
Contributed by Hans Clevers, December 15, 2020 (sent for review October 26, 2020; reviewed by Olivier Delattre and Frank Speleman)
Author contributions: E.S.H., T.M., M.v.d.W., and H.C. designed research; E.S.H., T.M., H.A.-S., and M.v.d.W. performed research; F.L.B. contributed new reagents/analytic tools; E.S.H., T.M., K.S., T.C., M.M.v.N., F.A.G.M.-W., M.v.d.W., F.C.P.H., and H.C. analyzed data; E.S.H., T.M., M.v.d.W., and H.C. wrote the paper; K.S., M.M.v.N., and F.A.G.M.-W. advised on survival analysis; and F.C.P.H. supervised single-cell analysis.
Reviewers: O.D., Institut Curie; and F.S., Institute of Human Genetics.
1E.S.H. and T.M. contributed equally to this work.
- Altmetric
Neuroblastoma is a childhood malignancy that originates from neural crest cells. We present a single-cell transcriptome and localization atlas of the developing adrenal gland and identify seven different cell clusters that make up the adrenal medulla. Their transcriptomic profiles were used to generate gene signatures that were compared to neuroblastoma samples. The neural crest-derived “SCP” gene signature score anticorrelates with disease severity based on staging and poor prognosis (molecular) markers, while a high score was associated with higher overall survival rates.
Neuroblastoma is the most common extracranial solid tumor and accounts for ∼10% of pediatric cancer-related deaths. The exact cell of origin has yet to be elucidated, but it is generally accepted that neuroblastoma derives from the neural crest and should thus be considered an embryonal malignancy. About 50% of primary neuroblastoma tumors arise in the adrenal gland. Here, we present an atlas of the developing mouse adrenal gland at a single-cell level. Five main cell cluster groups (medulla, cortex, endothelial, stroma, and immune) make up the mouse adrenal gland during fetal development. The medulla group, which is of neural crest origin, is further divided into seven clusters. Of interest is the Schwann cell precursor (“SCP”) and the “neuroblast” cluster, a highly cycling cluster that shares markers with sympathoblasts. The signature of the medullary SCP cluster differentiates neuroblastoma patients based on disease phenotype: The SCP signature score anticorrelates with ALK and MYCN expression, two indicators of poor prognosis. Furthermore, a high SCP signature score is associated with better overall survival rates. This study provides an insight into the developing adrenal gland and introduces the SCP gene signature as being of interest for further research in understanding neuroblastoma phenotype.
Neuroblastoma is an extracranial solid tumor that accounts for 7% of pediatric cases and 10% of all pediatric cancer deaths (1). Diagnosis is mostly made in early childhood with a median age of diagnosis of 18 mo, but neuroblastoma is even seen in neonates. The early age of diagnosis and the heterogeneity of the disease has been suggested to imply that neuroblastoma results from deregulated neural crest (NC) development (2, 3). Familial neuroblastoma can be caused by mutations in Phox2b and ALK (4). High levels of ALK expression, caused by the presence of mutation or amplifications, is associated with poor prognosis (5).
Neuroblastoma patients can be divided into categories with different prognoses, called stages. The international neuroblastoma staging system was introduced roughly 30 y ago (6). This system is based on the extent of surgical excision and presence of metastases at diagnosis. Stage 1 and 2 neuroblastomas are defined by localized tumor. Their primary treatment is surgery alone, with a 4-y event-free and overall survival rate of 98 to 100% (78–9). This does not hold true for stage 1 or 2 patients that are ≥2 y old with unfavorable histopathology or carrying a MYCN amplification. Stage 3 patients have a 5-y overall survival rate of roughly 70%, and stage 4S has a 5-y overall survival rate of 84%. Stage 4 neuroblastoma patients present with metastases at diagnosis and have a 5-y survival rate of roughly 43%, as observed in a Dutch cohort (9).
This staging system has been further revised, with the new system, International Neuroblastoma Risk Group (INRG) staging, being more compatible with the INRG classification system (10). The INRG classification system takes seven prognostic risk factors into account for risk assessment. High level of Schwannian stroma is associated with a good prognosis, while MYCN amplification, 11q aberrations, and diagnosis ≥18 mo are criteria associated with bad prognosis (6, 10, 11). The other prognostic risk factors are grade of tumor differentiation, ploidy, and INRG stage (10).
Primary tumors are found along the sympathetic nervous system, with over 50% arising in the adrenal medulla. The adrenal gland is surrounded by a capsule, which consists of stromal-like cells, fibroblasts, and myofibroblasts. The adrenal gland can be further divided into two parts, the adrenal cortex and the adrenal medulla (12). In humans, the adult adrenal cortex is composed of three different zones, the zona glomerulosa (ZG), zona fasciculata (ZF), and zona reticularis (ZR) (13). In mice, the adult adrenal cortex is made up of only the ZG and ZF, while in young mice there is an additional cortical zone called the X-zone (14).
Adrenal medulla cells are hormone-producing cells of NC origin. NC cells that migrate from the trunk regions form the adrenal medulla as well as the sympathetic ganglia (15). Trunk NC cells start migrating away from the NC, situated at the margin of the neural tube, at developmental stage embryonic day 9 (E9) to E10 in mice, and Carnegie stage 12 to 14 in humans (16). They migrate toward distant organs and differentiate into, among others, melanocytes, sympathetic neurons, and chromaffin cells. Since half of the neuroblastomas arise in the adrenal gland, a detailed understanding of its development may provide information for neuroblastoma research.
A recent study suggests that Schwann cell precursors (SCP), NC cell derivatives, may represent an alternative origin for the adrenal medulla. In this study, sympathoadrenal anlagen isolated from Wnt1-Cre; R26RTomato and Sox10-Cre; R26RTomato mice embryos were investigated using various experimental approaches (17). Among these, the transcriptome of NC derivatives at E12.5 and E13.5 was analyzed using single-cell mRNA sequencing (scRNA-seq). The study revealed four main clusters, termed “SCP”, “bridge” cells, “chromaffin” cells (that make up the adrenal medulla), and “sympathoblasts” (that make up the suprarenal sympathetic ganglion). The study led to the elucidation of SCP and its role in populating the adrenal medulla. This approach did not include nontraced adrenal cells. Here, we present an unbiased scRNA-seq study describing murine adrenal development from E13.5 to postnatal day 5 (P5) and provide a comparison of the adrenal medulla gene signatures to neuroblastoma.
Results
Identification of Cell Clusters of the Developing Murine Adrenal Gland.
To identify the different cell types in the developing adrenal gland, scRNA-seq was performed. At six different developmental time points (E13.5, E14.5, E17.5, E18.5, P1, and P5), mouse adrenal glands were isolated and dissociated into single cells (Fig. 1A). The cells were then sorted into 384-well plates and processed using the SORT-seq protocol (18). We analyzed a total of 2,229 cells across the six developmental time points. Unsupervised clustering of the nonerythroid cells defined 23 cell clusters (Fig. 1B). These 23 clusters formed five groups. A total of 471 cells were assigned to the medulla group, 235 cells to the cortex group, 298 to the stroma group, 618 to the endothelial group, and 175 to the immune group. The clusters were annotated using single-cell type identification analysis based on gene expression correlation to 358 bulk RNA-seq profiles of fluorescence-activated cell sorting (FACS)-sorted mouse cell populations (19) [SI Appendix, Fig. S1A/SingleR v2 (20)]. To further refine the annotation, cell type-specific marker expression levels were taken into account (SI Appendix, Fig. S1B).

