
Edited by Lucio Cocco, University of Bologna, Bologna, Italy, and accepted by Editorial Board Member Solomon H. Snyder December 5, 2020 (received for review June 3, 2020)
Author contributions: P.-G.S. and J.-Y.J. designed research; S.-H.K., S.Y., K.-H.L., H.-J.J., and J.-Y.J. performed research; E.K., M.K., P.-G.S., and J.-Y.J. contributed new reagents/analytic tools; K.-H.L., M.K., and J.-Y.J. analyzed data; and S.-H.K., S.Y., K.-H.L., M.K., P.-G.S., and J.-Y.J. wrote the paper.
1S.-H.K., S.Y., and K.-H.L. contributed equally to this work.
- Altmetric
DNA mutation within gene bodies contributes to abnormal translation and can lead to neurodegenerative disorders. High-throughput analysis is suitable for initial detection of gene mutations with details. Deep learning-based RNA splicing analysis facilitates accurate and precise predictions of genetic variants in DNA bodies. Although deep learning-based prediction methods have improved the screening of genetic variations for target diseases, there have been no reports showing a direct comparison with genetic information of an AD model. This study identified the gene mutations and abnormal splicing of PLCγ1 gene in AD using both high-throughput screening data and a deep learning-based prediction. Our findings provide insight for improvement in prediction and diagnosis of AD pathology.
Exon splicing triggered by unpredicted genetic mutation can cause translational variations in neurodegenerative disorders. In this study, we discover Alzheimer’s disease (AD)-specific single-nucleotide variants (SNVs) and abnormal exon splicing of phospholipase c gamma-1 (PLCγ1) gene, using genome-wide association study (GWAS) and a deep learning-based exon splicing prediction tool. GWAS revealed that the identified single-nucleotide variations were mainly distributed in the H3K27ac-enriched region of PLCγ1 gene body during brain development in an AD mouse model. A deep learning analysis, trained with human genome sequences, predicted 14 splicing sites in human PLCγ1 gene, and one of these completely matched with an SNV in exon 27 of PLCγ1 gene in an AD mouse model. In particular, the SNV in exon 27 of PLCγ1 gene is associated with abnormal splicing during messenger RNA maturation. Taken together, our findings suggest that this approach, which combines in silico and deep learning-based analyses, has potential for identifying the clinical utility of critical SNVs in AD prediction.
Alternative splicing (AS) occurs in most eukaryotic species and regulates gene expression, giving rise to diverse phenotypes (1, 2). Genetic variants arising due to RNA splicing are more frequently found in individuals having neurodevelopmental disorders, with variable mutation rates between the pre-messenger RNA (pre-mRNA) and mature RNA processing stages (3, 4). These events in genetic mutation reflect marked differences in the balance of transcriptional regulation fidelity among neural networking models (5). A correlation between splicing-mediated mutation and the likelihood of response to neurodegenerative disorders (NDs), together with the identification of associations between gene mutations and clinical outcomes of NDs, can provide comprehensive information on AS for diagnosis. Therefore, detection and evaluation of genetic variations in individuals are crucial for prediction and diagnosis of NDs (6).
Alzheimer’s disease (AD) is an ND affecting different brain regions, ranging from the cerebral cortex to hippocampus (7). Clinically, abnormalities, such as amyloid plaque (known as amyloid beta [Aβ] aggregation) and neurofibrillary tangle, are usually first observed in brain tissues of patients with AD. The progression of these abnormalities to other areas of the cortex is gradual and shows considerable differences in the rate of occurrence among individuals (8). Although biochemical research has contributed to important advancements in the diagnosis of AD pathogenesis, studies on single-nucleotide variants (SNVs) in AD are scarce due to the lack of informative genetic information (9). Therefore, identification of gene mutations in AD remains challenging, and a comparative genomics approach is required.
The products of PLC genes are activated by extracellular signaling factors, such as neurotransmitters and hormones, which trigger intracellular signaling receptors such as G protein-coupled receptors and receptor tyrosine kinases (RTKs) (10). Phospholipase c gamma-1 (PLCγ1), which encodes a signal transducer of RTKs, has been suggested as a candidate for neuronal development (11). Mutation in PLCγ2, a member of the PLC gene family, has been evaluated in diverse diseases, such as AD, myelodysplastic syndromes, and chronic lymphocytic leukemia (1213–14). However, the analysis of mutations in PLCγ1 has been limited to T cell lymphomas and angiosarcoma (15, 16). Although PLCγ1 is known to be associated with AD pathogenesis (1718–19), SNVs arising due to posttranscriptional splicing or frameshift remain unknown.
High-throughput screening-based deep learning has been used to predict splicing from primary sequences (4). However, high-throughput screening-based deep learning has not yet been extended to identify SNVs in specific target genes associated with AD. Therefore, we reasoned that genome-wide association study (GWAS)-derived deep learning would be an effective model system for predicting AD and its progression. We used a GWAS platform to analyze query expression patterns and mutations of the PLCγ1 gene, and found gradual increase and accumulation of acetylation in the gene during brain development. In addition, we found that exon splicing of PLCγ1 gene in mature mRNA was caused by single-nucleotide insertion in AD. The regions of PLCγ1 gene mutations are evolutionally conserved across various species, including human. The findings of this study provide insight regarding a previously uncharacterized AD-related gene that may have clinical significance for AD prediction.
Results
Genome-wide Analysis of Gene Expression in Cortex Using High-Throughput Total RNA Sequencing.
