
Edited by Fred J. Sigworth, Yale University, New Haven, CT, and approved January 6, 2021 (received for review June 22, 2020)
Author contributions: J.B., S.B., A.J.R.P., and H.S. designed research; J.B., S.B., A.J.R.P., and H.S. performed research; J.B. contributed new reagents/analytic tools; J.B., S.B., A.J.R.P., and H.S. analyzed data; and J.B., A.J.R.P., and H.S. wrote the paper.
- Altmetric
AMPA-type glutamate receptors (AMPARs) make the final step in a relay of excitability from one brain cell to another. The receptor contains an integral ion channel, which, when opened by neurotransmitter, permits sodium and other cations to cross the cell membrane. We investigated permeation of sodium, potassium, and cesium at the atomistic level using a computational molecular dynamics approach and obtained ion transit rates similar to those in wet experiments over tens of microseconds of simulations. We determined that the region selecting between cations is the simplest of any channel of this type. Distinct from ion channels that select single ion species, sodium and potassium remain partly hydrated and have only one major binding site in the channel.
Fast excitatory synaptic transmission in the central nervous system relies on the AMPA-type glutamate receptor (AMPAR). This receptor incorporates a nonselective cation channel, which is opened by the binding of glutamate. Although the open pore structure has recently became available from cryo-electron microscopy (Cryo-EM), the molecular mechanisms governing cation permeability in AMPA receptors are not understood. Here, we combined microsecond molecular dynamic (MD) simulations on a putative open-state structure of GluA2 with electrophysiology on cloned channels to elucidate ion permeation mechanisms. Na+, K+, and Cs+ permeated at physiological rates, consistent with a structure that represents a true open state. A single major ion binding site for Na+ and K+ in the pore represents the simplest selectivity filter (SF) structure for any tetrameric cation channel of known structure. The minimal SF comprised only Q586 and Q587, and other residues on the cytoplasmic side formed a water-filled cavity with a cone shape that lacked major interactions with ions. We observed that Cl− readily enters the upper pore, explaining anion permeation in the RNA-edited (Q586R) form of GluA2. A permissive architecture of the SF accommodated different alkali metals in distinct solvation states to allow rapid, nonselective cation permeation and copermeation by water. Simulations suggested Cs+ uses two equally populated ion binding sites in the filter, and we confirmed with electrophysiology of GluA2 that Cs+ is slightly more permeant than Na+, consistent with serial binding sites preferentially driving selectivity.
Glutamate receptor ion channels are found at synapses throughout the vertebrate nervous system, where they convert submillisecond glutamate signals into cation currents. Advances in structural biology have provided molecular scale maps of their ion pores, permitting comparison with a burgeoning menagerie of structures from related ion channels. It has been comparatively difficult to obtain candidate open pore structures of glutamate receptors, with the notable exceptions being from single-particle cryo-electron microscopy (Cryo-EM) of complexes between GluA2 and Stargazin (1, 2), the prototypical transmembrane AMPA receptor regulatory protein (TARP). However, it is unclear from simple inspection of these structures whether 1) the ion pore is conductive or 2) it is open to its fullest extent or to the highest conductance level. One structure has the unedited form of GluA2 (Q), that is, GluA2 lacking the common RNA editing of the Q586 residue to Arginine (3), while the other structure is of the edited form (Q586R), which should have a low conductance (4). Previous electrophysiological work suggests that the radius of the narrowest part of the AMPA channel [either with or without Stargazin (5, 6)] is 4 Å, and neither the upper gate nor the selectivity filter (SF) are this wide in either candidate open structure. However, these estimates are based on the geometric mean radius of large, elongated cations. A substantial body of electrophysiological work provides good benchmarks for how permeation should proceed in an open AMPA receptor pore. For example, canonical measurements of reversal potentials show that alkali earth cations from sodium (Na+) up to cesium (Cs+) should permeate GluA1 (7) and GluA2 (Q) (8) approximately equally well and that the single channel conductance of the full level of GluA2 (Q) should be considerable [∼30 pS (9)].
Most computational work on ion permeation through channels has been focused on simple, selective potassium (K+) channels like KcsA (1011–12), being the first reported crystal structures of ion channels (13). Thanks to their minimal sequences, these channels demand little computational overhead. Their key structural features are two membrane-spanning helices and a reentrant loop forming a narrow SF for permeant ions. This core motif defines a superfamily of tetrameric and pseudotetrameric channels that encompass selective, semiselective, and nonselective cation conductances. In common with many eukaryotic channels, the AMPA-type glutamate receptor (AMPAR) has a pore domain whose gating state is controlled by substantially larger domains outside the membrane (amino terminal domain [ATD] and ligand-binding domain [LBD], Fig. 1A), which account for about 75% of the protein mass. The large size presents a challenge for conventional molecular dynamic (MD) simulations, with the AMPA receptor being about six-times bigger than KcsA.

![AMPA receptor and the simulation setup. (A) The activated open state of the AMPA receptor from Cryo-EM [PDB ID: 5WEO (1)] with Stargazin molecules removed. The receptor is composed of ATD, LBD, and TMD. (B) The TMD and linkers to the LBD layer were included the MD simulations. The sites where the linkers were truncated and physically restrained (see Methods) are marked with red balls. Two out of the four subunits are shown. (C) The SF region of the AMPA receptor pore, with key residues labeled. Again, only two diagonally opposed subunits are drawn. (D) The computational electrophysiology setup was composed of two tetrameric AMPA channels, each embedded in a separate POPC lipid bilayer, solvated with water molecules and ions. A small cation imbalance between the two compartments α and β was maintained during the simulations. The resulting gradient gave a transmembrane potential to drive ion permeation.](/dataresources/secured/content-1765980780543-24197fdf-dafc-457f-9446-2e6b9f18a555/assets/pnas.2012843118fig01.jpg)
AMPA receptor and the simulation setup. (A) The activated open state of the AMPA receptor from Cryo-EM [PDB ID: 5WEO (1)] with Stargazin molecules removed. The receptor is composed of ATD, LBD, and TMD. (B) The TMD and linkers to the LBD layer were included the MD simulations. The sites where the linkers were truncated and physically restrained (see Methods) are marked with red balls. Two out of the four subunits are shown. (C) The SF region of the AMPA receptor pore, with key residues labeled. Again, only two diagonally opposed subunits are drawn. (D) The computational electrophysiology setup was composed of two tetrameric AMPA channels, each embedded in a separate POPC lipid bilayer, solvated with water molecules and ions. A small cation imbalance between the two compartments and
It remains to be seen to what extent key features of ion permeation elucidated in prokaryotic channels (selectivity, discrete sites, desolvated ions, and block by divalent ions) are widely applicable in channels with more substantial architectures. Recent experimental and MD work on nonselective prokaryotic channels like NaK and NaK-CNG as well as a mutant of the human hERG1 channel suggests that ion permeation in these channels differs substantially from classical K+-selective ion permeation (141516–17). The SF of nonselective cation channels is much more flexible, with fewer ion binding sites leading to distinct conduction mechanisms and hydration states for Na+ and K+ when passing through the filter. In the AMPA receptor, in one activated open Cryo-EM structure of GluA2 (1), density for a presumptive hydrated sodium ion was observed adjacent to the unedited Q586 residue. In the structure of RNA-edited GluA2 (Q586R) with Stargazin (2), ions were absent. The closed state structure of a GluA1/A2 heteromer featured strong density of unknown identity adjacent to C589 (18). Whether the paucity and heterogeneity of resolved ions is due to a lack of order in the filter region, the lack of detail in the coulomb potential density, or a true deficit of ions remains unclear. However, observations in these AMPA receptor structures are in marked contrast to the Cryo-EM structures of several other nonselective cation channels, such as hyperpolarization-activated cyclic nucleotide-gated (HCN) (19) and cyclic nucleotide-gated (CNG) channels (20), where two or three bound ions were visible. Further context comes from canonical K+-selective channels, which feature up to four ions in a row (21), and crystal structures of Ca2+-selective transient receptor potential (TRP) channels where two Ca2+ ions were readily resolved (22).
