
Contributed by Steven L. McKnight, January 2, 2021 (sent for review October 30, 2020; reviewed by Lynette Cegelski, Tatyana Polenova, and Natasha Snider)
Author contributions: R.T. and S.L.M. designed research; X.Z., Y.L., M.K., E.M., L.S., V.O.S., and D.T.M. performed research; X.Z., M.K., D.T.M., and R.T. analyzed data; G.L., R.T., and S.L.M. wrote the paper.
Reviewers: L.C., Stanford University; T.P., University of Delaware; and N.S., University of North Carolina.
1Present address: Tsinghua-Peking Center for Life Science, IDG/McGovern Institute for Brain Research, School of Life Sciences, Tsinghua University, 100084 Beijing, China.
- Altmetric
Assembly of intermediate filaments (IFs) is reliant upon amino-terminal head domains. These head domains are of low sequence complexity and are assumed to function in the absence of structural order. Herein, we provide evidence that the head domains of the desmin and neurofilament light (NFL) IF proteins self-associate via the formation of labile but structurally specific cross-β interaction. Disease-causing mutations in the head domains of both proteins cause enhanced cross-β interactions. By assembling desmin and NFL IFs bearing isotopically labeled head domains, we provide evidence of structural order in properly assembled biological filaments. We propose that these observations on IF head domains may be instructive to the function of low complexity domains operative in other aspects of cell biology.
Low complexity (LC) head domains 92 and 108 residues in length are, respectively, required for assembly of neurofilament light (NFL) and desmin intermediate filaments (IFs). As studied in isolation, these IF head domains interconvert between states of conformational disorder and labile, β-strand–enriched polymers. Solid-state NMR (ss-NMR) spectroscopic studies of NFL and desmin head domain polymers reveal spectral patterns consistent with structural order. A combination of intein chemistry and segmental isotope labeling allowed preparation of fully assembled NFL and desmin IFs that could also be studied by ss-NMR. Assembled IFs revealed spectra overlapping with those observed for β-strand–enriched polymers formed from the isolated NFL and desmin head domains. Phosphorylation and disease-causing mutations reciprocally alter NFL and desmin head domain self-association yet commonly impede IF assembly. These observations show how facultative structural assembly of LC domains via labile, β-strand–enriched self-interactions may broadly influence cell morphology.
Mammalian genomes encode 70 to 80 unique proteins that assemble into a variety of intermediate filaments (IFs). These filaments play cyto-architectural roles that vary considerably in skin, muscle, and nerve cells (1). Fluorescence recovery after photobleaching (FRAP) experiments have shown that IFs can be surprisingly dynamic, allowing insertion of new polypeptide subunits internal to the longitudinal axis of existing filaments (2). Despite access to a wealth of information pertinent to the form and function of IFs, important questions regarding the processes controlling their assembly/disassembly and its regulation remain unresolved.
Our interest in the assembly of IFs came by accident in studies of the aliphatic alcohol 1,6-hexanediol. This agent had long been known to compromise the permeability barrier of nucleopores and melt nuclear and cytoplasmic puncta not surrounded by investing membranes (34–5). We and others had noticed that 1,6-hexanediol also melts phase separated liquid-like droplets and hydrogels formed in test tubes via self-association of the low complexity (LC) domains associated with various RNA-binding proteins (6, 7). As a control experiment, we asked whether 1,6-hexanediol might melt cellular structures in a specific or nonspecific manner. The agent did not disassemble either actin filaments or microtubules upon administration to cultured mammalian cells. Much to our surprise, however, 1,6-hexanediol caused rapid and dramatic disassembly of IFs specified by either vimentin or keratin (6).
We quickly made note of extensive literature showing that IF assembly is reliant upon N-terminally localized “head domains” universally specified by low complexity sequences. We proceeded to confirm that the isolated head domains of six different IF proteins display the ability to become phase separated in a manner indistinguishable from prototypic LC domains associated with RNA-binding proteins (6). These observations raised the possibility that IF head domains might facilitate filament assembly in a manner analogous to LC domain function by RNA-binding proteins in the assembly of RNA granules.
IFs are defined by centrally located α-helical segments 300 to 350 residues in length. These central, α-helical segments are flanked on either end by head and tail domains thought to be devoid of structural order (8, 9). Assembly of IFs begins with the parallel association of two α-helical segments to form a coiled coil. Pairs of coiled-coil dimers, in turn, associate in an antiparallel orientation to form tetramers. Dimer and tetramer formation of IF proteins can proceed in the absence of their intrinsically disordered head and tail domains. By contrast, the end-to-end joining of tetramers into long protofilaments, as well as the assembly of eight protofilaments around one another to form mature, 10-nm IFs, cannot proceed with variants missing the N-terminal head domain (10111213–14).
Past studies of the isolated head domains of vimentin, peripherin, internexin, and the light, medium, and heavy neurofilament isoforms gave evidence of phase separation in the form of gel-like condensates (6). We proceeded to postulate that the molecular interactions leading to phase separation of IF head domains might be instructive as to how they assist in the assembly of IFs. Here, we describe experiments designed to investigate this hypothesis.
Results
The heavy, medium, and light neurofilament isoforms are important for the morphological integrity of axons where they interact intimately with longitudinally oriented microtubules and exist at an abundance 10-fold greater than microtubules or actin filaments (15). The studies reported herein were initially focused on the neurofilament light (NFL) isoform. The head domain of the NFL isoform, 92 residues in length, was prepared as a green fluorescent protein (GFP) fusion protein containing an amino-terminal 6×His-tag, expressed in bacterial cells, purified, and incubated at high concentration under physiologic conditions of salt and pH (Materials and Methods). As described previously, these procedures led to the formation of a gel-like condensate composed of uniform, amyloid-like polymers (6).
GFP:NFL head domain hydrogel droplets 5 mm in diameter were formed on parafilm and incubated for 2 h at 4 °C with a soluble extract prepared from mouse brain tissue (Materials and Methods). Following binding, hydrogel droplets were recovered by centrifugation, washed twice with phosphate-buffered saline, and melted by exposure to binding buffer supplemented with 6 M guanidine-HCl. Soluble material was passed through a nickel affinity resin to remove the 6×His-tagged GFP:NFL head domain fusion protein. Unbound material was recovered and subjected to shotgun mass spectrometry in order to identify the hydrogel-bound proteins listed in Fig. 1.


