4 °C, a night temperature of 23.9 °C, and a relative humidity of 50%. Soil consisted of a 1:1 mixture of Turface MVP and LM111. The maize inbred B73 was used for scRNA-seq and in situ hybridization analyses. The gras32 and gras33 alleles were obtained from the Maize Genetics Co-Op Center (Urbana, IL) in the W22 inbred background. Crosses were performed at a field site in Aurora, NY. Kn1-O/+ and WT sibling material along with seedlings and mature plants for ZmWUS1/2 RT-PCR experiments were grown at the Gutermann Greenhouse Facility in Ithaca, NY. Well-introgressed Kn1-O/+ and lg1-R mutants in the B73 genetic background were utilized for genetic interaction analysis.
In Situ Hybridization.
RNA was isolated by TRIzol (Thermo Fisher Scientific) extraction from liquid nitrogen-ground 2-wk-old maize seedling shoot apices. Total RNA was DNase I (Promega) treated and cDNA was prepared using polyT-primed SuperScript III reverse transcriptase (Thermo Fisher Scientific) according to the manufacturer’s instructions. cDNA was then used as a template to amplify probe sequences, which were TA cloned into the pCR4-TOPO vector backbone encoding flanking T3 and T7 polymerase promoters (Thermo Fisher Scientific) (see Dataset S12 for PCR primers). Following the verification of probe sequence and orientation by Sanger sequencing, antisense RNA probes were generated using a DIG-labeling kit and the resulting probes hydrolyzed as previously described (Roche Diagnostics) (42). A locked nucleic acid (LNA) probe was used for the WOX9c in situ presented in Fig. 3C and was ordered directly from Qiagen. LNA probe hybridization was carried out using 10 μM probe concentration and a 55 °C hybridization temperature according to published methods (43).
Tissues for in situ hybridization were prepared and processed as previously described (42). Briefly, 2-wk-old maize shoot apices were fixed overnight at 4 °C in FAA solution (3.7% formalin, 5% acetic acid, and 50% ethanol in water) and dehydrated through an ethanol series, cleared through increasing concentrations of Histo-Clear II (National Diagnostics), and then embedded in paraplast. Sections of 10 μm were prepared using a Leica RM2235 microtome (Leica Biosystems) and adhered to Probe-on-Plus microscope slides (Thermo Fisher Scientific). Sectioned tissues were then deparaffinized and rehydrated through a reverse ethanol series prior to Proteinase K (Sigma-Aldrich) treatment, refixation, and acetic anhydride treatment. Dehydrated tissues were then hybridized with probe overnight, washed, RNase A (Roche Diganostics) treated, and incubated with AP-conjugated anti-DIG antibody (Roche Diagnostics). A colorimetric nitro-blue tetrazolium/5-bromo-4-chloro-3′-indolyphosphate (Roche Diagnostics) reaction was then allowed to proceed until sufficient signal developed at which point the reaction was stopped in Tris(hydroxymethyl)aminomethane–ethylenediaminetetraacetic acid (EDTA), slides were dehydrated, washed in Histo-Clear II, and mounted with Permount (Thermo Fisher Scientific). Images were obtained using an Axio Imager Z10 (Carl Zeiss Microscopy) microscope equipped with an AxioCam MRc5 camera.
RT-PCR of ZmWUS1/2 Expression.
Total RNA was isolated (as in in situ hybridization) from B73 shoot apices consisting of the SAM and the three most recently initiated primordia, immature ∼2-mm ears containing inflorescence meristems, and mature seedling leaf 3 blade tissue. A total of 1 µg of total RNA from each tissue was DNase-I treated and used as input for reverse transcription using SuperScript III reverse transcriptase according to the manufacturer’s instructions. A total of 1 µL of the 25 µL RT reaction was used for PCR analysis (35 cycles). PCR products were run on a 2% agarose gel and visualized with ethidium bromide under ultraviolet (UV) light. The genomic DNA (gDNA) control was derived from B73 seedling leaf tissue prepared as previously described (44).