To examine transcriptional regulation of AD specific gene expression, we performed high-throughput total RNA sequencing (RNA-seq) analysis of cortices from 6-mo-old wild-type (WT) and 5xFAD mice. Since the expression levels of many genes are programed and regulated in tissues and blood of AD mouse models (2021–22), genes with increased or decreased expression levels in AD might be functionally important in the brain. We isolated the cortex region from mouse whole brain and extracted RNA for total RNA-seq (Fig. 1A). Data from the cortices of WT and 5xFAD mice were obtained to identify differentially expressed genes.

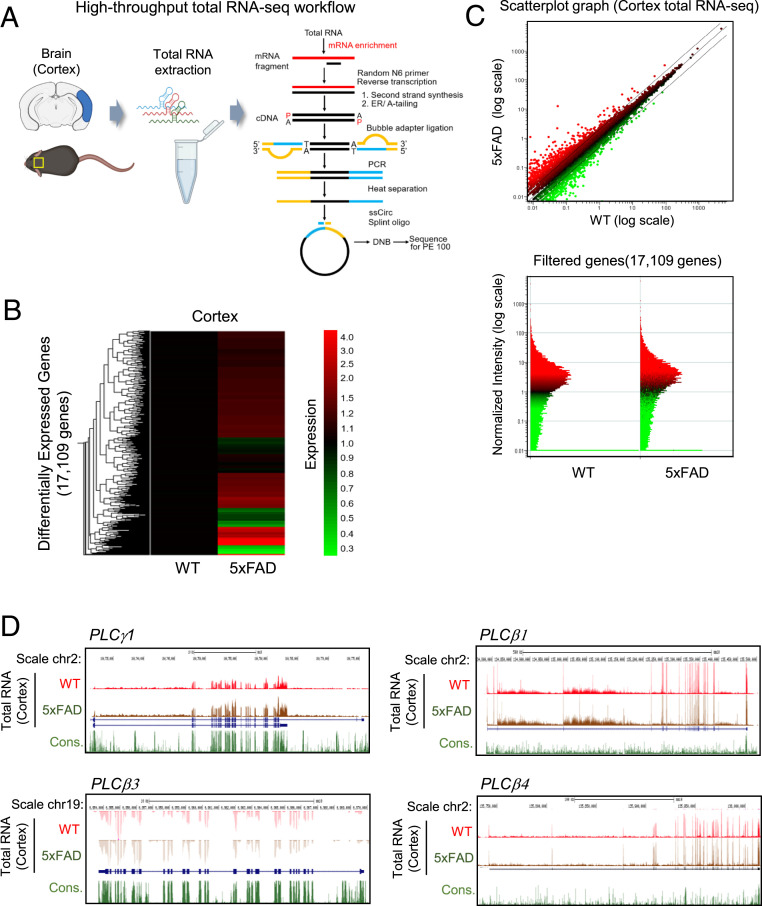
High-throughput total RNA-seq profile for PLCγ1 and PLCβ subfamily gene expression in the cortex. (A) High-throughput total RNA-seq analysis workflow. (B) Heat map and hierarchical clustering of total RNA-seq analysis from mouse cortex. The 17,109 genes are DEGs between WT and 5xFAD. (C) Normalization of total RNA-seq using scatterplot matrices from WT and 5xFAD model mouse cortex. The x axis indicates gene expression of WT; y axis indicates gene expression of AD. (D) Genome browser view of the PLCγ1, PLCβ1, PLCβ3, and PLCβ4 genes genomic locus and expression display with total RNA-seq data in cortex. ER, End Repair; ssCirc, single-strand Circle; DNB, DNA NanoBall; PE 100, Paired end 100; Cons, Consensus.
We confirmed that the expression levels of 17,109 genes were 5xFAD specific (Fig. 1B). Overall, 1,472 genes were significantly up-regulated, whereas 653 genes were down-regulated in the 5xFAD mouse cortex (Fig. 1C and SI Appendix, Fig. S1). Previous studies have reported that dysregulations of PLC genes are closely associated with many brain disorders, such as schizophrenia, bipolar disorder, Huntington’s disease, depression, and AD (10). Although the importance of the correlation between PLC genes and AD has been suggested, supporting evidence such as genes exhibiting differential expression or variation between healthy individuals and those with AD have not been identified. To demonstrate the expression levels of PLCβ1, PLCβ2, PLCβ3, PLCβ4, and PLCγ1 genes in the brains of WT and 5xFAD mice, we performed GWAS for PLCβ1, PLCβ2, PLCβ3, PLCβ4, and PLCγ1 genes using high-throughput total RNA-seq data to measure their transcription levels. The expression levels of PLCβ1, PLCβ2, PLCβ3, PLCβ4, and PLCγ1 were slightly up-regulated in the 5xFAD cortex, while PLCβ2 expression was up-regulated by ∼2.4 times in 5xFAD (Fig. 1D and SI Appendix, Fig. S2B). However, endogenous expression level of PLCβ2 was significantly lower than those of PLCβ1, PLCβ3, PLCβ4, and PLCγ1 in the 5xFAD cortex (SI Appendix, Fig. S2). In addition to the analysis of PLCβ1, PLCβ2, PLCβ3, PLCβ4, and PLCγ1 transcription levels, Western blot analysis was performed to confirm changes at the translational level, revealing that there were no significant differences among the levels of the proteins encoded by these genes in WT and 5xFAD cortices (SI Appendix, Fig. S3). Notably, PLCβ2 protein was not detected in the Western blot analysis because its endogenous level in neurons was very low (23).