Binding of mouse brain proteins to hydrogel droplets formed from the NFL head domain. Hydrogel droplets were formed from a fusion protein linking 6×His-tagged GFP to an N-terminal fragment specifying the first 92 residues of the NFL protein (Materials and Methods). Droplets 5 mm in size were formed on parafilm, incubated with a soluble lysate prepared from mouse brain tissue, washed, and eluted by melting gel droplets in gelation buffer supplemented with 6 M guanidine-HCl. The melted GFP:NFL head domain protein was removed by Ni-affinity beads, allowing bound proteins to then be identified by shotgun mass spectrometry. The intact, endogenous NFL protein yielded a higher number of spectral counts than any other mouse brain protein bound to GFP:NFL hydrogel droplets. All peptides derived from the first 92 residues of the NFL protein were disregarded from the mass spectrometry data. Many other hydrogel-bound proteins are known from previous studies to interact with neurofilaments. (Scale bar: 2 mm.)
Evidence of Specificity in IF Head Domain Self-Interaction.
As deduced by quantitation of spectral counts, the top protein retrieved from mouse brain extracts by hydrogel droplets composed of NFL head domain polymers was the NFL protein itself. The majority of peptides identified in gel-bound samples corresponded to regions of NFL specifying its coiled-coil and tail domains. Since the hydrogel itself was prepared from a protein specifying the NFL head domain alone (residues 2 to 92), it can be concluded that the NFL protein prominently bound by the hydrogel droplets was derived from the mouse brain extract.
Other than capturing the mouse brain NFL protein itself, NFL head domain hydrogel droplets retained a number of axonal proteins, including the α and β chains of nonerythrocyte spectrin, the α and β subunits of tubulin, three microtubule-associated proteins (MAPs) designated MAP1A, MAP1B, and MAP2, the heavy chain of cytoplasmic dynein, and the large plectin protein known to bridge association of assembled neurofilaments to microtubules (Fig. 1). The means by which these additional proteins might be trapped by GFP:NFL head domain hydrogels, as well as the potential biological implications of these observations, will be considered in Discussion.
Our simplistic interpretation of these observations is that the NFL head domain present in GFP hydrogel droplets is able to specifically bind the head domain of the native NFL protein present in a soluble brain tissue lysate. This binding would be expected to secure retention of the entire NFL polypeptide, thus accounting for the abundant occurrence of tryptic peptides from the coiled-coil and tail domains of the protein. We further offer that the heterotypic trapping of other proteins known to interact with neurofilaments reflects the fact that these proteins may remain associated with NFL endogenous to the brain lysate through our conditions of binding, washing, and elution.
These same hydrogel-trapping experiments were repeated and expanded to include GFP hydrogels formed from polymerized LC domains derived from the fused in sarcoma (FUS) RNA-binding protein, ataxin-2, synaptophysin, lamin, and Nup54. We again observed that the GFP:NFL head domain hydrogel trapped endogenous NFL as the top protein. None of the other five hydrogels retained the endogenous brain NFL polypeptide among their top 20 hydrogel-trapped proteins (SI Appendix, Table S1). As such, we tentatively hypothesize that the selectivity of brain NFL trapping by GFP:NFL head domain hydrogels might reflect specific, self-associative interactions engendered by the NFL head domain itself—perhaps the same cross-β interactions that allow for head domain polymerization.
The aforementioned interpretation was tested using recombinant NFL protein samples linked to GFP. One fusion protein linked GFP to intact NFL, a second linked GFP to an NFL variant deleted of its head domain, and a third linked GFP to a variant deleted of the NFL tail domain. Each of these fusion proteins was expressed, purified, and incubated with hydrogel droplets composed of NFL head domain polymers. As shown in Fig. 2B, NFL hydrogel droplets bound the GFP fusion proteins linked to either the intact NFL protein or the variant missing the unstructured tail domain. By contrast, the GFP fusion protein missing the unstructured NFL head domain was not trapped by hydrogels composed of the NFL head domain. As an extension of these experiments, GFP was fused to the isolated head, coiled-coil rod, or tail domains of NFL. Among the latter fusion proteins, only the GFP variant linked to the NFL head domain was retained by mCherry:NFL head domain hydrogel droplets (Fig. 2B).

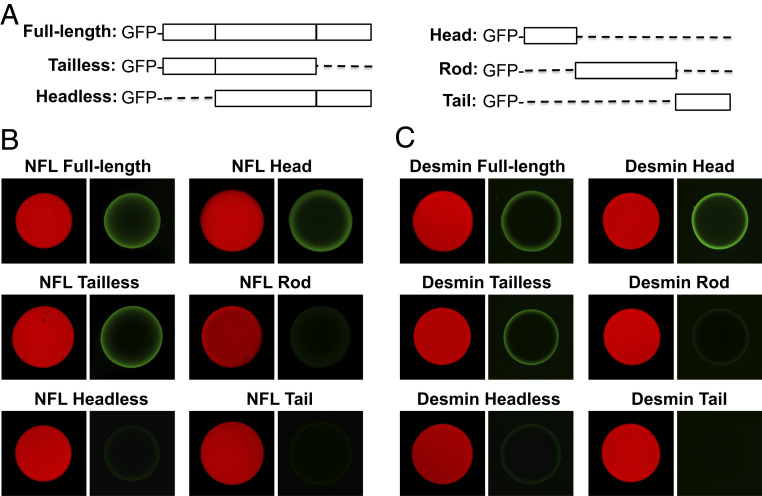
Hydrogel droplets formed from the NFL and desmin head domains bind cognate GFP-tagged proteins only if they retain the cognate head domain. (A) Schematic diagram of GFP-fusion proteins used in binding assays with mCherry:NFL and mCherry:desmin hydrogel samples. Hydrogel droplets, 1 mm in diameter, formed from fusion proteins linking mCherry to the head domains of NFL (B) or desmin (C), were exposed to GFP-tagged test proteins linked to the full-length versions of NFL or desmin, as well as versions missing the low complexity tail or head domain. GFP fusion proteins linked to the head domain alone, coiled-coil rod domain alone, or tail domain alone of either NFL or desmin were also tested for binding to hydrogel droplets composed of mCherry fused to the cognate head domain.
Similar experiments were conducted to study self-interaction of the head domain of desmin IFs. Desmin represents a canonical IF protein that has been extensively studied in the context of skeletal and cardiac muscle tissue (16). As shown in Fig. 2C, hydrogel droplets composed of a fusion protein linking mCherry to the head domain of desmin were observed to bind a test protein that linked the intact desmin protein to GFP. A desmin variant deleted of its head domain failed to bind the hydrogel droplets, yet removal of the disordered tail domain of desmin did not impede gel binding. When fused in isolation to GFP, only the head domain of desmin allowed for GFP binding to mCherry:desmin head domain hydrogel droplets.