Phenotyping.
For gras allele phenotyping, genomic DNA was extracted from the leaf tissue of F2 seedlings segregating for exonic gras32 and gras33 Mutator transposon insertion alleles. DNA extraction was performed as previously described (44). Plants were genotyped by PCR (see Dataset S12 for primers) and paraffin-embedded FAA-fixed shoot apex tissues (see in situ hybridization) were longitudinally sectioned to 10 μm and adhered to Probe-on-Plus slides. Tissues were deparaffinized and rehydrated through an ethanol series and then equilibrated in 1% sodium borate (wt/vol). Tissues were then stained in a 0.5% solution of o-toluidine (TBO) in 1% sodium borate for 5 min followed by an ethanol dehydration series, washing in Histo-Clear II, and mounting in Permount. Samples were then imaged (see in situ hybridization). SAM width and height were determined in medial sections using ImageJ. For analyses of Kn1-O/+ seedlings, leaf sheath and blade lengths were measured by hand with a meter stick when seedling leaves were fully mature. For Kn1-O/+ and lg1-R genetic interaction analysis, F1BC1 populations were generated, and greenhouse-grown Kn1-O/+; lg1 double mutants were selected based on phenotypic analysis of the blade/sheath boundary, where lg1-R mutants remove ligule and auricle tissues (SI Appendix, Fig. S10). Fully adult leaves were imaged, selecting the leaf above the uppermost ear, with a Canon Rebel digital single-lens reflex camera.
Cell-Cycle Quantification.
Medial SAM sections probed by in situ hybridization for expression of the S phase and G2/M phase up-regulated H3 and CYC1 genes, respectively, were imaged and imported into ImageJ. Images were converted to 16-bit format and processed using the thresholding tool such that stained areas could be differentiated from nonstained areas. For each SAM, five sections of equal height were measured and the percent ratio of above threshold (stained) area relative to the total area of each bin was calculated.
Generation and Collection of Protoplasts.
Two-week-old maize seedlings were hand dissected to either an ∼3-mm portion of the stem including the six most recently initiated leaf primordia (SAM + P6) or the SAM and the two most recently intiated leaf primordia (SAM + P2). Dissected tissue was briefly macerated and placed immediately in protoplasting solution, which consisted of 0.65 M mannitol, 1.5% Cellulase Onozuka R-10 (Research Products International [RPI]), 1.5% Cellulase Onozuka RS (RPI), 1.0% Macerozyme R-10 (RPI), 1.0% hemicellulase (Sigma-Aldrich), 10 mM 3-(N-morpholino)propanesulfonic acid (MOPS) pH 7.5, 10 mM l-arginine HCl pH 7.5, 1 mM CaCl2, 5 mM β-mercaptoethanol, and 0.1% bovine serum albumin (BSA) (Sigma-Aldrich). Prior to the addition of CaCl2, β-mercaptoethanol, and BSA, the solution was heated to 55 °C for 10 min to facilitate enzyme solubilization. All tissue collection was completed within 30 to 45 min. Tissue digestion was carried out with gentle shaking at 29 °C for 2 h. After digestion, fluorescein diacetate (FDA) was added to the cell suspension at a concentration of 5 μg/mL and cells were allowed to incubate in darkness for 5 min. The cell suspension was then filtered using a 40-μm nylon filter and the cells were centrifuged at 250 × g for 3 min at 4 °C. Cells were resuspended in washing buffer consisting of 0.65 M mannitol, 10 mM MOPS pH 7.5, and 10 mM l-Arginine pH 7.5 and washed three times using the same centrifugation conditions. Cell viability and concentration were assessed using a hemocytometer and a fluorescent Axio Imager Z10 microscope equipped with a 488-nm laser for FDA staining detection. If cell/debris clumping was observed, the suspension was again passed through a 40-μm nylon filter.