Here, we used MD-based computational electrophysiology to examine ion permeation through the mammalian AMPA receptor ion channel. We determined that the published structure is stably open, suitably detailed for MD simulations, and most likely represents a native fully open state. We identified a minimal SF consisting of a single major ion binding site that does not fully dehydrate ions. In simulations, Cs+ co-opted a secondary binding site. Consistent with multiple sites promoting ion selectivity, electrophysiology of AMPARs in human embryonic kidney (HEK293) cells showed that Cs+ is slightly more permeant than Na+.
Results
Monovalent Cation Permeation.
Our starting structure for MD simulations was the AMPAR channel in its activated conformation (Protein Data Bank [PDB] ID: 5WEO) (1), which we embedded into a palmitoyloleoyl phosphatidylcholine (POPC) lipid bilayer (Fig. 1D). To reduce the number of atoms in the simulations, we removed the extracellular ATD and LBD and the four Stargazin molecules decorating the periphery of the channel. To retain the open channel conformation, we held the transmembrane domain (TMD) of AMPAR open by physically restraining the end of the truncated linkers (Fig. 1B and see Methods for details). This approach, while facile, was highly reproducible, and the channel did not lose its overall stability (SI Appendix, Fig. S1) or close spontaneously during any simulation run (Fig. 2A). Simulations of ion conduction were performed with the computational electrophysiology method (23, 24). In this setup, the simulation box contains two membranes that define two compartments, and a voltage difference across each membrane is created by an ion gradient that is maintained alchemically during the simulations (Fig. 1D). From an analysis of the temporal evolution of the root mean square deviation (rmsd) (SI Appendix, Fig. S2), the different transmembrane voltages (up to 550 mV) induced by these ion gradients caused no systematic distortions to the receptor structure during each simulation run.

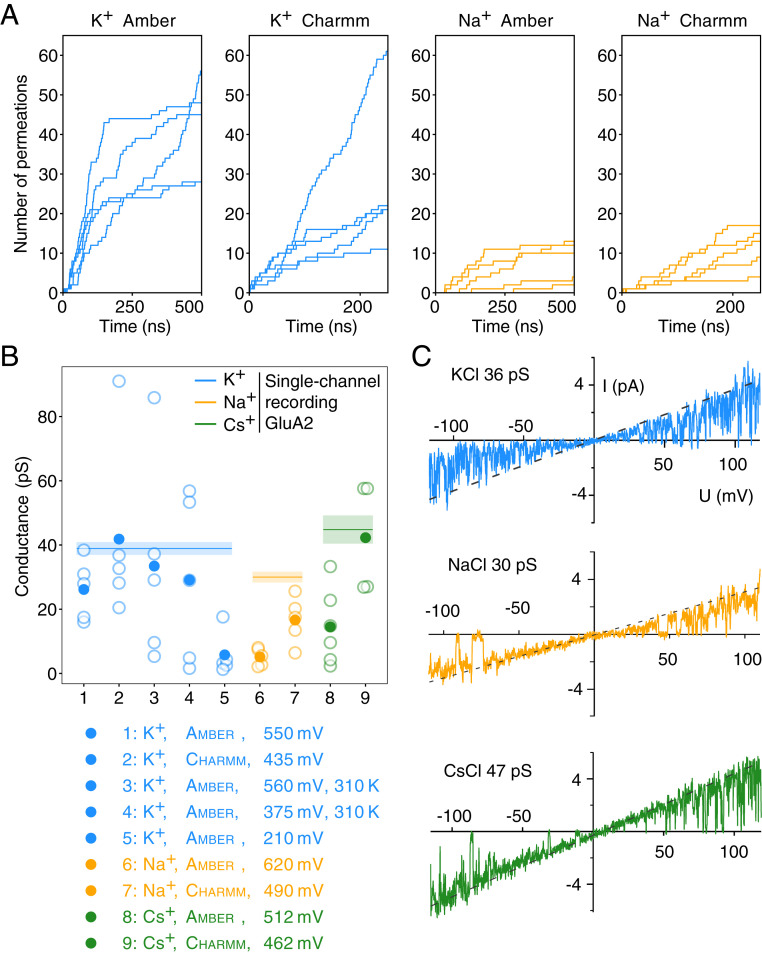
Ion conductances in GluA2. (A) Cumulative ion permeation events in the GluA2 pore for individual trajectories of K+ and Na+ simulations with the respective force fields. The simulations were performed at 303 K using an ion imbalance of 6e− between two compartments
Classical electrophysiology experiments suggest AMPARs are broadly nonselective over different alkali metal cations, although most measurements were not made on GluA2 (7, 25). We began by performing a series of ion permeation simulations with the principal biological ion species, Na+ and K+. To ensure the robustness of our observations, we undertook simulations at different temperatures, with different force fields and different driving forces on the ions (see Discussion and Methods). The conductances of the AMPA channel pore were directly calculated from the ion permeations observed in these simulations. We counted cations that traversed the entire narrow region of the pore domain delineated by the 586QQGCDI591 sequence, summarized in Table 1 and Fig. 2B. Outside of this region, ion movements lacked order and made only rare interactions with the channel, leading to a largely flat energy landscape (SI Appendix, Fig. S3). In all trajectories, the channel pore remained conductive for Na+ or K+ during the entire 500-ns run, as exemplified by the ion track plots in Fig. 2A and SI Appendix, Fig. S4. In a typical 500-ns trajectory with a transmembrane potential of 450 mV, we recorded over 40 K+ permeation events, corresponding to a conductance of 32 pS.