Single-nucleotide polymorphisms (SNPs) in APOE or TREM2 have been proposed to play pathogenic roles in AD. Moreover, somatic mutations in brain have been suggested to be associated with genetic architecture in AD (2425–26). RNA-seq data are benefitted by the detection of unidentified alternative variations and specific gene expression (27). Though the expression levels of PLC subfamily proteins showed no significant differences between healthy individuals and those with AD, the evidence led us to analyze the genomic variations of PLC genes in AD. To obtain the profiles of PLC subfamily genes based on SNV data from total RNA-seq, we proposed a pipeline for RNA-seq−based SNV and insertion/deletion (InDel) analyses (Fig. 2). We performed gene expression and SNV profiling of cortex from brains of WT and AD mice using total RNA-seq to obtain unknown information on SNVs in PLCγ1, PLCβ1, PLCβ2, PLCβ3, and PLCβ4, and provide insight into genetic variation in the pathogenesis of AD.

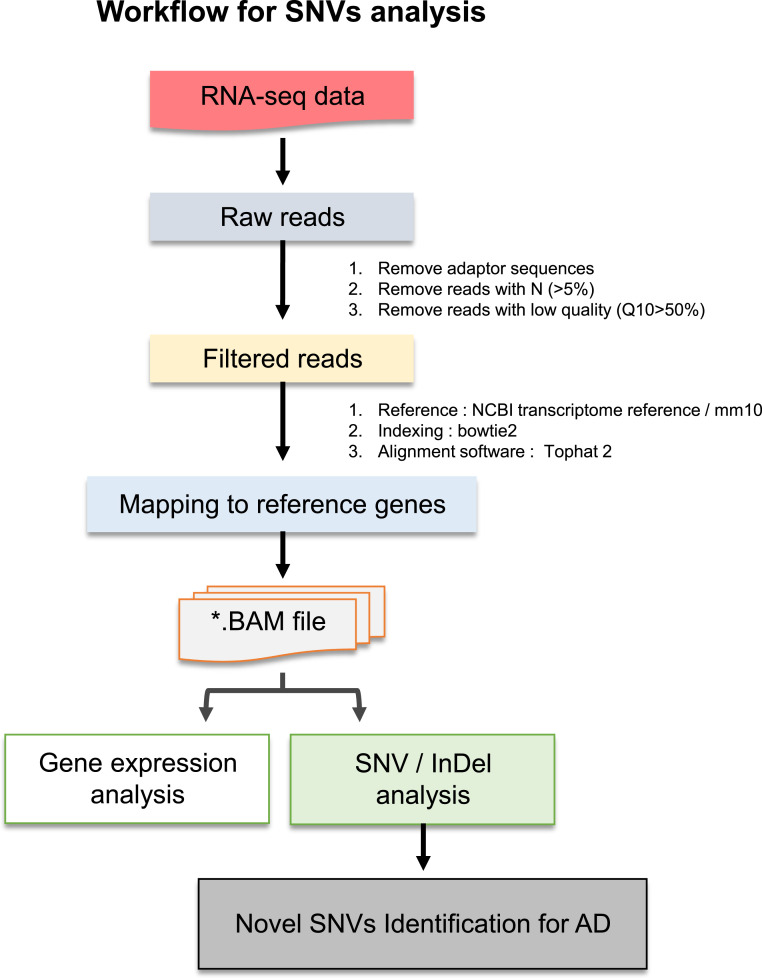
Total RNA-seq−based SNV identification workflow for AD. Single and paired-end reads are calculated from NGS (Next-generation sequencing). Mapping was performed with reference genes and each of bam files was indexed with SAMtools. Preprocessing formed Ras sequencing data. SAMtools mpileup was performed with UCSC (University of California Santa Cruz) mm10 genome as reference, and then analyzing the SNV/InDel for AD. VCF files were generated with vcfutils.pl varFilter, and functional annotation of each variant was performed with ANNOVAR (ANNOtate VARiation) software.
Identification and Characterization of AD-Specific Mouse PLCγ1 and PLCβ SNVs in the Cortex.
High-throughput total RNA-seq is one of the most potent techniques for determining SNVs and genetic variants of target genes. A total of 163 variations (132 SNVs and 31 InDels) were identified in five genes. Of these, most were located in introns (152 variants), and only five variations were located in exons; the remaining six variants were located in splicing regions. In the 5xFAD cortex, PLCγ1 had 13 variants, while PLCβ1, PLCβ2, PLCβ3, and PLCβ4 had 66, 6, 3, and 75 variants, respectively (SI Appendix, Tables S1–S5). In this study, we mainly focused on variations in the exons of PLCγ1. There were five functional variations in the exons, including three synonymous SNV, one frameshift insertion in exon 27 of PLCγ1, and one stop gain in exon 11 of PLCβ3 (SI Appendix, Tables S1 and S4). A frameshift insertion, wherein a single “A” nucleotide was inserted at position 160,759,682 in chromosome 2, gave rise to an abnormal codon that substituted the isoleucine at the 970th amino acid position to asparagine (Fig. 3A) in PLCγ1 protein of the AD mouse model. Stop-gain mutation occurred due to the substitution of the reference “G” nucleotide with “A” at position 6,963,413 in chromosome 2, encoding a terminal codon due to a change of glutamine at the 352nd amino acid of PLCβ3 in the 5xFAD cortex (SI Appendix, Table S4).

