
Edited by Stephen G. Young, David Geffen School of Medicine at UCLA, Los Angeles, CA, and approved November 2, 2020 (received for review July 12, 2020)
Author contributions: M.R.d.l.S. and P.M.K. designed research; M.R.d.l.S., M.R., F.S.D., A.S., J.P., D.T., L.M.V., A.V., K.S., D.M., M.L., G.V., A.K., B.F.-Z., L.W., B.T., P.N.R., D.H., S.M., U.K., A.J.B., D.S., Y.W., and P.M.K. performed research; B.F.-Z., L.W., B.T., S.M., U.K., A.J.B., D.S., and Y.W. contributed new reagents/analytic tools; M.R.d.l.S., M.R., F.S.D., A.S., J.P., and K.S. analyzed data; and M.R.d.l.S., M.R., F.S.D., J.P., P.N.R., D.H., and P.M.K. wrote the paper.
- Altmetric
Inherited GPI-anchor biosynthesis deficiencies (IGDs) explain many cases of syndromic intellectual disability. Although diagnostic methods are improving, the pathophysiology underlying the disease remains unclear. Furthermore, we lack rodent models suitable for characterizing cognitive and social disabilities. To address this issue, we generated a viable mouse model for an IGD that mirrors the condition in human patients with a behavioral phenotype and susceptibility to epilepsy. Using this model, we obtained neurological insights such as deficits in synaptic transmission that will facilitate understanding of the pathophysiology of IGDs.
Pathogenic germline mutations in PIGV lead to glycosylphosphatidylinositol biosynthesis deficiency (GPIBD). Individuals with pathogenic biallelic mutations in genes of the glycosylphosphatidylinositol (GPI)-anchor pathway exhibit cognitive impairments, motor delay, and often epilepsy. Thus far, the pathophysiology underlying the disease remains unclear, and suitable rodent models that mirror all symptoms observed in human patients have not been available. Therefore, we used CRISPR-Cas9 to introduce the most prevalent hypomorphic missense mutation in European patients, Pigv:c.1022C > A (p.A341E), at a site that is conserved in mice. Mirroring the human pathology, mutant Pigv341E mice exhibited deficits in motor coordination, cognitive impairments, and alterations in sociability and sleep patterns, as well as increased seizure susceptibility. Furthermore, immunohistochemistry revealed reduced synaptophysin immunoreactivity in Pigv341E mice, and electrophysiology recordings showed decreased hippocampal synaptic transmission that could underlie impaired memory formation. In single-cell RNA sequencing, Pigv341E-hippocampal cells exhibited changes in gene expression, most prominently in a subtype of microglia and subicular neurons. A significant reduction in Abl1 transcript levels in several cell clusters suggested a link to the signaling pathway of GPI-anchored ephrins. We also observed elevated levels of Hdc transcripts, which might affect histamine metabolism with consequences for circadian rhythm. This mouse model will not only open the doors to further investigation into the pathophysiology of GPIBD, but will also deepen our understanding of the role of GPI-anchor–related pathways in brain development.
The glycosylphosphatidylinositol (GPI) anchor is essential for connecting a remarkable number of proteins (GPI-linked proteins) to the cell membrane. GPI-linked proteins are essential for signal transduction, cell–cell adhesion, axonal outgrowth, synapse formation, and plasticity, as well as for regulation of the complement system (1, 2). Paroxysmal nocturnal hemoglobinuria (PNH) was the first disorder to be characterized as a GPI-anchor biosynthesis deficiency (GPIBD) (3). However, PNH is exceptional in two regards: First, it is the only GPIBD that is acquired and it is due to somatic mutations that cause complete loss of function. In inherited GPIBDs, also referred to as inherited GPI-anchor biosynthesis deficiencies (IGDs), residual GPI-anchor synthesis and maturation activities persist. Second, the prevalence of inherited GPIBDs is at least 10-fold higher than that of PNH. To date, recessive phenotypes have been reported for 21 genes of the GPI-anchor pathway. Bellai-Dussault et al. discussed the clinical variability in detail for the first 19 GPIBDs (4). However, most patients, including recently described cases due to GPIBD20 and GPIBD21, exhibit intellectual disability, psychomotor delay, and epilepsy (5, 6). Furthermore, due to the residual GPI-anchor synthesis and maturation, patient-derived fibroblasts have a reduced number of GPI-linked proteins on the cell surface (7).
Prior to the discovery of IGD, mouse models of GPI-anchor deficiency (891011–12), which mainly employed chimeric and conditional knockouts in which GPI-anchor biosynthesis was abolished in specific tissues, demonstrated that a complete loss of GPI anchors is embryonic lethal (9). Interestingly, the resultant phenotypes were often still so severe that the mutant mice died early, suggesting essential functions of GPI-anchor proteins in the skin, development of white matter, and dendritic arborization of Purkinje cells in the cerebellum (8, 10). In recent years, mice with constitutional GPIBDs were identified in mutation screens; these animals were viable probably because the mutations were only hypomorphic or affected isoforms that are limited to certain tissues, and therefore only explain some aspects of most inherited GPIBDs (13, 14). Lukacs et al. (13) showed that a missense mutation in Pgap2, p.(M1V), compromised transcription of this gene particularly in neural crest cells, resulting in a craniofacial phenotype in mutant mice. In contrast to most other genes involved in GPI-anchor biosynthesis and maturation, Pgap2 has a tissue-specific expression pattern that changes over embryonic development, and the existence of multiple isoforms in humans further complicates phenotypic comparisons between humans and mice. It is likely that the facial abnormalities described in human patients with a similar mutation, p.(M1R), are due to a similar mechanism; however, the other phenotypic features seem to be milder in humans, either because the substituted amino acid is different or because the isoforms resulting in p.(M52R) and p.(M58R) do not exist in mice. In addition, McKean and Niswander observed a holoprosencephaly-like phenotype in two mouse models with a frame-shift mutation in Pign and an in-frame deletion in Pgap1 (14).
Because most of the existing mouse models die at an early stage due to the severe phenotype, it has not been possible to use these models to characterize cognitive deficits, which represent the main challenge in individuals with IGDs. For this purpose, we used CRISPR-Cas9 to engineer a mouse model with the missense mutation c.1022C > A, p.A341E in Pigv, one of the most frequently encountered pathogenic alleles in humans (15). PIGV encodes mannosyl transferase II, which is essential for the attachment of the second mannose to the GPI anchor (16). Due to the residual function of Pigv341E, mutant mice are viable with a normal life span, making it possible to complete behavioral experiments that test motor, social, and cognitive abilities, and study the brain tissue, as well as cells of these mice.
Results
Patients with IGDs exhibit a heterogeneous spectrum of symptoms, including neurologic findings, movement disorders, and intellectual disability (7, 1718–19). In general, IGDs caused by pathogenic mutations in genes that catalyze the early steps of GPI-anchor synthesis, such as PIGA, tend to have more severe clinical features such as status epilepticus. In patients with mutations in later steps of synthesis, such as PIGV, epilepsies also occur in a substantial proportion of cases; however, they often disappear later in life, and intellectual disability becomes the key clinical feature. In contrast to acquired GPIBDs, such as PNH, all patients with IGDs have some residual function of GPI-anchor synthesis, implying that null mutants are not viable and that at least one hypomorphic allele must be present. When a novel mutation is encountered in a suspected IGD, flow cytometry with two or more different markers serves to confirm a GPIBD. To analyze the effect of Pigv341E on a cellular level, we used fluor-proaerolysin (FLAER), which can recognize all GPI-anchored proteins, and CD90, a GPI-linked protein that is highly expressed in human fibroblasts. The mean fluorescence intensity (MFI) of FLAER was reduced in hom-Pigv341E mouse embryonic fibroblasts (MEFs) (Fig. 1A). Likewise, CRISPR-Cas9–engineered mouse embryonic stem (mES) cell clones (hom-Pigv341E, hom-Pgap3107L) exhibited partial reductions in FLAER and GPI-anchored CD90 (SI Appendix, Fig. S1E). Unlike clones with homozygous hypomorphic mutations, the null mES clone [Pigv (−/−)] exhibited almost no cell-surface expression of FLAER and CD90 (SI Appendix, Fig. S1E). Therefore, we concluded that Pigv341E is hypomorphic in mice, as it is in humans.

