
Edited by Gebhard F. X. Schertler, Paul Scherrer Institute, Villigen, Switzerland, and accepted by Editorial Board Member Axel T. Brunger January 11, 2021 (received for review August 2, 2020)
Author contributions: W.S., H.K., K.I., and O.N. designed research; A.H., W.S., M.K., and K.I. performed research; A.H., W.S., M.K., T.I., and K.I. analyzed data; and A.H., W.S., H.K., K.I., and O.N. wrote the paper.
1A.H. and W.S. contributed equally to this work.
- Altmetric
We present a high-resolution structure of schizorhodopsin (SzR), a new rhodopsin family identified in Asgard archaea. SzRs work as light-driven inward H+ pumps as bacterial xenorhodopsins. Although SzRs are phylogenetically located at an intermediate position between type-1 microbial rhodopsins and heliorhodopsins, the structure of SzR resembles that of bacteriorhodopsin. Notably, the cytoplasmic parts of the transmembrane helices in SzR are shorter than those in other microbial rhodopsin, and thus the putative H+ acceptor E81 is located near the cytosol. Thus, we propose a model of untrapped inward H+ release through a water-mediated transport network, which is different from xenorhodopsins, suggesting essential insights into the convergent evolution of the same molecular function in Asgard archaea and bacteria.
Schizorhodopsins (SzRs), a new rhodopsin family identified in Asgard archaea, are phylogenetically located at an intermediate position between type-1 microbial rhodopsins and heliorhodopsins. SzRs work as light-driven inward H+ pumps as xenorhodopsins in bacteria. Although E81 plays an essential role in inward H+ release, the H+ is not metastably trapped in such a putative H+ acceptor, unlike the other H+ pumps. It remains elusive why SzR exhibits different kinetic behaviors in H+ release. Here, we report the crystal structure of SzR AM_5_00977 at 2.1 Å resolution. The SzR structure superimposes well on that of bacteriorhodopsin rather than heliorhodopsin, suggesting that SzRs are classified with type-1 rhodopsins. The structure-based mutagenesis study demonstrated that the residues N100 and V103 around the β-ionone ring are essential for color tuning in SzRs. The cytoplasmic parts of transmembrane helices 2, 6, and 7 are shorter than those in the other microbial rhodopsins, and thus E81 is located near the cytosol and easily exposed to the solvent by light-induced structural change. We propose a model of untrapped inward H+ release; H+ is released through the water-mediated transport network from the retinal Schiff base to the cytosol by the side of E81. Moreover, most residues on the H+ transport pathway are not conserved between SzRs and xenorhodopsins, suggesting that they have entirely different inward H+ release mechanisms.
Microbial rhodopsins are a large family of heptahelical photoreceptive membrane proteins that use retinal as a chromophore (1). They are found in diverse microorganisms such as bacteria, archaea, algae, protists, fungi, and giant viruses (23–4). The retinal chromophore in the microbial rhodopsins undergoes all-trans to 13-cis isomerization upon light illumination, leading to a photocyclic reaction in which the proteins exert their various biological functions. Ion transporting rhodopsins are the most abundant microbial rhodopsins and are classified into light-driven ion pumps and light-gated ion channels. Whereas light-driven ion pumps actively transport ions in one direction, light-gated ion channels passively transport them according to the electrochemical potential. Ion transporting rhodopsins are used as important molecular tools in optogenetics to control neural firing in vivo. Microbial rhodopsins evolved independently from animal rhodopsins, which are also retinal-bound heptahelical proteins and a subgroup of G protein–coupled receptors. The third class of rhodopsin, heliorhodopsin (HeR), was recently reported (56–7). It has an inverted protein orientation in the membrane, as compared with microbial and animal rhodopsins (5).
Bacteriorhodopsin (BR) is the first ion pump rhodopsin found in the haloarchaeon (8) Halobacterium salinarum, and it transports protons (H+) outward. An inward chloride (Cl−) pump, halorhodopsin, was subsequently identified in the same species (9, 10) (SI Appendix, Table S1). Although an outward sodium pump rhodopsin was not found for several decades after the discovery of BR, it was eventually identified in the marine bacterium Krokinobacter eikastus in 2013 (11). These ion-pumping rhodopsins hyperpolarize the membrane by their active ion transport against the electrochemical potential of the membrane. However, the bacterial xenorhodopsins (XeRs) reportedly work as light-driven inward H+ pumps (12). Thus, the membrane potentials are not exclusively hyperpolarized via active transport by ion pumping.
Asgard archaea are the closest prokaryotic species to ancestral eukaryotes (13) and have many genes previously thought to be unique to eukaryotes. Recently, a new microbial rhodopsin group, schizorhodopsin (SzR), was found in the assembled genomes of Asgard archaea and the metagenomic sequences of unknown microbial species (14, 15) (SI Appendix, Table S1). A molecular phylogenetic analysis suggested that SzRs are located at an intermediate position between typical microbial rhodopsins, also called “type-1 rhodopsins” (16), and HeR (5). Thus they were named “schizo- (meaning “split” in Greek)” rhodopsin. Especially the transmembrane helix (TM) 3 of SzR is more similar to that of HeR than type 1. By contrast, TM6 and 7 of SzR and type-1 rhodopsins share many identical residues (e.g., W154, P158, W161, D184, and F191), which are not conserved in HeR (15). SzRs heterologously expressed in Escherichia coli and mammalian cells displayed light-driven inward H+ pump activity (15). As SzRs are phylogenetically distant from XeRs (∼18% identity and ∼44% similarity), these two rhodopsin families with similar functions are thought to have convergently evolved.
In both SzR and XeR, an H+ is released from the Schiff base linkage connecting the retinal and a conserved lysine residue (retinal Schiff base, RSB) in TM7 to the cytoplasmic side. The transiently deprotonated RSB shows a largely blue-shifted absorption peak, and this blue-shifted state was named the M-intermediate. In the case of XeR from the marine bacterium Parvularcula oceani (PoXeR), the H+ is transferred to the cytoplasmic aspartate (PoXeR D216, H+ acceptor) in TM7 and then released to the cytoplasmic bulk phase (12). By contrast, the H+ acceptor of SzR was considered to be E81 in TM3 since the mutation of E81 to glutamine abolished the inward H+ transport (15). However, the H+ is not metastably trapped in E81, probably for a kinetic reason: The rate of H+ release from E81 to the cytoplasmic bulk phase might be faster than that of H+ transfer from RSB to E81. The reason why SzR and PoXeR exhibit different kinetic behaviors in H+ release has not been elucidated. Subsequently, another H+ is taken up from the extracellular side and directly transferred from the extracellular bulk phase to the RSB during the M-decay to the initial state.
