
Edited by Rene Bernards, The Netherlands Cancer Institute, Amsterdam, The Netherlands, and approved December 25, 2020 (received for review August 21, 2020)
Author contributions: N.K., Z.Q., R.D.M., and J.B.R. designed research; N.K., Z.Q., B.C.P., M.N.W., L.B., K.C.B., and X.C. performed research; J.G., J.N.R., R.D.M., and J.B.R. contributed new reagents/analytic tools; N.K., Z.Q., B.C.P., M.N.W., A.M., S.S., and R.D.M. analyzed data; and N.K. and Z.Q. wrote the paper.
1N.K. and Z.Q. contributed equally to this work.
2R.D.M. and J.B.R. contributed equally to this work.
- Altmetric
Consistent sex differences in incidence and outcome have been reported in numerous cancers including brain tumors. GBM, the most common and aggressive primary brain tumor, occurs with higher incidence and shorter survival in males compared to females. Brd4 is essential for regulating transcriptome-wide gene expression and specifying cell identity, including that of GBM. We report that sex-biased Brd4 activity drives sex differences in GBM and renders male and female tumor cells differentially sensitive to BET inhibitors. The observed sex differences in BETi treatment strongly indicate that sex differences in disease biology translate into sex differences in therapeutic responses. This has critical implications for clinical use of BET inhibitors further affirming the importance of inclusion of sex as a biological variable.
Sex can be an important determinant of cancer phenotype, and exploring sex-biased tumor biology holds promise for identifying novel therapeutic targets and new approaches to cancer treatment. In an established isogenic murine model of glioblastoma (GBM), we discovered correlated transcriptome-wide sex differences in gene expression, H3K27ac marks, large Brd4-bound enhancer usage, and Brd4 localization to Myc and p53 genomic binding sites. These sex-biased gene expression patterns were also evident in human glioblastoma stem cells (GSCs). These observations led us to hypothesize that Brd4-bound enhancers might underlie sex differences in stem cell function and tumorigenicity in GBM. We found that male and female GBM cells exhibited sex-specific responses to pharmacological or genetic inhibition of Brd4. Brd4 knockdown or pharmacologic inhibition decreased male GBM cell clonogenicity and in vivo tumorigenesis while increasing both in female GBM cells. These results were validated in male and female patient-derived GBM cell lines. Furthermore, analysis of the Cancer Therapeutic Response Portal of human GBM samples segregated by sex revealed that male GBM cells are significantly more sensitive to BET (bromodomain and extraterminal) inhibitors than are female cells. Thus, Brd4 activity is revealed to drive sex differences in stem cell and tumorigenic phenotypes, which can be abrogated by sex-specific responses to BET inhibition. This has important implications for the clinical evaluation and use of BET inhibitors.
Until recently, most basic and clinical research focused on investigating factors that influence disease susceptibility and progression without regard to biologic sex. However, mounting evidence has revealed sex differences in the incidence, age of onset, and outcome of numerous human diseases, including cardiovascular diseases, metabolic diseases, asthma, autoimmune diseases, birth defects, neurological diseases, psychiatric disorders, and cancers (123–4). The preponderance of sex differences in disease incidence and outcome led to the implementation of new guidelines by the NIH regarding inclusion of sex as a biological variable in all research.
Glioblastoma (GBM), the most common, aggressive, and incurable form of primary brain cancer (5, 6), is more prevalent in males, regardless of race or region of the world (male to female incidence of 1.6:1) (789–10). This sex difference extends across species; a male prevalence also occurs in spontaneous GBM in large dogs, suggesting a fundamental effect of sex on GBM risk (11). In addition to the sex difference in GBM incidence, Ostrom et al. documented a survival advantage for female GBM patients (12). The reasons for these sex differences in GBM incidence and outcome are largely unknown. We hypothesized that studying sex differences in GBM biology will inform the mechanisms underlying cancer risk and progression, with the ultimate goal of incorporating sex-informed approaches to treatment to improve survival of all patients.
While disease-related sex differences are often mediated through acute sex hormone actions, sex differences in the rates of multiple brain tumors are evident at all ages as well as in neutered and nonneutered dogs (8), suggesting that factors other than circulating sex hormones underlie this skewing (13). Such factors may include the organizational or epigenetic effects of transient in utero sex hormones, the effects of X chromosome alleles that escape inactivation, or the extragonadal expression of nonpseudoautosomal Y chromosome–encoded genes (141516–17).
We previously discovered that a genetically engineered model of GBM, involving combined loss of neurofibromin (NF1) and p53 function in murine neocortical postnatal day 1 (p1) astrocytes (male and female GBM astrocytes), exhibits sex differences in vivo (18, 19), including proliferation, clonogenic stem-like frequency and in vivo tumorigenesis, cell cycle regulation (18, 19), gene expression (18), and chemotherapy response (18), that mimic those observed in GBM patients (18, 19). Sex differences in the tumorigenic phenotype were validated in a second CRISPR-in utero electroporation murine model of GBM. In this model, guide sequence inserts targeting Nf1 and p53 were injected into the lateral ventricles of embryonic pups of male and female mice, and progenitor cells were targeted via bioelectroporation. Both male and female mice developed tumors; however, male mice exhibited an accelerated tumorigenic phenotype with a shorter median survival (18). Accordingly, data generated from both our in vitro and in vivo murine GBM models with combined loss of neurofibromin and p53 function further affirm that sex differences in tumorigenesis are evident in different mouse strains and independent of the method by which p53 function is abrogated. Together, these human and mouse data suggest that male and female GBM cells are differentially vulnerable to oncogenic events and to the cytotoxic effects of chemotherapy.
What are the factors that control these differences between male and female cells? In a recent multi-institutional study, we found that longer survival for male and female GBM patients after standard surgery, radiation, and chemotherapy is dependent upon different transcriptional programs (20). We discerned multiple sex-specific transcriptional subtypes of GBM that correlated with survival in a sex-specific manner. We hypothesized that these sex-specific transcriptional states arise through sex-specific epigenetics, analogous to mechanisms that drive normal sexual differentiation (21, 22).
Bromodomain and extraterminal (BET) family proteins are epigenetic readers of histone lysine acetylation and function as coactivators or corepressors of gene expression by recruiting specific transcriptional complexes to target genes (23). The BET family member Brd4 is essential for regulating transcriptome-wide gene expression and specifying cell identity, including that of GBM (2425–26). Additionally, Brd4 is an emerging drug target for epigenetic interventions (2324252627282930313233–34). Here, we show that sex differences in tumor phenotype are dependent on differential Brd4-bound enhancer regulation of stem cell–like phenotypes in male and female murine and human GBM cells. In vitro and in vivo studies of genetic or pharmacological Brd4 inhibition, as well as data mining of the Cancer Therapeutics Response Portal (CTRP), indicate that male and female murine and human GBM cells are differentially sensitive to inhibition of Brd4, resulting in decreased in vitro clonogenicity and in vivo growth of male tumors and opposite effects on female cells and tumors. Understanding the extent to which sex differences in GBM are mediated by epigenetic mechanisms will be imperative to understanding the biology of sex differences in GBM, stratifying patients for treatment with epigenetic agents, and anticipating potential sex-biased toxicities following systemic disruption of epigenetics.
Results
Male and Female GBM Cells Utilize Different Sets of Brd4-Bound Enhancers.
Previously, we reported correlations between sex differences in tumorigenicity and gene expression in our murine model of GBM (male and female GBM astrocytes lacking function of both NF1 and p53, see SI Appendix, Supplementary Materials and Methods for full details) (18, 19). Of the sex-biased differences in gene expression observed in this model, 50% were concordantly sex-biased in human GBM expression data (18). This concordance demonstrated that our murine model recapitulates important transcriptional pathways that govern clinically relevant sex differences in human GBM. In addition, we found correlations between sex differences in survival and gene expression in patients with GBM (20). We therefore determined whether these distinct GBM transcriptional states were the result of sex-specific enhancer activity and whether such states could be perturbed with small molecules that target epigenetic regulators that bind at these enhancers. We prioritized Brd4-bound enhancers for analysis because these enhancers play key roles in establishing cell identity (25, 30313233–34) and may regulate the cell-intrinsic sex differences observed in GBM. Brd4 is an epigenetic reader that binds acetylated histones H3 and H4 throughout the entire cell cycle and is deregulated in numerous cancers (35). Brd4 promotes epithelial-to-mesenchymal transition, stem cell–like conversion, and pluripotency (26, 28, 36). Brd4 pharmacological inhibition has shown therapeutic activity in a number of different cancer models (37383940–41).