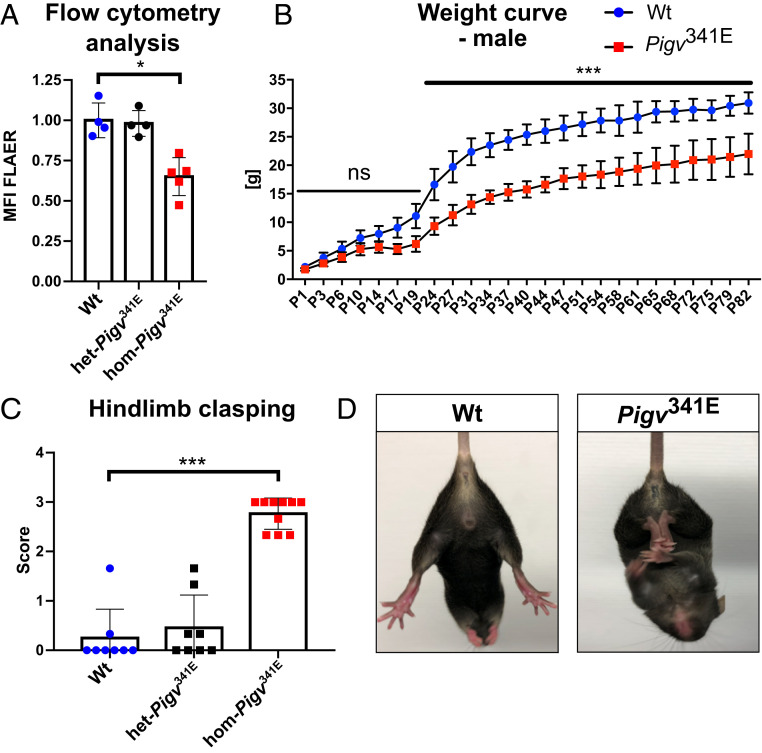
Features of Pigv341E mice. (A) Flow cytometry analysis of hom-Pigv341E MEFs isolated from embryos (E13.5) revealed a reduced MFI of FLAER. MFI was normalized against the wild type. (B) Male Pigv341E mice had a reduced weight on postnatal days (P) 1 to 82. (C) Hom-Pigv341E mice exhibited hindlimb clasping behavior. (D) Representative posture of hindlimb clasping behavior in hom-Pigv341E mice. By contrast, wild-type mice spread their hindlimbs when picked up by their tail. Hom-Pigv341E = homozygous for Pigv p.Ala341Glu; het-Pigv341E = heterozygous for Pigv p.Ala341Glu. Animals used for the weight curve: WT(male = 3), hom(male = 4). Animals used for the hindlimb clasping test were 6 wk old: WT(female n = 3, male n = 5), het-Pigv341E(female n = 4, male n = 4), hom-Pigv341E(female n = 4, male= n = 6). Data from the weight curve were analyzed by two-way ANOVA followed by Bonferroni’s multiple comparisons test. The data from flow cytometry and the hindlimb clasping test were analyzed by nonparametric t test (Mann–Whitney). *P < 0.05, ***P < 0.001. ns, not significant.
The results section is structured as follows: We will start with a description of the findings from behavioral experiments. Some of the dysfunctional behavior that we encountered, motivated further histopathological and electrophysiological analysis of the hippocampus that pointed to a synaptopathy. In the end, we present the results of a single cell transcriptome screen that we conducted to identify differentially expressed genes that might be involved in the observed pathophysiology.
Characteristic Features and Alterations of Sleep Patterns in Pigv341E Mice.
The most prominent differences that we observed first between Pigv341E and wild-type (WT) mice were reduced weight (Fig. 1B and SI Appendix, Fig. S1F) and hindlimb clasping behavior (Fig. 1 C and D). Due to the intellectual disability and psychomotor delay that are the key clinical features of IGD, patients are impaired in their everyday lives. Therefore, we sought to determine which spontaneous behaviors our mouse model exhibited while living undisturbed in their home cage (singly or group housed). Using HomeCageScan (HCS), we monitored singly housed Pigv341E mice for 23 h at two different time points (8 and 16 wk). Among the 19 behaviors accurately detected by analysis of the HCS data, three behaviors were consistently altered in Pigv341E mice at both time points. Pigv341E mice hung less often to the top of the cage (total occurrences) and for shorter durations (total duration and duration per hour) than wild-type mice; groomed more often (total occurrences) and for longer durations (total duration and duration per hour); and slept less (total duration and duration per hour) (Fig. 2 A and B and SI Appendix, Fig. S2A). At the earlier time point, Pigv341E mice spent more time walking (duration per hour) during the dark phase of the day than wild-type mice (Fig. 2B, Top graph). Furthermore, at both time points, hanging behavior was an important variable for differentiating genotypes along the dimensions of a principal component analysis (PCA) (SI Appendix, Figs. S3 A–F and S4 A–F).