Recently, a new SzR subgroup, AntR, was identified in metagenomic data obtained from Antarctic freshwater lake samples (17). Although SzR and AntR share substantial similarities (identity: ∼33%; similarity: ∼56%), and most of the SzR residues essential for the inward H+ pump function are conserved in AntR (e.g., SzR R67, F70, C75, E81, D184, and K188), they have several differences. While the SzR E81Q mutant cannot transport H+, as mentioned above, the H+ transport efficiency of AntR E81Q is close to that of AntR wild type (WT) (17), suggesting the diversity of H+ transport mechanisms. To understand the inward H+ pump mechanism of SzR as well as the similarities and differences between SzR, XeR, and AntR we present a three-dimensional structure of an SzR.
Results
Functional Characterization of SzR4.
For the structural analysis, we screened multiple SzRs and identified SzR AM_5_00977 (GenBank accession number: TFG21677.1; hereafter called SzR4) as a promising candidate (Fig. 1A). We purified and crystallized the full-length SzR4 using in meso crystallization. Eventually, we determined the 2.1 Å resolution structure of SzR4 by molecular replacement using BR as the search model (SI Appendix, Table S2).

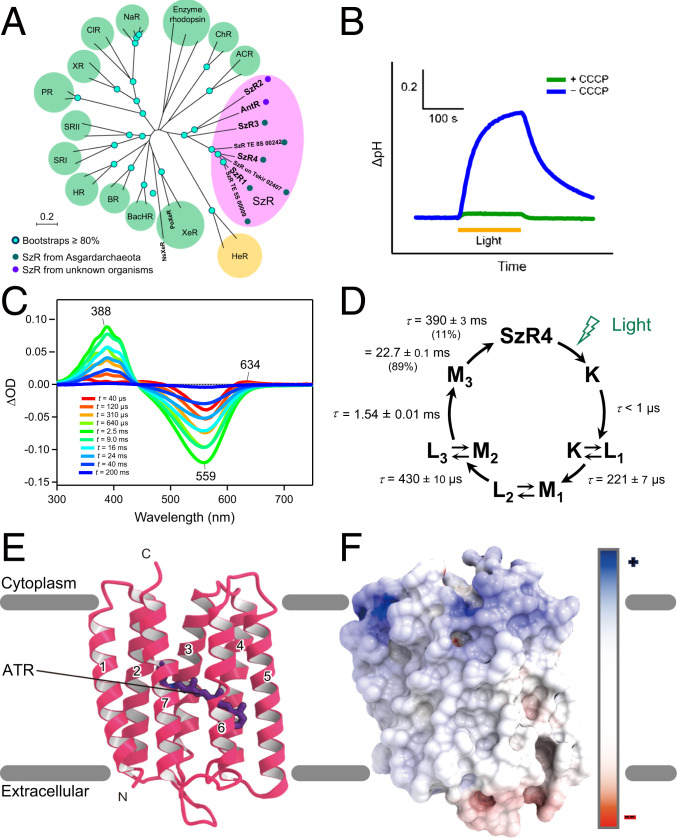
The molecular properties and overall structure of SzR4. (A) The phylogenetic tree of microbial rhodopsins. (B) H+ transport activity assay of SzR4 in E. coli cells suspended in 100 mM NaCl. Blue and green lines indicate the results in the absence and presence of carbonyl cyanide m‐chlorophenylhydrazone (CCCP), respectively. (C) Transient absorption spectra of SzR4 in 100 mM NaCl, 20 mM Tris HCl, pH 8.0, and POPE/POPG (molar ratio 3:1) vesicles with a lipid to protein molar ratio = 50. (D) The photocycle of SzR4 based on the fitting shown in SI Appendix, Fig. S1D and a kinetic model assuming a sequential photocycle. The lifetime (τ) of each intermediate is indicated by mean ± SD. The numbers in parentheses indicate the fraction of the M-intermediate decayed with each lifetime in its double exponential decay. (E) Ribbon diagram of the SzR4 structure, viewed from the membrane plane. (F) Electrostatic surface viewed from the membrane plane. Red and blue correspond to potentials of −8 kT e−1 and 8 kT e−1, respectively.
We first characterized the biochemical properties of SzR4. The phylogenetic tree of microbial rhodopsins indicated that SzR4 belongs to the SzR family, which is far from XeR, and it is close to the previously characterized SzR1 (Fig. 1A). To investigate the ion transport function of SzR4, we exposed SzR4-expressing E. coli to visible light and observed alkalization of the external solvent (Fig. 1B). The alkalization was largely eliminated by the addition of a protonophore, 10 μM carbonyl cyanide m‐chlorophenylhydrazone, suggesting that SzR4 functions as an inward H+ pump, as reported previously (15). The purified SzR4 showed a maximum absorption wavelength (λmax) at 557 nm, identical to that of SzR1 (15) (SI Appendix, Fig. S1A). The absorption peak in the visible wavelength region was decreased at higher pH, and another peak appeared in the ultraviolet (UV) region (λmax = 388 nm) (SI Appendix, Fig. S1 B and C). The latter represents the deprotonation of the RSB (15), and its acid dissociation constant (pKa) was 12.5 ± 0.2 (mean ± SD). This is one unit smaller than that of SzR1, suggesting that the protonated RSB is less stabilized in SzR4.
To investigate the photocycle of SzR4, we performed a laser flash photolysis experiment with SzR4 in lipid vesicles. Transient absorptions representing the accumulations of K, L, and M-intermediates were observed, as in the photocycle of SzR1 (15) (Fig. 1C and SI Appendix, Fig. S1D). The sum of five exponential functions effectively reproduced the time evolution of the transient absorption change of SzR4. The absorption spectra of the initial state and four photointermediates (K/L1, L2/M1, L3/M2, and M3) and the photocycle of SzR4 were determined (SI Appendix, Fig. S1E and Fig. 1D). The overall photoreaction cycle of SzR4 is similar to that of SzR1 (15). A large accumulation of the M-intermediate was observed in the millisecond region, representing the deprotonated state of the RSB. An equilibrium exists between M1 and M2, with L at different equilibrium constants, and it is more biased toward the M for L3/M2 than for L2/M1. Notably, the absolute spectra of the three M states were substantially different. Specifically, the vibrational structure observed in M2 was less pronounced in M1 and M3 (SI Appendix, Fig. S1D). This spectral change would originate from a large conformational change of the protein around the retinal chromophore and be associated with the conversion from the inward opened to outward opened state. The H+ release to the cytoplasmic side is not finished until the M3 formation, and thus a new H+ is taken up from the extracellular side during the M3 decay (13). A similar spectral change in the vibrational structures between two M states was also reported for XeR from Nanosalina (NsXeR) (18), suggesting that a comparable conformational change also occurs between the H+ release and uptake processes in SzR and XeR.