Solid-State NMR Spectra of NFL and Desmin Head Domain Polymers Give Evidence of Structural Order and β-Strand Secondary Structure.
Incubation of the isolated NFL and desmin head domains at a high protein concentration has been shown to yield amyloid-like polymers of uniform morphology (6). Unlike pathogenic, prion-like amyloids, polymers formed from the NFL or desmin head domains are labile to disassembly. Structural studies of these polymers were initiated by labeling the NFL and desmin head domains uniformly with 15N and 13C, allowing polymerization, and examining the samples by solid-state NMR (ss-NMR) spectroscopy. Whereas two-dimensional (2D) 15N-13C spectra of certain samples were recorded, these spectra did not add to the information available from the 2D 13C-13C spectra that are presented in Fig. 3.
Fig. 3 shows 2D 13C-13C spectra of NFL head domain polymers (Fig. 3 A, Left, blue contours) and desmin head domain polymers (Fig. 3 B, Left, blue contours), acquired respectively at 13 °C and 5 °C as described in Materials and Methods (SI Appendix, Table S2). Crosspeak signals in these spectra are not sufficiently sharp to allow signals from individual residues to be resolved and assigned, but certain crosspeaks can be assigned to amino acid types, including Thr, Ser, Val, Pro, Ala, and Ile. With the exception of proline, the average 13C chemical shifts for these residues are consistent with a predominance of β-strand secondary structure in both NFL and desmin head domain polymers. This interpretation rests upon the observations that 13CO and 13Cα chemical shifts are smaller than random-coil values and that 13Cβ chemical shifts are larger than random-coil values (SI Appendix, Table S3). The overall similarity of 2D 13C-13C ss-NMR spectra of the NFL and desmin head domain polymers reflects the similarity of their amino acid compositions (SI Appendix, Fig. S1).

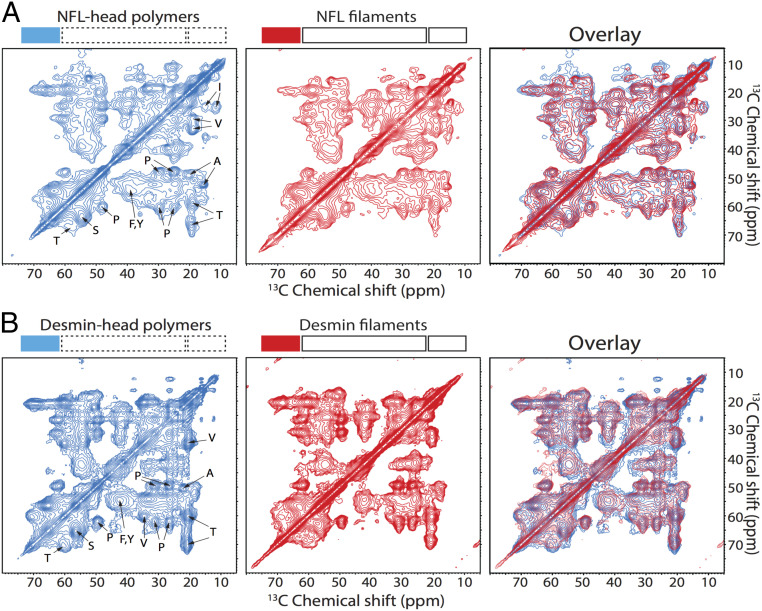
The 2D 13C-13C ss-NMR spectra of NFL and desmin head domain-only polymers as compared with segmentally labeled NFL and desmin IFs. (A) The 2D spectrum of uniformly 15N,13C-labeled NFL head domain-only polymers (Left, blue) adjacent to a spectrum of the segmentally 15N,13C -labeled NFL head domain within IFs (Middle, red). Overlay of the two spectra reveals extensive overlap (Right). Sample temperatures were 13 °C for head domain-only polymers and −23 °C for IFs. (B) The 2D 13C-13C spectrum of uniformly 15N,13C-labeled desmin head domain-only polymers (Left, blue) adjacent to a spectrum of the segmentally 15N,13C-labeled desmin head domain within IFs (Middle, red). Overlay of the two spectra reveals extensive overlap (Right). Sample temperatures were 5 °C for the head domain-only polymers and −23 °C for IFs. The 2D spectra were obtained with 25-ms dipolar-assisted rotational recoupling (DARR) 13C-13C mixing periods (64). Contour levels increase by factors of 1.4.
The observation that crosspeak signals from individual residues cannot be resolved in the 2D ss-NMR spectra of head domain polymers suggests that the level of structural order within these polymers is lower than in cross-β polymers formed by the low-complexity domains of FUS and hnRNPA2 (17, 18) or in certain pathogenic amyloid fibrils (1920–21) where better-resolved crosspeaks were observed. The precise level of order in NFL and desmin head domain polymers cannot be determined. However, it is notable that ss-NMR spectra of well-structured peptides and proteins in frozen solutions typically show 13C line widths in the 1 to 3 parts per million (ppm) range (22, 23), attributable to small-amplitude fluctuations around an average structure that are not averaged out by thermal motions. Similar structural variations among protein molecules within the head domain polymers, corresponding to a root-mean-squared deviation of the atomic coordinates on the order of 2 to 4 Å, may account for the observed level of spectral resolution.
Most amyloid-like, cross-β protein assemblies contain in-register parallel β-sheets (1718–19, 21). Exceptions to this rule include β-solenoidal fibrils (20), worm-like metastable “protofibrils” (24), and fibrils formed by certain short peptides (25). To test for in-register parallel β-sheets in NFL and desmin head domain polymers, we prepared samples with 13C labels only at backbone carbonyl sites of Val, Leu, or Phe residues and used the PITHIRDS-CT ss-NMR technique (26) to quantitatively measure 13C-13C dipole-dipole couplings (which are inversely proportional to the cube of internuclear distances). The PITHIRDS-CT data (SI Appendix, Fig. S2) indicate 13C-13C distances greater than 6 Å, inconsistent with in-register parallel β-sheets, for which 13C-13C distances of 4.8 Å are expected.
NFL and Desmin Head Domains Display Evidence of Molecular Structure in Their Respective Assembled IFs.
Having observed that the isolated NFL and desmin head domains adopt a preferred molecular structure upon polymerization, the question arose as to whether this structure might be of biological relevance. If so, one would anticipate formation of much the same structure in fully assembled NFL IFs. Recall that the coiled-coil domains of IFs become paired at an early step in filament assembly. This coiled-coil pairing positions two head domains in close proximity. Subsequent assembly of tetramers, elongated protofilaments, and fully mature IFs positions 16 head domains in immediate, cylindrical proximity. These head domains ring the circumference of mature filaments and are disposed at roughly 45-nm intervals along the axial filament length (1). The local concentration of NFL and desmin head domains must be high in these circumferential rings, perhaps sufficiently high to facilitate the weak, cross-β interactions formed when isolated head domains are incubated at a sufficiently high concentration to affect polymerization.