| Cation | Force field | ∆q (e–) | [C] (mM) | Vm (mV) | T (K) | Replicates | Ion permeations | γ (pS) |
| K+ | Amber | 6 | 273.8 | 550 | 303 | 5 | 220 | 26 ± 8 |
| K+ | Amber | 2 | 273.8 | 210 | 303 | 5 | 18 | 6 ± 6 |
| Na+ | Amber | 6 | 273.8 | 620 | 303 | 5 | 50 | 5 ± 2 |
| K+ | Charmm | 6 | 212.1 | 435 | 303 | 5 | 144 | 41 ± 25 |
| Na+ | Charmm | 6 | 212.1 | 490 | 303 | 5 | 63 | 17 ± 6 |
| Cs+ | Amber | 6 | 273.8 | 512 | 303 | 6 | 133 | 15 ± 11 |
| Cs+ | Charmm | 6 | 212.1 | 462 | 303 | 4 | 121 | 42 ± 15 |
| K+ | Amber | 6 | 273.8 | 560 | 310 | 5 | 300 | 33 ± 29 |
| K+ | Amber | 4 | 273.8 | 375 | 310 | 5 | 163 | 29 ± 23 |
| K+ | Amber | 0 | 121.5 | 77 | 303 | 5 | 0 | 0 |
| K+, closed | Amber | 6 | 284 | 433 | 303 | 3 | 0 | 0 |
| K+ | Single-channel recording | — | 150 | −120 to +120 | 296 | 3 patches | — | 39 ± 2 |
| Na+ | Single-channel recording | — | 150 | −120 to +120 | 296 | 3 patches | — | 30 ± 2 |
| Cs+ | Single-channel recording | — | 150 | −120 to +120 | 296 | 3 patches | — | 45 ± 4 |
Simulations of the GluA2 TMD for Na+, K+, and Cs+ permeation are shown. In addition to changing cations, we also varied temperature (T), charge imbalance (∆q), and the force field employed. C is the salt concentration. Each simulation replicate was 500 ns, except for charmm runs, in which the duration was 250 ns. These simulations represent an aggregate time of 20.5 µs. amber refers to amber99sb and charmm to charmm36. The single-channel conductance of GluA2 (Q) in Na+, K+, and Cs+ each determined from three recordings is also included.
Although K+ permeated readily in the simulations with amber99sb force field (26), we observed substantially fewer permeations of Na+ under the same simulation condition. The simulated conductances of both K+ and Na+ were higher in the charmm36 force field (27), allowing direct observation of Na+ permeation at close to physiological rates (Table 1 and Fig. 2B). Higher conductance using charmm36 compared to amber99sb was previously reported in simulations of K+-selective and nonselective NaK-CNG channels (14). We also performed five unbiased K+ simulation runs without ion gradient. Within each 500-ns run, several K+ ions spontaneously entered the SF from both intracellular and extracellular sides and found stable binding sites (SI Appendix, Fig. S5). Nevertheless, no complete permeation through the filter could be observed in 2.5 µs of simulation time. A comparison of ion occupancy showed that the major ion binding site within the SF (see below, SI) was rarely occupied during these simulations at nominal zero transmembrane voltage (SI Appendix, Fig. S6).
Binding Sites for Permeant Ions.
SF architectures differ widely among tetrameric ion channels. While K+ channels generally have long, straight filters, nonselective cation channels tend to have shorter filters that display more plastic or eccentric geometries (28, 29). To understand how the AMPAR SF supports cation-selective ion conduction, we determined one- and two-dimensional ion occupancies in the AMPA channel, which were sampled from cumulative simulations of K+ and Na+ with the respective force fields (Figs. 3B and 4A and SI Appendix, Fig. S7). For each ion species, these plots reveal only one principal binding site (which we call SI) in the SF, which is located around 15 Å along the pore axis (Fig. 3). At this site, the primary coordination of permeant ions is by backbone carbonyls of Q586 that replace some but not all of waters of hydration (SI Appendix, Fig. S8). Secondary, less frequent interactions occur with the mobile side chain of Q586 and the backbone carbonyls of Q587 (Fig. 1C). In some trajectories using amber99sb force field, solvated Na+ ions stayed at the SI longer than 100 ns (Fig. 4B), which may explain the slower permeation. During outward permeation, ions leaving the SI site frequently paused above the tip of the SF (25 Å at the pore axis in Fig. 4A) and then rapidly entered bulk water as they traversed the upper reaches of the channel. Before reaching the major binding site SI, multiple ions simultaneously entered the inner mouth of the channel, classically expected to be part of the SF (2, 30) and composed by the residues G588, C589, and D590 (Fig. 1C). K+ and Na+ paused frequently but briefly at several secondary sites in the lower SF region (for example, the SII site around 10 Å into the pore, cytoplasmic to the SI site, Fig. 3B). Compared to the primary site, the secondary sites were weakly populated (SI Appendix, Fig. S4) with shorter residence times (Fig. 4B). Therefore, we conclude that the true SF region of the AMPA is the shortest seen in any tetrameric channel to date, comprising essentially only Q586 and Q587 that form the SI site.

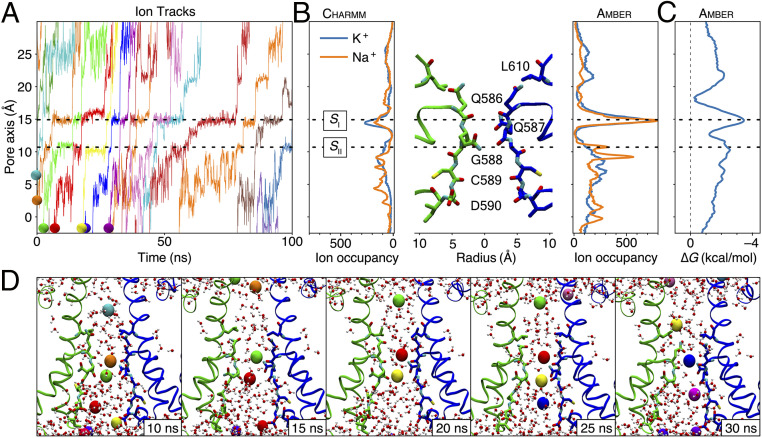
Simulated Na+ and K+ permeation of GluA2. Simulations were performed at 303 K using an ion imbalance of 6 e− between two compartments

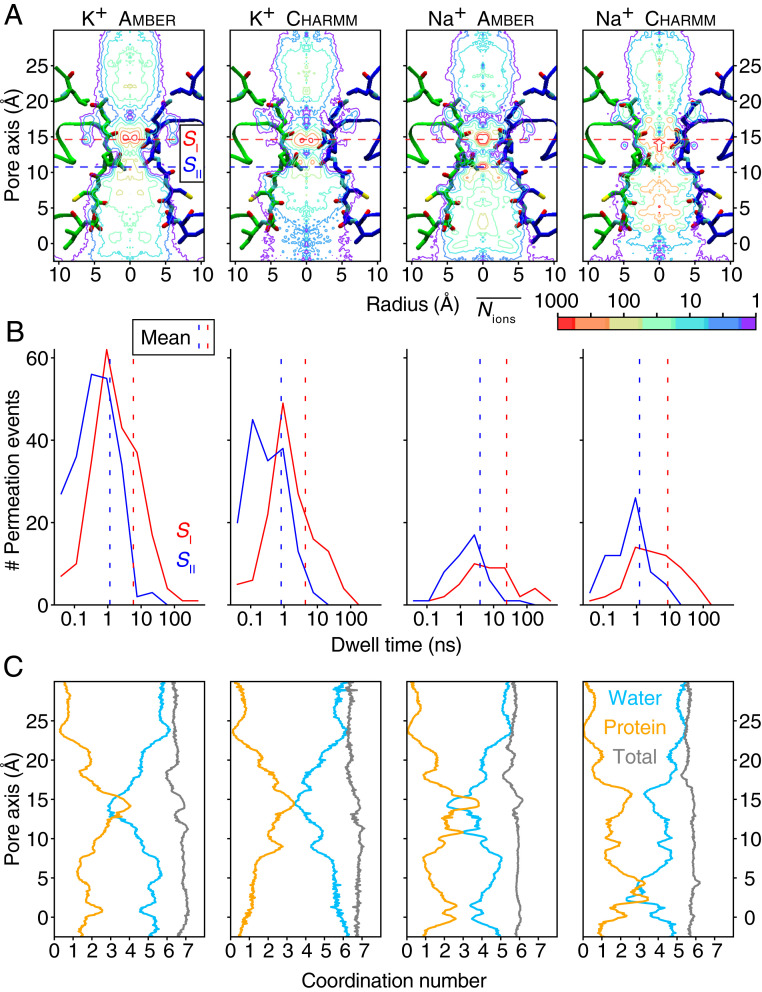
Ion occupancy, dwell time distribution, and hydration states. (A) Two-dimensional ion occupancy within the SF resolved radially and along the pore axis (z-axis) as a contour plot calculated from MD simulations of GluA2 with different ion types (K+ and Na+) and force fields (amber99sb and charmm36). The simulations were performed at 303 K using an imbalance of six cations between the two compartments, α and β. The ion occupancy in number of ions per 0.001 Å3 per 500 ns was normalized according to the volume change along the radius. The center of mass of the SF backbone atoms corresponds to the 10 Å point along the pore axis. Approximate centers of the binding sites are drawn as dashed lines (SI, red and SII, blue). (B) Dwell time histograms for SI and SII binding sites. Vertical dotted lines are means of the respective dwell times. (C) The number of oxygens within the first hydration shell of the according ion type along the pore axis. The number of coordinating water oxygens is in blue, the number of coordinating protein oxygens is in orange, and their sum is in gray.