Overall Structure of SzR4.
The crystallographic asymmetric unit contains three molecules (molecules A, B, and C) (SI Appendix, Fig. S1 F and G). The overall architectures of these three molecules are almost identical, and thus we focused on the molecule A structure. SzR4 consists of seven TMs and six loops (extracellular loops 1 through 3 and intracellular loops 1 through 3) (Fig. 1E). Three residues after H199 are disordered in the crystal structure. Extracellular loop 1 (residues G51 through Y64) contains two short antiparallel β-strands. All-trans retinal (ATR) is covalently bound to K188, forming the RSB as in other microbial rhodopsins.
A previous immunostaining analysis revealed that the C terminus of SzR1 is oriented toward the cytoplasmic side (15), as in the type-1 rhodopsins, which is opposite to the HeRs. Many positively and negatively charged residues are present on the cytoplasmic and extracellular faces, respectively, in the SzR4 structure (Fig. 1F). This electrostatic surface is consistent with the positive-inside rule (19) and also supports its topology.
The three SzR4 molecules in the asymmetric unit form a trimer in the crystal structure, in excellent agreement with the previous high speed atomic force microscope observation (15). TM1 and TM2 of one protomer interact with TM4′ and TM5′ of the adjacent protomer, creating the trimer interface (Fig. 2A and SI Appendix, Fig. S1H). The interface comprises mainly hydrophobic residues (SI Appendix, Fig. S1 I and J), and several hydrogen-bonding interactions are observed on the cytoplasmic side. The residues at the interface are conserved among SzRs (SI Appendix, Fig. S1K), suggesting that most of SzRs function as trimers.

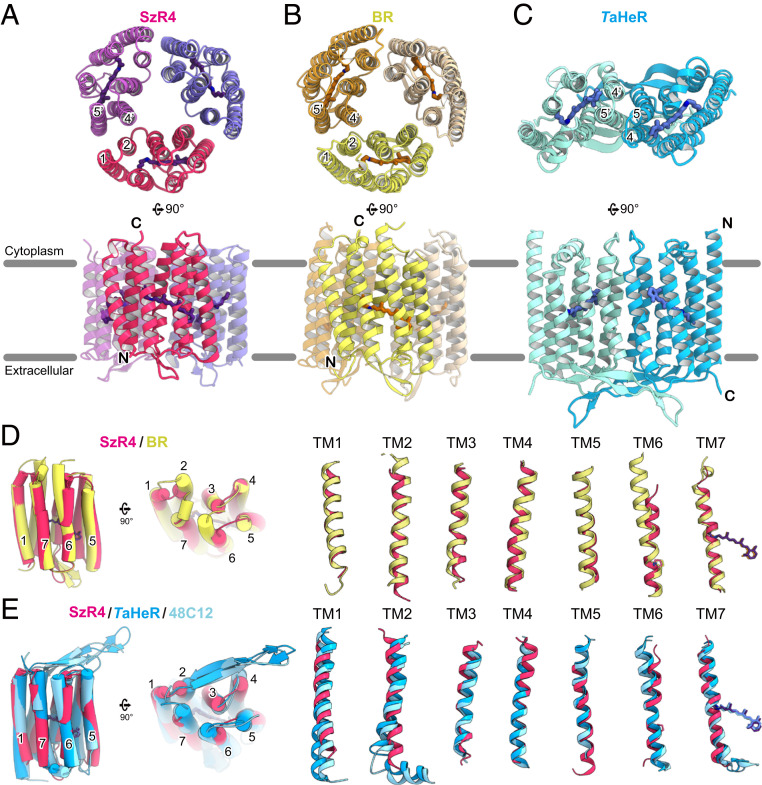
Comparison of SzR4, BR, and HeR. (A–C) Monomer and oligomeric structures of SzR4 (A), BR (PDB ID code 1M0L) (B), and TaHeR (PDB ID code 6IS6) (C) colored magenta, yellow, and dark turquoise. (D) Superimpositions of the SzR4 and BR structures. Individual TM helices are shown after superimposition of the two rhodopsins. (E) Superimpositions of the SzR4, TaHeR, and HeR 48C12 (PDB ID code 6SU3) structures. Individual TM helices are shown after superimposition of the two rhodopsins.
To determine whether SzRs are classified as either type-1 rhodopsins or HeRs, we compared the structures of SzR4, BR, and TaHeR (20, 21). SzR4 and BR similarly form trimers, while TaHeR forms a dimer (Fig. 2 A–C). The SzR4 and BR structures also have the same configuration of TMs forming trimeric binding interfaces, with TM1 and TM2 of one monomer creating a binding interface with TM4′ and TM5′ of the adjacent monomer. SzR4 and BR also share a common orientation relative to the membrane. Moreover, the monomer structure of SzR4 superimposes well on that of BR [Protein Data Bank (PDB) identification (ID) code 1M0L (22), rmsd of Cα atoms = 1.27 Å] (Fig. 2D). By contrast, the orientations of SzR4 and TaHeR are reversed in the membrane. When the N and C termini of SzR4 and TaHeR are aligned and their monomeric structures are superimposed, the slope and length of each TM do not overlap well [PDB ID code 6IS6 (20), rmsd of Cα atoms = 1.96 Å] (Fig. 2E). This is similar in comparison with the structure of bacterial HeR 48C12 [PDB ID code 6SU3 (23), rmsd of Cα atoms = 1.93 Å]. Overall, although SzR4 has ∼20% sequence identity to both BR and HeR, it is structurally more similar to BR. Hence, we suggest that SzRs belong to the type-1 rhodopsins.
We next compared the SzR4 and BR structures in detail. Each TM overlaps relatively well, and their ECL1 similarly contain antiparallel β-strands (Fig. 2D and SI Appendix, Fig. S2). However, there is a striking difference on the cytoplasmic side. The C terminus of BR contains a short α-helix and is directed toward the center of the protein, while the C terminus of SzR4 is disordered. Moreover, TM2 and TM6 of SzR4 are shorter than those of BR by one and two α-helical turns, respectively. Notably, the length between the conserved Pro and the cytoplasmic end of TM6 in SzR4 is 13 residues, while those in other type-1 rhodopsins are about 21 residues. Thus, the cytoplasmic part of TM6 in SzR4 is the shortest among the microbial rhodopsins (SI Appendix, Fig. S3 A and B). The sequence alignment of SzRs revealed that the shorter length of TM6 is highly conserved (SI Appendix, Fig. S2), and thus it is a unique structural feature of SzRs.
Retinal Binding Site.