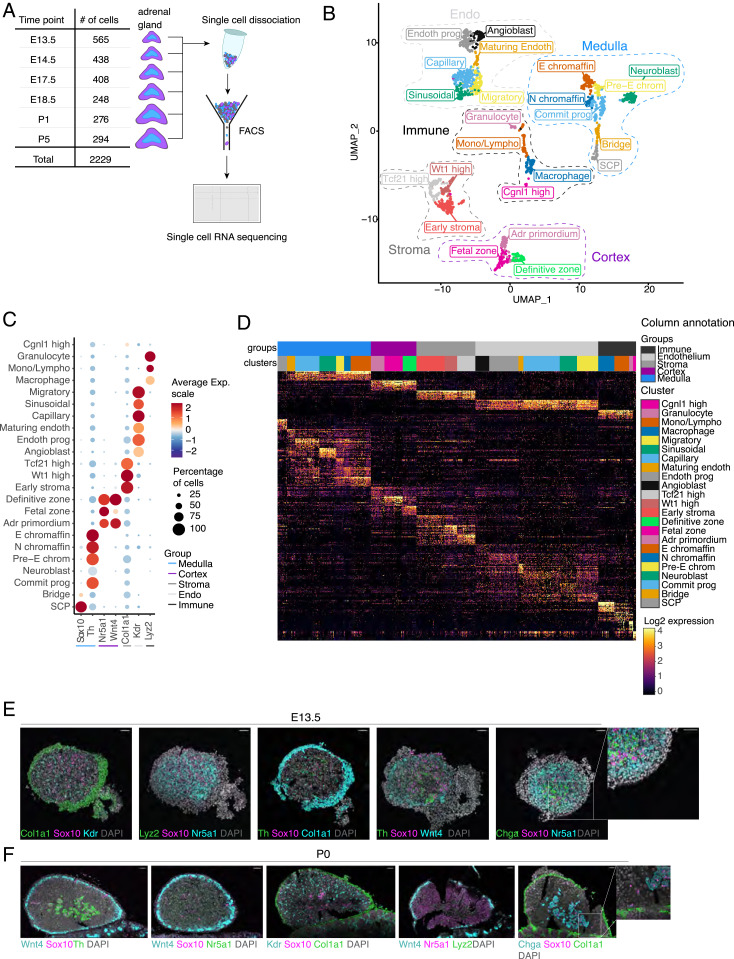
Identification of clusters present in the development of the murine adrenal gland. (A) An unbiased approach was used to analyze adrenal glands isolated at six different time points. (B) UMAP plot showing the clustering of the different cells based on their transcriptional programming. The cells are color-coded based on the 23 clusters they belong to. Cluster can be divided into five main groups included in dashed lines. (C) Dot plot of markers of the five main groups, Chga for the medulla, Nr5a1 for the cortex, Col1a1 for the stroma, Kdr for the Endo, and Lyz2 for the immune group. (D) Heatmap showing the top 20 differentially expressed genes for the main groups, followed by the top differentially expressed genes for each cluster. Color code of cluster and cell is the same as in Fig. 1B and shown in the column annotation. (E and F) In situ hybridization results of an E13.5 and P0 adrenal gland (respectively) using markers Sox10, Th, Chga, Nr5a1, Wnt4, Kdr, Col1a1, and Lyz2. (Scale bars, 45 μm.) Adr primordium, adrenal primordium; Commit prog, committed progenitor; E chromaffin, epinephrine chromaffin; Mono/lympho, monocytes or lymphocytes; N chromaffin, norepinephrine chromaffin; pre-E chrom, pre-epinephrine chromaffin.
Well-established markers for known adrenal cell types identified the indicated groups. Examples of such markers were Chga for the medulla group, Nr5a1 for the cortex group, Col1a1 for the stroma group, Kdr for the endothelial group, and Ptprc for the immune group (Fig. 1C and SI Appendix, Fig. S2A). Lyz2 was a prominent marker for most of the immune clusters and is a known marker for white blood cells. The top 20 genes of the groups and the clusters were then used to investigate potential overlap and/or the uniqueness of the individual clusters. Based on the heatmap, the clusters can be readily resolved (Fig. 1D). Among the top genes, apart from the established markers of the adrenal gland, new markers were evidently identified (SI Appendix, Fig. S2).
The cortex group consisted of three different clusters (Fig. 1B). Cells of all three clusters expressed genes that are typical markers of the cortex, including Nr5a1 encoding Steroidogenic factor 1 (Sf1), and steroidogenic enzymes such as Cyp11a1 and Hsd3b1 (SI Appendix, Figs. S1B and S3B) (21, 22). The cluster that was classified as the “adrenal primordium” positioned at the center of all cortical cells in the uniform manifold approximation and projection (UMAP) plot was mainly present at E13.5 after which it gradually disappeared (Fig. 1B and SI Appendix, Fig. S1E). In addition, this cluster expressed markers such as Wnt4, Shh, and Nr0b1 (Dax-1), and had the highest percentage of cells in G2/M phase of the cell cycle (SI Appendix, Fig. S1D). A developmental trajectory analysis using Monocle 3 was performed to assess the differentiation route taken by the adrenal primordium cells. The analysis predicted that the cells in the adrenal primordium first differentiate into “fetal zone” cells, and then a subset differentiates to “definitive zone” cells (SI Appendix, Fig. S1F) (23, 24). Of all cortical clusters, the fetal zone cluster most highly expressed genes such as Cyp11b1, Star, and the Mc2r accessory protein (Mrap), which implied that this cluster actively produced steroid hormones (SI Appendix, Fig. S1B). The definitive zone cluster highly expressed Wnt4, Shh, and Nr0b1, and accounted for the majority of adrenal cortex cells at the postnatal time points. The expression programs of the two later cortical clusters were already present in the adrenal primordium (Fig. 1D and SI Appendix, Fig. S2C), but to a much lower expression level. Such observations pointed to mutual exclusion of the differentiation potential of the developing cortex across the two cell fates.
The immune group consisted of four clusters identified using automatic cell type annotation (20). Upon comparison, the clusters could be identified as “macrophages,” “monocytes/lymphocytes,” and “granulocytes” cells (SI Appendix, Fig. S1A). The fourth cluster, named “Cgnl1high,” was identified by the expression of Cgnl1 (SI Appendix, Fig. S1B). In contrast, the automatic cell type annotation analysis could not distinguish between the stroma clusters or the endothelial clusters due to lack of corresponding cell types in the reference dataset. The stroma clusters were named based on developmental timing and expression of specific and general markers (SI Appendix, Fig. S1 B and E). The “early stroma” cluster was found predominantly at E13.5, while the two other clusters could be identified by Tcf21 and Wt1 expression, two markers expressed in the adrenal capsule (25, 26). The endothelial group consisted of six clusters, namely the “angioblast,” “endothelial progenitor,” “maturing,” “capillary,” “sinusoidal,” and “migratory” clusters. The clusters were so named based on expression of genes reported to be markers (SI Appendix, Table S1 and Fig. S1B).