Cation Conduction Mechanism and Selectivity.
During ion conduction, we observed multiple ions in the canonical SF region of 586QQGCDI591 (Fig. 3D and Movies S1 and S2), with an average occupancy of 2.4 ± 0.6 and 2.3 ± 0.7 for K+ and Na+, respectively, in amber simulation runs (SI Appendix, Fig. S9). Ions followed a loosely coupled knock-on mechanism, where in most cases, the exit of an ion to the upper cavity was closely followed by the entry of an ion to the SI site (Fig. 3D and Movies S1 and S2). From the free-energy profiles (Fig. 3C and SI Appendix, Fig. S7), we conclude that the energy barriers bracketing SI (at around 13 Å and 18 Å along the pore axis) are the major obstacles for ion conduction in the SF. The ion entering the SI site was normally supplied from the adjacent secondary sites, presumably aiding permeant ions to overcome the main energy barriers before and after the SI site (10).
To understand better where cation selection occurs, we examined the paths of chloride (Cl−) ions during our simulations. Consistent with a very short selective region, Cl− ions occasionally entered the cone-shaped space below the SF and readily approached the QQ filter from the bundle crossing side. Surprisingly, Cl− ions found stable binding sites immediately above the QQ filter (SI Appendix, Fig. S10) in all simulations. We also observed one Cl− permeation event during the K+ simulations (Movie S3) but not in the Na+ simulations, providing a crude estimate of the Cl−:K+ permeability ratio at around 1:220. In practical terms, this relative selectivity would correspond to an ∼15 µV shift in the reversal potential (Goldman-Hodgkin-Katz [GHK] equation, see Methods) compared to no Cl− permeation at all. In other words, at this level of selection between cations and chloride, any reversal potential shifts are too small to be measured experimentally. Further context comes from the observation that R-edited homomeric GluA2 channels are anion permeable (5). The observation of an infrequent yet stable anion approach to the face of the QQ filter favored for anions by the electric field provides a simple explanation as to how this point mutation can switch selectivity simply by changing local electrostatics.
Ion Hydration States during Permeation.
To determine the ion hydration states during permeation, we calculated the number of ion-coordinating oxygens from both water molecules and protein residues within each ion’s first solvation shell. Hydration shells are dynamic; here, we defined the waters of hydration using the radii corresponding to the minimum in the radius of gyration profiles: 3.1 Å for Na+, 3.4 Å for K+, and 3.8 Å for Cs+ (31, 32). All monovalent ions were principally coordinated by waters in the lower SF region (Fig. 4C), while the contribution of protein oxygens (backbone oxygens of Q586 and backbone and side-chain oxygens of Q586 and Q587, Fig. 1C and SI Appendix, Fig. S8) increased substantially at the major ion binding site and surpassed the water oxygens for K+ and Cs+. The average number of ion-coordinating oxygens in the first hydration shell (gray line) remained remarkably stable along the pore axis, being six, seven, and eight for Na+, K+, and Cs+, respectively (Figs. 4C and 5C). This total number of ion-coordinating oxygens was in very good agreement with the coordination number for hydrated ions determined experimentally (33). The simulations indicate a qualitatively similar mechanism for each ion species. Cations were never fully dehydrated, but we also observed permeation by water that was independent of ion conduction (SI Appendix, Table S1). The retention of water is probably a strong factor in determining the lack of selectivity across the alkali metal series.

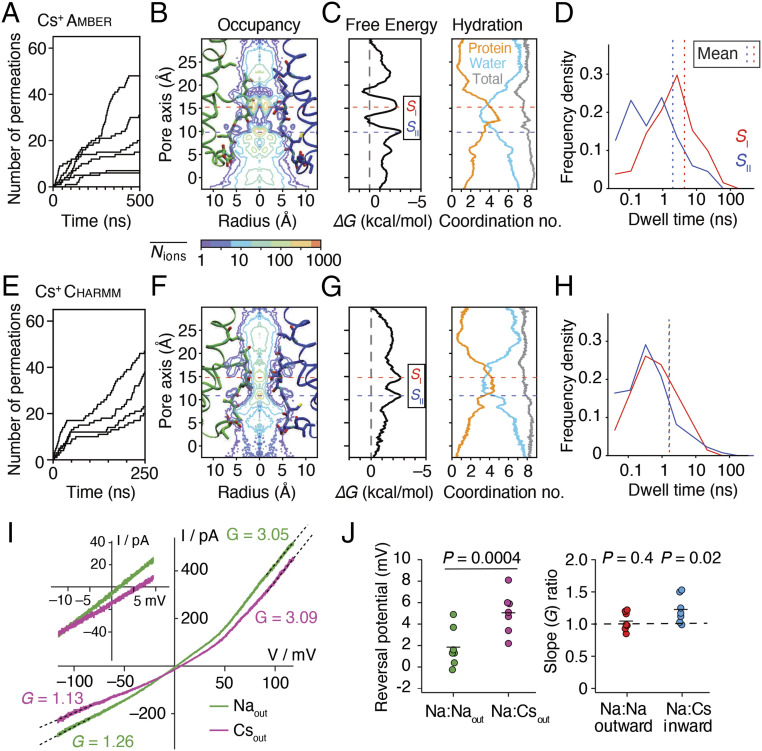
Cs+ permeation of GluA2. (A) Permeation events for Cs+ in the amber99sb force field. (B) Two-dimensional ion occupancy plots. The center of mass of the SF backbone atoms corresponds to the 10 Å point along the pore axis. Approximate centers of the binding sites are drawn as dashed lines (SI, red and SII, blue) that extend to C. (C) Free energy and hydration profiles for Cs+ along the pore axis. (D) Dwell time histograms for Cs+ in the SI and SII sites, with mean dwell time for each site indicated with a dashed line. (E–H) Same as for A–D but for the charmm36 force field. (I) Outward rectifying responses to voltage ramps in Na+ (green) and Cs+ (purple) measured with patch clamp electrophysiology. The outward rectification (ratio of chord conductances at +100/−100 mV) was 2.7 ± 0.3 for Na+ and 3.1 ± 0.3 for Cs+ (n = 7 patches). The inset shows the typical shift in reversal potential on exchange of external Cs+- and Na+. The pipette solution contained Na+. (J) Reversal potential of AMPAR currents in external NaCl and CsCl solution and conductance ratios of Na+ versus Cs+ for inward and outward permeation. Probability of no difference in the slope ratio was versus a ratio of 1 with paired Student’s t test.