We next describe the counterion interaction in SzR4 (Fig. 3A). In typical type-1 rhodopsins, two negatively charged residues (e.g., D85 and D212 in BR) function as counterions, and a water molecule (e.g., water402 in BR) bridges the RSB and the counterions via hydrogen-bonding interactions (SI Appendix, Fig. S3C). In SzR4, F70 and D184 are homologous to D85 and D212 in BR, respectively. D184 forms a direct salt bridge with the RSB and functions as a single counterion (Fig. 3A and SI Appendix, Fig. S3D), stabilizing the high pKa of the RSB (12.5, SI Appendix, Fig. S1C). The single counterion is similar in XeRs and HeRs, while the relative positions are different (SI Appendix, Fig. S3 E and F). The counterion D184 of RSB in SzR4 would have a large color-tuning effect on the absorption of the retinal chromophore, as in the other type-1 rhodopsins. However, the D184N mutation induced the loss of the color of the protein and H+ transport activity, indicating the deprotonation of RSB occurred (SI Appendix, Fig. S5 A and B). Since D184 is the only counterion in SzR4 in contrast to the double counterions in BR, the mutation of this residue to a noncharged amino acid probably no longer stabilizes the protonated state of Schiff base linkage, and the color shift by the mutation could not be determined.

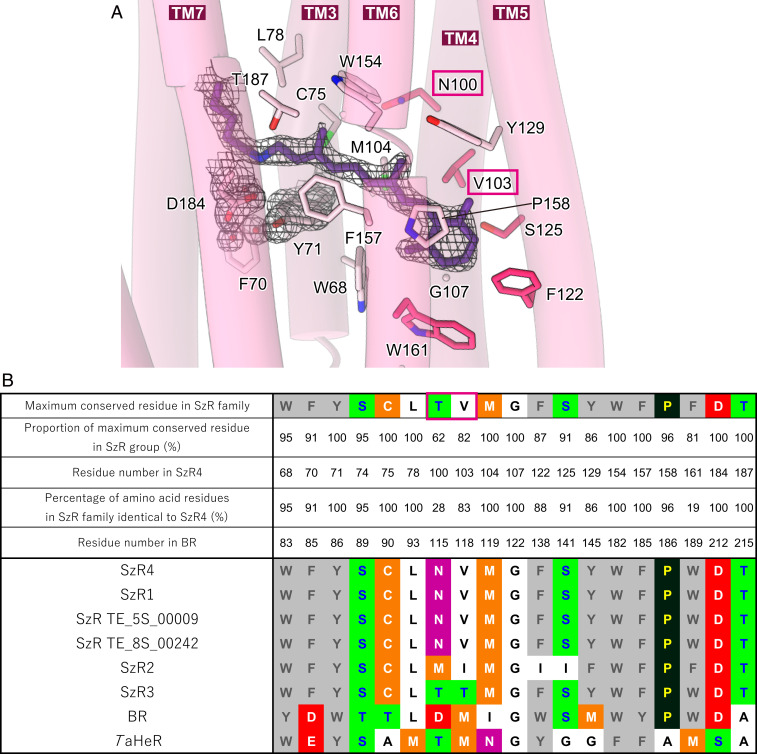
Conservation of retinal binding site. (A) The retinal chromophore and the residues within 4.5 Å involved in retinal binding. 2Fo-Fc map around the counterion is overlayed, contoured at 2.0σ. Black dashed lines indicate hydrogen-bonding interactions. Red boxes indicate the residues that play a critical role in color tuning. (B) Maximally conserved residues around retinal and their percentages in SzR family members, with residue numbering according to SzR4. The variations of the amino acid types in six SzRs, BR, and TaHeR are shown in the lower part.
In the current SzR4 structure, there is no electron density corresponding to water402 in BR. Around the RSB–counterion complex, three water molecules are present in the space opened by the flipping of the F70 side chain (Fig. 3A and SI Appendix, Fig. S3D). While these water molecules form an extensive polar interaction network, the RSB complex does not form any hydrogen-bonding interactions with them. This is in excellent agreement with the previous Fourier transform infrared (FTIR) analysis of SzR1, which indicated the presence of several water molecules around the chromophore that are not strongly hydrogen bonded (15). This strongly hydrogen-bonded water molecule is observed in all outward H+ pumping rhodopsins (24) and the XeRs, PoXeR (25) and NsXeR (18). Thus, the absence of the strongly hydrogen-bonded water molecule is a unique structural feature of SzR4 among the H+ pumping rhodopsins and might be associated with its function.
The other residues in the retinal binding pocket are mainly hydrophobic. Notably, the aromatic residues Y71 and W154 closely contact the C10-C13 moiety of the retinal from below and above, respectively, allowing the all-trans to 13-cis isomerization (15). These residues are completely conserved in 85 SzR homologs (Fig. 3B). The equivalent residues are two tryptophan residues in type-1 rhodopsins and tyrosine and phenylalanine residues in HeRs. From this viewpoint, SzR is in between BR and HeR. Among the residues constituting the retinal binding pocket, residues 7 and 3 of SzR4 are conserved in BR and TaHeR, respectively. Thus, the retinal binding pocket of SzR is similar to that of type-1 rhodopsins rather than HeRs,
Color-Tuning Mechanism among SzRs.
The environment around the retinal chromophore is closely associated with the absorption wavelength of rhodopsins. The purified SzR4 displayed the λmax at 557 nm, which is identical to that of SzR1, and 555 nm when expressed in E. coli cells (SI Appendix, Figs. S1A and S4A). Notably, SzR2 and SzR3 showed blue-shifted absorptions at 542 and 540 nm, respectively (SI Appendix, Fig. S4 B and C). To investigate the color-tuning mechanism in SzRs, we compared the residues constituting the retinal binding pocket between the six homologs of SzRs (Fig. 3B). These residues are entirely conserved in SzR4, SzR1, SzR TE_5S_00009, and SzR TE_8s_00242. Comparing SzR4, SzR2, and SzR3, the residues around the polyene chain are entirely conserved, whereas those around the β-ionone ring are more diverged. V103, F122, S125, and W161 in SzR4 are replaced in SzR2, and V103 is replaced in SzR3. Moreover, in the vicinity of the β-ionone ring, N100 in SzR4 is replaced with M and T in SzR2 and SzR3, respectively. In BR, D115 is present at the homologous position. BR D115N and D115A showed 2 and 11 nm blue shifts as compared with the WT, respectively (26, 27), and thus a different amino acid at this position would generate distinct λmax values among SzR4, SzR2, and SzR3.