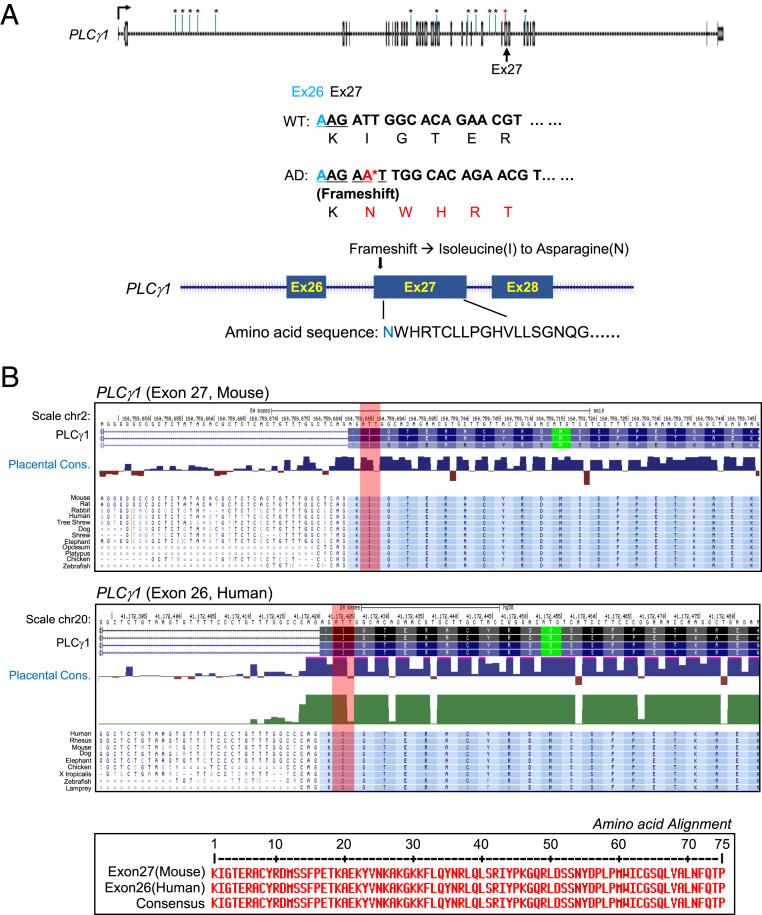
Identification and characterization of AD-specific sequenced mouse PLCγ1. (A) Mapping for AD model mouse-dependent frameshift at exon 27 of PLCγ1 gene body. One single-nucleotide insertion was caused by abnormal codon, by breaking 970th amino acid, isoleucine, into asparagine. (B) UCSC genome browser sequence alignment of various species, including human for PLCγ1 gene. Both exon 27 of PLCγ1 for mouse and exon 26 of human PLCγ1 was well conserved in isoleucine site, and full length of amino acid sequence was completely matched. Red vertical bars indicate the locations of the isoleucine for both human and mouse PLCγ1 gene.
However, we found no differences in the translation and termination of PLCγ1 and PLCβ3 between WT and the 5xFAD cortex (SI Appendix, Fig. S3). Eukaryotic AS events are closely related to adult tissue functions, organ development, and tissue homeostasis (28, 29); alterations in any of these can affect cell proliferation, methylation, and migration of cancer cells. Aberrant splicing is implicated as an important factor contributing to diseases (30). To determine whether exon 27 of PLCγ1 affects AS after mRNA processing, we performed RT-PCR using various primer sets targeting the region near this exon. Exons 26 to 30 of PLCγ1 were deleted in the mature mRNA of 5xFAD cortex. Conversely, exons 27 to 32 of PLCγ1 were conserved in the pre-mRNA of both WT and 5xFAD cortex (SI Appendix, Fig. S4), suggesting that exon skipping or AS occurs at the PLCγ1 SNV region in AD. Single amino acid substitutions have been linked to many human diseases; these disease-causing mutations tend to cause changes in hydrogen-bonded linkages or bridges, resulting in harmful amino acid mutations (31).
The frequency of AS differs among species. For example, although both human and mouse genomes have similar numbers of conserved motifs, the average alternative pre-mRNA splicing rates of human (>95 to 100%) and mouse (∼63%) genes are different (32). This comparison between human and mouse genes has also revealed that alternative exons of the reading frame are conserved in human and mouse, indicating that genetic changes are caused by AS (32). We further examined whether the 970th amino acid, isoleucine, at exon 27 of mouse PLCγ1 was conserved in other species. We obtained the PLCγ1 sequences of various species using the UCSC genome browser. Sequence alignment of PLCγ1 from various species, including human, revealed that exon 27 of mouse PLCγ1 was completely conserved across species. Moreover, exon 26 of human PLCγ1 was well conserved with respect to the isoleucine site. Surprisingly, the full-length amino acid sequences encoded by exon 26 of human and exon 27 of mouse PLCγ1 matched completely (Fig. 3B). Collectively, our findings suggest that the SNV in exon 27 of mouse PLCγ1 plays an important role in AS during mRNA processing, and may have potential as a biomarker for prediction of AD.
High-Resolution Profiling of Histone Modification at PLCγ1 in the Forebrain during Brain Development.