To evaluate a potential role for Brd4 in mediating sex differences in GBM, we mapped Brd4 genomic localization in male and female GBM astrocytes (highly active Brd4-bound enhancers and typical Brd4-bound enhancers) using transposon calling cards (42, 43) to identify enhancers differentially bound by Brd4. To do so, we fused the piggyBac (PB) transposase to the C terminus of the Brd4 protein, endowing the transposase with the ability to direct the insertion of the PB transposon into the genome close to Brd4-binding sites. Five biological replicates were carried out, and the correlation between replicates was r > 0.9 for all pairwise comparisons (SI Appendix, Fig. S1A). Using this protocol, we mapped ∼1.25 million unique insertions directed by the Brd4-PBase fusion for each male and female sample. As Brd4 is an epigenetic reader of the acetylated lysine 27 residue of histone H3 (H3K27ac), we confirmed that these newly identified Brd4-bound enhancers were enriched for H3K27ac. We performed H3K27ac chromatin immunoprecipitation sequencing (ChIP-seq) in male and female GBM cells to identify the genomic regions enriched for this well-known marker of active enhancers (44). Three biological replicates were analyzed, and the correlation between replicates was r > 0.9 for all pairwise comparisons (SI Appendix, Fig. S1B). As expected, data collected from calling cards and H3K27ac ChIP-seq revealed a high concordance between Brd4-bound enhancers and H3K27ac enrichment. A representative example of concordant Brd4 binding and H3K27ac in male and female GBM astrocytes at high- and low-enriched regions is depicted in Fig. 1A. We next analyzed the distances from Brd4-binding sites to the nearest H3K27ac-enriched regions. Brd4-binding sites were significantly enriched at H3K27ac candidate enhancer regions (Fig. 1B) and display concordant gene expression levels (Fig. 1C) indicative of active Brd4-bound enhancers (P < 0.01). More specifically, 82 and 81% of Brd4-bound enhancers were localized within 200 base pairs (bp) of H3K27ac peaks in male and female GBM cells, respectively.

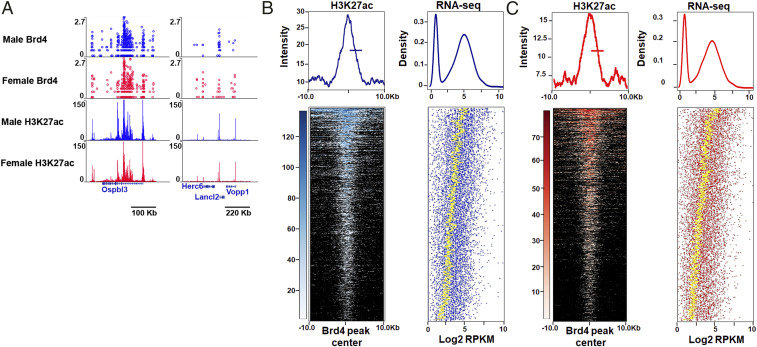
Concordance between Brd4-bound enhancers, H3K27ac enrichment, and gene expression in mouse GBM astrocytes. (A) A representative example of concordant Brd4 and H3K27ac binding at high- and low-enriched regions in male and female GBM astrocytes. (B and C) Heatmaps and intensity plots of H3K27ac signal at sex-biased Brd4-enriched peaks (+/−10 kb from the Brd4 peak center) in male (B) and female (C) GBM cells. For each sex-biased Brd4-enriched peak, the expression values of the nearest gene are presented in a dot plot in which the yellow dotted line depicts the average gene expression levels of 50 genes (Density refers to the fraction of genes). These analyses revealed that Brd4-binding sites are significantly enriched at H3K27ac candidate enhancer regions and display concordant gene expression levels indicative of active Brd4-bound enhancers (P < 0.01).
Having established that our genomic data were reproducible and that Brd4-binding sites occur at genomic regions enriched for H3K27ac, we next sought to identify the genomic loci that were differentially bound by Brd4 in male and female cells. We identified 2,679 enhancers (20% of all Brd4-bound enhancers in male GBM cells) that bound more Brd4 protein in males and 2,778 enhancers (21.2% of all Brd4-bound enhancers in female GBM cells) that bound more Brd4 protein in females (Fig. 2A). The Brd4 signal intensity for these male-biased and female-biased enhancers in male and female GBM astrocytes is depicted in Fig. 2B. To validate these putative male- and female-biased enhancers using an orthogonal method, we analyzed their H3K27ac status and found that the loci that bound Brd4 in a sex-biased manner displayed sex-biased enrichment of H3K27ac (Fig. 2C). Chromosome location analysis for these sex-biased Brd4-bound enhancers revealed that only 0.13 and 3.11% of male-biased Brd4-bound enhancers were located on the Y and X chromosomes, respectively, and that only 4.29% of female-biased Brd4-bound enhancers were located on the X chromosome, indicating that the observed differences in Brd4-bound enhancers are not simply due to differential Brd4 enhancer enrichment on sex chromosomes. The majority of enhancers exhibited a sex bias in Brd4 occupancy, while a subset appeared to be sex specific. We will collectively refer to these enhancers as sex biased. Representative examples of sex-specific, Nkx2.1, and sex-biased, Zic1/4, enhancers are depicted in Fig. 2 D and E. Nkx2.1 functions both as a “lineage survival” oncogene and tumor suppressor in lung adenocarcinomas (45, 46), whereas Zic1 acts as a tumor suppressor in breast and thyroid cancers (47, 48), supporting the potential context-dependent dual role of Brd4 in oncogenesis. This is the first demonstration of differential Brd4-bound enhancer usage by male and female cells of any kind, suggesting that these enhancers may function in sexual differentiation in a manner similar to their role in determining cell identity and fate (25, 3031–32, 49). A violin plot illustrating the full distribution of the enrichment signal for male- and female-biased loci and shared peaks is depicted in Fig. 2F.