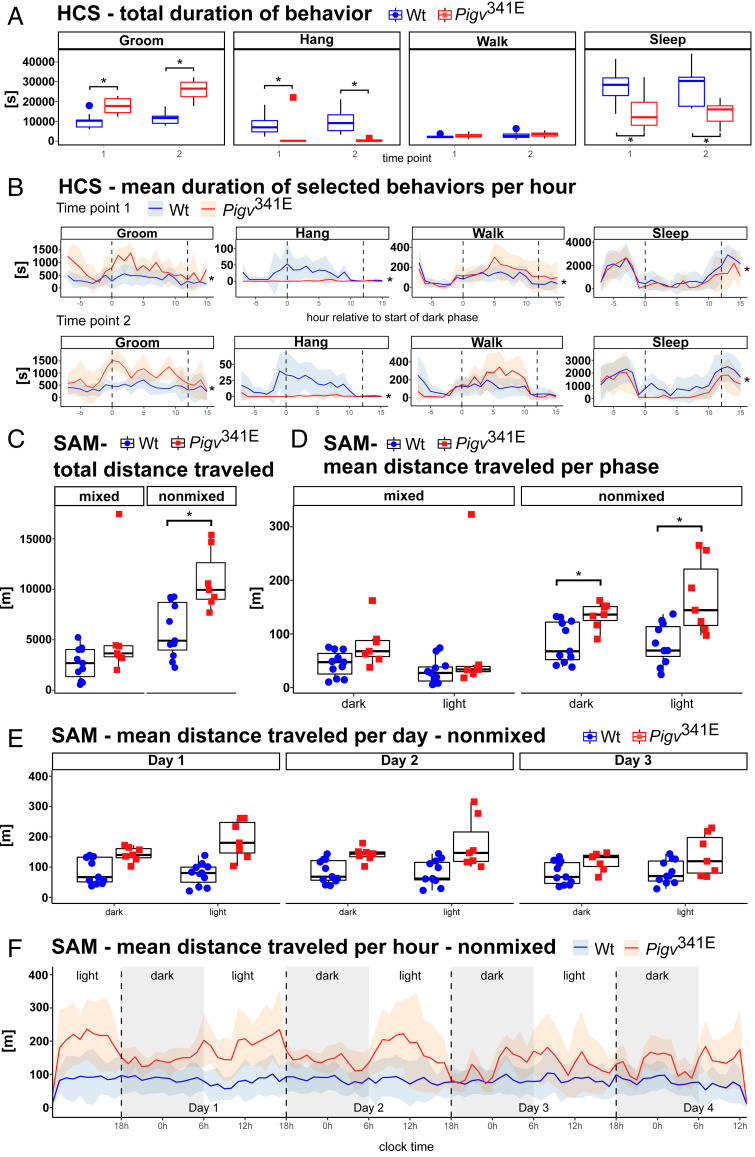
Pigv341E mice exhibit a behavioral phenotype in HCS and an elevated activity level in the SAM (second approach). (A) Duration of selected behaviors from the HCS. Pigv341E mice revealed elevated grooming and reduced hanging and sleeping behavior at both time points. (B) Duration of selected behaviors per hour, as determined by HCS. The dark phase took place between the two dashed vertical lines. Pigv341E mice exhibited elevated grooming and hanging and reduced sleeping at both time points. Pigv341E mice exhibited elevated walking at time point 1. (C) The SAM revealed an elevation in total distance traveled by Pigv341E mice when animals were separated by genotype (nonmixed). When housed in mixed genotypes (mixed), total distance traveled did not significantly differ between genotypes. (D) Pigv341E mice exhibited an increase in distance traveled per phase when separated by genotype (nonmixed). When housed in mixed genotype (mixed), total distance traveled per phase did not significantly differ between genotypes. (E and F) Pigv341E mice traveled longer distances per day and per hour when separated by genotype (nonmixed). Pigv341E = homozygous for Pigv p.Ala341Glu. Animals used for the HCS (WT: female n = 8, male n = 4; Pigv341E: female n = 4, male n = 2) were phenotyped at two different consecutive time points: tp1 at the age of 8 wk, and tp2 at the age of 16 wk. Animals used for the SAM: WT(female n = 4, male n = 6) Pigv341E(female n = 2, male n = 5). Data from the HCS (total duration) and SAM (total distance, mean distance traveled per phase and per day) were analyzed by Wilcoxon rank-sum test (nonparametric). Data from the HCS (duration per hour) and SAM (distance traveled per hour) were analyzed with a glmm using a MCMC. *P < 0.05.
We also used the social activity monitor (SAM) to assess spontaneous home-cage activity in Pigv341E mice while living in a group setting. For this test, we implanted a radio-frequency identification (RFID) transponder into the mice and put the home cage with mixed genotypes on a grid box that could locate individual animals and their position in the cage at all times (continuous 24 h/day recording). Because the animals were undisturbed in their home cage, SAM analysis could be performed several times without the animals noticing. For the first two time points (9 and 17 wk), SAM analysis revealed no difference between genotypes in total distance traveled over 14 d (SI Appendix, Fig. S4G), but an unexpected switch in diurnal/nocturnal activity for both genotypes was observed. The mice were more active during the light (normal sleeping time) than the dark phases of their days (SI Appendix, Fig. S5 A and C; confirmed by Markov chain Monte Carlo generalized linear mixed-effects models [MCMCglmm], pMCMC = 0.001). This observation supported the HCS data that Pigv341E mice slept less, which is a feature of some patients with IGDs who suffer from sleep disturbances (7, 20). We performed a second experiment in which we evaluated the spontaneous activity of the Pigv341E and wild-type mice separately (nonmixed vs. mixed-genotype cages). When Pigv341E and wild-type mice lived together in the same cage (mixed genotype), we reproduced our previous results: Mice of different genotypes exhibited no difference in total distance traveled over 4 d (Fig. 2C, Left graph), per phase (Fig. 2D, Left graph), per day (SI Appendix, Fig. S2C), and per hour (SI Appendix, Fig. S2D), but they still exhibited higher activity levels (larger variability) during the light vs. dark phase (SI Appendix, Fig. S2D, confirmed by MCMCglmm, pMCMC = 0.001). However, in nonmixed housing conditions Pigv341E mice walked greater distances over 4 d (Fig. 2C, Right graph), per phase (Fig. 2D, Right graph), per day, and per hour (Fig. 2 E and F) than wild-type mice, and again, Pigv341E mice were more active during the light than the dark cycle on days 1 and 2 (Fig. 2E).
Pigv341E Mice Exhibit Motor Dysfunction and Alterations in Sociability.
Extensive testing of various motor functions in Pigv341E mice revealed a clear and elaborated dysfunctional motor phenotype. Pigv341E mice had reduced balance and motor coordination, reflected by a reduced latency of falling off the rotarod (Fig. 3A). This was confirmed by an elevated latency of traversing an elevated beam (Fig. 3B). In the rope grip test, Pigv341E mice exhibited an elevated latency of climbing on the rope, and had a lower hanging score than wild-type mice (Fig. 3C and SI Appendix, Fig. S6A, Right graph). Furthermore, Pigv341E mice had reduced grip strength (Fig. 3D). Next, we evaluated the walking pattern of Pigv341E mice using the footprint test (Fig. 3E). Pigv341E mice had a larger distance between forepaw and hindpaw (S) in paw placement of the stride (Fig. 3F); this is remarkable because walking mice usually place their hind paws in the same positions as their forepaws (Fig. 3E, see WT).

![Pigv341E mice exhibit motor dysfunctions. (A and B) Pigv341E mice exhibited a reduction in motor coordination, reflected by a reduced latency of falling off the rotarod and elevated latency of traversal of the beam (diameter, 15 mm). (C) Pigv341E mice exhibited a reduction in climbing performance, reflected by a decrease in the time spent climbing the rope. (D) Pigv341E mice exhibited a reduced grip strength, as determined by grip strength meter. [g] was normalized to the weight of the animal. (E) Representative image of walking pattern between genotypes. SL–FL = stride length–forelimb, SL–HL = stride length–hindlimb, FBW = fore–base width, HBW = hind–base width, S = distance between forepaw and hindpaw. (F) Pigv341E mice had an altered walking pattern, reflected by an increase in the distance between forepaw to hindpaw and a decrease in fore–base width. (G) Pigv341E mice exhibited enhanced social approach behavior, reflected by a decrease in “rear up” behavior and an increase in the number of nose-to-anogenital contacts in the social proximity test. (H) The three-chamber test (first 5 min) indicated that Pigv341E mice exhibited an enhanced social approach behavior relative to wild-type mice, reflected by spending more time with the stranger mouse than with the empty. Pigv341E = homozygous for Pigv p.Ala341Glu. Animals used in the motor tests were 7 wk old: WT(female n = 8, male n = 4) Pigv341E(female n = 4, male n = 2). Animals used in the social proximity test: WT(female n = 9, male n = 11), Pigv341E(female n = 4, male n = 7). Animals used in the three-chamber test: WT(female n = 9, male n = 11), Pigv341E(female n = 4, male n = 6). The data from the motor tests and the social proximity test were analyzed by nonparametric t test (Mann–Whitney). The data from the three-chamber test were analyzed by two-way ANOVA followed by Bonferroni’s multiple comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001. ns, not significant.](/dataresources/secured/content-1765818034270-f44ec551-953a-4eff-bfe1-aa47e23b310b/assets/pnas.2014481118fig03.jpg)
Pigv341E mice exhibit motor dysfunctions. (A and B) Pigv341E mice exhibited a reduction in motor coordination, reflected by a reduced latency of falling off the rotarod and elevated latency of traversal of the beam (diameter, 15 mm). (C) Pigv341E mice exhibited a reduction in climbing performance, reflected by a decrease in the time spent climbing the rope. (D) Pigv341E mice exhibited a reduced grip strength, as determined by grip strength meter. [g] was normalized to the weight of the animal. (E) Representative image of walking pattern between genotypes. SL–FL = stride length–forelimb, SL–HL = stride length–hindlimb, FBW = fore–base width, HBW = hind–base width, S = distance between forepaw and hindpaw. (F) Pigv341E mice had an altered walking pattern, reflected by an increase in the distance between forepaw to hindpaw and a decrease in fore–base width. (G) Pigv341E mice exhibited enhanced social approach behavior, reflected by a decrease in “rear up” behavior and an increase in the number of nose-to-anogenital contacts in the social proximity test. (H) The three-chamber test (first 5 min) indicated that Pigv341E mice exhibited an enhanced social approach behavior relative to wild-type mice, reflected by spending more time with the stranger mouse than with the empty. Pigv341E = homozygous for Pigv p.Ala341Glu. Animals used in the motor tests were 7 wk old: WT(female n = 8, male n = 4) Pigv341E(female n = 4, male n = 2). Animals used in the social proximity test: WT(female n = 9, male n = 11), Pigv341E(female n = 4, male n = 7). Animals used in the three-chamber test: WT(female n = 9, male n = 11), Pigv341E(female n = 4, male n = 6). The data from the motor tests and the social proximity test were analyzed by nonparametric t test (Mann–Whitney). The data from the three-chamber test were analyzed by two-way ANOVA followed by Bonferroni’s multiple comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001. ns, not significant.