To determine the residues responsible for the color tuning, we comprehensively swapped the residues around the β-ionone ring between SzR4 to SzR2 and SzR4 to SzR3 and measured the λmax values of the swapped mutants (SI Appendix, Fig. S4 A–C). The mutants of SzR4 to SzR2 type (SzR4 N100M) and SzR3 type (SzR4 N100T) showed 11 and 4 nm blue shifts, respectively. By contrast, 1 and 3 nm red-shifted absorptions were observed for SzR2 M101N and SzR3 T103N. These results suggest that the difference in the amino acid at the SzR4 N100 position is one of the color-tuning factors, as in type-1 rhodopsins. Moreover, the mutation of SzR4 V103 to the SzR2-type residue (I) induced a 4 nm blue shift, while the λmax of SzR2 I104V was 3 nm longer as compared to SzR2 WT. Hence, V103 near the β-ionone ring in SzR2 also contributes to the absorption difference from SzR4. A methionine is present at this position in BR (M118) and most type-1 rhodopsins. The mutation of this residue to a smaller amino acid allows the rotation of the C6-C7 bond of retinal, connecting the β-ionone and polyene chain, and causes blue-shifted λmax values in channelrhodopsin and archaerhodopsin-3 (28). This result suggests that the replacement of the smaller valine with the larger isoleucine at this position in SzRs would generate blue-shifted λmax values as in type-1 rhodopsins.
Overall, the structure-based mutagenesis study demonstrated that the amino acid differences in N100 and V103 are essential factors for color tuning in SzRs (Fig. 3A). N100 and V103 are conserved in 61 and 82% of the 85 SzR homologs (Fig. 3B), respectively, and are less conserved as compared to the other residues in the retinal binding site. Thus, these differences create the diversity of the absorption spectra in SzRs.
SzR4 P158 in TM5 and T187 in TM7 are highly conserved in 96 and 100% of the SzRs (Fig. 3B), and the homologous residues in type-1 rhodopsins play a color-tuning role (29, 30). Mutating the former to threonine or the latter to alanine makes λmax longer for many type-1 rhodopsins. To determine whether these color-tuning rules also apply in SzR4, we constructed the SzR4 P158T and T184A mutants (SI Appendix, Fig. S4A). SzR4 P158T showed a 2 nm shorter λmax than that of SzR4 WT, suggesting that the color-tuning rule at this position is different between SzR and type-1 rhodopsins. By contrast, SzR4 T184A displayed a 7 nm red-shifted λmax as compared to that of SzR4 WT. A similar red shift by the mutation of an −OH-bearing residue at the same position was reported in several type-1 rhodopsins (21, 22), and SzR4 T184 has a similar effect on the excitation energy of the retinal π-electron.
Insight into Inward H+ Transport.
To investigate the mechanism of inward H+ transport, we compared the SzR4 and BR structures. In the outward H+ pumping BR, an H+ is transferred from RSB to the H+ acceptor D85 in the early stage of the photocycle at ∼10−5 sec, and this H+ is finally released to the extracellular milieu via a H+ release group, consisting of E194, E204, and a hydrating water between them (31) (Fig. 4A). However, D85 is replaced with the hydrophobic residue F70 in SzR4. The F70 side chain is directed toward the membrane environment and not involved in the interaction with the RSB (Fig. 4 B and C and SI Appendix, Fig. S3D). Moreover, the extracellular H+ acceptors E194 and E204 in BR are replaced with A168 and T176 in SzR4, respectively. There is no other specific extracellular H+ acceptor in SzR4. The SzR4 counterion D184 forms salt bridges with the RSB and R67 (Fig. 4B and SI Appendix, Fig. S3D), maintaining the low pKa of D184 and preventing its protonation. These structural features prove that SzR cannot work as an outward H+ pump.

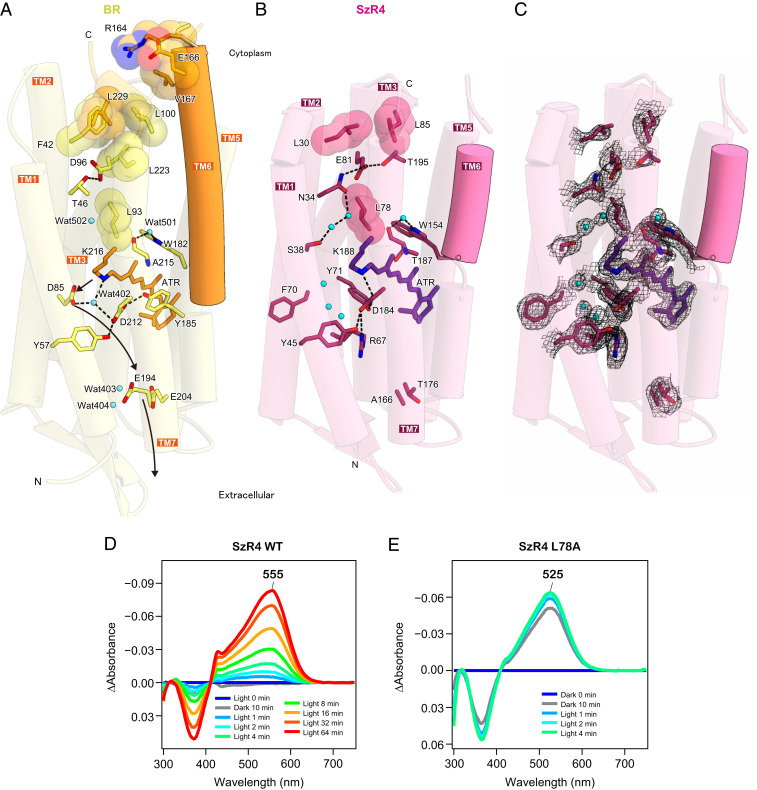
Essential residues for inward H+ uptake. (A and B) Essential residues for H+ transfer in BR (A) and SzR4 (B). Waters are shown as cyan spheres. Black dashed lines indicate hydrogen-bonding interactions. (C) 2Fo-Fc map for the essential residues in SzR4, contoured at 1.0σ. (D and E) The difference in absorption spectra before and after HA bleaching reactions of SzR4 WT (D) and SzR4 L78A (E) in solubilized E. coli membranes. The λmax of each SzR and mutant was determined by the positions of the absorption peaks of the original proteins indicated in each panel, and the absorption of retinal oxime produced by the hydrolysis reaction of RSB and HA was observed as peaks around 360 to 370 nm. The reaction was first performed in the dark for 10 min and then exposed to light for up to 64 min. Whereas no detectable bleaching of the visible region was observed for SzR4 WT in the first 10 min of the reaction in the dark, ca. 70% protein was bleached for SzR4 L78A during the same time period.