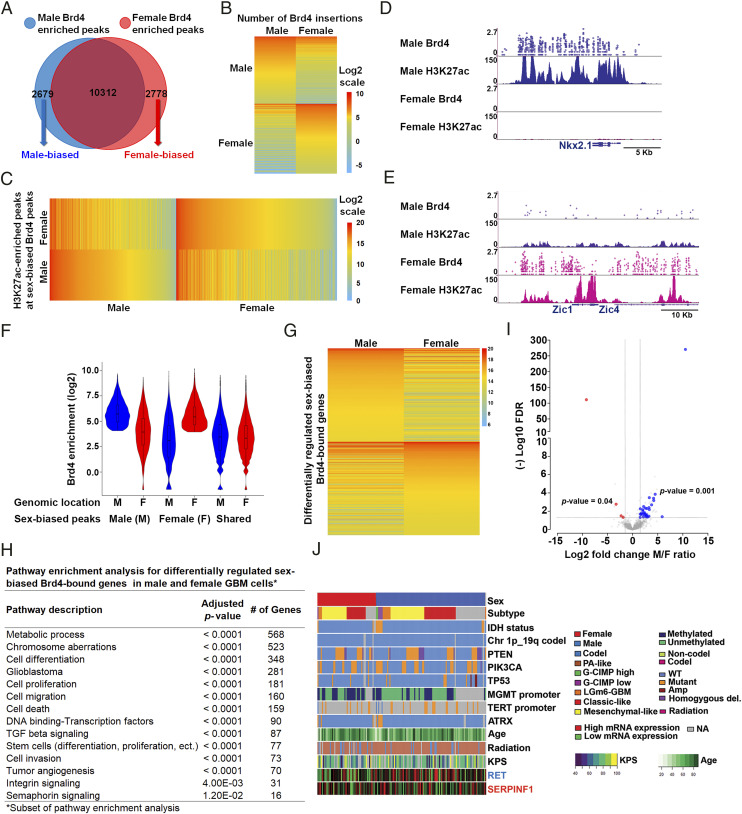
Male and female GBM cells have sex-biased Brd4-bound enhancers. (A) Venn diagram showing the number of unique male (2,679), unique female (2,778), and overlapping (10,312) Brd4-enriched peaks identified in GBM astrocytes. (B) A heatmap depiction of the number of Brd4 insertions at male- and female-biased Brd4-enriched peaks in GBM astrocytes (normalized data). (C) A heatmap analysis of H3K27ac signal intensity (read depth) at male and female-biased Brd4-enriched peaks in GBM astrocytes. (D) Nkx2.1 and (E) Zic1/4 illustrate male-specific and female-biased genes, respectively, associated with differential Brd4-binding affinity and H3K27ac enrichment. The x-axis (blue arrows) of all tracks corresponds to genomic location of the gene. The y-axis of calling card tracks represents the log10 scale of sequencing reads for each insertion as indicated by circles. The y-axis of ChIP-seq tracks represents the number of uniquely mapped reads. (F) Violin plot illustrating the full distribution range of the enrichment signal for male- and female-biased loci and shared peaks at different genomic locations. (G) RNA abundance (RNA sequencing) in male and female GBM cells for differentially regulated sex-biased Brd4-bound genes (n = 3). (H) Pathway enrichment analysis for differentially regulated sex-biased Brd4-bound genes in male and female GBM astrocytes. (I) Concordance in sex-specific gene expression patterns between murine GBM lines and human GSCs. Volcano plot depicting all mouse sex-biased significantly expressed genes with color-coded concordant human sex-biased genes (blue circles, male-biased genes; red circles, female-biased genes; log2FC 1.5 and FDR 0.05) genes. The x-axis represents mouse log2 fold change of male/female ratio for each significantly expressed sex-biased gene with its corresponding (−) log10 FDR value on the y-axis. (J) Gene expression pattern, whole-exome sequencing, and clinical phenotype data from the TCGA GBM datasets for two representative examples of concordant human and mouse male (blue) and female (red) sex-biased genes.
We next sought to identify the genes regulated by these sex-biased Brd4-bound enhancers. It is challenging to reliably link enhancers to the genes they regulate (50), so we used conservative criteria: we linked enhancers to the nearest gene within a 5 kb genomic distance. This analysis revealed 1,740 male-biased genes and 1,604 female-biased genes. Pathway enrichment analysis using the Genomatix Pathway System (GePS) of male-biased enhancer-gene pairs revealed functional enrichment for glioblastoma, cell proliferation, cell cycle, regulation of transcription, tumor angiogenesis, and cancer stem cells. Similar analysis on female-biased enhancer-gene pairs showed an enrichment in pathways involved in cell differentiation, glioblastoma, regulation of transcription, cell migration, and neural stem cells (SI Appendix, Table S1).
Based on our observation of sex-biased Brd4-bound enhancer usage between male and female GBM cells, we profiled male and female GBM cells with RNA sequencing (RNA-seq). For each condition, three biological replicates were performed; the data were highly reproducible (Pearson r ≥ 0.96 for all pairwise comparisons) (SI Appendix, Fig. S1C) and indicated differential expression of 3,490 transcripts (false discovery rate [FDR] < 0.05). We next integrated the male- and female-biased Brd4-regulated genes with the differentially expressed genes between male and female GBM astrocytes and narrowed the list of differentially regulated sex-biased Brd4-bound genes to 1,296, of which 52.4% were male-biased and 47.6% were female-biased Brd4-regulated genes. A heatmap depiction of the top 400 differentially regulated sex-biased Brd4-bound genes is presented in Fig. 2G. Pathway enrichment analysis for the 1,296 differentially regulated sex-biased Brd4-bound genes was performed using a combination of the Kyoto Encyclopedia of Genes and Genomes pathway and GePS. Classification of these genes according to function revealed a significant number of relevant and important pathways, including cell differentiation, glioblastoma, cell proliferation, cell migration and invasion, tumor angiogenesis, stem cell functions, and DNA-binding transcription factors (Fig. 2H and SI Appendix, Table S2). Thus, these sex-specific transcriptomic differences in core cancer pathways likely arise from male- and female-biased Brd4-bound enhancer activity.
We next sought to determine whether these sex-specific transcriptome differences are also present in human GBM samples. To do so, we reanalyzed published RNA-seq data of 43 human glioblastoma stem cells (GSCs) and segregated them by sex (51, 52). We found 246 differentially regulated genes between male and female GSCs, of which 223 were up-regulated in males and 23 were up-regulated in females (log2FC 1.5 and FDR 0.05). We then compared the gene lists of sex-biased differentially regulated genes in human GSCs with those of the murine GBM astrocytes at the same cutoff parameters (log2FC 1.5 and FDR 0.05; 981 and 771 up-regulated genes in male and female GBM cells, respectively) and found an overlap in gene identity and direction of change for both males (17.5% of human male-biased differentially expressed genes, P value 0.001) and females (17.4% of human female-biased differentially expressed genes, P value 0.04) (SI Appendix, Table S3). P values were calculated using the cumulative hypergeometric distribution. A volcano plot depicting all mouse sex-biased significantly expressed genes with color-coded concordant human sex-biased genes (blue circles show male-biased genes and red circles show female-biased genes; log2FC 1.5 and FDR 0.05) is presented in Fig. 2I. The expression patterns of two representative examples of concordant male-biased (RET) and female-biased genes (SERPINF1) are illustrated in Fig. 2J, with gene expression (RNA-seq), whole-exome, and clinical phenotype data collected from TCGA GBM data (53). This concordance of sex-biased gene expression patterns between our murine mouse model and the human GSCs validates the sex-specific tumorigenic phenotype observed in our mouse model. Pathway enrichment analysis for the human and mouse concordant genes identified functional enrichment for the MAPK family signaling cascade, cell adhesion, mesenchymal and pluripotent stem cells, neurofibromatosis, neuroendocrine tumors, and neuron differentiation. Altogether, the mouse and human data suggest the presence of sex-specific transcriptional programs in tumorigenesis that are regulated by Brd4-bound enhancers in a sex-biased manner.
Depletion or Inhibition of the BET Family Protein Brd4 has Differing Effects on Clonogenicity in Mouse and Human Male and Female GBM Cells.
Our genomic analyses of mouse and human GBM cells suggested to us that Brd4-bound enhancers might regulate a large number of genes and transcriptional networks in a sex-biased fashion. To test this hypothesis, we performed genetic depletion of individual BET family members in our murine model of GBM. Under basal conditions, male and female GBM cells expressed Brd3 and Brd4 messenger RNA (mRNA) at similar levels, whereas Brd2 was expressed at higher levels in female cells (*P = 0.02) (Fig. 3A). We evaluated the potency of five short hairpin RNAs (shRNAs) specific to each of the Brd2, Brd3, or Brd4 genes and selected the shRNAs that achieved the most robust knockdown of each gene for downstream functional experiments of the tumorigenic phenotype. Brd2, Brd3, and Brd4 mRNA levels were partially depleted in male and female GBM cells after infection with the corresponding lentiviral shRNAs (Fig. 3B). Knockdown of Brd2 did not affect clonogenic frequency in either male or female cells as measured by the extreme limiting dilution assay (ELDA) (Fig. 3C). Male GBM cells with Brd4 knockdown exhibited a decrease in clonogenic frequency, whereas female cells displayed an increase in clonogenic frequency. A similar effect on clonogenic frequency was observed following Brd3 knockdown in male cells, although it was not as robust as the effect of Brd4 knockdown. Strong Brd4 effects were observed despite the more modest knockdown of Brd4 compared to Brd3.