UMAP analysis identified seven distinct clusters in the medulla group (Fig. 1 B and C). Two clusters of the medulla were classified as the SCP and “neuroblast” cluster. The SCP cluster expressed the well-known SCP markers Foxd3, Erbb3, and Sox10 (17). We annotated the cluster with the highest level of Ccnd1as the neuroblast cluster (Fig. 2C and SI Appendix, Figs. S2B and S5E). Neuroblasts are described as the proliferative NC derivatives in the adrenal gland; this cluster expressed Tfap2b, Tln2, Gap43, and Eya1, all genes previously associated with neuroblasts (SI Appendix, Fig. S2B) (27).

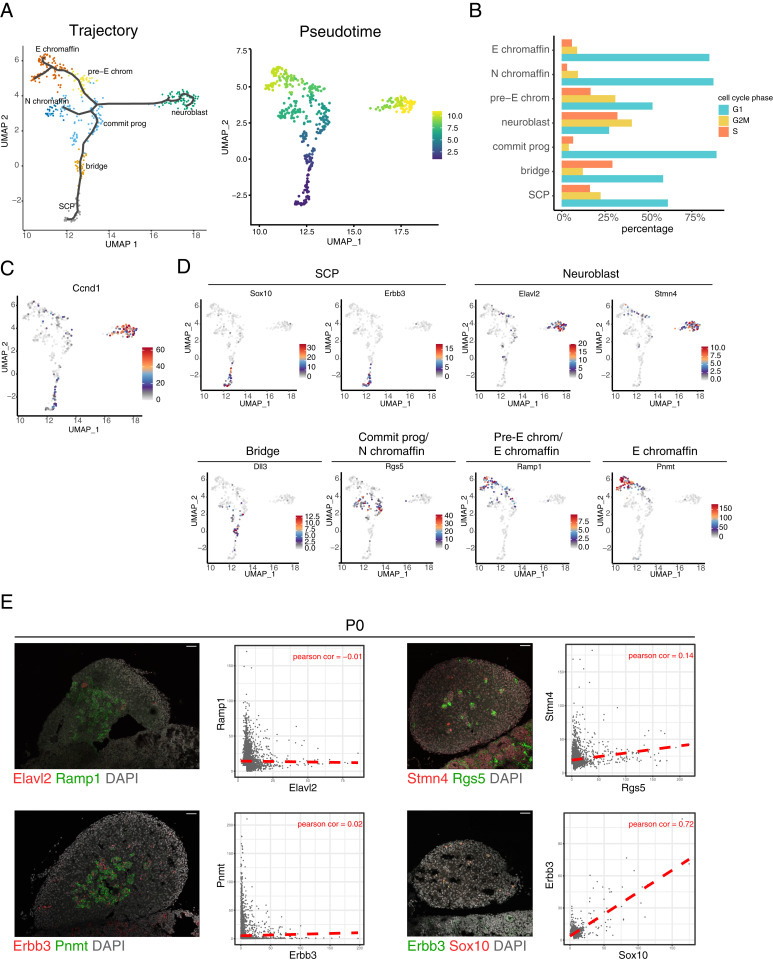
Identification of seven medulla clusters present during development. (A) UMAP plot of the developmental trajectory of the medulla and the pseudotime generated using Monocle 3. Cell cycle dynamics can be conferred by the (B) percentage of cells in the medulla allocated to a cell cycle phase and (C) the Ccnd1 expression level among the differing clusters. (D) The level of expression of genes marking the subclusters shown in UMAP plots, and (E) visualized using RNAscope in P0 adrenal glands. The average channel intensity per cell segment for the staining and the PCCs are represented in scatter plots to the Right of the image. (Scale bars, 45 μm.) Commit prog, committed progenitor; E chromaffin, epinephrine chromaffin; N chromaffin, norepinephrine chromaffin; pre-E chrom, pre-epinephrine chromaffin.
The bridge cluster was named following the Furlan nomenclature (17). To assess the overlap between the top 20 genes of the medulla clusters, a hypergeometric test was performed (Dataset S1). There was significant overlap of the SCP cluster to Furlan’s SCP cluster at the differentiation start. Sixteen of the top 20 genes overlapped as shown in Dataset S2 and SI Appendix, Fig. S3A. Fifteen of the top 20 bridge signature genes overlapped with the Furlan cluster that formed the bridge between their SCP and chromaffin clusters. This overlap was similar for the neuroblast top 20 gene signature. Furlan’s chromaffin cluster overlapped with the rest of our medulla clusters, namely the “committed progenitors,” “pre-epinephrine,” “norepinephrine,” and “epinephrine” chromaffin clusters. Based on the expression levels of the catecholamine biosynthesis enzymes combined with the developmental trajectory analysis, the chromaffin clusters were named pre-epinephrine, epinephrine, and norepinephrine chromaffin cluster (SI Appendix, Figs. S1C and S2B). The norepinephrine chromaffin cluster lacked Pnmt, an enzyme required for the catalysis of norepinephrine to epinephrine (28).
Apart from automatic computational analysis, the cells making up the developing murine adrenal gland could also be identified based on the expression of established markers. Visualizing the clusters would confirm that the cells were indeed localized within the adrenal gland and were present at different points during development.
Visualization of Adrenal Gland Clusters.
To further validate the scRNA-seq and provide spatial information of the clusters, RNAscope, a single-molecule fluorescent in situ hybridization technique, was performed. The adrenal gland was analyzed at time points E13.5 and P0 using the markers Th, Chga, and Sox10 for the medulla; Nr5a1 and Wnt4 for the cortex; Col1a1 for the stroma; Kdr for the endothelial; and Lyz2 for the immune group. These markers were chosen as they distinguish the scRNA-seq groups and are known markers for the different adrenal compartments (Fig. 1C) (29, 30).
At both time points, all marker genes were readily detectable. No overlap between these markers was observed, in concordance to the scRNA-seq analysis (Fig. 1 E and F). Nr5a1+ and Wnt4+ cells were found abundantly in both the E13.5 and the P0 adrenal gland. Nr5a1+ cells were found throughout the adrenal glands but not in the capsular region, whereas Wnt4+ cells were found adjacent to the capsular region. Staining of Col1a1 was observed in stromal cells surrounding the adrenal gland, i.e., the capsule (Fig. 1 E and F). The Kdr+ cells formed a web-like pattern throughout the adrenal gland at both time points. Islands of Chga+ and Th+ cells were in close proximity to Sox10+ cells in the E13.5 and P0 adrenal gland. Lyz2+ cells were sparsely observed at the two time points, in agreement with the kinetics of the development of the peripheral hematopoietic system (31), and in agreement with the scRNA-seq results.