Cesium Permeation.
Cs+ is impermeant or a blocker of most potassium channels (34) and also blocks nonselective HCN channels (35). In this context, it is somewhat surprising that classical electrophysiological measurements of reversal potentials for cloned AMPA receptors show, at most, minor differences in the permeation of Cs+ (van der Waals radius 343 pm) from that of Na+ and K+ (radii 227 pm and 280 pm, respectively) (7). The hydrated radius of Cs+ in bulk water (at least 3.1 Å) is expected to be substantially greater than that of Na+ and K+ (2.4 Å and 2.8 Å, respectively) (36). We reasoned that to achieve similar permeation statistics despite its greater size, Cs+ should either alter SF geometry, occupy distinct binding sites, or have a different hydration state during permeation. To resolve this point, we simulated Cs+ permeation in the same conditions as for K+ and Na+. From both amber and charmm simulations, we observed similar conductances for Cs+ and K+ (Fig. 2B and Table 1) that were higher than for Na+. However, the geometry at the tip of the pore loop did not change. The hydration profile of Cs+ as it passes through the pore is more similar to K+ than Na+, with protein oxygens outnumbering the water oxygens at the SI and SII sites in the QQ SF. However, in amber simulations, the major binding site (SI) for Cs+ is axially displaced by about 0.5 Å toward the bundle crossing gate compared to those of K+ and Na+ (Fig. 5B and SI Appendix, Fig. S7), with a concomitant longer distance to the SII site. Most critically, in both amber and charmm simulations, the mean residence time for Cs+ at the SI site was much less (∼1 ns) than for Na+ or K+ and matched that at the SII site (Figs. 4B and 5 D and H). Based on observations that NaK could be converted into a potassium-selective channel by introducing consecutive K+ binding sites (37), serial binding to two effectively equivalent sites suggested that Cs+ may be selected over Na+ and K+. In other words, the simulations suggest that Cs+ might be more permeant than Na+ or K+ and have a higher conductance than Na+, with the caveat that permeability and conductance are not related in a simple way (38).
Extensive studies of GluA1 (7) suggest a small shift in reversal potential between Cs+ and Na+, corresponding to selection, but the permeation of cations in GluA2 (Q) might be different. We performed patch clamp electrophysiology experiments on unedited GluA2 channels expressed in HEK293 cells to reexamine the reversal potentials and slope conductances for Na+ and Cs+ in the same recording. We readily detected a small but consistent shift in the reversal potential in external Cs+ of 3.7 ± 0.5 mV (n = 7 patches, Fig. 5 I and J), meaning Cs+ was slightly more permeant than Na+ (PCs:PNa = 1.16 ± 0.02), like in GluA1. The slope conductance for the inward arm of the current–voltage (I–V) relation in Cs+ was similar to that of Na+ (82 ± 6%; n = 7 patches). Taken together with our single-channel recordings (Fig. 2), these observations are consistent with the AMPAR pore being mildly selective for Cs+ over Na+ and Cs+ permeating with a higher conductance despite using a subtly different permeation mechanism on the same overall scaffold.
Conformational Dynamics in the Vicinity of the SF.
To identify the major conformational states of the SF during ion permeation, we performed a conformational clustering analysis on Na+, K+, and Cs+ simulations, respectively. Interestingly, only one mainly populated state was found for the respective simulations across different cations (SI Appendix, Table S2), which in each case was highly similar (Fig. 6A). A comparison of these representative structures from clustering analysis with the original Cryo-EM structure revealed no large structural differences, but the top part of the SF (Q586 and Q587) is slightly more open in the Cryo-EM structure (Fig. 6A).

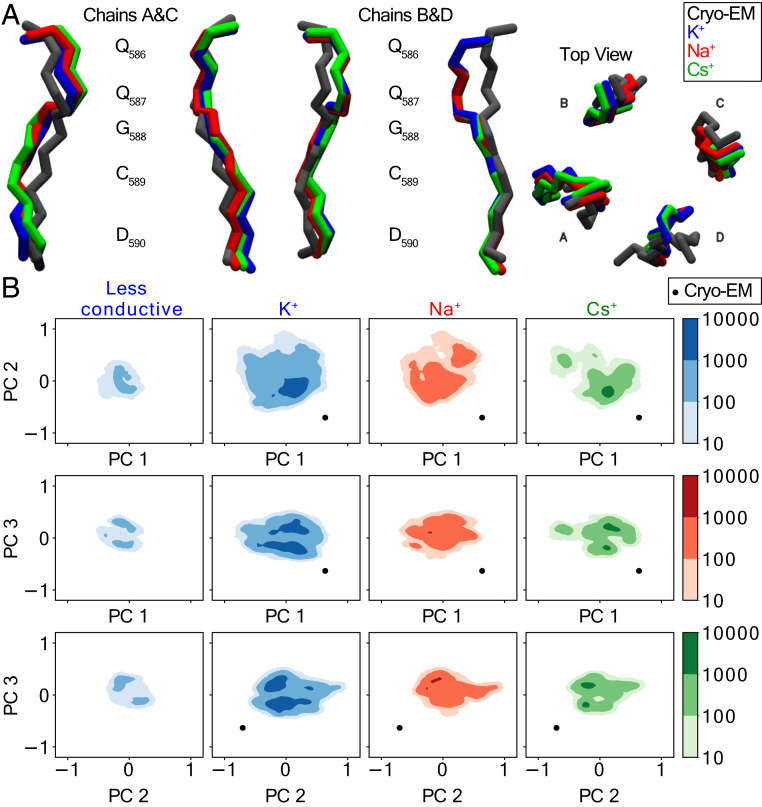
PCA and clustering analysis. (A) Comparison of the SF in Cryo-EM structure and the representative structures of the K+, Na+, and Cs+ simulations using amber99sb force field. The representative structure is the middle structure of the most populated cluster from clustering analysis (SI Appendix, Table S1). An rmsd cutoff of 0.05 nm was used for cluster determination. A total simulation time of 2.5 μs was obtained for K+ and Na+ simulations, respectively, while for Cs+, the total simulation time is 3.0 μs. (B) Results of the PCA analysis on the SF part, where trajectories of K+, Na+, and Cs+ simulations were combined for the determination of the principal components. Conformational dynamics of the SF in K+, Na+, and Cs+ simulations were projected individually onto the first three principal modes. Less conductive states were combined from K+ simulation run 1 (300 to 450 ns), run 3 (200 to 300 ns), and run 5 (200 to 350 ns) of the lower channel (SI Appendix, Fig. S4), where only one permeation event was observed during the entire simulation period.