To test whether head domains adopt structures or structural distributions within assembled IFs that are similar to structures in head domain-only polymers, we acquired 2D 13C-13C spectra of segmentally labeled IFs (formed by full-length desmin or NFL, but with 13C labeling localized only within the respective head domains via intein chemistry) (SI Appendix, Fig. S3). These 2D spectra are shown with red contours in the middle panels of Fig. 3 and are superimposed upon the spectra of head domain-only polymers in the right panels. Crosspeak signal patterns of head domain polymers and segmentally labeled IFs are strikingly similar, providing strong evidence for structural similarity.
As a more quantitative measure of conformational similarity between head domain-only polymers and the same head domains in segmentally labeled IFs, we used Cα/Cβ crosspeak volumes from Ser and Ala residues in the 2D 13C-13C spectra (SI Appendix, Fig. S4A). Ser and Ala were chosen because their Cα/Cβ crosspeaks are resolved from those of other residues and because their crosspeak positions are known to be dependent on local secondary structure. From the measured crosspeak volumes for β-strand–like and non-β-strand–like signal components, we estimate that roughly 50% of isotopically labeled Ser and Ala residues have β-strand conformations, both in uniformly 15N,13C-labeled NFL head domain-only polymers and in segmentally labeled NFL IFs (SI Appendix, Table S4). Roughly 80% of Ser and 40% of Ala residues have β-strand conformations in uniformly 15N,13C-labeled desmin head domain-only polymers. In segmentally labeled desmin IFs, β-strand contents of both Ser and Ala are reduced by a factor of two. These estimates are necessarily approximate, with error bars on the order of ±15%. Error bars are derived from uncertainties in the volumes of crosspeaks that arise from residues in different secondary structure elements, as shown in SI Appendix, Fig. S4, due to uncertainties in the precise boundaries of the 2D crosspeak regions. The 2D difference spectra (SI Appendix, Fig. S5A) also confirm the similarity of crosspeak intensities between NFL head domain-only polymers and segmentally labeled NFL IFs. Intensity differences are somewhat greater in 2D spectra of the corresponding desmin samples (SI Appendix, Fig. S5B). Nonetheless, in both cases, the 2D spectra of head domain-only polymers and segmentally labeled IFs have nearly the same crosspeak positions (Fig. 3). A 2D 13C-13C spectrum of the uniformly 15N,13C-labeled desmin head domain in an amorphous, precipitated state shows larger differences in crosspeak positions and intensities (SI Appendix, Fig. S5C).
Although crosspeaks of individual residues are not resolved, the 2D ss-NMR spectra of head domain polymers and assembled IFs are strongly indicative of the presence of structural order. To probe this interpretation more rigorously, we prepared head domain-only polymers with 13C-15N labeling restricted to either Val and Ile residues (NFL) or Val and Thr (desmin). The spectra of these polymers were compared with the same protein samples dissolved in trifluoroacetic acid (TFA) precipitated by cold ether. The latter treatment yielded amorphous aggregates not expected to exist in a structurally ordered state. The 2D 13C-13C spectra of the Val/Ile (NFL) or Val/Thr (desmin) labeled head domain polymers and ether precipitates are shown in Fig. 4. In these spectra, crosspeak signals of head domain polymers are clearly shifted away from the corresponding signals of amorphous head domain precipitates. The directions of shift correspond to expectations of a transition from a predominantly random coil structure in the amorphous material (red vertical lines in Fig. 4) to a β-strand-enriched structure in the head domain polymers (blue vertical lines in Fig. 4).

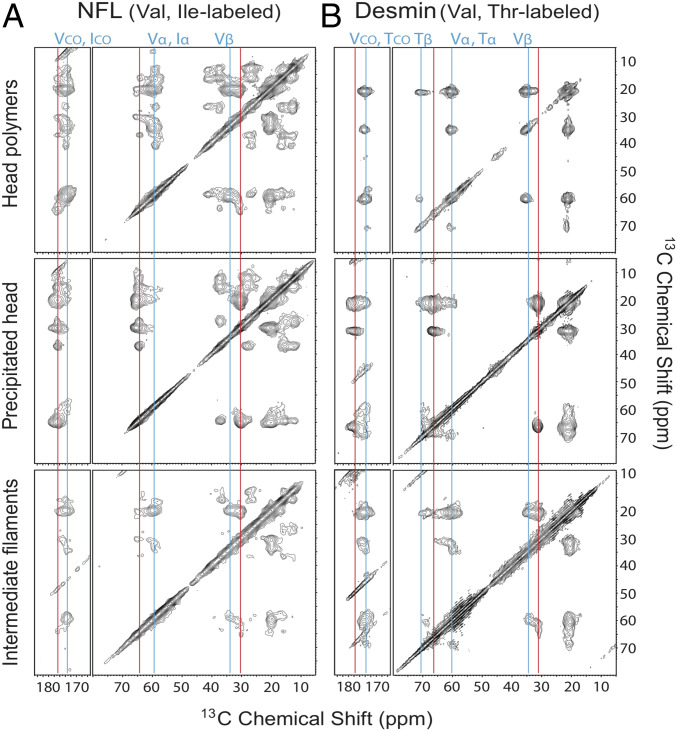
Two dimensional 13C-13C ss-NMR spectra of head domain-only polymers compared with IFs segmentally labeled with specific amino acids. (A) The 2D spectra of 15N,13C-Val and Ile-labeled NFL head domain-only polymers (Top), an ether-precipitated sample of the 15N,13C-Val and Ile-labeled NFL head domain protein (Middle), and the segmentally Val,Ile-labeled NFL head domain within IFs (Bottom). Sample temperatures were 13 °C for the head domain-only samples and −23 °C for IFs. (B) The 2D spectra of 15N,13C-Val and Thr-labeled desmin head domain-only polymers (Top), an ether-precipitated sample of the 15N,13C-Val and Thr-labeled desmin head domain alone protein (Middle), and the segmentally Val,Thr-labeled desmin head domain within IFs (Bottom). Sample temperatures were 5 °C for the head domain-only samples and −23 °C for IFs. Contour levels increase by factors of 1.4. Vertical blue lines indicate peak positions for β-strand–like crosspeak signals in spectra of head domain-only polymers. Vertical red lines indicate non-β-strand–like signals in spectra of ether-precipitated samples. VCO, ICO, TCO: carbonyl carbon of valine, isoleucine, and threonine. Vα, Iα: α carbon of valine and isoleucine. Vβ, Tβ: β carbon of valine and threonine.