On the cytoplasmic side, E81 forms hydrogen bonds with N34 and T195, stabilizing its low pKa and negative charge (Fig. 4 B and C). In BR, the equivalent residue D96 works as a cytoplasmic H+ donor, supplying an H+ to the deprotonated RSB in the M-intermediate during the outward H+ pump cycle (1) (Fig. 4A). In SzRs, E81 plays a critical role in the H+ release process upon M-formation during the inward H+ pump cycle. The E81Q mutant of SzR4 lost the H+ transport activity, whereas the E81D mutant retained (SI Appendix, Fig. S5 A and B), suggesting that the negative charge of E81 plays an essential role in the H+ transport activity, as in SzR1.
However, a previous FTIR analysis indicated that the H+ is not metastably trapped by E81 in the L/M-intermediate of SzR1, unlike PoXeR (12). Instead, it is directly released into the cytoplasmic milieu in SzR and does not interact with the protein in the L/M state (15). In the current SzR4 structure, E81 is closer to the cytosol, since the cytoplasmic parts of TMs 2, 6, and 7 are shorter than those in the other type-1 rhodopsins, as described above (Fig. 2D and SI Appendix, Fig. S3 A and B). E81 is separated from the solvent by only two leucine residues, L30 and L85, and easily exposed to the solvent by the light-induced structural change. An H+ would be attracted to the negative charge of E81 and released to the cytoplasm through the solvent water molecules.
What light-induced structural changes enable the inward H+ release? A recent time-resolved study of BR with millisecond time resolution (32) has shown that the rotation of L93 opens the hydrophobic barrier between the RSB and D96 (Fig. 4A), creating space for the three water molecules that connect them. This structural change allows the H+ transfer to the RSB. In SzR4, the homologous residue L78 also forms the hydrophobic barrier between the RSB and E81 (Fig. 4B). Around L78, three hydrating waters exist in the SzR4 structure, as in BR (Fig. 4A). Thus, a similar rotation of L78 would create a water-mediated transport network from the RSB to the cytosol, with the H+ released to the cytoplasm through the network.
To investigate the importance of L78 for inward H+ transport, we constructed the L78A mutant. No pH change was observed upon light illumination of E. coli cells expressing SzR4 L78A (SI Appendix, Fig. S5 A and B), suggesting that L78 plays a critical role in the inward H+ transport function. A homologous leucine is conserved in BR (L93), and the BR L93A mutant results in two orders of magnitude longer photocycle compared to the WT (33). Hence, we suggest that the loss of function we observe for the L78A mutant in SzR4 is due to a similar interruption of the photocycle. It would be interesting to confirm this point in future time-resolved measurements that are beyond the scope of the current study. Furthermore, the RSB in SzR4 WT was not hydrolyzed by hydroxylamine (HA) in the dark (Fig. 4D), whereas that in SzR4 L78A was breached by HA even without light exposure (Fig. 4E). In this mutant, the RSB would be more accessible to external solvents on the cytoplasmic side and small hydrophilic molecules such as HA. This result supports the solvent access to E81 during the photocycle.
Working Model of Untrapped Inward H+ Release.
We mutated the residues on a putative H+ transport pathway in SzR4, and all of the mutations reduced the H+ transport activity (Fig. 5A and SI Appendix, Fig. S5 A and B). Notably, the mutants of the residues in TM3 completely lost the transport activity (S74A, C75A, C75S, C75T, L78A, and E81Q), while those in TM 2, 4, 6, and 7 retained the transport activity itself, except for the counterion mutant (D184N). These results suggest the functional importance of TM3 on the inward H+ transport. Moreover, previous time-resolved studies of BR with femtosecond time resolution (34, 35) reported a movement of TM3 that coincides with the deprotonation of the RSB in BR. Given the structural similarity between BR and SzR4, light-induced structural rearrangements in TM3 could trigger the large structural rearrangement of SzR4, allowing the inward H+ transport.

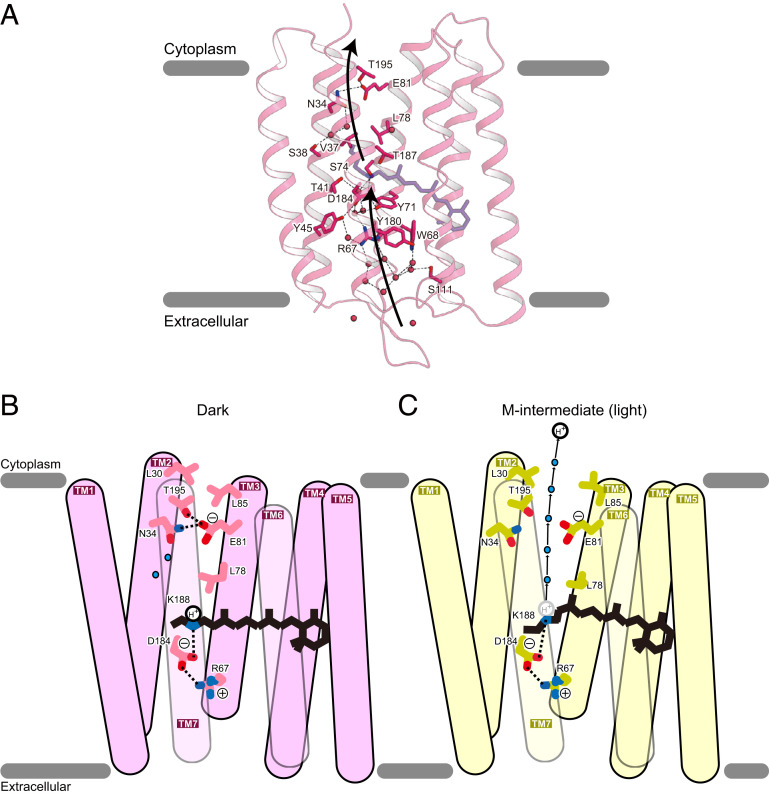
The working model of inward H+ release by SzR. (A) Putative ion translocation pathway in SzR4. Black dashed lines indicate hydrogen bonds. (B and C) Models of dark (B) and M-intermediate (C) of SzR4. Water molecules are shown as cyan spheres. Black dashed lines indicate hydrogen-bonding interactions.
Integrating these findings, we propose a structure-based working model of inward H+ release (Fig. 5 B and C). During rise of the M-intermediate, the protein moieties, including TM3, undergo structural changes, disrupting the hydrogen-bonding network around E81 and the two hydrophobic barriers above and below E81. Thus, a water-mediated transport network is formed between the RSB to the cytosol. Then, the RSB is deprotonated, and the H+ is released to the solvent through the network, attracted by the negative charge of E81. We refer to this mechanism as “untrapped inward H+ release.”