The distribution of RNAscope signals of the markers were quantified to better assess colocalization. Individual cells were computationally segmented based on nuclear DAPI staining. The average intensities of each of the markers were measured per segment making use of the EzColocalization plug-in available on Fiji (32). The values were then plotted, and the Pearson correlation coefficient (PCC) was calculated. For E13.5 adrenal glands, Sox10 was stained together with markers for the nonmedulla clusters, namely Nr5a1 (cortex), Lyz2 (immune), Kdr (endothelial), Col1a1 (stroma), and Wnt4 (cortex). The PCC values for Sox10 and the above-mentioned markers ranged from −0.01 to −0.2, suggesting lack of correlation or even anticorrelation (SI Appendix, Fig. S4A). The PCC values for the costaining of Sox10 with Th and Chga ranged from 0.14 to 0.19 (SI Appendix, Fig. S4A). These PCC values suggested that correlation of Sox10 with the other medulla markers was negligible, probably related to the minor overlap in expression of the corresponding markers observed (SI Appendix, Fig. S1B). To sum up, the SCP cluster as indicated by the expression of Sox10, exhibited a small overlap with the Th+ and Chga+ medulla clusters, but no colocalization whatsoever with the other groups. The scRNA-seq findings for E13.5 were therefore validated.
For the P0 adrenal gland, Sox10 was stained together with Wnt4, Kdr, Col1a1, and Nr5a1. The PCC values were close to zero and negative for comparisons between Sox10 and Wnt4, Kdr, Col1a1, and Nr5a1, ranging from −0.01 to −0.12. For the costaining of Sox10 with the other medulla markers, Th and Chga, the PCC values were 0.11 and −0.02, respectively (SI Appendix, Fig. S4B). The P0 adrenal glands were interpreted as Sox10 positivity having negligible correlation to the other average marker intensities. This confirmed the scRNA-seq findings for P0, where low colocalization was observed between the markers chosen for the RNAscope experiment. Therefore, the clusters observed from scRNA-seq could be localized in the adrenal gland. This was observed for both the earliest timepoint analyzed, and at P0.
In summary, the scRNA-seq was able to identify all major cell type groups in the adrenal gland, as well as different clusters within the groups. This was validated by RNAscope, which showed that this dataset is a reliable representation of the developing murine adrenal gland.
Seven Clusters of the Medulla Cell Cluster Identified.
The seven different time points that were used to analyze the adrenal gland allowed for the changes in clusters across development to be mapped (Fig. 1B and SI Appendix, Fig. S1E). To better understand the developmental relationship between the clusters of the medulla, pseudotime analysis was implemented (Fig. 2A). The lineage trajectory of medulla cells starts with the SCP cluster. The cells then appeared to follow the differentiation trajectory by down-regulating the SCP genes and up-regulating the transcriptome observed in the cells of the bridge cluster, as shown by the lack of overlap of genes between the two clusters (Fig. 1D). The cells would then further differentiate into committed progenitors and afterward are fated to become either neuroblast, pre-epinephrine chromaffin cells, or norepinephrine chromaffin cells.
The pseudotime analysis was in part supported by the expression of transcription factors related to adrenergic specification. The expression of transcription factors as annotated by Gene Ontology term “DNA-binding transcription factor activity” was compared to the medulla cluster marker genes (SI Appendix, Fig. S5A). Among the lineage-specific transcription factors, numerous known and new transcription factors were identified. Eighty-four transcription factors overlapped, of which nine were associated with adrenergic specification and further implicated in neuroblastoma (33, 34). Phox2a, Phox2b, Gata3, and Hand1 were expressed in almost all cells in the medulla clusters with SCP cluster as the exception (SI Appendix, Fig. S5C). Ascl1, another important regulator of chromaffin differentiation, was found expressed highly by the bridge and epinephrine chromaffin cluster (35). Isl1, Sox11, Tfap2b, and Gata2 were listed as transcription factors associated with the adrenergic cell fate that neuroblastoma cells could present with. There was little to no overlap of the transcription factors involved with the mesenchymal cell fate in neuroblastoma (SI Appendix, Fig. S5 B and D) (34).
An aspect that distinguished the clusters was the cell cycle profile. When the cells in the cluster were categorized as either being in cell cycle phases G1, G2/M, and S, the neuroblast cluster had a majority of cells in the G2/M phase (Fig. 2B and SI Appendix, Fig. S1D). Following the developmental trajectory from the SCP and bridge to the committed progenitors cluster, the cell cycle phase shifted from S and G2/M toward G1 phase. Exceptions to this trend were the neuroblast and pre-epinephrine chromaffin clusters, which had a higher number of cells in the G2/M phase, while the chromaffin clusters had more cells in the G1 phase.
The high number of cells in G2/M phase was partially supported by the high number of cells expressing Ccnd1. When compared to the medulla clusters (Fig. 2B), and the rest of the adrenal gland clusters (SI Appendix, Fig. S5E), Ccnd1 was expressed, but not to the same high level as in the neuroblast cluster. This would suggest an association of Ccnd1 to the neuroblast cluster that extends beyond the proliferative state. In previous studies, high expression of Ccnd1 was found in neuroblastoma when compared to other malignancies (36, 37). Mycn was highly expressed in the medulla, with higher levels in the neuroblast and pre-epinephrine chromaffin clusters (SI Appendix, Fig. S5E). Alk expression was restricted to the neuroblast cluster. Both genes are associated with neuroblastoma, making the neuroblast cluster interesting for further research into neuroblastoma.
When examining the top 20 genes from each medulla cluster, the genes for the SCP cluster displayed the least coexpression with other medulla clusters genes (Fig. 1D and SI Appendix, Fig. S2B). For the rest of the clusters, some similarities were observed. Genes were chosen that represented their medulla clusters as markers (Fig. 2D). For the SCP cluster, Sox10 and Erbb3 were expressed by majority of the cells. The rest of the markers were expressed to some degree in multiple clusters. Stmn4 and Elavl2 were expressed at a high level in the neuroblast cluster, with low expression found in a few cells of the bridge and pre-epinephrine chromaffin clusters. Dll3 was expressed in the bridge cluster and a few cells of the committed progenitors cluster. Rgs5 was expressed in the committed progenitors and norepinephrine chromaffin clusters. Expression of Ramp1 was found in the pre-epinephrine chromaffin cluster, whereas Pnmt was expressed by all the epinephrine chromaffin cluster at a high level. The markers expression was a further confirmation of how unique the transcriptome of the SCP cluster is, and how the rest of the medulla clusters have some overlap in their gene programs.
The gene list provided by Dong et al. (38), a study that performed single-cell RNA sequencing on four human fetal adrenal glands, was used for comparison of the top 20 differentially expressed genes of the medulla clusters. This study also identified SCP and “sympathoblast” clusters, as did Furlan, and provides an interesting database for comparison. Their SCP gene list overlaps highly significantly with the medulla SCP signature (hypergeometric test, P value of 5.5*10−19) without any overlap with the chromaffin signature genes (SI Appendix, Fig. S3B). Their chromaffin and sympathoblast gene list overlapped with the neuroblast and chromaffin signature, respectively (SI Appendix, Fig. S3 C and D).