We evaluated social behavior in Pigv341E mice in the social proximity and three-chamber tests. In the social proximity test, Pigv341E mice exhibited a reduced number of “rear up” behaviors and an elevated number of nose-to-anogenital contacts with the stranger mouse (Fig. 3G). However, no differences in the number of nose-tip-to-nose-tip, nose-to-head-contact, “crawl over,” or “crawl under” behaviors were observed between genotypes (SI Appendix, Fig. S7A). In the three-chamber test, Pigv341E mice spent more time with the stranger mouse than in the vicinity of the empty cage (Fig. 3H). Furthermore, the discrimination ratio (stranger vs. empty cage) was higher in Pigv341E than in wild-type mice (SI Appendix, Fig. S7D, Right graph).
Pigv341E Mice Exhibit Cognitive Deficits in Spatial Long-Term Memory and Species-Specific Hippocampus-Dependent Functions.
To characterize the cognitive and affective profile of Pigv341E mice, we performed a battery of tests to assess aspects of spatial learning and memory, species-specific functions (Barnes maze, y-maze, marble burying, and nest building behavior), and spontaneous response to novel, open, and elevated or bright environments (open field, elevated plus maze, and dark/light box). In the Barnes maze test, Pigv341E mice were delayed in spatial learning, as indicated by an elevated latency to escape during days 1 to 3 (Fig. 4A). Despite this delay in spatial learning during days 1 to 3, Pigv341E mice had learned the location of the escape box by day 4 and exhibited normal short-term spatial memory at day 5. However, Pigv341E mice had impaired long-term spatial memory at day 12, reflected by an elevated latency to escape (Fig. 4A). Similar to the results that measured latency to escape, Pigv341E mice had an elevated path length for days 1 to 4 and 12, but not day 5 (Fig. 4B). Furthermore, Pigv341E mice spent less time in the target quadrant than wild-type mice at day 12, confirming deficits in long-term spatial memory (SI Appendix, Fig. S8B, Right graph). In the y-maze test, similar spontaneous alternation behavior between genotypes suggested normal short-term spatial working memory in Pigv341E mice (SI Appendix, Fig. S8A). In the species-specific and hippocampus-dependent tests of marble burying and nest construction (21), Pigv341E mice buried fewer marbles and had lower-quality nests (Fig. 4 C and D).

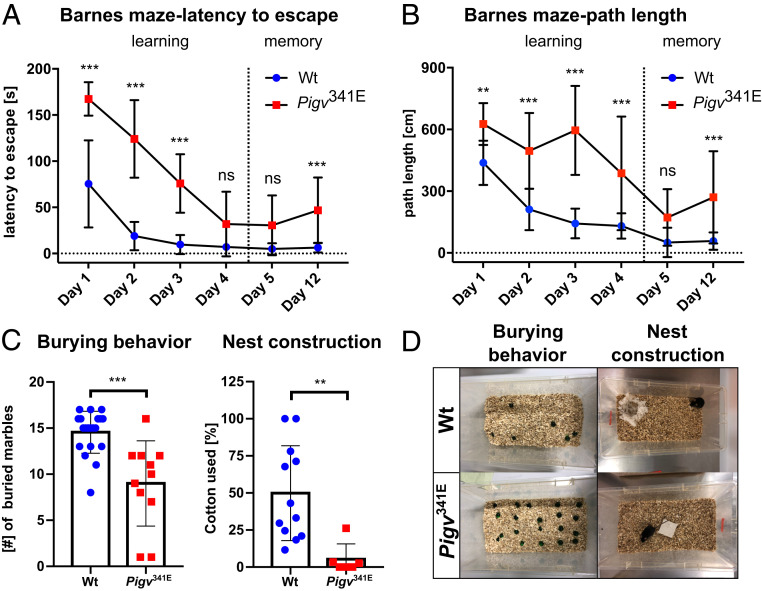
Pigv341E mice exhibit cognitive deficits in learning and memory. (A and B) Pigv341E mice exhibited cognitive deficits in learning and long-term memory, reflected by increases in latency to escape and path length on days 1 to 3 (learning) and day 12 (long-term memory in the Barnes maze). (C) Pigv341E mice exhibited a decrease in burying and nest construction behavior. (D) Representative image of burrowing behavior (Left) and nest construction behavior (Right) of wild-type and Pigv341E mice. Pigv341E = homozygous for Pigv p.Ala341Glu. Animals used in the Barnes maze: WT(female n = 8, male n = 11), Pigv341E(female n = 4, male n = 6). Animals used in the marble-burying test: WT(female n = 9, male n = 11), Pigv341E(female n = 4, male n = 7). Animals used in the nest construction test were 7 wk old: WT(female n = 8, male n = 4), Pigv341E(female n = 4, male n = 2). The data from the nest construction and marble-burying tests were analyzed with a nonparametric t test (Mann–Whitney). The data from the Barnes maze (latency to escape, path length) were analyzed by two-way ANOVA followed by Bonferroni’s multiple comparisons test. *P < 0.05 **P < 0.01, ***P < 0.001. ns, not significant.
Pigv341E mice behaved similarly to wild-type mice in the dark/light box and elevated plus maze (SI Appendix, Fig. S9 A and B). In the open field test, Pigv341E mice spent less time in the center and more time in the periphery than wild-type mice (SI Appendix, Fig. S9C). Moreover, the path length and number of visits to the center were reduced in Pigv341E mice (SI Appendix, Fig. S9 D and E).
Pigv341E Mice Exhibit Defects in Synaptic Transmission.