To inwardly uptake an H+, the deprotonated RSB should be reprotonated from the extracellular milieu. In PoXeR, the branched thermal isomerization of retinal from the 13-cis–15-anti to all-trans–15-anti and 13-cis–15-syn configurations is the rate-limiting process for the reprotonation of RSB during the M-decay (36). The 13-cis–15-anti to all-trans–15-anti isomerization changes the inward-directed orientation of the lone pair on the nitrogen atom of RSB toward the outward-directed one, and the H+ can access the RSB from the extracellular side. An extensive hydrogen-bonding network, including seven hydrating water molecules, exists between the RSB and the extracellular side (Fig. 5A). Since there is no specific extracellular H+ donor, as suggested by the comprehensive mutations of SzR1 (15), the H+ is directly taken up from the extracellular milieu simultaneously with the thermal isomerization of the retinal chromophore in the M-decay through this hydrogen-bonding network as in PoXeR (36). Because the configuration of the retinal in SzR in the M state is unknown, it is not known whether thermal isomerization to the all-trans–15-anti form occurs before the reprotonation or at the same time, as in PoXeR, and further studies are needed.
Comparison of SzR4 and NsXeR.
SzRs and XeR similarly function as inward H+ pumps (12, 15) despite their distant sequence similarity. To explore their common structural features as inward H+ pumps, we compared the SzR4 and NsXeR structures. As described above, SzR4 has the shortest TM6 (Fig. 2D and SI Appendix, Fig. S3 A and B), and it enables the “untrapped” inward H+ release. However, TM 2, 5, and 6 of NsXeR are longer than those of SzR4 by over two turns (SI Appendix, Fig. S6 A and B). Unlike SzR4, the N terminus and ICL1 in NsXeR contain characteristic α-helices. SzR4 superimposes well on BR [PDB ID code 1M0L (22), rmsd of Cα atoms = 1.26 Å] rather than NsXeR [PDB ID code 6EYU (18), rmsd of Cα atoms = 1.56 Å]. At the secondary structure level, SzR4 and NsXeR do not share common structural features as inward H+ pumps.
We also compared the H+ transport pathways in SzR4 and NsXeR (SI Appendix, Fig. S6 C and D). F70 and D184 in SzR4 are replaced with D76 and P209 in NsXeR, respectively. Thus, SzR4 and NsXeR similarly have a single counterion, although its relative position in the structure is different. The protonation of BR D85 was suggested to catalyze the 13-cis to all-trans isomerization during the N → O → BR transition (37). In SzR and AntR, a phenylalanine is completely conserved at this position, and while the mutation of F70 in SzR1 to alanine did not alter the inward H+ transport efficiency, the mutations to an aspartic acid or glutamic acid completely eliminated the inward H+ transport (15). This implies the electric neutralization at this position would be essential for the fast cycling and high H+ transport efficiency of SzR and AntR.
The smaller negative charge on the extracellular side of the RSB, as compared to the outward H+ pumps with double counterions, would increase the directionality of H+ transfer to the cytoplasmic H+ acceptor relative to the extracellular counterion. NsXeR has the H+ acceptors H48 and D220 on the cytoplasmic side, and the H+ is trapped by these residues in the M state. However, these residues are not conserved in SzR4, and the H+ is not trapped in the M state. Moreover, the other residues on the H+ transport pathway are not conserved between SzR4 and NsXeR, suggesting that they have entirely different inward H+ release mechanisms. Nevertheless, SzR4 and NsXeR similarly have the extensive water-mediated hydrogen-bonding network in their extracellular halves, and the H+ is easy to access from the extracellular milieu to the RSB.
Discussion
Our SzR4 structure offers numerous insights into the structure–function relationships and color-tuning mechanisms of the SzR family members. Although SzRs are phylogenetically located at an intermediate position between type-1 rhodopsins and HeRs, SzRs are structurally similar to type-1 rhodopsins, and we classified SzRs with them. Since the cytoplasmic part of TM6 is the shortest among the microbial rhodopsins (Fig. 2D and SI Appendix, Fig. S3 A and B), E81 is located near the cytosol and plays a critical role in the inward H+ transport (Fig. 4B). Since E81 is homologous to BR D96, which is protonated in the dark state, the protonation state of E81 is needed to be carefully considered. In many type-1 rhodopsins, it is known that the position of infrared absorption peak of the stretching vibration of C = O group of protonated aspartic acid or glutamic acid sensitively shifts upon the conformational change of the protein, even without the change in the protonation state (38, 39). However, no appearance and/or shift of the IR peak of protonated carboxylic acid in the different FTIR spectra of all photointermediates of SzR1 suggests that E81 is deprotonated both in the dark and during the photocycle (15). Given that the H+ is not trapped in E81, light-induced structural changes would displace the rotamer of L78, releasing the H+ to the solvent water molecules, attracted by the negative charge of E81 (Fig. 5C).
By contrast, in AntR, the H+ transport rate of E81Q is reportedly similar to that of the WT (17), indicating that the negative charge of E81 is not essential for its function. To understand the H+ uptake mechanism of AntR, we constructed a homology model of AntR based on the SzR4 structure (SI Appendix, Fig. S7). In this model, similar to D96 in BR, E81 does not form any polar interactions and is surrounded by hydrophobic residues. Thus, E81 would be protonated and not associated with the proton transport in AntR. Notably, D30 forms a hydrogen-bonding network with R84 and Y193, which are located somewhat closer to the cytoplasmic side as compared with E81. These residues are unique in AntR (SI Appendix, Figs. S2 and S7). A previous study demonstrated that the R84A mutant retains the H+ transport activity, whereas M-formation is absent in the photocycle of the Y193F mutant. These observations suggest that the hydrogen-bonding interaction between D30 and Y193 is critical in the photoactivation of AntR. Instead of E81, D30 might be deprotonated and negatively charged and thus play an essential role in the inward H+ release by AntR. The homology model of AntR represents the diversity of the inward H+ transport mechanisms among the SzR/AntR family members.
Materials and Methods
Expression and Purification.