Histone acetylation facilitates transcriptional activity and plays an important role in the development of human brain diseases through epigenetic modification events, such as altering chromatin structure or accessibility of transcription factors (33, 34). To determine whether epigenetic changes may occur in the PLCγ1 gene during mouse brain development, we performed H3K27ac enrichment profiling using chromatin immunoprecipitation sequencing (ChIP-seq) dataset to generate high-resolution histone modifications. Histone acetylation gradually accumulated in PLCγ1 gene in mouse forebrain regions ranging from E11.5 to P56. Histone acetylation increased in the introns of PLCγ1, and AD-associated SNVs were also concentrated in this region (Fig. 4). In addition, AD-specific nucleotide alternation sites were distributed in the noncoding region of PLCγ1, and we found several transcription factor binding motifs at AD-specific AS sites (SI Appendix, Fig. S5). These results suggest that SNVs in PLCγ1 are concentrated in the introns in AD, indicating that they may interfere with the epigenetic role associated with AD.

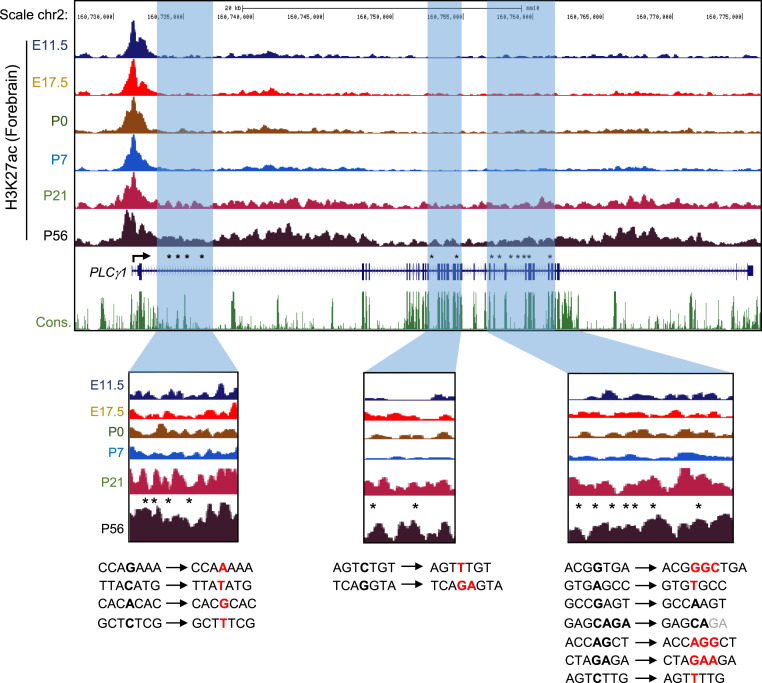
High-resolution binding profiles of histone acetylation in the forebrain during the brain development. H3K27ac binding profiles throughout the mouse forebrain development. Histone acetylation was highly accumulated at PLCγ1 gene body in P56 mouse forebrain. AD-dependent expressed SNVs were concentrated in the intron region of the PLCγ1 gene, which is enrichment of histone acetylation. Blue vertical bars indicate the locations of the AD-dependent SNVs region of the PLCγ1 gene. Black bold letters indicate the nucleotides for WT, red bold letters indicate the insertion or substitution nucleotide for AD, and gray letters indicate the deleted nucleotide for AD.
Prediction of AD-Specific Nucleotide Alteration Sites in the Human Genome Using Deep Learning.
Dysregulation of AS has been implicated in AD (35, 36). Recently, a new deep learning-based tool called SpliceAI, which can predict splice junctions of target genes from pre-mRNA nucleotide sequences with high accuracy, was introduced (4). Total RNA-seq−based SNV analysis confirmed the profile of AD-specific SNVs in PLCγ1 in 5xFAD mouse cortex. Then, we further examined whether a single-nucleotide alteration in human PLCγ1 may be correlated with the 5xFAD cortex. After SpliceAI analysis predicted a total of 14 splicing sites in the human PLCγ1 gene, accurate delta scores and positions were analyzed (Fig. 5), and a novel splicing site in exon 26 of human PLCγ1 was identified. The single nucleotide “G” at position 41,172,421 in chromosome 20 was substituted with “A,” “C,” or “T” at position 41,172,423, which was completely correlated with exon 27 of mouse PLCγ1 in the AD model cortex. SpliceAI analysis accurately revealed that the SNVs in exon 26 of human PLCγ1 and exon 27 of mouse PLCγ1 were correlated. These AD-associated SNVs occurred at the same position in human and the 5xFAD cortex. Taken together, these results clearly demonstrated the prediction accuracy of SpliceAI in humans with respect to genetic mutations and splicing in the AD model.