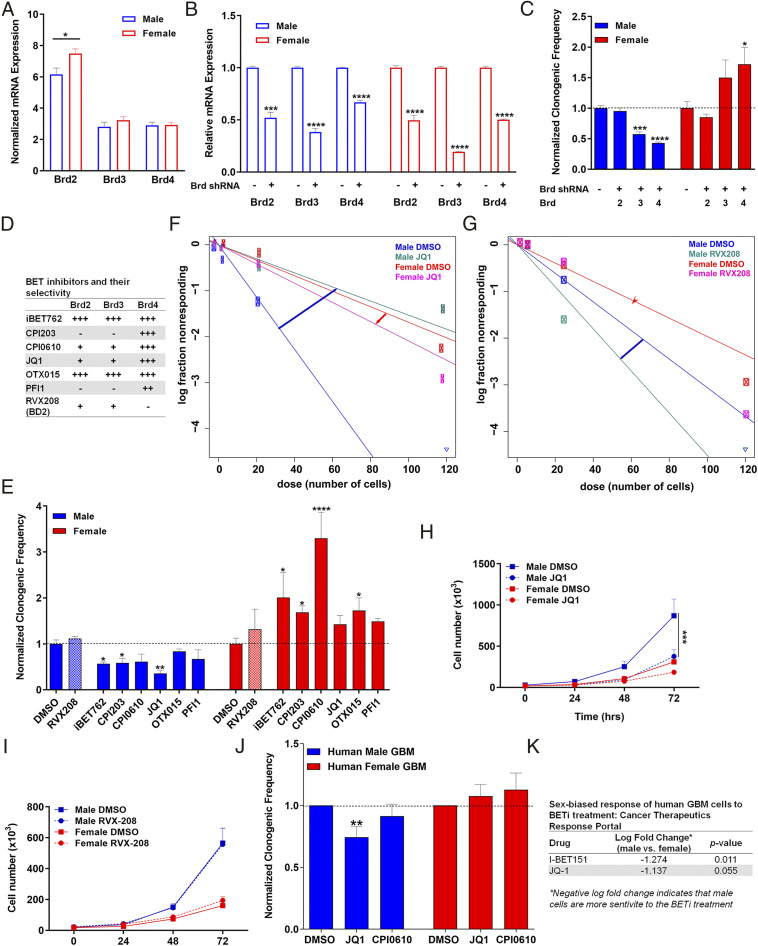
Brd4 inhibition has differing effects on clonogenicity in mouse and human male and female GBM cells. (A) Normalized Brd2, Brd3, and Brd4 mRNA expression in male and female GBM cells under basal conditions. Brd3 and Brd4 are expressed at equivalent levels between male and female GBM cells, while Brd2 is expressed at higher levels in female cells. (B) Brd2, Brd3, and Brd4 mRNA expression following lentiviral shRNA infection. mRNA levels in knockdown samples were normalized to their respective control. (C) Normalized clonogenic cell frequency as determined by ELDA assay in control and Brd2, Brd3, and Brd4 knockdown male and female GBM cell lines. Knockdown of Brd4 and Brd3 suppresses clonogenic frequency in male GBM cells, while female cells showed a significant increase in clonogenic frequency following Brd4 knockdown only. Brd2 depletion was without effect. All treatment groups were normalized to control clonogenic frequency levels. (D) Tabular representation of the BET inhibitors currently in clinical trials and their selectivity to the three BET family members. (E) Normalized clonogenic cell frequency as determined by ELDA assay in male and female GBM cells treated with DMSO (control) or the indicated BET inhibitor. BET inhibitors significantly reduced clonogenic cell frequency in male cells and significantly increased clonogenic cell frequency in female cells. Shaded bars are used to indicate the effect of RVX208, a Brd2/3 inhibitor, as a control for Brd4 inhibition. (F) Frequency of clonogenic stem-like cells as determined by ELDA assay in male and female GBM cells treated with DMSO or JQ1 (Brd2/3/4 inhibitor). Male cells exhibited greater clonogenic cell activity than female cells under control conditions. JQ1 significantly reduced clonogenic cell frequency in male cells to levels almost equivalent to female cells under control conditions, while female cells exhibited an increase in their clonogenic cell frequency. (G) Frequency of clonogenic stem-like cells was measured by ELDA in male and female GBM astrocytes following RVX-208 treatment, a BD2 inhibitor with selectivity to Brd2 and Brd3. No significant change in clonogenic frequency was observed in male and female GBM astrocytes following RVX208 treatment. (H) Proliferation assays were performed in mouse GBM cells treated with DMSO or JQ1. Male cells exhibited greater proliferation than female cells under control conditions. JQ1 significantly reduced growth in male cells to female basal growth level. (I) Proliferation rates of male and female GBM astrocytes were unaffected by RVX-208 treatment. (J) Normalized clonogenic cell frequency as determined by ELDA assay in male and female human primary GBM cells treated with DMSO (control), JQ1, or CPI0610. JQ1 treatment significantly reduced clonogenic cell frequency in human primary GBM male cells and slightly increased clonogenic cell frequency in human primary GBM female cells. CPI0610 treatment had a similar response pattern as JQ1 in male and female human GBM primary cells. (K) Tabular representation of the sex-biased growth response of human GBM cells to BETi treatment. Male GBM cells are more sensitive to BETi treatment than female GBM cells (CTRP). (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 as determined by two-tailed t test or one-way ANOVA).
Small molecule inhibitors of the BET family of proteins (BETi) are a novel class of epigenetic compounds that selectively target BET proteins and have been shown to have promise as cancer therapeutics, decreasing cell proliferation and invasion in many cancer types, including GBM (24, 27, 29). However, almost all of the American Type Culture Collection human cell lines used in the preclinical studies are of male origin and are therefore potentially uninformative with regard to sex differences in drug effects. Therefore, we evaluated whether there was a sex difference in the treatment response to some of the BET inhibitors currently in clinical trials for cancer in our mouse GBM cells. We treated male and female GBM astrocytes with a panel of BETi (Fig. 3D), which are being tested in clinical trials (24 clinical trials listed in https://www.clinicaltrials.gov/ and https://wwwselleckchem.com), and then performed ELDAs to measure clonogenic cell frequency (5455–56). Treatment with BETi reproducibly decreased clonogenic frequency in male GBM cells while increasing clonogenic frequency in female cells (Fig. 3 E–G and SI Appendix, Fig. S2). This striking abrogation of sex differences, driven by sex-specific response to Brd4 inhibition, is illustrated in response to the Brd4 antagonist JQ1 (Fig. 3F). JQ1 is a thieno-triazolo-1,4-diazepine that displaces Brd4 from chromatin by competitively binding to the acetyl-lysine recognition pocket (37, 38). Treatment of acute myeloid leukemia cells with JQ1 causes a rapid release of Mediator 1 (MED1) from a subset of enhancer regions that were co-occupied by Brd4, leading to a decrease in the expression of neighboring genes (57). We treated male and female GBM cells with either 0.05% dimethyl sulfoxide (DMSO) or 500 nM JQ1 and then performed ELDAs to measure clonogenic cell frequency. Male cells exhibited greater clonogenic cell activity than female cells under control conditions. Treatment with JQ1 reproducibly abrogated the basal differences in clonogenic frequency between male and female GBM cells by decreasing the clonogenic cell fraction in male cells and increasing the clonogenic cell fraction in female cells, rendering the male JQ1–treated cells equivalent to the female control cells (Fig. 3F). To confirm that these effects were mediated by Brd4-bound enhancers, we performed similar experiments using RVX-208, a BD2 inhibitor with high selectivity to Brd2/3 (58). Consistent with the Brd2 and Brd3 knockdown results, treatment with 5 µM RVX-208 for 24 h did not have an effect on the clonogenic frequency of male and female cells (Fig. 3 E and G). Next, we examined the effect of drug treatment on cell growth and proliferation. Male cells exhibited decreased proliferation and a growth phenotype equivalent to control female cells following JQ1 treatment (Fig. 3H), whereas treatment with RVX208 did not alter proliferation of male and female cells (Fig. 3I). Taken together, these results demonstrate that the sex differences in the tumorigenic phenotype we observe in our murine GBM cells are mediated by differential Brd4-bound enhancers and that the response to BET inhibition is sex dependent.