The gene encoding SzR4 (GenBank ID: TFG21677.1), with codons optimized for an E. coli expression system, was synthesized (Genscript) and subcloned into the pET21a(+) vector with the resulting construct encoding a TEV cleavage site followed by a GFP-His10 tag at the C terminus. The protein was expressed in E. coli C41(Rosetta). Protein expression was induced by 1 mM isopropyl β-D-thiogalactopyranoside for 20 h at 25 °C, and then the culture was supplemented with 10 μM all-trans retinal (Sigma Aldrich). The harvested cells were disrupted by sonication in buffer containing 20 mM Tris HCl (pH 7.5) and 20% glycerol. The crude membrane fraction was collected by ultracentrifugation at 180,000 g for 1 h. The membrane fraction was solubilized for 1 h at 4 °C in buffer containing 20 mM Tris HCl (pH 7.5), 150 mM NaCl, 1% n-dodecyl-β-d-maltoside (DDM), and 10% glycerol. The supernatant was separated from the insoluble material by ultracentrifugation at 180,000 g for 20 min and incubated with TALON resin (Clontech) for 30 min. The resin was washed with 10 column volumes of buffer containing 20 mM Tris HCl (pH 7.5), 500 mM NaCl, 0.03% DDM, and 15 mM imidazole. The protein was eluted in buffer containing 20 mM Tris HCl (pH 7.5), 500 mM NaCl, 0.03% DDM, and 200 mM imidazole. The eluate was treated with TEV protease and dialyzed overnight. The cleaved GFP and the protease were removed with Co2+-NTA resin. The flowthrough was concentrated and loaded onto a Superdex200 10/300 Increase size-exclusion column and equilibrated in buffer containing 20 mM Tris HCl (pH 7.5), 150 mM NaCl, and 0.03% DDM. Peak fractions were pooled, concentrated to 30 mg ⋅ mL−1 using a centrifugal filter device (Millipore 50 kDa molecular weight cutoff), and frozen until crystallization.
Crystallization.
As previously described (40), the protein was crystallized in lipidic cubic phase (LCP). The protein was reconstituted into monoolein at a weight ratio of 1:1.5 (protein:lipid). The protein-laden mesophase was dispensed into 96-well glass plates in 30 nL drops and overlaid with 800 nL precipitant solution using a Gryphon robot (ARI). Crystals of SzR4 were grown at 20 °C in precipitant conditions containing 20% PEG500DME, 100 mM Na-acetate, pH 4.75, 250 mM MgSO4, and 10 mM ZnCl2. The crystals were harvested directly from the LCP using micromounts (MiTeGen) or LithoLoops (Protein Wave) and frozen in liquid nitrogen without adding any extra cryoprotectant.
Data Collection and Structure Determination.
X-ray diffraction data were collected at the SPring-8 beamline BL32XU with an EIGER X 9M detector (Dectris) using a wavelength of 1.0 Å. In total, 148 small-wedge (10° per crystal) datasets were obtained using a 15 × 10 μm2 beam. The collected images were processed with KAMO (https://github.com/keitaroyam/yamtbx/blob/master/doc/kamo-en.md) (41). Each dataset was indexed and integrated with XDS (42) and then subjected to a hierarchical clustering analysis based on the unit cell parameters using BLEND. After outlier rejection, 127 datasets were finally merged with XSCALE (42). The SzR4 structure was determined by molecular replacement with PHASER (43) using the structure of BR (PDB code: 1M0K) (22). Subsequently, the model was rebuilt and refined using COOT (44) and phenix.refine (45). Figures were prepared with CueMol (www.cuemol.org/ja/).
Laser Flash Photolysis.
For the laser flash photolysis measurement, SzR4 was purified and reconstituted into a mixture of POPE (Avanti Polar Lipids) and POPG (sodium salt, Avanti Polar Lipids) (molar ratio = 3:1), with a protein to lipid molar ratio of 1:50, in buffer containing 20 mM Tris HCl (pH 8.0) and 100 mM NaCl. The absorption of the protein solution was adjusted to 0.8 to 0.9 (total protein concentration: ∼0.25 mg ⋅ mL–1) at an excitation wavelength of 532 nm. The sample was illuminated with a beam of the second harmonics of a nanosecond-pulsed Nd3+-YAG laser (λ = 532 nm, 3 mJ pulse–1, and 1 Hz) (INDI40; Spectra-Physics). The transient absorption spectra were obtained by monitoring the intensity change of white-light from a Xe-arc lamp (L9289-01, Hamamatsu Photonics) passed through the sample, with an ICCD linear array detector (C8808-01, Hamamatsu). To increase the signal-to-noise (S/N) ratio, 90 spectra were averaged, and a singular value decomposition analysis was applied. To measure the time evolutions of transient absorption changes at specific wavelengths, the light from the Xe-arc lamp (L9289-01, Hamamatsu Photonics) was monochromated with a monochromater (S-10, SOMA OPTICS), and the change in the intensity after photoexcitation was monitored with a photomultiplier tube (R10699, Hamamatsu Photonics) equipped with a notch filter (532 nm, bandwidth = 17 nm, Semrock) to remove the scattered pump pulses. To increase the S/N ratio, 100 signals were averaged.
Measurement of Absorption Maximum Wavelength by HA Bleaching.
The λmax values of the WT and mutants of SzR4, SzR2, and SzR3 were determined by bleaching the protein with HA, according to the previously reported method (30). E. coli cells expressing rhodopsins were washed three times with buffer containing 50 mM Na2HPO4 (pH 7.0) and 100 mM NaCl. The washed cells were treated with 1 mM lysozyme for 1 h at room temperature and then disrupted by sonication. To solubilize the rhodopsins, 3% DDM was added and the samples were stirred overnight at 4 °C. The rhodopsins were bleached with 500 mM HA in the dark or under yellow light illumination (λ > 500 nm) from the output of a 1 kW tungsten halogen projector lamp (Master HILUX-HR, Rikagaku) passed through a glass filter (Y-52, AGC Techno Glass). The absorption change upon bleaching was measured by a UV-visible spectrometer (V-730, JASCO) equipped with an integrating sphere (ISV-922, JASCO).
Acknowledgements
The diffraction experiments were performed at SPring-8 BL32XU (proposals 2018B2544 and 2019A0153). We thank the members of the Nureki laboratory and the beamline staff at BL32XU of SPring-8 for technical assistance during data collection; Drs. O. Béjà, R. Ghai, S.P. Tsunoda, and S. Hososhima for useful discussions; and R. Nakamura and Y. Yamauchi for technical assistance. This research was partially supported by the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research) from Japan Agency for Medical Research and Development (AMED) under Grant Number JP19am0101070 (Support Number 1627). This work was supported by JSPS KAKENHI Grants 16H06294 (O.N.), 20H05437, 20K15728 (W.S.), 25104009, 15H02391, 18H03986 (H.K.), and 17H03007 (K.I.), and by Japan Science and Technology Agency PRESTO (JPMJPR15P2) and CREST (JPMJCR1753 and JPMJCR17N5).
Data Availability
The crystal structure of schizorhodospin 4 reported in this paper has been deposited in the Protein Data Bank (PDB, ID code 7E4G).
References
1
2
3
4
5
6
8
9
10
13
14
15
16
17
18
19
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
44
45
46
47
48
 Crystal structure of schizorhodopsin reveals mechanism of inward proton pumping
Crystal structure of schizorhodopsin reveals mechanism of inward proton pumping