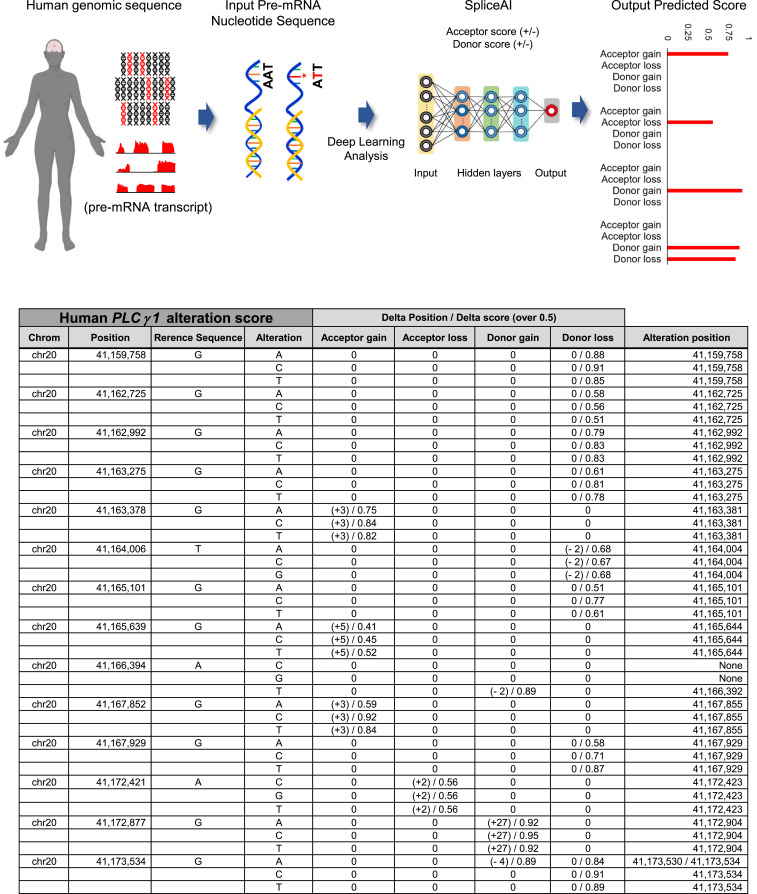
Prediction of AD specific nucleotide alteration sites from human genomic sequence with deep neural network. (Top) The schematic diagrams of the deep neural network procedure from human genomic sequence database through the SpliceAI analysis. (Bottom) The total 14 accurate prediction splicing sites in the human PLCγ1 gene, with the delta scores and position information.
Discussion
Given the fact that genetic variants exist in mammals at different stages of development, it is likely that these variants play roles in determining phenotypes and adaptation to environments as response to unexpected stimuli (37). It frequently has been questioned whether genomic information is directly associated with the expression to determine phenotypes. In addition, comparison of expression at transcriptional and translational levels in the same tissue may not be appropriate (38). Although gene copy number variations and gene copy number alterations (CNAs), which usually lead to changes in mRNA levels, are believed to contribute toward the development of several diseases such as tumors, CNAs leading to transcriptional changes do not cause translational changes (39). A previous study evaluated genetic variation leading to changes at the transcriptional level and in ribosome profile using RNA-seq and GWAS (40). However, correlations between genomic variation and protein expression in NDs, such as AD, are not fully understood yet. Therefore, we performed careful measured mature RNA and analyzed splicing patterns in genetic variants using GWAS-based deep learning to provide insight into individual SNVs in AD.
This study has at least two major implications. First, genetic variants arising due to gene mutations and RNA splicing are respectively conserved and observed in AD. Second, these variants could be predicted using deep learning-based tools to identify therapeutic and diagnostic targets for AD. GWAS of AD brain tissues described here was instrumental in defining genetic variations. This study evaluates a deep learning-based analysis for the detection of gene variants in AD, and examines the phenotypes of these variants and the association of these gene variants with disease progression. The marked increase in the level of histone acetylation in introns during development presumably enables highly specific activation of long noncoding RNAs as enhancer RNA, which may be needed to mediate the expression of some proteins (41, 42). H3K27ac is a well-known marker of active enhancers and is associated with enhancer activity (43, 44). Superenhancers, which are large clusters of enhancers, play an important role in biological regulation (transcription and translation) and various human diseases. Indeed, many diseases are associated with variations in superenhancer-enriched region (45). Previous studies have identified SNPs associated with various human diseases, such as AD, Type 1 diabetes, white blood cell distribution, fasting insulin level, and so on (45). For example, two SNPs in the superenhancer region of BIN1 gene are associated with AD (46). Moreover, trait-associated SNPs occur in noncoding regions within enhancers, where H3K27ac accumulate (45). AD-specific nucleotide alternation sites are distributed in noncoding regions of the PLCγ1 gene, and we identified several transcription factor binding motifs in AD-specific alternative sites (SI Appendix, Fig. S5). These variations can also be attributed to RNA maturation and splicing (Fig. 4). Consistent with these findings, epigenetic changes of target genes are also correlated with AD.
Among the unresolved questions regarding molecular mechanisms of genetic variations, including RNA splicing in AD progression, the potential of deep learning for application in prediction of diagnostic targets for diseases remains obscure. For example, although the genetic risk factors associated with AD progression mainly support the importance of Aβ aggregation, it has been indicated that Aβ is necessary but not sufficient for AD progression, and other unknown factors may play a key role (47). However, identifying such unknown factors is difficult without specific genetic analysis. Therefore, deep learning, including genome footprinting, has now become an alternative method to predict diseases (48). Through genetic analysis and exon mapping, we found that mature RNA was modified through splicing in an AD model (SI Appendix, Fig. S4), suggesting that genetic mutations might be conserved in human and that they can be analyzed with deep learning of pre-mRNA nucleotide sequences. Surprisingly, predictions of genetic mutations in human PLCγ1 matched exactly with the genome variant data of the AD model (Figs. 3 and 5). Although a wide range of clinical phenotypes is observed in AD patients, a liquid biopsy of bone marrow, blood, and urine is required for the diagnosis of AD. We also analyzed the SNVs in PLC genes and their expression in the blood of the AD model; the result indicated that AD-specific novel PLCβ1, PLCβ2, PLCβ4, and PLCγ1 SNVs in the blood are different within the cortex (SI Appendix, Table S6). PLCγ1 expression was enriched in AD patients, according to peripheral blood mononuclear cells microarray data, and this was consistent with the results of GWAS analysis in the AD mouse model (SI Appendix, Fig. S6). Collectively, GWAS-based deep learning results may be applied to predict the clinical outcome of AD patients and could contribute to both diagnosis and clinical treatment of AD.