Because Pigv341E mice exhibited cognitive impairments in spatial learning and memory, we hypothesized a hippocampal defect, and this idea was supported by the reduced burrowing and nest building behavior (hippocampus-dependent) in Pigv341E mice. Hence, we decided to analyze the hippocampus in more detail. Nissl staining revealed no morphological abnormalities in the hippocampus of Pigv341E mice (SI Appendix, Fig. S10C). Because many GPI-linked proteins play crucial roles in synapse formation and plasticity, causing deficits that can be detected in cell culture (22), we performed immunostaining to visualize synaptophysin, a presynaptic marker. We observed a decreased immunoreactivity for synaptophysin in cornu ammonis 1–stratum radiatum (CA1–SR) of Pigv341E mice (Fig. 5A). By contrast, we observed no significant differences between genotypes in cornu ammonis 3–stratum radiatum (CA3–SR) or cornu ammonis 1–molecular layer of dentate gyrus (CA1–ML) (SI Appendix, Fig. S10 A and B). To determine whether the behavioral abnormalities were accompanied by deficits in synaptic transmission, we conducted electrophysiology recordings in the CA1–SR region, where synaptophysin levels were reduced in Pigv341E mice. We tested input–output functions and observed lower amplitudes of excitatory postsynaptic potential (EPSPs) at different stimulation intensities in Pigv341E mice (Fig. 5B). In paired pulse facilitation (PPF), Pigv341E mice exhibited an elevated paired pulse ratio (PPR) (Fig. 5C). Moreover, posttetanic potentiation (PTP) exhibited elevated facilitation in Pigv341E mice (Fig. 5D).

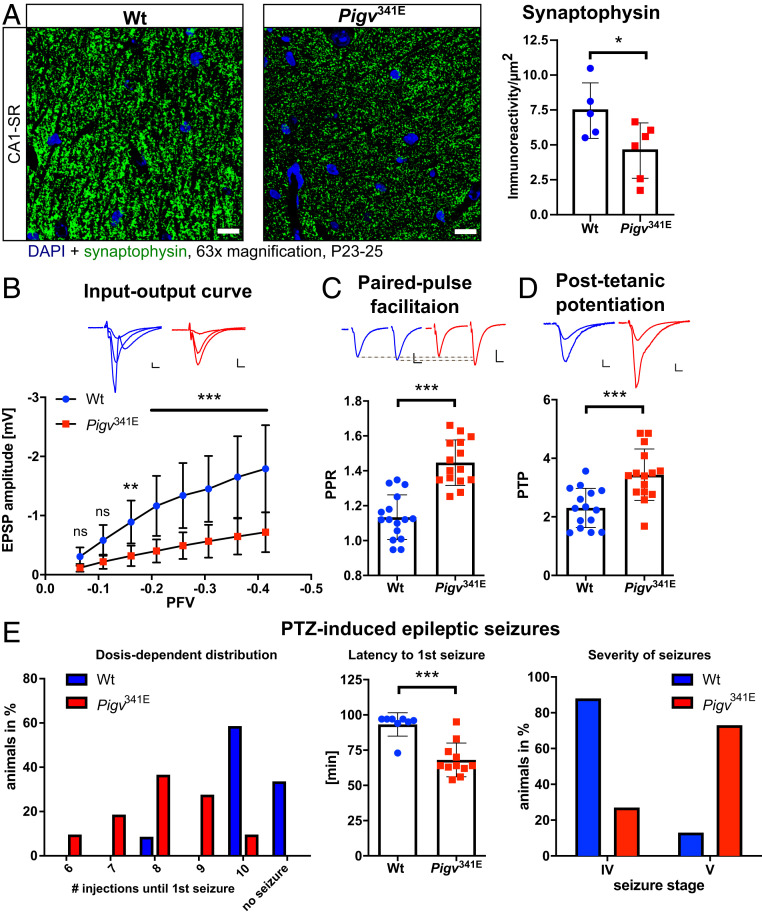
Pigv341E exhibit a hippocampal synaptic defect. (A) Representative images of immunofluorescence staining show less synaptophysin immunoreactivity (green) in CA1–SR of Pigv341E mice. 4′,6-diamidino-2-phenylindole (DAPI) staining is shown in blue. (Scale bar: 10 µm.) Synaptophysin immunoreactivity per µm2, quantified using FIJI ImageJ, was significantly reduced in CA1–SR in Pigv341E mice (Right). (B) Input–output: EPSP amplitude was reduced in Pigv341E mice at different presynaptic fiber volleys (PFVs). (Scale bar: L indicates 0.2 mV/5 ms.) (C) PPF revealed an elevated PPR in Pigv341E mice. (Scale bar: L indicates 0.2 mV/10 ms.) (D) PTP was elevated in Pigv341E mice. (Scale bar: L indicates 0.2 mV/5 ms.) (E) In the PTZ kindling model Pigv341E mice had a significantly different seizure threshold compared to that of wild-type control littermates (Left graph). In Pigv341E mice the latency to the first convulsive seizure was significantly reduced (Middle graph). Furthermore, the seizure phenotype appears more severe (Right graph). Pigv341E = homozygous for Pigv p.Ala341Glu. Animals used for the synaptophysin-staining were 3 wk old: WT(female n = 3, male n = 2) Pigv341E(female n = 2, male n = 4). Animals used for the electrophysiology recordings: WT(female n = 2, male n = 2) Pigv341E(female n = 2, male n = 2). Animals used for the PTZ model: WT(female n = 3, male n = 9) Pigv341E(female n = 5, male n = 6). The data from the synaptophysin immunofluorescence staining and electrophysiology recordings (PPF, PTP) were analyzed by parametric Student’s t test. Furthermore, the data from the input–output curve were analyzed by two-way ANOVA followed by Bonferroni’s multiple comparisons test. In the PTZ model, group comparisons (dose-dependent distribution, severity of seizures) were analyzed by χ2 test and latency to first seizure by unpaired Student’s t test with Welch’s correction. *P < 0.05 **P < 0.01, ***P < 0.001.
Increased Susceptibility of Pigv341E Mice to Chemically Induced Acute Seizures.
Considering the altered electrophysiological properties in Pigv341E mice, we examined for differences in the seizure threshold between Pigv341E and corresponding littermate wild-type mice in an acute epilepsy model, the pentylenetetrazole (PTZ)-induced kindling model (23). To this end, mice were repetitively exposed to PTZ (10 mg/kg, i.p.) every 10 min until the occurrence of a first focal-to-bilateral tonic–clonic seizure occurred. Intriguingly, Pigv341E mice exhibited a significantly lower seizure threshold, manifesting as a reduced latency to first seizure than wild-type mice, which exhibited convulsive seizures after PTZ exposure. Wild-type animals manifested generalized seizures after 93.3 min, whereas in Pigv341E mice, the first seizure manifested already after 63.3 min (Fig. 5E, Middle graph). Furthermore, the observation that four wild-type animals did not exhibit any seizure after 10 injections, whereas all Pigv341E mice did, underlines the higher susceptibility of Pigv341E mice for seizure induction via PTZ (χ2 = 105.3, df = 4, P < 0.0001) (Fig. 5E, Left graph). In addition, Pigv341E mice exhibited more severe seizures than wild-type mice (χ2 = 74.21, df = 1, P < 0.0001) (Fig. 5E, Right graph).
Shift in Relative Cell Count of Hippocampal Cellular Subgroups in Pigv341E Mice.
The synaptic defect in the CA1–SR region of Pigv341E mice could have been responsible for the observed impairments in spatial learning and memory. To identify the cell types most affected by GPIBD, we performed single-cell RNA sequencing on freshly isolated hippocampal cells after cognitive behavioral tests. The isolation of hippocampal cells was performed as previously described (24). Based on the gene expression profiles of 8,800 single cells from Pigv341E mice and 7,100 cells from wild-type animals, we defined 17 cellular subgroups (Fig. 6A). Cells from both Pigv341E and wild-type mice were present in all subgroups, but the distributions differed between genotypes (χ2 = 306.49, df = 16, P < 2.2 × 10−16). While the fractions of granule cells, oligodendrocytes, and a microglia subpopulation (microglia 3) were reduced in pooled samples from Pigv341E mice, the proportions of subicular neurons (neurons subiculum 1), GABAergic (inhibitory) interneurons, and fibroblast-like cells were higher than in the pooled wild-type samples (Fig. 6B).