Intein-mediated chemistry was used to introduce the same Val/Ile isotopes into the head domain of the full-length NFL and the same Val/Thr isotopes into the head domain of the full-length desmin. Following purification, the segmentally labeled samples were assembled into mature IFs and analyzed by ss-NMR. The same patterns of crosspeak signal shifts were observed in the assembled NFL and desmin IFs as had been seen in the head domain-only polymers. Although not all Val/Ile (NFL) or Val/Thr (desmin) crosspeak signal intensity in 2D spectra of the assembled IFs aligned exactly with signal intensity of 2D spectra of the head domain-alone polymers, spectra of the Val/Ile- and Val/Thr-labeled IFs clearly agree more closely with spectra of head domain polymers than with spectra of amorphous, ether-precipitated material.
To estimate secondary structure contents, we measured volumes of conformation-dependent Cα/Cβ, Cα/Cγ, and Cα/Cβ crosspeaks for the labeled amino acids (SI Appendix, Fig. S4 B and C). As summarized in SI Appendix, Table S4, the crosspeak volumes indicate that more than 50% of Val and Ile residues have β-strand conformations in both NFL head domain-only polymers and segmentally Val/Ile-labeled NFL IFs. β-strand conformations are below 20% in the TFA/ether precipitated Val/Ile-labeled NFL sample. For desmin, Val and Thr residues have β-strand contents above 60% in head domain-only polymers, roughly 40% in segmentally Val/Thr-labeled IFs, and about 10% in the TFA/ether precipitated sample. These analyses support the structural similarity between head domain-only polymers and head domains in IFs, and the dissimilarity from an amorphous state.
NFL and Desmin Head Domains Are Partially Dynamic in IFs at Biologically Relevant Temperatures.
The 2D ss-NMR spectra of IFs in Figs. 3 and 4 were recorded at low temperatures (−23 °C) because spectra recorded above 0 °C showed significantly weaker crosspeaks (SI Appendix, Fig. S6). The reductions in crosspeak intensities at higher temperatures are attributable to molecular motions that partially average out nuclear magnetic dipole-dipole couplings, which drive the nuclear spin polarization transfers that produce crosspeaks. Apparently, the molecular structures and degree of conformational disorder of head domains in head domain-only polymers and IFs are similar, but the head domains are more highly dynamic (i.e., “mushy”) in IFs above 0 °C. Conformational variations in IFs are not necessarily larger in amplitude at higher temperatures but occur on shorter time scales. Once the motional time scales exceed roughly 1 ms at lower temperatures, motional effects on the 2D 13C-13C ss-NMR spectra become minimal.
To probe molecular motions of head domains in the IFs in more detail, we recorded one-dimensional (1D) 13C NMR spectra of the segmentally 13C-labeled IFs with two different methods. To observe signals from relatively rigid protein segments, we used 1H-13C cross-polarization (CP) (27), driven by nuclear magnetic dipole-dipole couplings, to prepare transverse 13C polarization and high-power 1H decoupling with two-pulse phase modulation (TPPM) (28) during signal acquisition. To observe signals from highly dynamic segments with nearly isotropic motions, we used the “insensitive nuclei enhanced by polarization transfer” (INEPT) (29) technique, driven by scalar couplings, to prepare transverse 13C polarization and low-power composite pulse decoupling (30, 31). As shown in Fig. 5 for both NFL IFs and desmin IFs, CP-based signals increase in intensity with decreasing temperature while INEPT-based signals decrease in intensity.

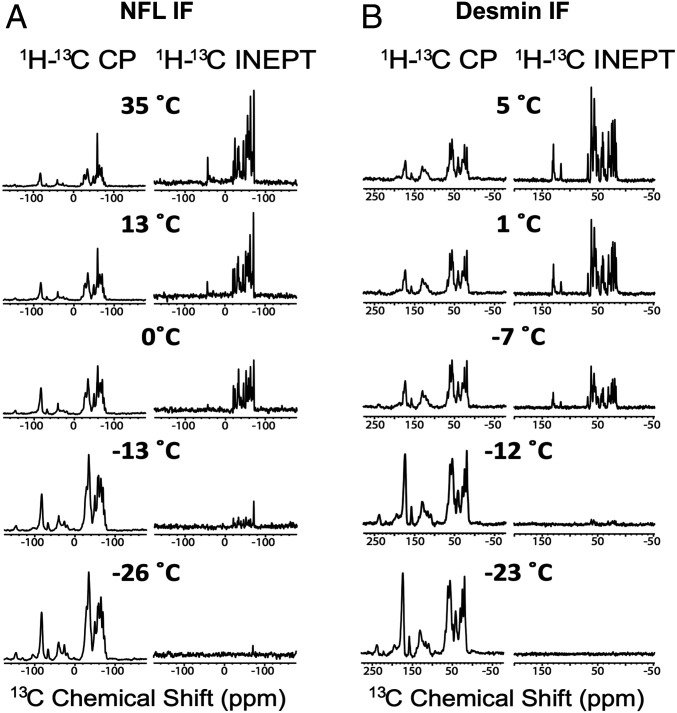
The 1D CP and INEPT spectra of segmentally labeled IFs. (A) Spectra of the segmentally 15N,13C-labeled NFL head domain as situated in situ in the context of assembled IFs. Spectra were recorded at temperatures ranging from 35 °C to −26 °C as indicated. (B) Spectra of the segmentally 15N,13C-labeled desmin head domain as situated in situ in the context of assembled IFs. Spectra were recorded at temperatures ranging from 5 °C to −23 °C as indicated. Spectra within each column are plotted on the same vertical scale.
We interpret the data in Fig. 5 and SI Appendix, Fig. S6 as evidence that both desmin and NFL head domains contain structured segments, with moderate-amplitude disorder, and unstructured segments. The structured segments have limited internal motion, occurring on submillisecond time scales at the higher temperatures in these experiments and >1-ms time scales at the lowest temperatures. The unstructured segments have large-amplitude motions (i.e., are dynamically disordered) at the higher temperatures, on submicrosecond time scales. Motions of the unstructured segments are reduced at lower temperatures, leading to inefficient INEPT polarization transfers and weak signals in INEPT-based spectra. Importantly, it is primarily the overall signal intensities, rather than the peak positions and line shapes, that change with temperature in both the CP-based and the INEPT-based spectra in Fig. 5. Therefore, these spectra should not be interpreted to be giving evidence of a conversion of disordered segments to ordered segments with decreasing temperature.