Next, we investigated whether the sex-specific responses to BETi observed in our murine GBM model were also present in human GBM. To do so, we treated male and female human GBM primary cell lines with 0.05% DMSO, 500 nM JQ1, or 1 µM CPI0610 and then performed ELDA to measure clonogenic cell frequency. Similar to the murine data, treatment with BETi decreased clonogenic cell frequency in male patient-derived GBM cells, while slightly increasing the clonogenic cell frequency in female patient-derived GBM (Fig. 3J and SI Appendix, Fig. S3). Although we observed the expected trend in response to BETi treatment in human male and female primary cell lines, our sample size was small with limited power. Additionally, human GBM samples are intrinsically heterogeneous with different genetic backgrounds, and hence the magnitude of treatment effects can be more variable compared to isogenic cell lines. To expand our sample size, we analyzed the CTRP to examine if human male GBM cells are indeed more sensitive to BETi than female cells (59). First, we segregated the CTRP human GBM samples by sex. We then analyzed the cell growth response of male and female GBM cells to BETi treatment. This analysis revealed that male GBM cells are more sensitive to BETi, displaying slower growth and more cell death, consistent with what we observed using our murine GBM model (Fig. 3K).
While the female stem cell response to BETi treatment data were variable, a clear difference in response to BET inhibition was present in the human GBM cell lines. This result was also supported by the sex difference in proliferation following BETi identified in the CTRP datasets. Thus, the sex-specific effects of Brd4 inhibition on tumorigenic phenotype and response to treatment observed in our murine GBM model extend to human GBM cells. Collectively, the concordance of data generated from three different GBM models provide strong evidence for the generalizability of our results generated from the murine GBM model. Taken together, these results demonstrate that sex differences in GBM cellular phenotypes are mediated by differential Brd4-bound enhancers and that reduced Brd4 function results in differing sex-dependent effects in both mouse models and human patients. These data further affirm the sex-dependent role of Brd4 in regulating tumorigenesis in GBM and demonstrate how clinically important it will be to consider sex in the design of clinical trials and the decision making for treatment of cancer patients.
Brd4-Bound Enhancers Regulate Sex Differences in GBM.
Having established that Brd4-bound enhancers mediate sex difference in our murine model of GBM, we next sought to determine the transcriptional pathways responsible for these basal sex differences. To do so, we used our Brd4-binding data in male and female GBM astrocytes to categorize Brd4-regulated genes as male biased (Fig. 4A), female biased (Fig. 4B), or shared (Fig. 4C). Although these genes were categorized using only Brd4-binding data, we found a high degree of concordance with H3K27ac intensity and gene expression within each category. When considered as a group, 95% of the male-biased Brd4-regulated genes displayed more H3K27ac signal in males, and 72% were more highly expressed in males (Fig. 4 A, P value < 10−6), while 93% of the female-biased Brd4-regulated genes displayed more H3K27ac signal in females, and 67% were more highly expressed in females (Fig. 4 B, P value < 10−6). These results demonstrate that sex-biased Brd4 binding correlated with sex-specific gene expression patterns.

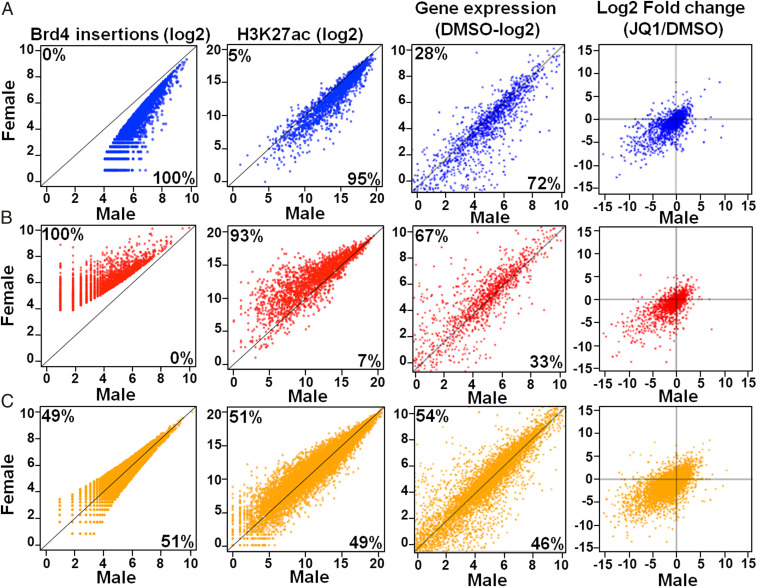
Male and female GBM cells have sex-biased Brd4-bound enhancers and concordant gene expression. Brd4 insertions, H3K27ac signal, and gene expression values (normalized reads per kilobase million) of the nearest gene(s) in male (x-axis) and female (y-axis) GBM astrocytes for male-biased (A), female-biased (B), and shared (C) genes are plotted. The percentage of genes enriched in each sex is indicated at the top left (female) and bottom right (male) of each graph. Log2 fold changes (Log2FC) of the gene expression value following JQ1 treatment are indicated in the far-right column and are plotted as above. Most genes are located in the bottom left quadrant, indicating down-regulation of the gene in both sexes following JQ1.
To determine which sex-biased genes were modulated after treatment with a BET inhibitor, we performed RNA-seq on male and female GBM cells treated with JQ1, one of the BET inhibitors that had a strong sex-biased effect on clonogenicity (SI Appendix, Fig. S1D). Briefly, cells were treated with either vehicle (0.05% DMSO or 500 nM JQ1) for 24 h prior to RNA isolation. Gene expression analysis of JQ1- and DMSO-treated cells revealed that JQ1 treatment predominantly down-regulated gene expression (Fig. 4, last column of scatterplots, and SI Appendix, Fig. S4A). In addition, genes proximal to Brd4-binding sites were significantly down-regulated compared to genes that were distal to Brd4-binding sites (P < 0.01), indicating that JQ1 had a specific and directed effect on genes whose expression was driven by Brd4-bound enhancers (SI Appendix, Fig. S4B). Pathway analysis of sex-biased Brd4-bound enhancer associated genes down-regulated following JQ1 treatment in male and female GBM cells revealed functionally important pathways, such as chromosome aberrations, integrin signaling, and stem cell proliferation in males and immune system process, cell cycle, and transforming growth factor-beta signaling in females (SI Appendix, Table S4).
To begin to understand the sex-specific stem cell response to BETi, we focused our attention on the functions of the sex-biased male and female Brd4-bound enhancer genes under basal conditions. Functional classification of these genes using pathway enrichment analysis revealed that male GBM cells were highly enriched for cancer, neoplastic, and pluripotent stem cell pathways, while female GBM cells were mainly enriched for neural stem cell and stem cell proliferation pathways (SI Appendix, Fig. S5A). Representative examples of these pathways are depicted in SI Appendix, Fig. S5 B and C. Male-biased genes in stem cell pathways included well-known oncogenes, such as Myc, Tet1, and Lif, while female-biased genes included tumor suppressors, such as Six2 and Six3. Based on these results and our previous observation of a sex-dependent differing response in clonogenicity following Brd4 inhibition with JQ1, we examined the effects of JQ1 treatment on the expression of stem cell pathway genes. As BETi reduced sex differences in clonogenicity, we hypothesized that sex differences in the expression of stem cell–related genes present in male and female GBM cells under basal conditions would be eliminated by JQ1 treatment, accounting for the abrogation of sex differences in the tumorigenic phenotype. We identified genes with sex differences in expression at baseline that became equivalent following JQ1 treatment and performed pathway analysis. As anticipated, the cancer stem cell pathway was affected by JQ1 treatment (SI Appendix, Fig. S5D). Concordant with differential growth responses, JQ1 down-regulated male-biased genes, while JQ1 up-regulated female-biased genes. The fold change in expression following JQ1 treatment (ratio of JQ1/DMSO) is depicted above or below each gene. Overall, there was concordance with the direction of change in gene expression and the decrease and increase in stem cell function in male and female ELDA assays, respectively. These results support the existence of Brd4-regulated sex-specific transcriptional programs that mediate key functional properties of GBM, including clonogenicity. Identifying the specific pathways critical to sex differences in GBM will require further functional studies.
Motif Analysis at Sex-Biased Brd4-Bound Enhancers Identifies Candidate Transcription Factor Drivers of Sex Differences in the Tumorigenic Phenotype.