In conclusion, we have demonstrated the importance of GWAS-based deep learning analysis for predicting genetic variation in AD. This study identifies the SNVs in the AD model through a combination of high-throughput screening and deep learning analysis. Given the DNA sequence of the target gene, our method enables the prediction of transcriptional events based on RNA splicing patterns. Although RNA-seq data are needed to analyze RNA splicing, deep learning with given referenced genomic sequences can be a novel alternative for accurate prediction of splicing. Furthermore, understanding of RNA splicing mechanisms using deep learning could provide novel ways to develop therapeutics and diagnostic procedures for AD.
Materials and Methods
Animals.
The 5xFAD (6-mo-old) transgenic mice were used as model of AD. The 5xFAD mice were obtained from the Jackson Laboratory. All animal experiments performed in this study were reviewed and approved by the Institutional Animal Care Use (IACUC) committee at KBRI (Korea Brain Research Institute) (IACUC-20-00018).
Total RNA-seq.
Total RNA-seq was performed as previously published (20). Total RNA extraction from mouse cortex was performed using commercial methods based on TRIzol (Invitrogen). Libraries construction was performed using TruSeq Stranded Total RNA LT Sample Prep Kit (Human Mouse Rat) by manufacturer’s instructions (Illumina). Firstly, to make a short fragment of mRNA, we added the fragmentation buffer. The oligo dT-primer was used to synthesize the first-strand complementary DNA (cDNA) to take short fragments as templates. Preparation of synthesis for second-strand cDNA was performed using buffer, 2′-deoxynucleoside 5′-triphosphate (dNTPs, containing 2′-deoxyuridine 5′-triphosphate instead of thymidine 5′-triphosphate), RNaseH, and DNA polymerase I, respectively. Double-stranded cDNA was purified with QIAQuick PCR extraction kit (Qiagen), and cDNA was eluted by elution buffer. Following the synthesis of second strand, end repair, addition of single A base, and adaptor ligation, cDNAs were connected with sequencing adaptors. The library concentration was measured by real-time PCR, and Agilent 2100 Bioanalyzer was used for profiling the distribution of insert size. Library constructions were sequenced by Illumina HiSeq 4000 based on the manufacturer’s instructions (Illumina), and were sequenced for 100 cycles. The HiSeq Control Software HCS (HiSeq Control Software) (v3.3) with RTA (Real-Time Analysis) (v2.7.3) was used to provide the management and execution of the HiSeq 4000 experiment runs.
Sequencing Data Analysis.
Total RNA-seq data analysis was performed as previously published (20). Images generated by HiSeq4000 were converted into nucleotide sequences by base calling and stored in FASTQ format utilizing Illumina package bcl2faseq (v2.16.0.10). To filter the dirty reads, which contain adaptors, unknown or low Phred quality-scored bases were obtained from raw reads, and clean reads were generated. Clean reads were mapped to reference UCSC hg19 genome and gene sequences using Tophat2 (v2.1.0). No more than five bases mismatched reads were allowed in the alignment. To annotate gene expression, fragments per kb per million reads values of each gene were calculated using Cufflinks package (v2.2.1). DEGs (differentially expressed genes), fold-change analysis was performed by Agilent GeneSpring (v7.3). Hierarchical clustering analysis for expression pattern was performed using Agilent GeneSpring (v7.3). Functional enrichment analysis was done using Gene Ontology functional classification system (geneontology.org/) and DAVID website (https://david.ncifcrf.gov/) as well. Raw reads and data are accessible in the Gene Expression Omnibus (GEO) (accession nos. GSE 151270 and GSE 147792).
SNV Analysis.
We performed SNV calls using the SAMtools (v.1.3) and bcftools (v.1.3) with bam files from two samples (WT and 5xFAD cortex). Each of bam files was indexed with SAMtools, and SAMtools mpileup was performed with UCSC mm10 genome as reference. To obtain variant calling format (VCF) files for specific target genes, option –r was used to set the position during mpileup. To perform variant calling for five PLC genes, five specific regions were set (in detail, chr2:160,731,310 to chr2:160,761,923 for PLCγ1, chr2:134,786,164 to chr2:134,988,192 for PLCβ1, chr2:118,728,319 to chr2:118,709,202 for PLCβ2, chr2:135,741,830 to chr:136,013,068 for PLCβ4, and chr19:6,969,643 to chr:6,954,299 for PLCβ3), and each of five VCF files were generated and merged for further analysis. After mpileup, VCF files were generated with vcfutils.pl varFilter using option –D100. Functional annotation of each variant was performed with ANNOVAR software.
SpliceAI Analysis.
The human gene, PLCγ1, was analyzed by SpliceAI to predict RNA splicing types on the genetic variations (4). SpliceAI, a residual convolutional neural network, predicts the probability of RNA splicing taking place in either donor or acceptor given DNA sequences. SpliceAI trained the complex patterns of RNA splicing from DNA sequences of 10 kbps around nucleotides of interests from the GENCODE (Genome ENCyclopedia Of DNA Elements) V24 databases (GRCh38/hg38) including more than 10,000 genes in chromosomes 2, 4, 6, 8, 10 to 22, X, and Y. We conducted the analyses using the pretrained SpliceAI, downloaded at https://github.com/Illumina/SpliceAI. We computed the probability of RNA splicing types (probabilities of donor, acceptor, or neither) for the alternate nucleotides on the starting locus of every exon of the gene, PLCγ1, based on the reference sequences (GRCh38/hg38). Then, the probabilities of the variant being splicing altering on acceptor and donor are computed by
where , and
cDNA Synthesis.