Cells isolated from SAM + P6 tissue were suspended at a concentration of ∼10,000 cells/mL and loaded onto the 10× Genomics Chromium Controller using v3 reagents following the manufacturer’s instructions to target ∼10,000 cells. Cells from the SAM + P2 tissue were resuspended in 1 mL of wash buffer and a 100- to 200-μL aliquot was transferred to the well of a clear-bottom CoStar plate kept over ice. A total of 200 μL of wash buffer was distributed to other wells on the plate. A Leica M205 FCA microscope equipped with a 488-nm laser was then used to transfer individual viable (FDA+) cells. Each cell was carried in 0.1-μL volumes through three wash buffer wells. Following washing, cells were transferred to a 96-well LoBind (Eppendorf) plate containing reagents for reverse transcription (see library construction and sequencing) and kept on dry ice. Cell collection was completed in <1 h. Plates were sealed with adhesive film and transferred to a −80 °C freezer prior to further processing.
Cell Viability Assays.
Protoplasts were obtained as described (see generation and collection of protoplasts) with the exception of the pH and concentration of buffer components used. For the pH 5.7 condition, 10 mM 2-(N-morpholino)ethanesulfonic acid buffer was used, whereas for the pH 6.5, 7.0, and 7.5 conditions, 10 mM MOPS buffer was used. To quantify cell viability, the ratio of fluorescing (living) protoplasts to dead (nonfluorescing) and large debris (>5 μm) were quantified in each of the four larger corners of a hemocytometer using a fluorescent Axio Imager Z10 microscope equipped with a 488-nm laser.
SAM + P2 Single-Cell RNA Isolation and Amplification.
Single-cell RNA-seq library construction was performed using an adaptation of the Cel-Seq2 protocol (45). The 96-well LoBind plates were prepared with each well containing 0.22 μL 25 ng/μL Cel-Seq2 RT primer (1s–96s primers, Dataset S12), 0.11 μL 10 mM deoxynucleoside triphosphates (dNTPs), and 0.77 μL nuclease-free H2O such that each well of the plate contained a unique cell barcoded RT primer. After cell collection and storage at −80 °C, plates were thawed briefly on ice and centrifuged at 2,000 × g for 2 min at 4 °C. Next, plates were incubated at 65 °C for 5 min and again centrifuged using the same settings. For reverse transcription, 0.54 μL First Strand Buffer, 0.27 μL 0.1 M dithiothreitol (DTT), 0.135 μL RNAseOUT (Thermo Fisher Scientific), 0.034 μL SuperScript III, and 0.52 μL nuclease-free H2O were added to each well and plates were incubated for 1 h at 42 °C followed by RT inactivation for 10 min at 70 °C. cDNA was pooled horizontally into the eight wells at the end of each plate. Then, 2.5 μL 10× Exonuclease I buffer and 2.1 μL Exonuclease I were added to each well and incubated at 37 °C for 15 min followed by heat inactivation at 80 °C for 15 min. cDNA was purified using Ampure RNAClean XP beads according to the manufacturer’s instructions and resuspended in 7 μL nuclease-free H2O. Second strand synthesis was performed by adding 2.31 μL Second Strand buffer, 0.23 μL dNTPs, 0.08 μL Escherichia coli DNA ligase, 0.3 μL E. coli DNA polymerase, and 0.08 μL RNase H and incubated at 16 °C for 2 h. Pooled dsDNA from each of the eight reactions was pooled and then purified using Ampure XP beads according to the manufacturer’s instructions followed by resuspension in 6.4 μL nuclease-free H2O. In vitro transcription was performed by the addition of 1.6 μL each of the A, G, C, U dNTPs, 10× T7 polymerase buffer, and T7 polymerase (Ambion) followed by incubation at 37 °C for 13 h. Amplified RNA was then treated with ExoSAP-IT (Thermo Fisher Scientific) for 15 min at 37 °C followed by RNA fragmentation by addition of 5.5 μL fragmentation buffer (200 mM Tris pH 8.0, 150 mM MgCl2) and incubation for 3 min at 94 °C. Fragmentation was stopped by transfer to ice and the immediate addition of 2.75 μL 0.5 M EDTA pH 8.0.