Our results suggest that male and female GBM cells reside in two distinct transcriptional states established by the actions of Brd4. Inhibiting Brd4 function abrogates the sex differences in these transcriptional states and the concomitant differences in tumorigenic phenotype. As Brd4 does not bind DNA directly, but instead is recruited to enhancers by transcription factors either directly or indirectly though histone acetylation, we sought to identify candidate transcription factors (TFs) responsible for the recruitment of Brd4 to sex-biased genomic loci. To do so, we searched for TF-binding motifs enriched at male-biased or female-biased Brd4-bound enhancers. TFs enriched at male-biased Brd4-bound enhancers included oncogenes and stem cell markers, such as Myc, Klf5, and Oct4, whereas female-biased Brd4-bound enhancers were co-occupied and enriched with TFs with tumor suppressor functions, such as p53 and Smad4 (Table 1). These results could explain the observed dual functions of Brd4 in male and female GBM cells. Future studies, including knock-in and knockout of transcriptional master regulators and partners of Brd4, are warranted to investigate the dual functions of Brd4 as an oncogene in male GBM cells and a tumor suppressor in female GBM cells.

| Pathway description | Genes | Adjusted P value | # of genes |
| Male GBM Cells: 20 genes/motifs | |||
| Transcription factor activity, sequence-specific DNA binding | Myc, Hoxb4, Max, Pou5f1, Fli1, Klf1, Bach1, Fosl2, Bach2, Nfe2l2, Mafk, Isl1, Nfe2, Jun, Klf5, Fosl1, Batf, Lhx3, Atf3 | <0.0001 | 19 |
| Neoplastic cell transformation | Myc, Hoxb4, Max, Pou5f1, Fli1, Klf1, Bach1, Fosl2, Bach2, Nfe2l2, Mafk, Nfe2, Jun, Klf5, Klf14, Fosl1, Batf, Lhx3, Atf3 | <0.0001 | 19 |
| Cell proliferation | Myc, Hoxb4, Fosl2, Isl1, Nfe2, Jun, Fosl1, Atf3 | <0.0001 | 8 |
| Pluripotent stem cells | Myc, Hoxb4, Max, Pou5f1, Klf1, Mafk, Isl1, Klf5, Egr1, Max, Fosl2, Jun, | <0.0001 | 8 |
| Angiogenesis | Nfe2l2, Isl1, Jun, Klf5 | <0.0001 | 4 |
| Mitogen-activated protein kinase signaling | Max, Fosl2, Jun, Fosl1, Atf3 | 1.10E-02 | 5 |
| Stem cell differentiation | Pou5f1, Nfe2l2, Isl1, Batf, Hoxb4 | <0.0001 | 5 |
| Female GBM Cells: 26 genes/motifs | |||
| Transcription factor activity, sequence-specific DNA binding | Mybl1, Rela, Gata4, Ets1, Atf4, Arnt, Maff, Irf1, Smad2, Irf2, Prdm1, Irf3, Ddit3, Ebf1, Smad4, Myod1, Ep300, Irf4, Cebpa, Pgr, Ctcf, Trp53 | <0.0001 | 22 |
| Growth arrest | Mybl1, Hoxb13, Atf4, Irf1, Smad2, Ddit3, Ebf1, Myod1, Cebpa, Trp53 | <0.0001 | 10 |
| Negative regulation of cell cycle | Mybl1, Ets1, Irf1, Ddit3, Ep300, Cebpa, Trp53 | 1.00E-03 | 7 |
| Histone modification and chromosome organization | Mybl1, Hoxb13, Atf4, Irf1, Smad2, Ddit3, Ebf1, Myod1, Cebpa, Trp53 | 1.00E-03 | 6 |
| Cyclin-dependent kinase inhibitor 1 signaling | Hoxb13, Irf1, Myod1, Trp53 | 1.00E-03 | 4 |
| Estrogen receptor signaling | Hoxb13, Arnt, Irf4, Pgr | 3.00E-03 | 4 |
* Subset of enriched pathways for enriched motifs in male and female GBM cells.
BET Inhibitors Have Sex-Specific Effects on In Vivo Tumorigenicity in Male and Female Mouse GBM Astrocytes.
The results described thus far suggested to us that Brd4-bound enhancers play an important role in maintaining sex differences in our murine GBM model. However, these experiments all measured in vitro surrogates for the tumorigenic phenotype. Therefore, we next sought to determine what role, if any, Brd4 plays during in vivo tumorigenesis. We treated male and female murine GBM astrocytes with BETi and used these cells to perform in vivo tumor flank implantation studies in female nude mice. Only females were used as recipients for tumor flank implantation because we have previously shown that the sex of the recipient mouse does not affect the in vivo growth of implants in this murine model (19). Based on our in vitro ELDA studies, we chose to use JQ1 and CPI0610, the two BETi with the most dramatic sex-specific effect in male and female GBM astrocytes, respectively. Each mouse underwent flank implantation of DMSO-, JQ1-, or CPI0610-treated male cells (5,000 cells) or DMSO-, JQ1- or CPI0610-treated female cells (1.5 million cells). The sex differences in cell dose were empirically determined to match the timeframe for in vivo tumor growth up to the threshold for euthanasia. We attribute the sex differences in tumor growth kinetics to differences in clonogenic cell frequency and rates of proliferation (18, 19). Tumor formation was monitored by observers blinded to group assignment for 7 to 16 wk. Flank implantation of JQ1-treated male GBM cells was less likely to result in tumor formation than implantation with control DMSO-treated male cells (Fig. 5 A and C). In contrast, flank implantation of CPI0610-treated female GBM cells was more likely to result in tumor formation than implantation with control DMSO-treated female cells (Fig. 5 B and C). These results are consistent with the sex-specific effects of JQ1 and CPI0610 on in vitro clonogenic cell frequency and cell growth. This effect was seen following only one dose of BETi prior to implantation, suggesting that this robust response is maintained and manifested at the epigenetic level. Both JQ1 and CPI0610 display a similar pattern in their selectivity to target the different Brd family members, with the highest selectivity for Brd4 (Fig. 3D). Although both drugs had a similar sex-specific directional effect on clonogenic frequency in male and female cells, the magnitude of their effect differed in both the clonogenic cell frequency and in vivo tumorigenic assays. A possible explanation for this discrepancy is that the BETi treatment could be affecting different or multiple Brd family members. To rule out this possibility and validate that the sex-biased response to treatment was driven by Brd4, we treated male and female control and Brd4 knockdown cells with either 0.05% DMSO, 500 nM JQ1, or 1 µM CPI0610 and then performed ELDA to compare clonogenic cell frequency. No significant difference in the effect on clonogenic cell frequency was observed between control and Brd4 knockdown cells treated with BETi, indicating that Brd4 is required for sex-specific responses to BETi (SI Appendix, Fig. 6). Although it is still unclear why we observe different magnitude effects of these two BETi, the fact that both drugs show a sex-specific effect in vivo is notable and predicted by our earlier observations. These results provide strong and critical evidence that the biology of sex affects cancer incidence and outcome and addressing these sex differences in cancer will be fundamental to improving outcomes and a better quality of life for cancer patients.

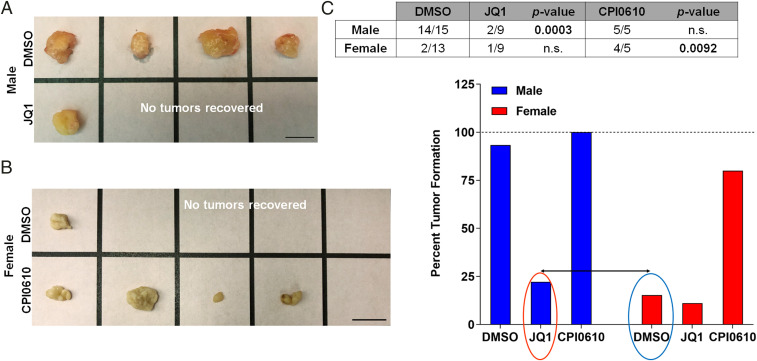
BET inhibitors have sex-specific effects on in vivo tumorigenicity in male and female GBM astrocytes. (A) Representative flank tumors from DMSO- or JQ1-treated male GBM astrocyte-initiated tumors. (B) Representative flank tumors from DMSO- or CPI0610-treated female GBM astrocyte-initiated tumors. (C) Fraction of male and female GBM cell implants that formed tumors following treatment with DMSO, JQ1 (500 nM), or CPI0610 (1 μM). JQ1-treated male GBM cells were significantly less likely to form tumors than DMSO-treated male GBM cells (Fisher’s exact test P < 0.0003), while CPI0610-treated female GBM cells were significantly more likely to form tumors than DMSO-treated female GBM cells (Fisher’s exact test P < 0.0092). The bar graph represents the percent tumor formation under each treatment condition. Notably, JQ1-treated male GBM cells show a decrease in their tumor formation capacity equivalent to the control DMSO-treated female GBM cells, whereas CPI0610-treated female GBM cells had an increase in tumor formation capacity almost equivalent to control DMSO-treated male GBM cells.