![Differences between Pigv341E and wild-type mice in the transcriptional landscape of single hippocampus cells. (A) Hippocampal cells from both Pigv341E and wild-type mice clustered into 17 cellular subgroups (B), with a skewed distribution of cells across subgroups in the Pigv341E sample relative to wild-type. Percentages below 3% are not shown for clarity. (C) Differential expression testing between Pigv341E and wild-type cells within each cellular subgroup yielded differentially expressed genes with absolute log2(fold change) ≥0.25 and adjusted P value ≤0.01. The most extensive changes in gene expression were observed for the third subgroup of microglia cells and the first subgroup of subicular neurons. Gene counts are not shown for sets with <20 differentially expressed genes. (D) Global comparison between all Pigv341E and wild-type cells highlights the hippocampus-wide effect on the genes Abl1, Hdc, Cyp4x1, and Gm14216, for which significant changes in expression were detected within each cellular subgroup. The dashed horizontal line is located at an adjusted P value of 0.01, the dashed vertical lines at an absolute log2(fold change) of 0.25, and the dotted vertical lines at an absolute log2(fold change) of 0.7. For genes with an absolute log2(fold change) >0.7, gene symbols are shown. Within the subgroups of microglia 3, neurons subiculum 1 and pyramidal neurons (CA3), GO enrichment analysis revealed a number of overrepresented biological process (E), cellular component (F), and molecular function terms (G) in the sets of genes differentially expressed between Pigv341E and wild-type cells within the cellular subgroups. For each set of genes that were up-regulated or down-regulated between genotypes, up to three terms with the lowest adjusted P value after removal of redundant GO terms are shown. The gene ratio, i.e., the number of genes annotated with a term divided by the respective number of differentially expressed genes, is encoded by point size. Cellular subgroups with no significantly enriched terms were omitted. GO terms enriched in differentially expressed genes with elevated or reduced expression within a subgroup are contracted into a single star-shaped symbol. OPC, oligodendrocyte precursor cells; DE genes, differentially expressed genes; FC, fold change; Padj, adjusted P value. Pooled samples [WT(male = 4), Pigv341E(male = 4)]. n = 1 per genotype.](/dataresources/secured/content-1765818034270-f44ec551-953a-4eff-bfe1-aa47e23b310b/assets/pnas.2014481118fig06.jpg)
Differences between Pigv341E and wild-type mice in the transcriptional landscape of single hippocampus cells. (A) Hippocampal cells from both Pigv341E and wild-type mice clustered into 17 cellular subgroups (B), with a skewed distribution of cells across subgroups in the Pigv341E sample relative to wild-type. Percentages below 3% are not shown for clarity. (C) Differential expression testing between Pigv341E and wild-type cells within each cellular subgroup yielded differentially expressed genes with absolute log2(fold change) ≥0.25 and adjusted P value ≤0.01. The most extensive changes in gene expression were observed for the third subgroup of microglia cells and the first subgroup of subicular neurons. Gene counts are not shown for sets with <20 differentially expressed genes. (D) Global comparison between all Pigv341E and wild-type cells highlights the hippocampus-wide effect on the genes Abl1, Hdc, Cyp4x1, and Gm14216, for which significant changes in expression were detected within each cellular subgroup. The dashed horizontal line is located at an adjusted P value of 0.01, the dashed vertical lines at an absolute log2(fold change) of 0.25, and the dotted vertical lines at an absolute log2(fold change) of 0.7. For genes with an absolute log2(fold change) >0.7, gene symbols are shown. Within the subgroups of microglia 3, neurons subiculum 1 and pyramidal neurons (CA3), GO enrichment analysis revealed a number of overrepresented biological process (E), cellular component (F), and molecular function terms (G) in the sets of genes differentially expressed between Pigv341E and wild-type cells within the cellular subgroups. For each set of genes that were up-regulated or down-regulated between genotypes, up to three terms with the lowest adjusted P value after removal of redundant GO terms are shown. The gene ratio, i.e., the number of genes annotated with a term divided by the respective number of differentially expressed genes, is encoded by point size. Cellular subgroups with no significantly enriched terms were omitted. GO terms enriched in differentially expressed genes with elevated or reduced expression within a subgroup are contracted into a single star-shaped symbol. OPC, oligodendrocyte precursor cells; DE genes, differentially expressed genes; FC, fold change; Padj, adjusted P value. Pooled samples [WT(male = 4), Pigv341E(male = 4)]. n = 1 per genotype.
Pigv341E Hippocampal Cells Exhibit a Deregulation in Gene Expression Related to Synapse Organization and Signaling Transduction.
Expression analysis of single-cell RNA sequencing data revealed multiple genes that were differentially expressed between Pigv341E and wild-type cells within each cellular subgroup (Fig. 6C). In the mutant mice, nonreceptor tyrosine kinase Abl1 was down-regulated, whereas histidine decarboxylase Hdc, cytochrome P450 member Cyp4x1, and predicted lncRNA Gm14216 were up-regulated, both within and across all cellular subgroups (Fig. 6D and SI Appendix, Figs. S16–S18). The most extensive change in gene expression within a cellular subgroup was observed in the first subgroup of subicular neurons and the third subgroup of microglia (Fig. 6C and SI Appendix, Figs. S16 and S17). We also performed Gene Ontology (GO) analysis of differentially expressed genes between genotypes, including terms in three categories: biological process, cellular component, and molecular function (Fig. 6 E–G). Among the differentially expressed genes in subicular neurons, the biological process term “regulation of synapse organization” was enriched in genes up-regulated in Pigv341E cells, whereas the biological process terms “cell morphogenesis” and “commissural neuron axon guidance” were enriched in down-regulated genes (Fig. 6E). Furthermore, the biological process term “cell–cell adhesion via plasma-membrane adhesion molecules” was enriched in genes with elevated and reduced expression (Fig. 6E).
Single-cell RNA sequencing revealed three hippocampal microglia populations in both genotypes (Fig. 6 A and B, microglia 1 to 3). All three subpopulations expressed the marker genes Csf1r and C1qa (SI Appendix, Fig. S15). Among the most significant differentially expressed genes between the three microglia subgroups and all remaining cells in both genotypes, “cell activation,” “migration,” “phagocytosis,” and “immune responses” were among the top 10 GO biological process terms in microglia 1 and 2 (Dataset S2). By contrast, the top 10 GO biological process terms in microglia 3 cells were “ribosome,” “ribonucleoprotein complex biogenesis,” and “cytoplasmic translation” (Dataset S2). Hence, we considered microglia 1 and 2 cells as potentially more phagocytic and migratory than microglia 3 cells. We identified 326 genes that were differentially expressed between genotypes in microglia 3 cells. Remarkably, 306 of these 326 genes were down-regulated in microglia 3 cells of Pigv341E mutants (Fig. 6C). GO analysis revealed that the down-regulated genes were enriched for the biological process terms “small GTPase-mediated signal transduction” and “regulation of microtubule cytoskeleton polymerization and depolymerization” (Fig. 6E). In addition, we identified 20 genes differentially expressed between genotypes that were up-regulated in Pigv341E microglia 3 cells. Among these were Rpl38, Rps21, and Rps28, which encode ribosomal proteins.
CA3 pyramidal neurons project their axons to the CA1 region, where we observed dysfunction in synaptic transmission in Pigv341E mice. Therefore, we were particularly interested in the cellular subgroup of CA3 pyramidal neurons, in which we identified 79 genes that were differentially expressed between genotypes (Fig. 6C). GO analysis revealed that down-regulated genes were enriched for the terms “adherens junction organization,” “regulation of cell-substrate junction assembly,” and “focal adhesion assembly” in Pigv341E CA3 pyramidal neurons (Fig. 6E). Interestingly, we also observed enrichment of genes with reduced expression related to “neuron–neuron synaptic transmission,” “regulation of synaptic vesicle exocytosis,” and “synaptic vesicle transport” (Dataset S3).