The neuroblast cluster expressed Cartpt, a marker of sympathoblasts, a cell type identified by Furlan et al. (17) to be located in the suprarenal sympathetic ganglion (SI Appendix, Fig. S2B). Expression of RAMP1 and PNMT was found in the sympathoblast cluster by Dong et al. (38). To ensure that the clusters of the medulla were situated in the adrenal gland, RNAscope stainings were performed. The staining for the SCP, neuroblast, committed progenitors, pre-epinephrine chromaffin, norepinephrine chromaffin, and epinephrine chromaffin clusters in the P0 adrenal gland was performed using their representative markers (Fig. 2E). The pattern of localization of the cells positive for the markers Rgs5, Ramp1, Pnmt, Elavl2, and Stmn4 (Fig. 2E) was similar to cells positive for Th and Chga expression (Fig. 1F). The scatter plots of the average intensity of the two markers for each cell segment can be seen with the PCC. For the double stainings of Elavl2 with Ramp1, Erbb3 with Pnmt, and Stmn4 with Rgs5, the PCC ranged from −0.01 to 0.14, which indicated no colocalization. For the double staining of Erbb3 and Sox10, two markers chosen that represent the SCP cluster, the PCC values were 0.72, which can be interpreted as colocalization. The RNAscope stainings showed that the markers of the medulla clusters are present in the adrenal gland. This validated the medulla clusters and elucidated the presence of the neuroblast cluster in the adrenal gland. The neuroblast contribution to the adrenal medulla would be worthy of further study.
Genes Expressed in the Medulla Group and Clusters Found Specifically in Neuroblastoma.
In order to assess whether the gene programs identified by scRNA-seq are relevant to neuroblastoma, bulk RNA-sequencing data were used from the TARGET (Therapeutically Applicable Research to Generate Effective Treatments) initiative (phs000218). The TARGET RNA-seq database allowed us to assess whether the top 20 genes of the medulla group and cluster were enriched in neuroblastoma tumor transcriptomes, and not in other cancer types. The dataset included acute myeloid leukemia (AML) (39), B-lymphoblastic leukemia/lymphoma (BLL) (40), rhabdoid cancer (41), Wilms’ tumor (42), and neuroblastoma (43). The top 20 genes of all adrenal gland groups and medulla clusters were used. The z-score for the 20 genes was combined using Stouffer’s Z-score method into a signature score.
The medulla score was highest in the neuroblastoma samples (Fig. 3A). The cortex score was slightly higher in a few neuroblastoma samples, which also had higher stromal and endothelial scores, pointing to sample-specific healthy tissue contamination. The stroma score was at a similar level in the solid cancers (Wilms’ tumor, rhabdoid cancer, and neuroblastoma) and a lower level in the hematological malignancies (AML and BLL). The levels of the endothelial score in the solid cancers were similar, while the hematological cancers displayed low score levels. The immune score was highest in the hematological malignancies, and lower in the solid cancers. Observing the scores to be higher in the malignancies that are derived from similar origins showed the analysis approach was sound.

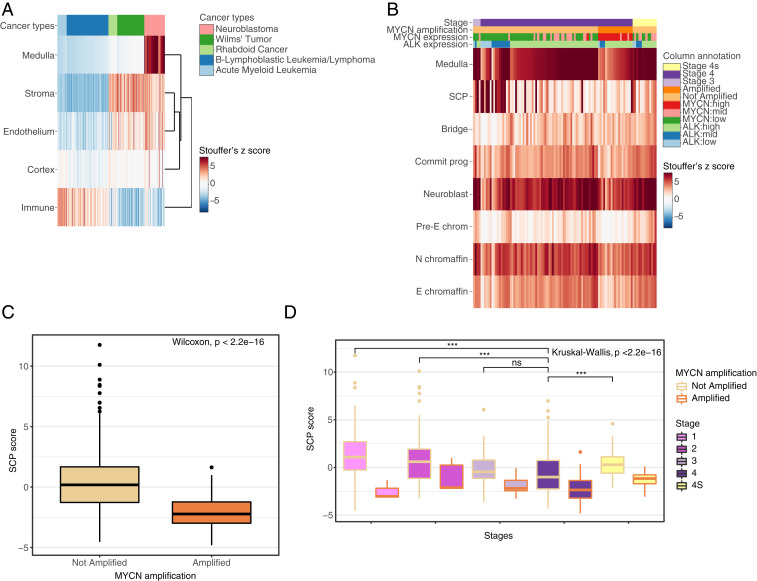
Comparison of the main cluster and medulla cluster signatures to neuroblastoma bulk transcriptomes. (A) Heatmap showing the z score of the medulla, cortex, stroma, endothelium, and immune group top 20 gene signatures compared to the TARGET RNA-seq dataset, which includes acute myeloid leukemia, B lymphoblastic leukemia/lymphoma, rhabdoid cancer, Wilms’ tumor, and neuroblastoma. (B) Heatmap showing the z scores of the signature of medulla group, and the medulla clusters compared to the TARGET RNA-seq dataset. Samples were ordered by stage, MYCN amplification, and by medulla z-score level. Box plot showing the z score of the SCP cluster signature compared to the SEQC dataset containing neuroblastoma patients of all stages either separated by (C) MYCN amplification status, or (D) staging, stages 1, 2, 3, 4, and 4s. For global comparison, the significance was calculated using the Kruskal–Wallis test, while the Wilcoxon test was used for pairwise comparisons. *P < 0.05, **P < 0.01, ***P < 0.001, or nonsignificant (ns), P ≥ 0.05. Commit prog, committed progenitor; E chromaffin, epinephrine chromaffin; N chromaffin, norepinephrine chromaffin; pre-E chrom, pre-epinephrine chromaffin.
The medulla cluster that yielded the highest scores in neuroblastoma belonged to the neuroblast followed by the norepinephrine chromaffin cluster (Fig. 3B). The presence of MYCN amplification in the genome, the stage, and the levels of MYCN and ALK expression did not lead to significant variation in the neuroblast score among the patients. This indicated that the neuroblastoma transcriptome was most similar to the neuroblast gene program when compared to the other hematological and solid malignancies. The medulla clusters correlated with the medulla group score, with the SCP cluster being the exception (SI Appendix, Fig. S6A). The majority of samples with a high SCP score was derived from stage 3 neuroblastoma patients. A high SCP score further anticorrelated with high ALK and MYCN expression and MYCN amplification. This clarified the low presence of SCP score in the stage 4S samples, as roughly 50% had high MYCN expression. Taken together, this indicated a high SCP score correlating with the most favorable disease phenotype present in the database.
Neuroblastoma Stages Exhibit Differing Levels of the Medulla Scores.