Phosphorylation of the NFL and Desmin Head Domains Disassembles IFs and Inhibits Head Domain Self-Association.
IFs have long been known to disassemble during mitosis (32, 33). In numerous instances, disassembly has been attributed to phosphorylation of IF head domains (15, 3435363738–39). Upon incubation of either NFL or desmin IFs with a combination of adenosine 5′-triphosphate (ATP) and protein kinase A (PKA), both filaments were observed to disassemble (Fig. 6). Exposure of NFL and desmin intermediate filaments to either ATP or PKA alone failed to affect filament disassembly. The combination of ATP and PKA also effected release of the GFP:NFL head domain fusion protein from mCherry:NFL head domain hydrogel droplets. Likewise, the GFP:desmin head domain fusion protein bound to mCherry:desmin head domain hydrogel droplets was released upon exposure to both ATP and PKA (Fig. 6). Reactions missing ATP or PKA left the respective GFP fusion proteins hydrogel-bound.

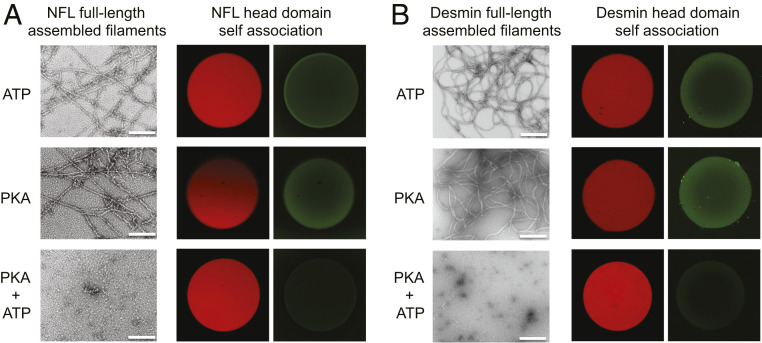
PKA-mediated disassembly of NFL and desmin IFs and release of GFP:head domain test proteins from cognate hydrogel binding. Assembled IFs prepared from full-length NFL and desmin were incubated with ATP alone (Top Images), PKA alone (Middle Images), or both ATP and PKA (Bottom Images). PKA-mediated phosphorylation led to disassembly of both types of IFs as deduced by transmission electron microscopy. Hydrogel droplets, 1 mm in diameter, formed from mCherry fused to the head domain of either NFL (A) or desmin (B), were prebound with GFP fusion proteins linked to the head domain of either NFL or desmin. Release of hydrogel-bound GFP-tagged protein was only observed upon coincubation with both ATP and PKA. (All scale bars: 200 nm.)
Recurrent Mutations in NFL and Desmin Head Domains Accelerate β-Strand–Enriched Self-Association.
Human genetic studies have identified perplexingly recurrent, autosomal dominant mutations localized to the NFL head domain. Studies of pedigrees tracing the genetic cause of neurological deficits, commonly described in the context of Charcot–Marie–Tooth (CMT) disease, have identified mutations in one of two proline residues located within the NFL head domain (40, 41). Independent familial mutations changing proline residue 8 to leucine, arginine, or glutamine are commonly understood to be causative of CMT disease. Surprisingly, independent familial mutations causative of CMT disease have also been found to change proline 22 within the NFL head domain to serine, threonine, or arginine. The reason why CMT disease-causing mutations are recurrently found in these two proline residues has been obscured by the assumption that the head domains of all IF proteins function in the absence of molecular order. If a protein segment is intrinsically disordered, why would its biological function care about the subtle change of but a single amino acid?
NFL variants carrying a P-to-Q, P-to-L, or P-to-R mutation at residue 8, or a P-to-S, P-to-T, or P-to-R mutation at residue 22, were expressed as recombinant protein, purified, and tested for the formation of IFs. All six mutations significantly impeded IF assembly, yielding much shorter filaments relative to the native NFL protein (Fig. 7A and SI Appendix, Fig. S7A) (42). In order to study these NFL variants more carefully, head domains bearing three different P8 mutations, and three different P22 mutations, were expressed in bacterial cells, purified, and assayed for their capacity to form labile, cross-β polymers. Our standard methods involve purification of the NFL head domain in 8 M urea, followed by sequential dialysis into gelation buffer supplemented by 4 M, 2 M, 1 M, 0.5 M, and no urea (Materials and Methods). Upon initiating studies of the six different CMT-causing mutations, it was noticed that the mutated variants became cloudy upon dialysis of the protein samples from 8 M to 4 M urea (Fig. 7C). Analysis of the cloudy samples by transmission electron microscopy revealed uniform, amyloid-like polymers indistinguishable from the labile, cross-β polymers formed by the native NFL head domain (Fig. 7C and SI Appendix, Fig. S7B) which do not form without complete removal of the urea denaturant.

![Effects of disease-causing mutations within NFL and desmin head domains on assembly of IFs (A and B), cross-β polymerization of isolated head domains (C and D), and ss-NMR spectra of isotopically labeled head domains (E and F). Full-length (FL) NFL and desmin proteins bearing indicated head domain mutations were incubated under conditions suitable for assembly of IFs. Compared with native (wild-type [WT]) proteins, disease-causing mutations in the NFL and desmin head domains led to the formation of either short (NFL) or tangled (desmin) IFs (A and B, and SI Appendix, Fig. S5A). Isolated head domain samples of native (WT) or six different NFL head domain mutants were tested for formation of cross-β polymers (C). Relative to the native NFL head domain, all six head domain mutants formed polymers prematurely in the presence of 4 M urea as revealed by opalescence. Transmission electron microscopy showed no polymers formed from the native NFL head domain in urea (Materials and Methods), but clear evidence of polymers formed from the P8Q and P22R head domain mutants. Isolated head domain samples of native (WT) and six desmin head domain mutants were also tested for formation of cross-β polymers in the presence of 3 M urea (D). Relative to the native desmin head domain, all six head domain mutants formed polymers prematurely in the presence of 3 M urea. Transmission electron microscopy revealed no polymers formed from the native desmin head domain in urea (Materials and Methods), but clear evidence of polymers formed from the S7F, S12F, and S46F head domain mutants. The 2D 13C-13C ss-NMR spectra of isotopically labeled head domains of NFL and desmin show no significant differences between native (WT) proteins and indicated head domain mutants (E and F). (All scale bars: 200 nm.)](/dataresources/secured/content-1765980604534-5f01a505-0ab0-48c2-a193-9bf1d5e7024a/assets/pnas.2022121118fig07.jpg)
Effects of disease-causing mutations within NFL and desmin head domains on assembly of IFs (A and B), cross-β polymerization of isolated head domains (C and D), and ss-NMR spectra of isotopically labeled head domains (E and F). Full-length (FL) NFL and desmin proteins bearing indicated head domain mutations were incubated under conditions suitable for assembly of IFs. Compared with native (wild-type [WT]) proteins, disease-causing mutations in the NFL and desmin head domains led to the formation of either short (NFL) or tangled (desmin) IFs (A and B, and SI Appendix, Fig. S5A). Isolated head domain samples of native (WT) or six different NFL head domain mutants were tested for formation of cross-β polymers (C). Relative to the native NFL head domain, all six head domain mutants formed polymers prematurely in the presence of 4 M urea as revealed by opalescence. Transmission electron microscopy showed no polymers formed from the native NFL head domain in urea (Materials and Methods), but clear evidence of polymers formed from the P8Q and P22R head domain mutants. Isolated head domain samples of native (WT) and six desmin head domain mutants were also tested for formation of cross-β polymers in the presence of 3 M urea (D). Relative to the native desmin head domain, all six head domain mutants formed polymers prematurely in the presence of 3 M urea. Transmission electron microscopy revealed no polymers formed from the native desmin head domain in urea (Materials and Methods), but clear evidence of polymers formed from the S7F, S12F, and S46F head domain mutants. The 2D 13C-13C ss-NMR spectra of isotopically labeled head domains of NFL and desmin show no significant differences between native (WT) proteins and indicated head domain mutants (E and F). (All scale bars: 200 nm.)