To further understand the conformational dynamics of the SF during ion permeation, we performed a principal component analysis (PCA), in which the most pronounced motion of the SF can be described by a few eigenvectors with large eigenvalues (39). For the PCA analysis, we used the combined trajectories of Na+, K+, and Cs+ simulations (with the amber forcefield) that were initially aligned on the SF (586QQGCD590). The first three eigenvectors (with the largest eigenvalues) account for >45% of all the motion in the combined trajectories (SI Appendix, Fig. S11). We have visualized the motion of these first three eigenvectors, which correspond to asymmetric deformation or distortion of one to two subunits in the SF (SI Appendix, Fig. S12). Conformational asymmetry is also reflected in the major states determined from clustering analysis (Fig. 6A). Furthermore, we projected the first three eigenvectors onto the individual Na+, K+, and Cs+ simulations, respectively. States of the SF sampled from the K+, Na+, and Cs+ simulations are highly similar (Fig. 6B), suggesting no significant difference in conformational dynamics during ion permeations with different ion types. Finally, we combined three parts of less-conductive trajectories from K+ simulations (SI Appendix, Fig. S4, lower channel, run 1: 300 to 450 ns, run 3: 200 to 300 ns, and run 5: 200 to 350 ns). Projection of the first three eigenvectors on the less-conductive simulations showed that they sampled the same conformational landscape as the rest of the simulations, ruling out structural changes for the variations in the instantaneous conduction rate observed during the simulations.
Examining the root mean square fluctuations (rmsf) of heavy atoms in residues in the traditional SF and its neighboring regions (585MQQGCDI591) reveals a clear split in dynamics (SI Appendix, Fig. S13). The backbone atoms at the pore loop (585MQQG588) are comparatively immobile, similar to the M3 backbone adjacent at L610. In the lower filter region, the backbone and side chains of CDI residues are comparatively unrestrained. The side chain of Q586 is more flexible than the Q587, as it points toward the cavity and thus has more freedom for movement. Similar to the results of PCA analysis, the dynamics of the SF were indistinguishable across the different ionic conditions, with the rmsf profiles of both backbone and side-chain atoms in the Na+, K+, and Cs+ simulations being highly similar (SI Appendix, Fig. S13). These observations further underline that the stable structure of the QQ filter is sufficient for selecting cations.
Discussion
In this study, we simulated the permeation of various monovalent cations through the AMPA channel pore using the computational electrophysiology approach. Our direct measurements of ion conduction confirm that the Cryo-EM structure reported by Twomey et al. (1) is bona-fide open state, with a conductance large enough to be the fully open state. The whole TMD of the AMPAR remained stable during the simulations, even at transmembrane voltages that are relatively high compared to normal physiology. Particularly, the upper bundle crossing in this structure, which we held open mechanically, provides almost no barrier to conduction. Our simulations provide an answer to how this channel can allow a range of cations to pass with similar rates. The simulated conductance shows a striking agreement with the single-channel recording data of monovalent cations in GluA2, suggesting that to a first approximation, the ion conduction we observe resembles physiological permeation. The simulations suggested that Cs+ permeates with a subtly different mechanism to Na+ and K+, and we could provide evidence for this using patch clamp electrophysiology.
Although in principle using the computational electrophysiology approach we should simulate both inward permeation through one channel and outward permeations through the other, we observed more outward ion flux (from intracellular to extracellular) through one AMPA channel pore in all simulation setups. As an example, in K+ simulations, the ratio of conduction for outward and inward permeation is 13 (amber99sb) and 6 (charmm36), respectively. Although electrophysiology revealed that the channel is indeed outward rectifying (Fig. 5) for all ion types, the magnitude of the effect (about threefold) is smaller compared to the simulations. Nevertheless, it should be noted that the experiment was carried out at a transmembrane voltage of about ±100 mV, while high membrane potential (about ±500 mV) was applied in the simulations. It is thus conceivable that the channel shows higher outward rectification at higher voltages. Similar directional flux has also been observed in previous computational electrophysiology simulations of K+-selective channels, such as KcsA (40). As inward permeation events were too few to be analyzed in a statistically representative manner, all simulated conductances listed in Table 1 and Fig. 2B were derived from the outward ion permeations, and all analyses were based on outward ion permeation.
For the major physiological ions Na+ and K+, we observed only one strongly populated ion binding site (SI) within the SF. This main ion binding site is remarkably congruent with the single ion density observed in the Cryo-EM structure (PDB ID: 5WEO, SI Appendix, Fig. S14). The centers of the ion densities from these two very different approaches match within 1 Å. This difference may, in part, be attributed to the simulations being performed under transmembrane voltages, while the Cryo-EM structure was determined at 0 mV in the absence of ion gradients. In contrast to the major binding site SI, the other ion binding sites in our simulations, such as SII, were rather short-lived, and ions pausing there were less ordered. This observation markedly contrasts with the four adjacent ion binding sites of canonical K+ channels that were determined by X-ray crystallography (21) and MD simulations (40). In several nonselective cation channels, such as NaK (41, 42), CNG (20), and HCN channels (19), the SF consists of either two or three consecutive sites. It has been suggested that a single ion binding site within the NMDA receptor SF is the key to the ion nonselectivity of these cation channels (43). Our finding from the AMPA simulations, revealing a single major ion binding site with twin energy barriers, is clearly in line with this mechanism. In other words, the AMPA receptor channel is minimally selective and has one of the simplest observed SFs comprising only two residues (Q586 and Q587). The mechanism of cation selection in the AMPA receptor also shows some similarity with the one of ryanodine receptor, where four Q4863 at the constriction zone act as a charge SF for selecting cations over anions, as suggested by previous MD simulations (44, 45).
Moreover, we found that during permeation, hydration states of the ions in the SF also differ from the ones in the K+-selective and several nonselective cation channels. Previous simulations using computational electrophysiology suggested K+ pass desolvated through the SF of potassium channels (14), while an entire hydration shell surrounding Na+ ions was observed while they traverse the SF (14, 15, 32). There is no narrow constriction in the AMPA receptor pore to match those in K+ and nonselective NaK channels, with the narrowest portion at Q586 (7.6 Å between beta carbon atoms in 5WEO). Therefore, all monovalent ions remain mostly hydrated in the SF and are only partially dehydrated at Q586. Dehydration of smaller Na+ ions requires much more energy (46, 47), and, consistent with this expectation, the Na+ ions were more hydrated at the QQ filter compared to K+ and Cs+. The very short SF has a permissive architecture that accommodates the different alkali metals in distinct solvation states.