Discussion
Sex differences in the incidence and severity of numerous human diseases, including cancer, are substantial and demand an understanding at a molecular level. Despite the abundant data in the literature supporting an important role for sex on incidence, prognosis, and mortality in cancer, there has been limited effort, until recently, to include sex as a biological variable in the design of clinical trials, data analysis, and treatment (3, 12, 20, 606162–63). New approaches to treatment could be revealed by dissecting the biological and molecular mechanisms that drive sex differences in phenotype. Similar to other cancers, GBM has a higher incidence in males compared to females with a ratio of 1.6:1 (63). Additionally, after developing GBM, males tend to have a shorter median survival of 17.5 mo compared to 20.4 mo for females (64). Recently, we found that longer survival for male and female GBM patients after standard surgery, radiation, and chemotherapy is associated with different transcriptional programs (20). Using a JIVE (joint and individual variation explained) algorithm, male and female patients with GBM cluster into five distinct male and female subtypes distinguished by gene expression and survival. Although, a recent study by Yuan et al. (65) assigned GBM as a weak sex-effect cancer, their analysis of the TCGA GBM human patient data was performed using a propensity score algorithm, a more standard approach compared to the specialized JIVE analysis we have previously adapted in our analysis of the TCGA GBM transcriptome datasets (20).
The epigenetic mechanisms involved in GBM initiation and treatment response have been heavily investigated in the past few years and include epigenetic readers, writers, erasers, and histone proteins (25, 26, 6667–68). The strong evidence for epigenetic dysregulation in GBM has led to clinical trials of multiple drugs targeting tumor epigenetics in hope of improving treatment response in patients (24, 27, 29, 69). However, sex differences in the epigenetic landscape in male and female cells have not been taken into account when investigating cancer risk or treatment response. As sexual differentiation is in large part an epigenetic phenomenon and sex differences are found in the epigenetic landscape of male and female cells, it will be critical to examine the efficacy of therapies in both sexes separately to avoid missing important clinical effects when data are compiled from both sexes. Studying disparities in threshold for transformation in male and female cells will provide a powerful tool to investigate epigenetic mechanisms that establish a dimorphic tumorigenic phenotype and better treatment and survival outcomes for cancer patients.
Our study establishes that sex-biased Brd4 activity drives sex differences in the GBM tumorigenic phenotype and renders male and female tumor cells differentially sensitive to BETi. Using our murine GBM model, in which male and female cells are syngeneic except in their sex chromosomes, we discovered transcriptome-wide sex differences in gene expression, H3K27ac marks, and Brd4-bound enhancers in male and female GBM cells. Brd4 is an epigenetic reader that promotes stem-like conversion and pluripotency (26, 28, 36), and its inhibition has shown therapeutic activity in a number of different cancer models (37383940–41). Sex-biased Brd4-bound enhancers, identified in our study, were also enriched for H3K27ac, indicative of sex-biased highly active enhancer regions. These sex-specific gene expression patterns were evident in human GSCs. validating our model to recapitulate the human GBM phenotype. Thus, sex is established as an intrinsic element of cellular identity that is driven by Brd4 activity in GBM.
To validate whether the phenotypic differences in male and female GBM cells were indeed mediated by Brd4, as suggested by our transcriptome-wide gene expression and Brd4 binding analyses, we performed Brd4 knockdown using shRNA in male and female GBM cells. Brd4 depletion decreased clonogenic stem cell frequency in male cells while increasing it in female cells, abrogating the sex differences observed in the tumorigenic phenotype. These results suggest that differential sex-biased Brd4 localization may be responsible for the sex differences in GBM. These data emphasize the importance of understanding and including the fundamentals of the biology of sex while investigating epigenetic mechanisms of tumorigenesis.
As bromodomain inhibitors are currently being evaluated in clinical trials, we hypothesized that this differential sex-biased Brd4 localization in male and female GBM cells would contribute to a differential response to BETi. Male and female murine and human GBM cells responded in differing ways to BETi. Consistent with our knockdown data, this dimorphism in Brd4 function was linked to the sex of the experimental cells and correlated with sex-specific patterns of Brd4 localization throughout the genome, which renders male and female cells differentially sensitive to BETi. Accordingly, BETi decreased the growth of male in vivo tumors but increased growth of female tumors. These results strongly indicate that sex differences in disease biology translate into sex differences in therapeutic responses. The sex-specific response to the inhibition of Brd4 in our GBM model could help explain previously published data in breast and prostate cancer, wherein ectopic expression of Brd4 in breast cancer cells decreased invasiveness and tumor growth, while Brd4 inhibition decreased viability of prostate cancer cells (40, 7071–72). These studies also revealed that in women with estrogen receptor positive breast cancer (70) or endometrial cancer (71), low Brd4 expression correlated with worse survival. This is in contrast to men with prostate cancer, in whom low levels of Brd4 are associated with improved survival (40, 71). The sex-specific response of primary human GBM cells to BETi corroborated data obtained from the CTRP that show human male GBM cells to be more sensitive to BETi, displaying slower growth and increased cell death compared to human female GBM cells.
The functions of Brd4 are determined by the action of master transcriptional regulators that bind directly or indirectly to Brd4 and dictate its localization in the genome. We performed a motif-based analysis to identify potential TFs that endow Brd4 with its pro- or anti-tumorigenic functions. This analysis revealed that Brd4 colocalized in a sex-biased manner with Myc and p53 in male and female GBM cells, respectively. These results are consistent with previously published data investigating sex differences in gene expression and regulatory networks in multiple human tissues. The study revealed that multiple TFs, although not differentially expressed between male and female cells, display sex-biased regulatory targeting patterns (2). These results warrant further functional studies to validate whether these master TFs drive the sex differences in Brd4-bound enhancer activity (sex-biased Brd4 genomic localization) and, therefore, the sex differences in tumorigenic phenotype and response to BETi. Given that Brd4 is shown to have pleiotropic functions in the regulation of gene expression, these results suggest that Brd4 can exhibit dual functions in GBM as an oncogene or a tumor suppressor, dependent upon cellular sex. Further functional and mechanistic experiments are warranted to ascertain this Brd4 dual function in GBM. We previously presented evidence for cell-intrinsic sex differences rendering male and female cells differentially vulnerable to malignant transformation. We showed that murine male and female GBM cells displayed differences in cell cycle regulation through differential RB inactivation (19). These data, in addition to our newly identified role for Brd4 in regulation epigenetic mechanisms involved in stem cell function, could further explain the sex differences in the male and female GBM cells tumorigenic phenotypes.
The consistency between our mouse and human data (GBM cell lines, GSCs, and CTRP) and published breast and prostate cancer studies provides strong evidence for context-dependent, sex-specific dual roles of Brd4 in regulating gene expression programs in oncogenesis. In future work, we will take advantage of this remarkable sex difference in Brd4 function in GBM to better define the molecular basis for Brd4 pleiotropy in cancer and mechanisms of resistance to Brd4 inhibitors. As BETi are currently being evaluated in a number of clinical trials, understanding this phenomenon is of critical importance, both for the interpretation of existing trials and to guide better application of these drugs. Increasing our knowledge of these sex-biased genetic and epigenetic mechanisms will lead to a greater understanding of cancer biology and its relationship to normal development. We have identified sex-biased Brd4-regulated genes and pathways, which could translate into new and promising therapeutic targets to enhance survival for all GBM patients and potentially other cancers that exhibit substantial sex differences in incidence or outcome.