To compare native and mutant polymers in greater detail, we isotopically labeled the P8Q and P22S CMT variants of the NFL head domain with 13C and 15N, allowed for polymerization, and examined the material by ss-NMR. As shown in Fig. 7E, 2D 13C-13C ss-NMR spectra of mutant NFL head domain polymers were highly similar to the wild-type spectrum (SI Appendix, Fig. S5D). We conclude from these observations that mutation of either proline residue 8 or proline residue 22 to any of a number of different amino acids enhances the ability of the NFL head domain to self-associate in the form β-strand–enriched polymers without grossly changing protein structure.
Human genetic studies have also led to the discovery of autosomal dominant mutations localized to the head domain of desmin. Pedigree studies have associated desmin head domain mutations with cardiomyopathy (43, 44). Surprisingly, all six of these desmin head domain mutations change individual serine residues, including S2I, S7F, S12F, S13F, S46F, and S46Y. The latter five mutations change a single serine residue to either phenylalanine or tyrosine. All of these disease-causing desmin variants were expressed as recombinant protein, purified, and tested for the formation of IFs. All six mutants impeded IF assembly, yielding tangled and aggregated filaments (Fig. 7B and SI Appendix, Fig. S7A) (45).
When expressed in the context of the isolated desmin head domain and assayed for formation of β-strand–enriched polymers, all six variant proteins, including S2I, S7F, S12F, S13F, S46F, and S46Y, aberrantly polymerized in the presence of 3 M urea, just as was observed for the six different CMT-causing P8 and P22 mutations within the NFL head domain (Fig. 7D and SI Appendix, Fig. S7B). The S7F, S12F, and S46F variants of the desmin head domain were labeled with 13C and 15N, allowed to polymerize, and evaluated by ss-NMR. As shown in Fig. 7F, all three mutants exhibited 2D 13C-13C ss-NMR spectra that are high similar to the spectrum of the native desmin head domain (SI Appendix, Fig. S5E).
Discussion
Here, we described experiments focused upon the amino-terminal head domains of the NFL and desmin IF proteins. The head domains of IF proteins are of low sequence complexity and have long been believed to function in a state of intrinsic disorder (8). Evidence is shown that the NFL and desmin head domains, when studied in isolation, can form labile β-strand–enriched polymers (6). Hydrogel preparations composed of NFL head domain polymers, upon incubation with total soluble lysate from mouse brain tissue, selectively bind the endogenous NFL protein (Fig. 1). Capture of the intact NFL protein in this hydrogel binding reaction was shown to be dependent upon the presence of the NFL head domain (Fig. 2). Similarly conceived experiments, focused on the desmin head domain, yielded similar observations. We conclude from these observations that the head domains of NFL and desmin are capable of some means of self-association that is of demonstrably significant specificity.
In addition to the selective capture of the mouse brain NFL protein itself, hydrogel droplets composed of the isolated NFL head domain also captured a number of neuronal proteins known to associate with neurofilaments, including tubulin, spectrin, and MAPs (Fig. 1). Having employed a cryo-mill method of lysate preparation designed to enhance the stability of subcellular assemblies (46), it was not surprising that GFP:NFL head domain hydrogels trapped brain proteins other than NFL itself. Many of the observed hydrogel-bound proteins other than NFL have long been known to interact with neurofilaments (15, 4748495051–52). As such, we suspect that most such proteins were bound to GFP:NFL head domain hydrogels indirectly via the endogenous NFL protein itself.
Surprisingly, two RNA-binding proteins were also captured by NFL hydrogels: hnRNPM and DDX17. Both hnRNPM and DDX17 are endowed with their own LC domains. Histological studies of cultured nerve cells have revealed hnRNPM at synaptic terminals, giving evidence that this RNA-binding protein may participate in the control of localized, synaptic translation (53). We offer that interaction between RNA granules, perhaps containing hnRNPM and/or DDX17 (54), microtubules, and neurofilaments may form a multivalent nexus responsible for assisting the transport of messenger RNAs (mRNAs) to specialized regions of neurons for the purpose of localized translation.
Previously reported electron microscopic studies have revealed binding of a GFP protein linked to the low complexity domain of the FUS RNA-binding protein to vimentin IFs (6). The reported binding was observed at 45-nm intervals along the axial length of vimentin filaments, corresponding to the repetitive, circumferential organization of the vimentin head domain as it helps organize filament architecture. Knowing that RNA granules in the eggs of both flies and frogs colocalize with IFs (55, 56), the observations reported herein may be offering mechanistic insight relevant to how cytoplasmic RNA granules come to be localized in eukaryotic cells.
ss-NMR methods were used to ask whether the NFL and desmin head domains might employ molecular structure to facilitate the specificity of self-association. Our studies give evidence of β-strand−enriched interactions that define the structural interface dictating specificity of NFL and desmin head domain self-association (SI Appendix, Table S3 and Figs. 3 and 4). By use of intein chemistry facilitating segmental labeling, intact NFL and desmin polypeptides were produced bearing several different forms of isotope labeling restricted to the respective head domains of the two proteins. Subsequent to assembly into mature IFs, the isotopically labeled head domains of NFL and desmin were evaluated by ss-NMR. As a function of sequential cooling, we observed the appearance of CP-based ss-NMR spectra highly similar to spectra found in polymeric samples formed from the NFL and desmin head domains alone (Figs. 3 and 4). From these observations we offer three conclusions. First, we propose that the observed structural interactions are responsible for the specificity of NFL and desmin head domain self-interaction. Second, we propose that these interactions are responsible for the augmentative role of the respective head domains in assembly of mature NFL and desmin IFs. Third, we propose that the observed structural interactions are inherently dynamic—with their diagnostic NMR spectra becoming enhanced upon cooling (Fig. 5 and SI Appendix, Fig. S6).