Based on these observations, we propose that the mechanism of ion conduction through the AMPA channel pore is remarkably different from that seen to date in other tetrameric channels. For K+ (40) and several other nonselective cation channels, such as NaK (15) and the NaK-CNG mutant (14), ions strictly follow a direct knock-on mechanism when passing through the SF. In contrast, although multiple cations generally occupied the SF region in the AMPA receptor pore, only one stable axial binding site could be resolved, resulting in a quasi-uncoupled knock-on for monovalent cation conduction. All ions retained a degree of hydration and water was copermeant. Furthermore, quite distinct from the nonselective NaK channel (15), all monovalent ions investigated in this study (K+, Na+, and Cs+) follow a similar path during permeation, with no requirement for conformational change for physiological permeation rates. The conduction mechanism in the AMPA channel displays some similarities with Na+ conduction in the voltage-gated sodium channels, where both loosely coupled knock-on and “pass-by” processes compete during Na+ permeation (32, 48).
Using extensive MD simulations combined with Markov state model analyses, Furini and colleagues suggested that ion-triggered conformational change within the SF determines ion selectivity in bacterial sodium channels (49). Along the same lines, in our previous study on nonselective NaK channels, we proposed that K+ and Na+ permeation is coupled to distinct SF conformations that are stable in the microsecond time scale and did not readily interconvert (15). In marked contrast to these findings, the short AMPA SF does not show ion-dependent conformations and, rather, achieves nonselectivity in a quite simple way. The AMPAR provides a minimal selection architecture that is insensitive to ion diameter; the ion hydration shells provide a compensatory variation. However, it is probably not correct to think of the AMPA receptor SF region as a single permissive structure. Instead, the SF of the AMPA receptor is flexible at the nanosecond time scale and allows different ions to permeate using distinct mechanisms but at similar rates. A comparison of the conductances derived from the simulations with experimental ones from electrophysiology suggests that the simulations sample the fully open state (Fig. 2B). Distinct from previous simulations of K+ channels performed at longer time scales (12, 17), the overall structure and the SF of the AMPAR remained stable during the simulations with different ion species (Fig. 6 and SI Appendix, Figs. S1 and S2). We did not observe relaxation from the permeation-competent conformation to other states.
It is also possible that the SF may be coupled to the upper gate movement or fluctuation as in KcsA (50) and that by holding the upper gate in a single stable conformation, we reduced the SF dynamics. Recent results suggesting that SF perturbations can change the desensitization lifetime are consistent with coupling to regions outside the pore domain (51). On this point, results from Cryo-EM are inconclusive, with some open and closed structures having similar SF architectures, whereas one open state structure (with the Q586 residue RNA-edited to R586) has marked twofold symmetry (2). Restraining the linkers was enough to keep the channel stably open, even though we removed flanking Stargazin molecules. Longer time scales may be required to see any putative relaxation into a TARP-free state. The stability of the structure with TARPs removed is consistent with the idea that the modulation of channel gating by Stargazin and other TARPs is principally outside the membrane, although our study does not distinguish between extracellular (52) or intracellular modulation (53). Future work will further clarify the relation between conduction properties, receptor conformation, and dynamics of the AMPAR pore (54).
Methods
Computational Electrophysiology.
To prepare the Cryo-EM structure of the AMPAR in an open state (PDB ID: 5WEO) (1) for MD simulations, we removed the four copies of the auxiliary protein Stargazin. We truncated the receptor linkers before I504 and G774 and after I633 and A820 and added N-methyl amide and acetyl caps at the newly created C and N termini. We used MODELER (55) to build missing loops (residues 550 to 564 of the intracellular loops of each subunit) as well as missing side chains (sidechains of S818 and R819 of subunits B and D and K505 of subunit D). To hold the truncated structure together and in an open state, we restrained protein at the heavy atoms of all four peptide chain termini with a harmonic potential of 10,000 kJ/mol for each subunit.
We performed MD simulations with both charmm36 (27) and amber99sb (26) force fields. In the amber setup, insertion of the AMPAR TMD into a POPC lipid bilayer was performed with the Gromacs internal embedding function, whereas in the charmm setup, this process was carried out in charmm–GUI (56). The concentration of KCl, NaCl, and CsCl was 274 mM in simulations with amber, except for one simulation setup of the closed channel in which the amber force field was 284 mM KCl. For charmm simulations, we used a salt concentration of 212 mM. Improved lipid parameters (57) and ion parameters (36) were used in the amber simulations. We used the TIP3P (transferable intermolecular potential with three points) water model (58) in all simulations.
All the MD simulations were carried out with the gromacs software package (versions 2016.1 and 2019.5) (59). Short-range electrostatic interactions were calculated with a cutoff of 1.7 nm, whereas long-range electrostatic interactions were treated by the particle mesh Ewald method (60). The cutoffs for van der Waals interactions were set to 1.7 nm. The simulations were performed at 303 K or 310 K with an enhanced Berendsen thermostat (gromacs v-rescale thermostat) (61). A surface-tension Berendsen barostat was employed to keep the pressure within the membrane plane (xy-axis) at 250 bar per nm per lipid surfaces and at 1 bar in z-axis direction (62). All bonds were constrained with the Linear Constraint Solver (LINCS) algorithm (63). Interatomic forces (van der Waals and Coloumb) were calculated with a 1.5-nm cutoff. Long-range electrostatic forces were computed with the Particle-Mesh-Ewald method. To decrease computational cost, we employed virtual sites for hydrogens in all amber simulations. amber simulations were calculated with an integration time step of 4 fs. All charmm-based simulations used an integration time step of 2 fs.
Before starting the simulation of the AMPAR with an ion gradient, the system was first energy minimized and equilibrated. After the systems energy was minimized to below 1,000 kJ/mol with gromacs “steepest descent” implementation, a 10-ns free equilibration without any restraints (except those for the linkers) was performed. For charmm-based simulations, the recommended energy minimization and equilibration steps from charmm-GUI were performed.
For the computational electrophysiology study (23), two copies of the equilibrated system of the AMPAR TMD in the lipid bilayer were included in a simulation box of typically 10 × 10 × 20 nm (Fig. 1D). A charge gradient and therefore transmembrane potential was generated by introducing an ion difference of 2, 4, or 6 cations between the two compartments separated by the two lipid bilayers. Since the instantaneous introduction of a charge gradient into the system can produce large perturbations in the system, we omitted the first 20 ns of every simulation. During MD simulations, the number of ions in each chamber was kept constant alchemically by an additional algorithm (23). The resulting membrane potential can be calculated by double integration of the charge distribution using the Poisson equation as implemented in the gromacs tool gmx potential (64). During the simulations, a permeation event was counted when an ion traversed the entire filter region. Individual simulation runs were 500 ns in the amber setups and 250 ns in the charmm setups (Table 1).
The majority of the production runs were conducted at 303 K using an imbalance of six cations (∆q of six elementary charges, e–) between the two compartments
To align results over different setups, we used the center of mass of the backbone atoms of the classical SF residues (586QQGCD590) as a reference point. All trajectories were analyzed with gromacs tools and python using mdanalysis (65). Molecular visualizations were made with Visual Molecular Dynamics (VMD) (66).