Materials and Methods
Please find in SI Appendix, Supplementary Materials and Methods detailed descriptions of mouse male and female GBM cells; ChIP-seq for H3K27ac; RNA sequencing; pathway analysis; GSCs gene expression data analysis; real-time qPCR; shRNA lentiviral infection and knockdown of Brd2, Brd3, and Brd4; ELDA analysis, growth assays, and CTRP data analysis, in vivo tumorigenesis: flank implantation, bioinformatic motif analysis, and statistical analysis.
Transposon Calling Cards Assay for Brd4 Binding.
For each sample (female and male murine GBM cells), 10 wells with an area of 9.5 mm2 were seeded at 50% confluency (200,000 cells per well of a 6-well plate). Each well represented a unique set of insertion events and was processed individually. Briefly, each well received 3 µg plasmid DNA containing a PB transposon (carrying a puromycin selection marker) with or without 3 µg transposase plasmid encoding Brd4 fused to the N terminus of hyperactive PB (HyPBase). Plasmids were delivered using Lipofectamine LTX/Plus Reagent (Invitrogen). Before transfection, cells were incubated in Dulbecco’s Modified Eagle Medium (DMEM) without fetal bovine serum (FBS) or antibiotics for 30 min and then replaced with fresh media containing DMEM/F12 with 10% FBS and 1% penicillin-streptomycin. Transfection complexes were applied to cells for 12 to 18 h at 37 °C and 5% CO2. Cells were allowed to recover in fresh medium for another 24 to 48 h. Each well was then separately expanded onto 10 cm dishes and placed in medium containing puromycin at a concentration of 2.5 µg/mL for 3 d. After 3 d, cells that received transposon DNA alone showed no growth, while those that also received transposase DNA showed robust growth. Each 10 cm dish of expanded puromycin-resistant cells were harvested and processed by a modified version of the transposon calling card protocol as previously described (43). The calling card library was prepared by assigning each well a unique Illumina P7 index so that each well of a sample could be demultiplexed as a set of unique barcoded insertions. All four sets of libraries, (Male_PB_helper, Male_PB_Brd4_fusion, Female_PB_helper, and Female_PB_Brd4_fusion) were then pooled and sequenced on an Illumina HiSeq 3000. A total of 40 Illumina P7 indexes were demultiplexed (10 replicates per library). Reads were mapped to the mm10 reference genome using NovoAlign (version 3.08.02; Novocraft Technologies Sdn Bhd). For each library, data from constituent replicates were pooled, and discrete insertions were exported in calling card library format for subsequent analysis. Insertions were visualized on the WashU Epigenome Browser (v46.1).
Sequencing Data Alignment and Analysis.
Raw reads from transposon calling cards were aligned to the murine genome build mm10 using Bowtie 2 (version 2.3.4.3) (73). Significant calling card peaks or Brd4-enriched enhancer sites were identified by a modified version of the previously described algorithm, which also has similarities to the MACS2 ChIP-seq peak caller (43). Briefly, transposon insertions were grouped into peaks using a hierarchical clustering algorithm with a maximum distance of 5 kb between insertions. Significant peaks were identified using Poisson distribution to test for enrichment over the background (unfused transposase) calling card data with an adjusted P value threshold less than 0.05. The expected number of transposon insertions per TTAA was estimated by considering the total number of insertions observed in a large region of 100 kb distance centered at the calling card cluster/insertion. We computed a P value based on the expected number of insertions and identified Brd4-enriched enhancer sites using Poisson distribution.
To identify the sites with an excess of Brd4 insertions in male GBM cells relative to female cells, we used the algorithm from ChIP-seq peaks caller MACS (version 2.1.0) (74) but modified for the analysis of calling card data. First, the Brd4-enriched enhancer sites from both male and female cells were merged if they overlapped by 1 bp. For each merged or unmerged enhancer site, the normalized insertions from the female samples were used to compute the lambda of Poisson distribution. We then computed a P value from the cumulative distribution function of the observed number of independent insertions in the male sample. Brd4-binding sites with a P value less than 0.05 and with more than 20 insertions were considered as Brd4-binding sites significantly enriched in male samples. To identify the Brd4 sites with an excess of insertions in female GBM cells relative to male cells, we performed the same analysis, substituting the male and female datasets.
Although male and female GBM cells have a different number of X chromosomes, one X chromosome is inactivated in female cells, and so the amount of accessible chromatin has been found to be similar in both cell types (75). For this reason, we treated the X chromosome in an identical manner to the autosomes for peak calling. However, to explore how sensitive our analysis was to this assumption, we recalculated P values for peaks on the X chromosome as we adjusted the insertions in male cells with a multiplier from 1 to 2 (using a step size of 0.1). The total number of male-biased peaks varied from 2,679 to 2,995 and the total number of female-biased peaks varied from 2,778 to 2,727, indicating that the vast majority of differential bound peaks in our analysis are robust to the actual percentage of accessible chromatin on the inactivated X chromosome.
ChIP-seq datasets for H3K27ac were aligned to the murine genome build mm10 using Bowtie 2 (version 2.3.4.3), and only uniquely aligning reads were used for downstream analyses (73). Regions of enrichment of H3K27ac over the background were calculated using the MACS version (2.1.0) peak finding algorithm (74). An adjusted P value threshold of enrichment of 0.01 was used for all datasets. The resulting peak files were used as inputs for DiffBind (version 3.5) to derive consensus peak sets (76, 77). The differential enrichment of H3K27ac signals between male and female analysis was carried out with DiffBind using DESeq2 (method = DBA_DESEQ2) with libraries normalized to total library size.
RNA-seq datasets were aligned to the transcriptome and the whole genome with Spliced Transcripts Alignment to a Reference (STAR) (version 2.7.0) (78). Genes or exons were filtered for just those that were expressed. The read-count tables for annotated genes in the mm10 gene transfer format (.GTF) file were derived from uniquely aligned reads using HTSeq (version 0.11.1) (79). The raw counts for each gene were converted into transcripts per million (TPM) that is more appropriate for comparisons of gene expression levels between samples (8081–82). TPMs from biological replicates were averaged for subsequent analysis. Differential gene expression between pairs of samples was computed using DESeq2 (version 1.28.1) and was filtered by FDR < 0.05 for differentially expressed genes (83). In in some cases, a twofold change was also applied as a filter for identifying differentially expressed genes between two conditions.
Acknowledgements
This work was supported by NIH Grants RO1 CA174737 (J.B.R.), RF1MH117070, U01MH109133 (National Institute of Mental Health) (R.D.M.), R01GM123203 (National Institute of General Medical Sciences) (R.D.M.), and R21 HG009750 (National Human Genome Research Institute) (R.D.M.); Joshua’s Great Things (J.B.R.); The Children’s Discovery Institute (J.B.R. and R.D.M.); and NIH Grants T32GM007200, T32HG000045, and F30HG009986 (A.M.). We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by National Cancer Institute Cancer Center Support Grant No. P30 CA91842 to the Siteman Cancer Center and by Institute of Clinical and Translational Sciences/Clinical and Translational Sciences Award Grant No. UL1 TR000448 from the National Center for Research Resources (NCRR), a component of the NIH, and NIH Roadmap for Medical Research. This publication is solely the responsibility of the authors and does not necessarily represent the official view of the NCRR or NIH.
Data Availability
All raw data and processed files have been deposited in the Short Read Archive/Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/). GSE156821 is the reference series for all data. The SubSeries that are linked to GSE156821 are GSE156678, GSE156819, and GSE156820. Source code for the python package used to map calling card reads and call peaks can found in GitLab at https://gitlab.com/rob.mitra/mammalian_cc_tools (see README_CCFTOOLS.pdf in the ccf tools directory).
References
1
2
3
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
75
76
77
79
81
82
 Brd4-bound enhancers drive cell-intrinsic sex differences in glioblastoma
Brd4-bound enhancers drive cell-intrinsic sex differences in glioblastoma