Library Construction and Sequencing.
For reverse transcription, 5 μL of the amplified RNA was added to 1 μL randomhexRT primer and 0.5 μL dNTPs, incubated at 65 °C for 5 min, and chilled on ice. Next, 2 μL First Strand buffer, 1 μL 0.1 M DTT, 0.5 μL RNAseOUT, and 0.5 μL SuperScript III were added and the samples incubated for 10 min at 25 °C followed by a 1-h incubation at 42 °C. Half of the completed RT reaction was subjected to PCR by addition of 5.5 μL nuclease-free H2O, 12.5 μL Phusion High-Fidelity PCR Master Mix with HF buffer (New England Biolabs), RNA PCR Primer1 (RP1), and 1 μL of RNA PCR Primer X (RPIX). PCR conditions were as follows: 30 s at 98 °C, 11 cycles of 10 s at 98 °C, 30 s at 60 °C, 30 s at 72 °C, and 10 min at 72 °C. The pooled cDNA from each plate received a unique RPIX. Samples were then purified and size selected via two rounds of AMPure XP bead treatment using a 1:1 ratio of beads-to-sample and the final library was resuspended in 10 μL nuclease-free H2O. A total of 1 μL of the library was submitted for fragment analysis using an Agilent Bioanalyzer to confirm a target library size between 200 and 400 bp. An additional 1 μL was used for concentration measurement using a Qubit. If the library concentration was suboptimal, the second unused half of the RT reaction was amplified using up to 15 PCR cycles. Libraries were then sequenced using a single flow cell on an Illumina NextSeq 500 instrument using the small RNA chemistry. Paired-end sequencing was performed with 15 and 77 bp obtained for read 1 and read 2, respectively. The libraries generated from the biological replicates of SAM + P6 cells were also sequenced using a NextSeq 500 instrument, with each replicate allocated a single flow cell of sequencing. For experiments using cells from sibling Kn1-O/+ and WT seedlings, barcoded libraries were pooled together prior to sequencing.
Single-Cell RNA-Seq Read Processing and Cell Filtering.
SAM + P2 FASTQ files were processed using the default settings in the celseq2 pipeline (https://github.com/yanailab/celseq2), which includes read trimming, alignment, and unique molecular identifier (UMI) counting steps to generate a UMI count matrix (45). Reads were aligned to version 3 of the B73 reference genome. SAM + P6 reads were trimmed, aligned, and UMI count matrices generated using the CellRanger version 3.1.0 pipeline under the default settings. Reads were aligned to version 3 of the B73 reference genome. The UMI count matrices for individual biological replicates were merged prior to further analysis. For the SAM + P2 dataset, cells with fewer than 500 genes detected were removed while in the SAM + P6 dataset; cells with fewer than 2,500 genes detected were removed. In both datasets, cells with over 1% of transcripts encoded by the mitochondrial genome were removed.
Dimensionality Reduction, Cell-Type Classification, and Differential Expression Analysis.