For clustering analysis, five runs, each comprising a 500 ns trajectory, were combined for the simulations of K+ and Na+, while six runs of 500 ns were concatenated for Cs+ simulations. All simulations were performed with amber99sb force fields. Clustering analysis was performed with gromacs clustering tool. To decrease the computational effort, we calculated the clusters based on the backbone heavy atoms of the SF (Q586-D590) only. The SF atoms were fitted for all frames of the trajectories. A clustering cutoff of 0.05 nm in rmsd was used to decide cluster affinity. Simulations involving K+, Na+, or Cs+ each yielded one dominant cluster and a number of minor ones (SI Appendix, Table S2). PCA was performed on the same selected atoms as the clustering analysis, where trajectories of K+, Na+, and Cs+ simulations were combined for the determination of the principal components. Proportion of variance for the derived principal modes from PCA analysis is shown in SI Appendix, Fig. S11. Conformational dynamics of the SF in K+, Na+, and Cs+ simulations were projected individually onto the first nine principal modes (SI Appendix, Fig. S15).
Patch Clamp Electrophysiology.
HEK293 cells were plated on glass coverslips in dishes and incubated for 20 to 44 h before calcium phosphate transfection with 3 µg cDNA. For macroscopic current recordings, the cDNA transfection was done with the Rat GluA2 (Q) pRK5 vector encoding enhanced green fluorescent protein after an internal ribosome entry site. Single-channel recordings from outside-out patches were performed 24 h after transfection. For single-channel recordings, we used a plasmid ratio approach to obtain sparse expression (67), whereby the rat GluA2 (Q) vector was cotransfected with enhanced green fluorescent protein and empty vector (pCDNA3.1+) in a ratio of 1:63:313. Standard extracellular solution contained 150 mM NaCl, 0.1 mM MgCl2, 0.1 mM CaCl2, 5 mM HEPES, and 10 µM EDTA and was titrated with NaOH to a pH of 7.3. We included EDTA to chelate trace contamination by divalent ions. The internal solution contained 115 mM NaCl, 1 mM MgCl2, 0.5 mM CaCl2, 10 mM NaF, 5 mM Na4BAPTA, 10 mM Na2ATP, and 5 mM HEPES, titrated to a pH of 7.3 with NaOH. Pipettes were mounted in an ISO holder (G23 Instruments) and had a resistance of 3 MΩ for macroscopic current recordings. For single-channel recording, pipettes were fire polished to a resistance of 10 to 25 MΩ and were coated with Sylgard. The junction potential between the pipette and Na+-bath solution (Ebath-Epip, considering Na+, Cl−, and F− mobilities) was 3.7 mV (68). For reversal potential experiments with Cs, we substituted NaCl in the extracellular solution with CsCl and titrated the pH to 7.3 with CsOH. For single-channel recordings in near-symmetrical K+ or Cs+, in external solutions, we substituted NaCl with KCl or CsCl, respectively, and titrated to pH 7.3 with KOH or CsOH, respectively. The Cs internal solution contained 115 mM CsCl, 1 mM MgCl2, 0.5 mM CaCl2, 10 mM CsF, 5 mM Cs4BAPTA, 10 mM K2ATP, and 5 mM HEPES, titrated to a pH of 7.3 with CsOH. The potassium internal solution contained 115 mM KCl, 1 mM MgCl2, 0.5 mM CaCl2, 10 mM CsF, 5 mM Cs4BAPTA, 10 mM K2ATP, and 5 mM HEPES, titrated to a pH of 7.3 with KOH. Cyclothiazide (Hello Bio) was prepared as a 100-mM stock solution in DMSO and used at 100 µM (giving a final concentration of 0.1% DMSO). EDTA stock solution was prepared in NaOH. Reagents were obtained from Carl Roth GmbH, Sigma Aldrich, or Toronto Research Chemicals, unless otherwise noted. For fast perfusion of outside-out patches, the perfusion tools were made with custom-designed four-barrel square profile glass (Vitrocom) mounted on a piezo electric stack (Physik Instrument). Currents were filtered at 10 kHz (−3 dB cutoff, 8-pole Bessel) with an Axopatch 200B amplifier (Molecular Devices). For analog–digital conversion, an InstruTECH ITC-18 digitizer (HEKA Elektronik Dr. Schulze GmbH) was used at 40-kHz sampling rate. Data were recorded and analyzed with AxoGraph X (AxoGraph Scientific).
For single-channel conductance measurements, in each record, we ran a ramp protocol (−120 to +120 mV, 1.2 V ⋅ s−1) both before and during glutamate application (10 mM) to the outside-out patch. The leak current recorded during the no-glutamate ramp was subtracted from the current recorded during glutamate application. Stretches of the recording corresponding to one open channel were selected and the open levels were fit with a linear relation to obtain the chord conductance.
For macroscopic measurements of reversal potential and conductance, we alternated washing each patch with Cs+ and Na+ external solution. Slope conductances were fit to the traces over the 30-mV ranges at the extreme positive and negative ends of the ramp. For the slope conductance, we compared the conductance ratios (Cs versus Na inward and Na versus Na outward) from the same patch against the null (ratio = 1) using a paired t test. We assumed that there was a similar junction potential (within 0.5 mV) in both cases because the junction potential from the pipette to the Cs+ solution (−1.7 mV) is cancelled by a second junction potential from Cs+ solution back to the bath electrode in Na+ (4.9 mV) (69). We added this junction potential to the measured mean reversal potential. The flow rate through the local perfusion tool was low (<200 µL/min), meaning the overall bath Cs+ concentration remained low.
We used the GHK equation (70) to calculate permeability ratios based on shifts in reversal potentials:
where X was Na+ or Cs+.
Likewise, to calculate the putative shift in Erev because of a minor chloride permeability, we took internal [X]i and external [X]O ion concentrations, where X was Na+, Cs+, or Cl− permeability ratios of PCs:Na of 1.16:1, as measured, and assumed PCl:Na of either 1:220 (an upper estimate based on the observation of a single anion permeation in our potassium simulations) or zero (no chloride permeability). Cesium terms were only used for the bi-ionic condition of [Na+]i to [Cs+]o:
Statistical analysis and data plotting was done in IGOR Pro (Wavemetrics).
Acknowledgements
This work was funded by the Deutsche Forschungsgemeinschaft (DFG) RU2518 DynIon to A.J.R.P. (P3; PL 619/5-1) and H.S. (P3; SU 997/1-1), the DFG under Germany’s Excellence Strategy—EXC 2008–390540038—UniSysCat to H.S., a DFG Heisenberg Professorship (PL 619/3-1 and PL619/7-1) to A.J.R.P., and the European Research Council Consolidator Grant “GluActive” (647895) to A.J.R.P. The computations were performed with resources provided by the North-German Supercomputing Alliance (HLRN). We gratefully acknowledge the Gauss Centre for Supercomputing e.V. (https://www.gauss-centre.eu/) for funding this project by providing computing time through the John von Neumann Institute for Computing on the GCS Supercomputer JUWELS at Jülich Supercomputing Centre.
Data Availability
All study data (including MD trajectories) are available from the authors on request.
References
1
2
3
4
5
6
7
8
10
11
12
13
14
15
16
17
18
19
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
 Nonselective cation permeation in an AMPA-type glutamate receptor
Nonselective cation permeation in an AMPA-type glutamate receptor