Neuroblastoma is well known for heterogeneity between clinical subtypes. Comparison of the genes to the cancer entities of the TARGET RNA-seq dataset confirmed the similarity of the medulla group and neuroblast cluster gene program to neuroblastoma. However, the samples included in the TARGET dataset were classified as stage 3, 4, and 4S, with an overrepresentation of stage 4 (84% of the total samples). A more suitable dataset to investigate the difference in the transcriptome of neuroblastoma stages when compared to the medulla group and clusters was generated by the SEQC Consortium (44). The SEQC dataset lacks expression data from other cancer entities but does include all neuroblastoma stages. By using this dataset, any association of the medulla group and cluster gene programs with disease phenotype was assessed.
The SCP score was higher in the samples lacking a MYCN amplification a confirmation of the finding of the TARGET dataset (Fig. 3C). This trend was observed in the medulla group and cluster scores as well (SI Appendix, Fig. S6B). This was an indication that carrying a MYCN amplification or having high MYCN expression, both markers of bad prognosis, changes the transcriptome further away from the medulla transcriptome. The samples with the highest SCP score were concentrated in stages 1 and 2, followed by stage 4S (Fig. 3D). The SCP signature showed the best separation between the stages. This was similar to the observation in the TARGET dataset and implied an association of the SCP signature to disease phenotype.
SCP Signature Can Be Associated with Disease Phenotype Severity.
To assess whether the SCP signature could be further associated with a more severe disease phenotype, survival analyses were performed. In univariate analyses, Cox regression showed that the SCP signature was associated with better overall survival (P = 9.0 × 10−11; hazard ratio = 0.694; 95% CI, 0.621–0.775). Patients were subsequently classified as having either high or low SCP signature based on an optimal cutoff value calculated with a receiver operating characteristic (ROC) curve. Survival analysis using the Kaplan–Meier product-limit method showed that patients with a high SCP signature (n = 318) had a significantly better prognosis than patients with a low SCP signature (n = 175; Fig. 4A). In the TARGET dataset, the SCP score was anticorrelated with MYCN amplification (Fig. 3B), and with high MYCN and ALK expression (Fig. 3B). In the SEQC dataset, the SCP score was also anticorrelated with ALK expression (Pearson correlation = −0.244, P = 4.35e-08), and MYCN amplification (Pearson correlation = −0.398, P = 3.60e-20). The SCP score also anticorrelated with age (Pearson correlation = −0.138, P = 0.002). This further confirmed that a high SCP score was associated with a less severe phenotype.

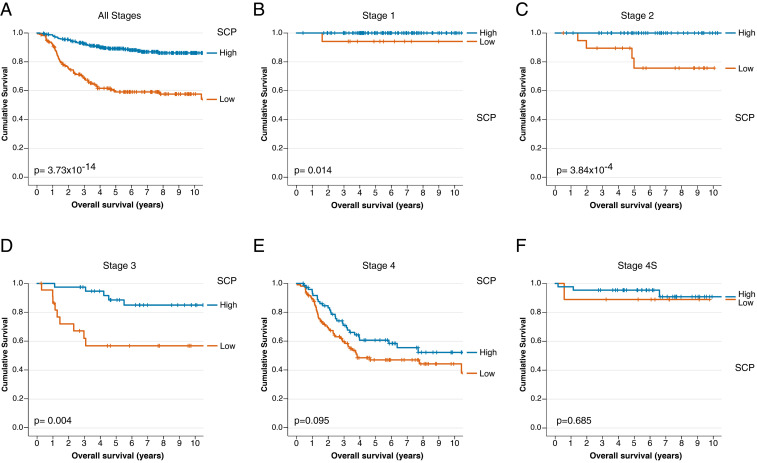
Survival analysis of the SEQC patients based on the SCP signature. (A) The 10-y overall survival rate of all stages classified to either have high levels (high, blue line, n = 318) or low levels (orange line, n = 175) of SCP signature. The 10-y overall survival rate of patients, categorized with either high or low levels of SCP signature, diagnosed with (B) stage 1 (high, n = 103; low, n = 17), (C) stage 2 (high, n = 58; low, n = 20), (D) stage 3 (high, n = 39; low, n = 23), (E) stage 4 (high, n = 75; low, n = 106), and (F) stage 4S neuroblastoma (high, n = 43; low, n = 9). Censored patients were represented as tick marks.
To investigate whether a cohort of patients with less severe disease phenotype could be classified based on the SCP score even within the same disease phenotype staging, the survival analysis was performed per stage. For stages 1, 2, and 3, samples characterized as having a high SCP signature showed a significantly better overall survival rate (Fig. 4 B–D). This trend was present in the stage 4 and 4S patients, while the difference in survival between high and low SCP signature level was not significant (Fig. 4 E and F). These findings were confirmed by a multiple Cox regression model, which showed that the SCP signature could predict overall survival independent of staging (P = 0.003; hazard ratio = 0.858; 95% CI, 0.760–0.947).
The consistency of having a better overall survival rate irrespective of the neuroblastoma stage, lends credence to the SCP signature identifying a cohort of patients with a different disease phenotype.
Discussion
In this study, we have identified cell clusters that make up the murine adrenal gland at multiple developmental timepoints. Single-cell RNA sequencing of the adrenal gland of mice during embryonic development has been performed earlier by Furlan et al. (17). With the use of Wnt1-Cre; R26RTomato E12.5 and E13.5 embryos and a biased sorting approach, the adrenal medullae and the suprarenal sympathetic ganglion were analyzed. The study revealed three main clusters. By taking six different developmental time points, two of which postnatal, and performing unbiased scRNA-seq, we identified seven clusters that have gene programs classifying them as medulla cells. Using the genes Furlan et al. associated with the NC derivatives of the adrenal medulla, and the catecholamine biosynthesis pathway, we identified the clusters as SCP, neuroblast, bridge, committed progenitors, pre-epinephrine chromaffin, norepinephrine chromaffin, and epinephrine chromaffin cluster. The norepinephrine chromaffin cluster consisted of chromaffin cells that are only capable of producing norepinephrine. The presence of these cells in rodents has been reported before (28). This cell type was identified by histological assessment of the chromaffin granules and by lack of Pnmt staining. The transcriptome of this cell type was not reported previously; therefore, having the gene program provides for more markers to distinguish these cells from the epinephrine chromaffin cell.
The proliferative neuroblast cluster is of particular interest. A dogma in the field is that neuroblastoma arises from a proliferating NC derivative that evades differentiation cues. This study provides a transcriptome by which the neuroblast cluster can be identified and distinguished from the other NC derivatives in the adrenal gland. Our study showed that healthy neuroblasts express high levels of MycN and Alk. Due to their expression, their loci are more open, and it can be hypothesized that this makes it easier for them to be amplified or mutated. Generation of mouse models for neuroblastoma could be supported by targeting oncogene expression to the clusters that we have identified. An example of such is the ectopic expression of MYCN under the control of genes that are markers for the neuroblast cluster, such as Stmn4 and Elavl2. Comparing the findings of the single-cell atlas to currently available neuroblastoma mouse models would be intriguing. Of note is Rgs5, the marker for the committed progenitor cluster. It is suggested to be involved in chromaffin differentiation, is negatively regulated by MYCN, and its genomic locus is reported to be affected in neuroblastoma (45, 46). It therefore would be intriguing to investigate what occurs to cells with the committed progenitors gene program in MYCN mouse models. The gene programs identified for the medulla clusters may also serve for future research on NC derivatives, in particular for in vitro modeling. This will allow for better defined characterization of these models.