The cDNA synthesis for mature mRNA was performed with PrimeScript first-strand cDNA synthesis kit (Takara) according to the manufacturer’s protocol. To synthesize cDNA, we used 1 µgof total RNA from each sample, and added the reaction mixture containing the Oligo dT Primer (2.5 µM), dNTP mixture (0.5 mM), 5× PrimeScript buffer, RNase Inhibitor (20 U) and PrimeScript RTase (200 U). The synthesized cDNA was heat incubated at 42 °C for 60 min and 95 °C for 5 min, amplified with a thermal cycler dice. RNA templates were reverse transcribed into cDNA, which corresponds to pre-mRNA transcript using a High-Capacity reverse transcription kit (Applied Biosystems) according to the protocol described. Following the reference component suggested, each reaction was composed of 10× RT buffer, 25× dNTP Mix (4 mM), 10× RT Random Primers, MultiScribe Reverse Transcriptase (50 U), RNase inhibitor, and adjusted total volume with nuclease-free water. Then, template RNA sample with equal volume was added to set the total reaction volume. The reverse transcription was performed as suggested optimal thermal cycler condition, heat incubated at 25 °C for 10 min, 37 °C for 120 min, and 85 °C for 5 min, amplified with a thermal cycler dice.
RT-PCR.
PCR was performed using the Phusion-HF DNA polymerase kit (NEB). Primers employed were PLCγ1 Ex27-29 forward, 5′-GATTGGCACAGAACGTGCTT-3′, reverse, 5′-CTCCGTCTTTTGCTTGGTGC-3′; PLCγ1 Ex27-28 forward, 5′-GATTGGCACAGAACGTGCTT-3′, reverse, 5′-GCAAATGACACAGGGTTCCA-3′; PLCγ1 Ex28-29 forward, 5′-GCAGATGAACCAGGCCC-3′, reverse, 5′-CTCCGTCTTTTGCTTGGTGC-3′; PLCγ1 Ex26-30 forward, 5′-CTGAGGGGAAGATGATGGA-3′, reverse, 5′-CAGGCCTTTTACTGGGAAAG-3′; GAPDH forward, 5′-CATGGCCTTCCGTGTTCCTA-3′, reverse, 5′-GCGGCACGTCAGATCCA-3′; PLCγ1 Ex27-32 forward, 5′-GATTGGCACAGAACGTGCTT-3′, reverse 5′-CAAATGGCTGCTGGTATCTG-3′. Each reaction mixture contains 5× phusion HF buffer, dNTP (0.2 mM), primer forward and reverse (0.5 μM), Phusion DNA polymerase (1 U), and template cDNA, and nuclease-free water was added. The amplification was performed by the recommended condition. PCR products were detected through the ChemiDoc (Bio-Rad) imaging system.
RT-qPCR.
Total RNA was isolated from mouse whole blood using the TRIzol method. The cDNA synthesis for RT-qPCR was performed by High-Capacity reverse transcription kit (Applied Biosystems). Primers for RT-qPCR employed were PLCγ1 coding forward, 5′-GGTGACCTCAGTCCTTTCAG-3′, reverse, 5′- GAAATCTTCAAATGGCTGCTG-3′. RT-qPCR amplification was performed for 10 min for initial denaturation, followed by 45 cycles with 10 s at 95 °C for denaturation, 30 s at 60 °C for annealing, and 1 min at 72 °C for extension. PCR products were analyzed by LightCycler 480 (Roche).
Western Blot and Antibodies.
Electrophoresis was performed with 8% polyacrylamide gels, and proteins were then electrotransferred to nitrocellulose membranes. Membranes were immunoblotted with mouse monoclonal anti-PLCγ1 antibody (B16-5), generated as described previously (49), and mouse monoclonal anti-PLCβ1 (sc-5291, Santa Cruz), rabbit polyclonal anti-PLCβ3 (sc-403, Santa Cruz), rabbit polyclonal anti-PLCβ4 (sc-404, Santa Cruz), and mouse monoclonal anti-β-actin (GT5512, GeneTex) antibodies in Tween 20/Tris-buffered saline containing 5% skim milk. After incubation with the appropriate peroxidase-conjugated secondary antibody, proteins were detected with the enhanced chemiluminescence system (ECL system, Amersham).
ChIP-Seq Data.
H3K27ac binding profiles throughout the mouse forebrain were adapted from the data generated by GSE52386 data. H3K27ac ChIP-seq data are available in GEO (https://www.ncbi.nlm.nih.gov/geo/) (50).
Acknowledgements
We thank Dr. S.-W. Lee for assistance with bioinformatics analysis. This work was supported by KBRI Basic research program through KBRI funded by the Ministry of Science and ICT (Grant 20-BR-02-13), and Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (Grants 2019R1F1A1059595 and 2017R1D1A1B03030741). Figures were created with Biorender.com.
Data Availability.
RNA sequencing data have been deposited in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/, GSE 151270 and GSE 147792).
References
2
3
4
5
6
8
9
11
12
13
14
17
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
46
47
48
49
 Prediction of Alzheimer’s disease-specific phospholipase c gamma-1 SNV by deep learning-based approach for high-throughput screening
Prediction of Alzheimer’s disease-specific phospholipase c gamma-1 SNV by deep learning-based approach for high-throughput screening