Self-associative interactions specified by the NFL and desmin head domains appear, by evolutionary design, to be inherently weak and readily disrupted by phosphorylation (Fig. 6). We proposed that these head domains, even in the context of assembled IFs, are continuously moving in and out of the structurally ordered state. If so, access to enzymes affecting posttranslational modification might be facile. PKA has been reported to phosphorylate the NFL head domain on six different serine residues, including S2, S12, S41, S49, S55, and S62 (34). The desmin head domain is reported to be phosphorylated by PKA on residues S45 and S60, protein kinase C on S13, S48, and S68, and cyclin-dependent kinase on S7, S32, and T76 (39). We as yet do not know the location of these sites with respect to the β-strand–enriched structural regions of either the NFL or desmin head domains. If proximal to transiently structured regions, phosphate groups might impart repulsive charge:charge interactions incompatible with the structured state, as has been observed for DNA-dependent protein kinase-mediated phosphorylation and disruption of cross-β structural interactions formed by the FUS LC domain (17).
Quite by contrast to the hypothetically destabilizing influences of head domain phosphorylation, we conclude that disease-causing mutations in the NFL and desmin head domains enhance self-association (Fig. 7). All such mutations are autosomal dominant (40, 41, 44) and are most easily understood to elicit gain-of-function phenotypic effects. CMT-causing mutations in the NFL gene recurrently localize to proline residues 8 or 22. All such mutations were observed to enhance head domain self-association (Fig. 7 and SI Appendix, Fig. S7). Paradoxically, enhancement of NFL head domain self-interaction was observed to be detrimental to proper assembly of neurofilaments. It was similarly observed that recurrent, disease-causing mutations in the desmin head domain also enhance head domain self-association and impede IF assembly (Fig. 7 and SI Appendix, Fig. S7). At present, we remain ignorant as to why enhanced self-association of NFL and desmin head domains interferes with IF assembly.
The studies reported herein combine human genetics, biochemistry, and NMR spectroscopy to study the head domains of NFL and desmin. As such, this work is reminiscent of similarly interwoven studies on RNA-binding proteins. Idiosyncratically recurrent mutations of an analogous aspartic acid residue present in the low complexity domains of the hnRNPA1, hnRNPA2, and hnRNPDL proteins lead to various forms of neurologic disease (57, 58). The relevant aspartic acid residue of the three hnRNP proteins is localized within self-associating cross-β regions that allow for formation of both liquid-like droplets and hydrogel polymers. It has been interpreted that these aspartic acid residues impart repulsive, charge:charge interactions that are structure-destabilizing. Upon mutational change, oftentimes to valine, the destabilizing interactions are removed, leading to enhanced self-association of the hnRNP LC domains and age-related neurodegenerative disease (18, 59). We offer that human genetic studies are helping us to understand that evolution has established a proper balance of “designed instability” to self-associative, β-strand–enriched interactions that are of biologic utility to both RNA-binding proteins and IF proteins.
Five of the six disease-causing mutations in the desmin head domain change individual serine residues to either phenylalanine or tyrosine (S7F, S12F, S13F, S46F, and S46Y). The importance of aromatic amino acids to LC domain self-association was proposed and functionally confirmed in the earliest studies of phase transition of both the FG repeats of nucleoporins (60) and the LC domain of the FUS RNA-binding protein (61, 62). We speculate that the substitution of hydrophilic serine residues by either phenylalanine or tyrosine somehow accelerates or enhances desmin head domain self-association. At the same time, we appreciate the possibility that S-to-F or S-to-Y mutations might eliminate sites of phosphorylation important to the regulation of desmin assembly in living cells.
We close by asking why proline residues 8 and 22 are so important for proper function of the NFL head domain? CMT-causing mutations in the NFL protein are restricted to P8 and P22, and disease can result from mutations that change either of these proline residues to any of a number of different amino acids. Proline is an unusual amino acid residue for two reasons. First, it is an imino acid extending a covalently closed ring of three CH2 groups extending off the peptide nitrogen and back onto the adjacent α-carbon to which the side chains of the other 19 amino acids are invariably attached. This chemical conformation eliminates the opportunity for the peptide backbone of proline residues to participate in the hydrogen-bonding network essential for β-strand interactions (63). Second, the peptide bond adjacent to proline residues can rotate between the cis and trans states. The peptide bond adjacent to all other amino acids specifying biological polypeptides is invariably restricted to the trans conformation. Mutation of proline to any of the other 19 amino acids would—at the same time—restore the opportunity for β-strand hydrogen bonding and confine the adjacent peptide bond to the trans state. Deeper experimental investigation into the structure and function of the NFL head domain may allow us to probe these perplexing observations.
Materials and Methods
See SI Appendix, Materials and Methods for detailed materials and methods that describe 1) methods for protein expression and purification; 2) preparation of labile cross-β polymers and hydrogels from the desmin and NFL head domains; 3) hydrogel-binding assays; 4) assembly of segmentally labeled, full-length desmin and NFL proteins into IFs; 5) negative staining and electron microscopic analysis of both labile, cross–β polymers and assembled IFs; 6) 13C and 15N isotopic labeling of desmin and NFL head domains and segmental assembly of full-length proteins via intein chemistry; 7) sample packing and ss-NMR spectroscopy; and 8) phosphorylation of desmin and NFL head domains and full-length proteins and assays of effects on hydrogel binding and IF stability.
Acknowledgements
We thank Yonghao Yu for extensive help with mass spectrometry; Deepak Nijhawan for thoughtful advice on human genetic mutations in the NFL and desmin head domains; and the Electron Microscopy Core Facility of the University of Texas Southwestern Medical Center (supported by NIH Grant 1S10OD021685-01A1) for technical support. This work was supported by Grant 5R35GM130358 from the National Institute of General Medical Sciences and by unrestricted funding provided to S.L.M. by an anonymous donor. This work was also supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, NIH.
Data Availability
All study data are included in the article and/or SI Appendix.
References
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
 Transiently structured head domains control intermediate filament assembly
Transiently structured head domains control intermediate filament assembly