Comparing the medulla cluster signature to the TARGET RNA-sequencing database demonstrated that the signatures were specific for neuroblastoma. A high SCP score was associated with a less severe phenotype in neuroblastoma in all stages. We can only speculate as to what cells are giving rise to the signature. One possibility is that there are indeed healthy SCP cells in the “good prognosis” tumors that confer a protective effect. Another is that the signature is derived from other cell types that are present exclusively in the tumors, like Schwannian stromal cells. Stages 3 and 4 have been described by Shimada et al. (11) as being Schwannian stroma poor, and yet in the SEQC dataset a subset of patients have a high SCP score. The SEQC dataset does not provide any information on the presence of Schwannian stroma. Direct comparison of the transcriptomes of Schwann cells and SCP would help addressing this question, as would ascertaining whether SCP cells persist postembryogenesis in the adrenal gland.
Of interest here, the development of the human NC was elucidated further by a recent study making use of single-cell RNA sequencing (38). Dong et al. analyzed NC cells upon specification and delamination from the neural tube, as well as NC cells found in the fetal adrenal gland. A link between clinical heterogeneity and the signatures of 10 cell types was made. Making use of the SEQC dataset, they observed an association with good prognosis when the signature of their most differentiated chromaffin cell type was present, and a poorer prognosis with the presence of their undifferentiated chromaffin cell type. No survival analysis was performed using their SCP signature and there was minimal difference in SCP levels observed across the patients. A discrepancy with their findings was that our SCP signature differed between the clinical subtypes. This could be due to the genes included in the signature list. By using the top 20 genes for our comparison to the SEQC dataset, only genes highly enriched in the SCP cluster were included in the analysis. Their SCP signature consisted of 239 genes, 2 of which we identified as being cortical markers in our single-cell atlas (Star and Cyp11a1). This could explain why we find a high SCP signature score leading to a more favorable overall survival rate than a low score.
In line with our findings, a recent study showed that high expression of Sox10 in the Schwannian stroma of neuroblastoma was associated with a good prognosis (47). This study focused on the Sox family transcription factors and made use of the same dataset as our study. Our study has therefore identified a list of independent genes that could be associated with a less severe disease phenotype. Further research into these genes and their relation to neuroblastoma could lead to new therapeutic strategies that might make use of the microenvironment to target the cancer cells. Stage 2 and 3 neuroblastoma patients can progress to stage 4 (48). Another interesting avenue of research is to further investigate the difference in level of the SCP signature in patients that progressed from intermediate stage to stage 4.
Since we included the entire adrenal gland in the scRNA-sequencing approach, the results obtained in this study provide insight not just into adrenal medulla development, but also into adrenal cortex development. Based on the expression of several adrenocortical markers, we were able to identify the three cortical clusters as adrenal primordium, fetal zone, and definitive zone in an unbiased fashion. The pseudotime trajectory analysis suggested that the adrenal primordium cells first turn into fetal zone cells, after which they reach a bifurcation where they either remain as fetal zone cells or turn into definitive zone cells. This is in line with the results of Zubair et al. (24), who show that fetal zone cells can act as precursors for the definitive zone cells, but only in a specific developmental time window. The fact that the NC-derived cells migrate through the developing adrenal cortex signals that factors present in the cortex could also influence medulla development. The single-cell atlas generated in this study represents a valuable tool to assess which pathways and ligands are required to establish in vitro models derived from either part of the adrenal gland.
In summary, the single-cell RNA sequencing of the murine adrenal gland has provided a tool to better understand the development of the adrenal gland. It opens up various research streams for better understanding of neuroblastoma pathobiology and provides a resource in the further possibilities of modeling neuroblastoma in vitro and in vivo.
Materials and Methods
Mice.
For the mouse experiments, a project license was granted by the Central Animal Testing Committee of the Dutch government. Ethical approval was granted by the Royal Netherlands Academy of Arts and Sciences–Hubrecht Institute Animal Welfare Body. The mice that were used were wild-type C57/BL6 mice. The day a plug was detected, and the day of birth were considered as E0.5 and P0, respectively.
Single-Cell RNA Sequencing.
Mice were collected at time points E13.5, E14.5, E17.5, E18.5, P1, and P5. Adrenal glands were isolated and dissociated into single cells. Adrenal glands were collected in PBS containing 1 mg/mL collagenase/dispase (Roche) and sheared using a 1-mL pipette on ice until single-cell suspension was observed. The enzymatic digestion was stopped by the addition of basic medium (Advanced DMEM/F12 [Gibco] containing N2 [1×; Gibco], B27 [1×; Gibco], Hepes [10 mM; Gibco], penicillin–streptomycin [100 U/mL; Gibco], Glutamax [1×; Gibco], N-acetyl cysteine [1.25 mM; Sigma]), and the suspension was centrifuged at 250 × g before being resuspended in PBS. All centrifugation steps were done at 4 °C, with the samples kept on ice for the duration of the experiment.
Single-cell suspensions were stained using 1 µM DAPI (Sigma). Viable single cells were sorted based on forward/side-scatter properties and DAPI staining (gating strategy shown in SI Appendix, Fig. S7) using FACS (FACSJazz; BD Biosciences) into 384-well plates (Bio-Rad) containing 10 µL of mineral oil (Sigma) and 50 nL of RT primers. Libraries were prepared using the Sort-seq method (18) and subjected to paired-end sequencing with 75-bp read length using the Illumina NextSeq500 sequencer.
Mapping.
Sequencing data were processed using the Sharq pipeline as described (49). Mapping was performed using STAR, version 2.6.1, on the Genome Reference Consortium Mouse Build 38. Read assignment was with featureCounts, version 1.5.2, using a gene annotation based on GENCODE, version M14.
Analysis.
Transcripts mapping to the mitochondrial genome and external RNA controls (ERCCs) were removed from all cells. A total of 410 cells forming two distinct clusters of erythroid lineages was identified based on high levels of hemoglobin complex genes and removed. To avoid contamination, all cells expressing more than 25% erythroid marker genes (SI Appendix, Fig. S8 and Table S2) were also removed. Finally, cells with mitochondrial-encoded transcripts over 50% of nuclear ones were removed, as well as cells with less than 1,000 nuclear-encoded transcripts or 334 genes. Subsequently, unique transcript counts were normalized using sctranform (50) and analyzed using the Seurat R package, version 3.1.5 (51). A Pearson residuals variance of 2 was used as a cutoff for identification of the variable genes. The following genes were removed from the list of variable genes in order to avoid biases in the cell clustering: genes associated with sex (Xist, Tsix, and Y chromosome-specific genes), cell cycle phase, dissociation stress (heat shock and chaperone proteins according to GO:0006986), and activity (ribosomal protein genes according to GO:0022626), as described before (SI Appendix, Table S3) (52). The first 25 principal components were used to calculate dimensionality reduction using the UMAP technique. Clustering was performed using the same principal components, a resolution of 2 and the Louvain algorithm.