Cell-type analysis and clustering were performed using Seurat v3.0 (46). The merged UMI count matrices were converted to Seurat objects. Normalization and variance stabilization were performed using SCTransform and the 3,000 genes with the highest expression variability were used for the calculation of principal components. UMAP was then used to embed cells in lower dimensional space for data visualization. For projection of SAM + P2 cells, UMAP was run using dimensions (dim) = 1:5, number of neighbors (n.neighbors) = 15, minimum distance (min.dist) = 0.1, and spread = 5. For the projection of SAM + P6 cells, UMAP was run using dim = 1:25, n.neighbors = 25, min.dist = 0.01, and spread = 1. For the SAM + P6 subset cells, cells belonging to clusters 5 and 0 were reclustered in isolation using the same parameters as for the full dataset. Cells were assigned to clusters using k-means hierarchical clustering. All differential expression analyses used to compare gene expression on a per-cluster basis were performed using Wilcoxon ranked-sum tests using the Seurat FindMarkers function. GO enrichment analysis was done using a Fisher’s exact test implemented in AgriGO v2 (47). Cell-cycle regression was used to reduce the effects of the cell cycle on cell clustering in the SAM + P2 dataset. Differentially expressed genes among cells belonging to cell clusters with S-phase and G2/M-phase marker gene expression were first identified (adjusted P value <0.05). Genes that were highly specific for these clusters were identified using a ratio of the number of cells expressing a given differentially expressed gene within the cluster to those in all clusters. Those with a ratio greater than 2 were deemed phase specific and their expression was used to calculate a numeric cell-cycle score and a cell-cycle stage (G1, S, and G2/M) implemented in Seurat (46). SCTransform was run again on the raw UMI count matrix with cell-cycle score as a variable to regress, followed by principal component analysis and UMAP dimensionality reduction.
Trajectory Inference and Pseudotime Analysis.
Trajectory inference and pseudotime analysis was performed using Monocle3 (https://cole-trapnell-lab.github.io/monocle3) (48). A principal graph was generated using the learn graph function and cells were assigned a pseudotime value using order_cells with a pseudotime start or “root” position manually selected. Genes that were differentially expressed along the inferred trajectories were identified using the graph_test function, which applies a Moran’s I test to detect spatial autocorrelation. For the SAM + P2 dataset, individual Moran’s I tests were performed on cell subsets to better identify genes with branch-specific expression patterns. This involved two tests on a common population of cells derived from the tip, meristem 1, and meristem 2 clusters merged with cells from the Primordia and Vasculature clusters or the epidermis 1 and 2 clusters. The significantly differentially expressed genes along both trajectories were then further analyzed. For the visualization and analysis of pseudotime-dependent gene expression patterns, cubic smoothing splines were fit to each gene using the R smooth.spline function with a spar parameter of 1.1. To compare the expression behavior of genes in the SAM + P2 and SAM + P6 datasets, smoothed expression profiles for each gene were averaged in 10 pseudotime bins and z-scaled values for each bin were calculated. The Fréchet distances between the curves of identical genes and all nonidentical genes were calculated using the SimilarityMeasures R package (49).
Phylogenetic Analysis.
For the phylogenetic analysis of GRAS proteins, amino acid sequences of all Arabidopsis GRAS proteins and HAM-LIKE homologs in rice (Oryza sativa) and maize were downloaded from Phytozome. Amino acid sequences were then aligned using Clustal Omega. Maximum likelihood phylogenetic tree construction was performed using PhyML. The Jones–Taylor–Thornton (JTT) amino acid substitution model was selected based on its Akaike information criterion (AIC) calculated using smart model selection (SMS) implemented in PhyML (50). Branch support values were calculated using the aLRT SH-LIKE fast likelihood-based method (51).
Acknowledgements
We thank the Cornell University Biological Resource Center for assistance in single-cell genomics and sequencing techniques, as well as J. Cammarata and A. Roeder for critical feedback on the manuscript. We thank S. Hake for discussions of Kn1 mutants. This work was funded by NSF grants IOS-2016021 and IOS-1238142 (to M.J.S.), a Schmittau-Novak small grant (to J.W.S.), and NSF Postdoctoral Research Fellowship in Biology grant IOS-1710973 (to J.S.).
Data Availability.
The sequence data have been deposited in the National Center for Biotechnology Information Short Reads Archive (PRJNA637882). Plant materials are available upon request.
References
1
2
3
4
5
6
7
9
10
12
13
14
15
16
17
18
20
21
22
23
24
25
26
27
28
29
30
31
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
 Plant stem-cell organization and differentiation at single-cell resolution
Plant stem-cell organization and differentiation at single-cell resolution