Discussion
Pigv341E is a mouse model for GPI-anchor deficiency with a hypomorphic mutation that is viable after weaning. The mice exhibited significant alterations in behavior that reflect key aspects of patients with IGD.
In these mice, we observed a severe motor phenotype that included deficits in motor coordination, grip or muscle strength, climbing, and hanging behavior (in HCS); alterations in walking pattern; and hindlimb clasping. Behavioral traits such as altered walking pattern, hindlimb clasping, and motor coordination deficits are usually observed in mouse models with ataxia-like behavior and cerebellar dysfunction (25, 26). In agreement with these findings, ataxia has been reported in some IGD patients, and an ataxia-like behavior was observed in a conditional Piga knockout mouse model (10, 19). Lukacs et al. analyzed the microscopic anatomy of cerebellum sections from their conditional Piga knockout mouse model and observed mild deficits in Purkinje cell arborization (10). However, in histologic analysis with various stainings, the cerebellum of our Pigv341E mutants did not exhibit any abnormalities (SI Appendix, Fig. S11 A–F), and the overall folial pattern appeared to be unchanged. In particular, calbindin staining exhibited no differences in Purkinje cell dendritic arborization between genotypes (SI Appendix, Fig. S11 E and F). Therefore, we hypothesize that deficits in dendritic arborization in this neuronal cell type, as reported by Lukacs et al. (10) require a more severe GPIBD than that induced by the hypomorphic mutation c.1022C > A in Pigv. This is consistent with the longer lifespan of our mouse model, which allowed us to analyze the associated cognitive deficits. In addition to the cerebellum, we focused on the hippocampus, where we performed histology and electrophysiology to achieve a deeper understanding of the memory and species-specific deficits. Although we did not observe significant morphological changes in the hippocampus, the input–output curve, PPR, and PTP were significantly altered in Pigv341E mice, indicating that electrophysiology is a sensitive functional assay for mouse models with mild GPIBD.
Because Pigv341E mice exhibited increased self-grooming, which is a repetitive, highly stereotyped pattern that is associated with autistic-like behavior in rodents (27), we suspected abnormalities in social behavior as well. Autistic features have been reported in a subgroup of patients with IGD due to pathogenic mutations in PGAP3 (7). By contrast, patients with PIGV deficiency are keen to interact socially despite their severe speech impairments. Interestingly, in Pigv341E mice we observed enhanced social approach behavior, reflected by an elevated number of nose-to-anogenital contacts and reduced “rear up” behavior in the social proximity test. The reduction in “rear up” behavior in Pigv341E mice suggested reduced social avoidance. The enhanced social approach behavior was confirmed in the three-chamber test: relative to wild-type controls, Pigv341E mice spent more time with the stranger mouse than in the vicinity of the empty cage. Comparable performance between genotypes in the buried food test ruled out compromised olfaction, a potential confounder in social behavior tests (SI Appendix, Fig. S7C). Taking into account the enhanced social approach behavior in Pigv341E mice, the positive social abilities of patients with PIGV deficiency seem to be characteristic of these individuals and should be considered during diagnosis. However, it remains unknown to what extent IGD patients who are affected in genes other than PIGV exhibit positive social abilities. In addition, because we did not observe social behavior characteristic of autism (28) in Pigv341E mice, and autistic features are seen only in a subgroup of patients with IGD, autism should not be considered as a specific feature of IGD.
Pigv341E mice exhibited a deficit in spatial long-term memory in the Barnes maze, correct short-term spatial memory, and short-term working memory (y-maze test). Furthermore, Pigv341E mice exhibited a delay in spatial learning relative to wild-type mice in the Barnes maze (days 1 to 3). In the Barnes maze, both latency to escape and path length were elevated in Pigv341E mice; therefore, we excluded the motor phenotype as a confounder. However, even though path length and latency to escape were elevated in Pigv341E mice, the number of visits to the wrong holes was not significantly altered on days 2 through 12 (SI Appendix, Fig. S8C). Analysis of the search strategy revealed that Pigv341E mice were targeting the correct hole without any errors (number of wrong holes) less often than wild-type mice (SI Appendix, Fig. S8D, direct strategy) (WT: 15.8%, Pigv341E: 3.3%). Moreover, we quantified two further search strategies: random choice of a hole without any order and a serial strategy that tests holes one after another in close proximity. Pigv341E mice used more often the random strategy (WT: 9.7%, Pigv341E: 56.7%), whereas wild-type mice used more often the serial strategy to find the correct hole (WT: 25.4%, Pigv341E: 20.0%) (SI Appendix, Fig. S8D). In contrast to random guessing, the serial strategy has the advantage that it minimizes the path length and by that means will result in a quicker escape. However, the serial strategy results also in a higher number of errors. This potentially explains why the latency to escape was significantly elevated in Pigv341E mice, whereas the number of wrong holes did not significantly differ between genotypes.
Furthermore, we observed no difference between genotypes in affective-related behavior (dark/light box, elevated plus maze) except in the open field test, in which Pigv341E mice spent more time in the periphery than in the center. However, this observation could also represent a confounder due to the motor phenotype, as the number of entries to the center and the path length were also reduced in Pigv341E mice.
Interestingly, electrophysiology recordings revealed reduced synaptic transmission at CA1–SR in Pigv341E mice, consistent with the observed impairment in long-term spatial memory and hippocampus-dependent species-specific behaviors (marble-burying and nest construction test). While PTP and PPR were elevated in Pigv341E mice, the input–output curve was reduced, indicating a decrease in synaptic release probability. Due to the increase in PTP and PPR, and the reduced immunoreactivity of synaptophysin, a presynaptic vesicle protein, we hypothesize that the pool of readily releasable vesicles in the presynapse is reduced, resulting in a damped input–output curve in the postsynapse. Notably in this regard, GO analysis of single-cell RNA sequencing data revealed that genes which were significantly down-regulated in Pigv341E-CA3 pyramidal neurons were enriched for GO biological process terms associated with “synaptic transmission” and “vesicle transport.” The impaired synaptic transmission in the hippocampi of Pigv341E mice was reflected in their lower threshold to the excitotoxin-induced epileptic events and aggravated seizures than wild-type mice.
Single-cell RNA sequencing data revealed the most prominent differences in gene expression in a subgroup of subicular neurons and microglia. In subicular neurons 1, up-regulated genes were associated with biological process terms such as “synapse organization.” Because we observed a synaptic defect in CA1–SR, as revealed by immunohistochemistry and as further supported by electrophysiological recordings, we hypothesized a synaptic defect in the subiculum as well. The subiculum, an area of the hippocampus that is important for memory retrieval, is linked through microcircuits with the CA1 (29). Ledergerber and Moser (29) described two distinct circuits for memory acquisition and retrieval: memory acquisition involves the CA1 and medial-entorhinal cortex, whereas memory retrieval involves the CA1, the medial-entorhinal cortex, and the subiculum. Future studies should seek to determine whether memory acquisition, memory retrieval, or even both conditions are affected in Pigv341E mice.
Strikingly, 306 genes were down-regulated in Pigv341E microglia 3 cells. Therefore, microglia might play more important roles in GPI-anchor deficiency than previously thought. These genes were enriched in GO biological process terms “protein localization to cell periphery,” “small GTPase-mediated signal transduction,” and “regulation of microtubule polymerization or depolymerization.” Small GTPases are important mediators of the cytoskeleton (30). Hence, we hypothesized that a GPI-anchor defect leads to down-regulation of small GTPase-mediated pathways, which has further consequences for cytoskeleton organization in this microglia subtype. In this regard, GPI-anchored ephrin A proteins could play an important role, as EphrinA1 regulates small GTPase (Rho)-dependent cytoskeleton rearrangement through Src/focal adhesion kinases (31).