The resulting 23 clusters were annotated on a single-cell basis using SingleR 1.2.4 (20) with the MouseRNAseqData reference dataset, consisting of 358 bulk RNA-seq samples of sorted cell populations (SI Appendix, Fig. S1A). Marker genes were used for further refining the cell annotations (SI Appendix, Fig. S1B). The 23 clusters were partitioned in five groups based on correlation. Differential expression analysis was performed with the FindMarkers Seurat (51) function for groups, clusters, and their combination using the Wilcoxon test with 1.5-fold change expression cutoff and 5% Bonferroni multiple testing corrected statistical significance cutoff. All genes passing the above criteria are shown in Dataset S3. Each marker gene was assigned to a unique group/cluster, based on higher overall expression.
Heatmaps were generated by heatmaply, version 1.1 (53). Columns represent cells ordered by cluster and developmental stage. Rows represent, in order, the top 20 differentially expressed genes of each group, followed by the top 20 differentially expressed genes in each of the clusters. The mean-centered expression of each gene–cell combination is represented in color according to the legend. Dot plots show gene expression changes across groups or clusters shown in the y axis. The size of the dot encodes the percentage of cells expressing each gene and the color encodes the log differential expression levels, as calculated by the DotPlot function in Seurat.
For the cortex and medulla group, trajectory graphs and pseudotime projections were generated with monocle3, version 0.2.2 (23), using code described in the SeuratWrappers package of the Seurat developers (https://github.com/satijalab/seurat-wrappers) and specifying E13.5 and E14.5 SCPs as the root node.
External Tumor Sample Data.
Public TARGET clinical and RNA-seq data were downloaded from cbiortal (https://www.cbioportal.org/datasets) and can be further found from https://portal.gdc.cancer.gov/projects. The results published here are in whole or part based upon data generated by the TARGET initiative, phs000218, managed by the National Cancer Institute (40). The data used for this analysis are available at https://portal.gdc.cancer.gov/projects. The SEQC microarray dataset, also known as the Fischer laboratory Agilent microarray (44), and clinical data (54, 55) were downloaded from the GEO database (GSE49710 and GSE62564). Samples with unknown MycN status were removed from all neuroblastoma datasets. In addition, samples with annotated tumor purity less than 80% were also removed.
Cell Type Signature Scoring in Bulk Expression Datasets.
For RNA-seq expression data, missing values were random forest imputed (56), mitochondrial genes were removed, and expression values were scaled to one million counts per sample. Subsequently, expression data were log base 2(log2) transformed using a pseudo count of 1. For both datasets, the normalized log2 data were quantile normalized using the R package limma (57). Finally, the normalized log2 data were median-centered per gene across all samples.
Mouse group- and cluster-specific genes were mapped to their human orthologs and to the genes present in each platform. After removing cell state-specific genes, the expression values were scaled and centered per gene (i.e., Z-scored) and each set of top 20 genes of each group and cluster 20 z-scores was combined using the Stouffer’s Z-score method to provide the signature score.
Heatmaps were generated by heatmaply, version 1.1 (53). Columns represent tumor samples ordered as described in the figure legend. Rows represent the signature scores of groups or clusters, as indicated. The sample-signature score combination is represented in color according to the legend. The expression of MYCN and ALK shown per sample is grouped in high (over 10-fold of the median), medium (between 4- and 10-fold), and low (less than 4-fold).
RNAscope In Situ Hybridization.
Adrenal glands were isolated at time points E13.5 and P0. Upon collection, they were rinsed twice with PBS, before fixation in 10% neutral buffered formalin (Sigma-Aldrich; HT501128) ranging from 16 to 32 h. Adrenal glands were further processed toward paraffin blocks, and sections were sliced 5 μm thick. RNA in situ was performed according to the instructions provided by RNAscope Multiplex Fluorescent Reagent Kit v2 Assay (Advanced Cell Diagnostics Bio). The RNAscope probes used are listed in Table 1.

| Probe target | Cluster | Catalog no. |
| Sox10 | Medulla | 435931 |
| Th | Medulla | 317621-C2 |
| Chga | Medulla | 447851-C3 |
| Nr5a1 | Cortex | 445731-C2 |
| Wnt4 | Cortex | 401101 |
| Col1a1 | Stroma | 319371-C4 |
| Kdr | Endothelial | 300031 |
| Rgs5 | Commit progenitor/N chromaffin | 430181 |
| Elavl2 | Neuroblast | 491961-C2 |
| Stmn4 | Neuroblast | 537541-C4 |
| Pnmt | E chromaffin | 426421-C3 |
| Erbb3 | SCP | 441801-C2 |
Imaging and Analysis.
The images were taken using a Leica TCS SP8 confocal microscope at 40× magnification. The adrenal gland was segmented based on DAPI signal in which the local maxima was determined, and a binary mask was created (58). The marker intensity was quantified using the Fiji plugin EzColocalization (32). All segments that had marker intensity values lower than half of the mode were designated as negative. The PCC was plotted along with the intensity values of the markers on the scatter plot. Segments that were double negative for the markers were not included in the analysis.
Survival Analyses.
Association of the SCP signature with survival was analyzed with the Cox proportional hazards model using univariate analysis. To divide the patients in two groups according to their SCP signature, the optimal signature cutoff value was calculated with a ROC curve. Patients with unfavorable outcome (death despite aggressive chemotherapy; n = 90) were included in the negative group, and patients with favorable outcome (survival 1,000 d after diagnosis with no chemotherapy; n = 180) in the positive group. The SCP signature value with the highest Youden’s index (sensitivity + specificity – 1) was selected as the cutoff value that was used for further analyses. The survival times were calculated using the Kaplan–Meier product-limit method, and the log-rank test was used to determine whether differences between groups were significant. Multiple Cox regression was performed using forward selection (Wald), and included the SCP signature and staging as variables. All survival analyses were performed using IBM SPSS Statistics, version 24 (IBM).
Acknowledgements
We thank Reinier van den Linden for help with flow cytometry; the Hubrecht Single Cell Core Facility for help with library preparation; Useq for NGS services; the Máxima Single Cell Genomics Facility for help with scRNA analysis; Yvonne M. A. Rijksen for help with RNAscope procedures; and Rijndert Ariese for help with imaging.
Data Availability.
Single-cell RNA-sequencing data have been deposited in the Gene Expression Omnibus database (accession no. GSE162238). The SEQC dataset containing microarray and clincal data was accessed from GSE49710 and GSE62564. The Public TARGET clinical and RNA-seq data were obtained from https://portal.gdc.cancer.gov/projects.
References
1
2
3
4
5
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
 Single-cell atlas of developing murine adrenal gland reveals relation of Schwann cell precursor signature to neuroblastoma phenotype
Single-cell atlas of developing murine adrenal gland reveals relation of Schwann cell precursor signature to neuroblastoma phenotype