Up to 0.5% of the eukaryotic proteins are GPI linked, with a broad range of functions including cell–cell adhesion, signal transduction, and antigen presentation (32). Therefore, it is surprising that the pathophysiology of acquired GPI-anchor deficiency PNH can be explained by the reduced expression of only two substrates, CD55 and CD59, which reduces the protection of cells against membrane attack complex (MAC), and can also be effectively treated by eculizumab, which inhibits complement activation (33). Likewise, analysis of mouse models of congenital forms of GPI-anchor deficiencies has aimed at identifying other lead targets. McKean and Niswander suggested a pivotal role of Cripto/TGFβ signaling in the development of holoprosencephaly (14), whereas Lukacs et al. discussed the role of GPI-anchored Folr1 in neural crest cells and the cranial neuroepithelium, and argued that compromise of Folr1 could be linked to the facial gestalt (13).
Interestingly, a considerable number of GPI-linked proteins are involved specifically in synapse formation and plasticity (1). Because Pigv341E mice exhibit a hippocampal synaptic defect, this subset of GPI-linked proteins, including GPI-linked EphrinA, may play pivotal roles in the development of the disease as well. Single-cell RNA sequencing analysis revealed that Abl1, which interacts on the protein level with several EphrinA receptors (SI Appendix, Fig. S6F) (34), was not only down-regulated in Pigv341E mice across all cellular subgroups, but also within each cellular subgroup. Our hypothesis is that the GPI-anchor defect in the hippocampus is especially critical for EphrinA signaling, and that defective GPI anchoring of EphrinA in turn reduces hippocampal EphrinA receptor and Abl1 activity. Notably in this regard, axon repulsion is EphrinA dependent and mediated through the Abl kinase family (35).
Along with Abl1, Hdc and Ptgds were also dysregulated in Pigv341E mice, with elevated expression across all cellular subgroups. Hdc encodes a histidine decarboxylase that catalyzes the conversion from histidine to histamine, an important neurotransmitter regulating circadian rhythm (36). In rodents, histamine levels are elevated during the dark phase to induce wakefulness and are reduced during the light phase to induce sleep (37). Furthermore, intracerebroventricular application of histamine triggers characteristic signs of wakefulness, such as elevated grooming and exploration behavior, which were also observed in Pigv341E mice (HCS and SAM) (37). In addition, during home-cage activity monitoring (group housed) and the HCS (individually housed), Pigv341E mice were more active during the light cycle and slept for shorter durations. Consequently, higher expression of Hdc may lead to higher production of histamine, thereby disturbing circadian rhythm and causing classical signs of wakefulness in Pigv341E mice during the light phase. However, it remains unknown how misregulation of Hdc is associated with GPI-anchor deficiency. Interestingly, in the conditional Piga knockout mouse model, bulk RNA sequencing of the cerebellum revealed an enrichment of deregulated genes associated with the circadian rhythm (10). Moreover, Ptgds encodes prostaglandin D2 synthase, which converts prostaglandin H2 (PGH2) into prostaglandin D2 (PGD2), which in turn induces sleep (38). PGD2 levels fluctuate with the circadian rhythm and are elevated in the cerebrospinal fluid when rats are sleep deprived (39). Therefore, up-regulation of Ptgds expression could be an indicator of sleep deprivation in Pigv341E mice. Because Pigv341E mice exhibit fewer resting phases during their classical inactive (light) phase, a careful analysis of sleep pattern in IGD patients is indicated. To date, sleep disturbances have mainly been reported in patients with PGAP3 deficiency (19).
In summary, we have performed a deep phenotypic characterization of a mouse model that mirrors the symptoms of human patients with IGD. In addition, we detected a hippocampal synaptic defect that may impair spatial long-term memory and important species-specific behaviors for the survival of the animal. We hope that our model, as well as our phenotyping approach, will be useful in future studies aimed at a detailed elucidation of the pathomechanism of IGD and the response to therapeutic interventions.
Materials and Methods
For full methods, see SI Appendix, Supplementary Material and Methods.
Animals.
Pigv341E mice were generated by diploid or tetraploid aggregation (40) (SI Appendix, Fig. S1A) and maintained by crossing with C57BL.6/J mice. Mice were genotyped by PCR using the primers mPigvEx4_fw and mPigvEx4_rv. PCR amplicons were digested with BcuI (Thermo Fisher Scientific) and subjected to agarose gel electrophoresis (SI Appendix, Fig. S1D). All animals were handled according to government regulations as approved by local authorities (LaGeSo Berlin and LANUV Recklinghausen). In addition, all experiments were carried out following the 3R (Replacement, Reduction and Refinement) guidelines for animal welfare. Mice were housed in groups with mixed genotypes in single ventilated cages with an enriched environment. The mice were housed in a pathogen-free animal facility with a 12 h dark/light cycle and had food and water ad libitum unless otherwise indicated. Mice used for experiments were 8 wk to 6 mo old unless otherwise indicated. Pigv341E and wild-type mice used in a given experiment were the same age. To avoid bias effects, littermates were assigned equally to both experimental groups, according to their genotype and sex. The experimenter was blinded except during behavioral testing, as Pigv341E mice were physically smaller. Moreover, experiments were randomized with respect to mouse genotype.
Statistical Analysis.
For all experiments, at least four animals per genotype were used, except for the weight curve, for which at least three animals per genotype were used. One animal was defined as one biological replicate and represented one data point, except for electrophysiology recordings (see SI Appendix, Schaffer collateral recordings). Means and SDs were calculated for each genotype group unless otherwise indicated. Data were statistically analyzed with GraphPad Prism (version 7) or R (version 3.6.3) (41), and results are expressed as means ± SDs. Statistical tests were performed for each experiment as indicated in Dataset S1. Results with P value <0.05 were considered significant unless otherwise indicated.
Acknowledgements
We thank the Core Unit for Bioinformatics Data Analysis from the University Hospital of Bonn for their support. We thank Andrea Christ from the Anatomisches Institut, Anatomie & Zellbiologie, Medical Faculty of the University of Bonn, for excellent technical support. Furthermore, we thank the Animal Outcome Core Facility of the NeuroCure center and Charité-Universitätsmedizin, as well as the animal facility, transgenic core, and the sequencing core facility from the Max Planck Institute for Molecular Genetics in Berlin for their support. This study was supported by the German Research Council (Deutsche Forschungsgemeinschaft, DFG), by grant KR3985/6-1, awarded to P.M.K. and grants BE 2078/5-1 and D-256.0154 (Sonderforschungsbereich (SFB) 1089) awarded to A.J.B.; project 327654276 [SFB 1315] awarded to D.S.; project 184695641 [SFB 958] awarded to D.S.; and under Germany’s Excellence Strategy [EXC-2049] 390688087 awarded to D.S.), by the European Research Council under the European Union’s Horizon 2020 Research and Innovation Programme (grant agreement 810580 awarded to D.S.), and by the Berlin-Brandenburg School for Regenerative Therapies (final year stipend awarded to M.R.d.l.S.).
Data Availability.
The single-cell RNA sequencing data are freely available from the Gene Expression Omnibus repository under accession number GSE147722.
References
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
38
39
40
41.
 A CRISPR-Cas9–engineered mouse model for GPI-anchor deficiency mirrors human phenotypes and exhibits hippocampal synaptic dysfunctions
A CRISPR-Cas9–engineered mouse model for GPI-anchor deficiency mirrors human phenotypes and exhibits hippocampal synaptic dysfunctions
