
- Altmetric
Skeletal muscle health is important for the prevention of various age-related diseases. The loss of skeletal muscle mass, which is known as sarcopenia, underlies physical disability, poor quality of life and chronic diseases in elderly people. The transcription factor NRF2 plays important roles in the regulation of the cellular defense against oxidative stress, as well as the metabolism and mitochondrial activity. To determine the contribution of skeletal muscle NRF2 to exercise capacity, we conducted skeletal muscle-specific inhibition of KEAP1, which is a negative regulator of NRF2, and examined the cell-autonomous and non-cell-autonomous effects of NRF2 pathway activation in skeletal muscles. We found that NRF2 activation in skeletal muscles increased slow oxidative muscle fiber type and improved exercise endurance capacity in female mice. We also observed that female mice with NRF2 pathway activation in their skeletal muscles exhibited enhanced exercise-induced mobilization and β-oxidation of fatty acids. These results indicate that NRF2 activation in skeletal muscles promotes communication with adipose tissues via humoral and/or neuronal signaling and facilitates the utilization of fatty acids as an energy source, resulting in increased mitochondrial activity and efficient energy production during exercise, which leads to improved exercise endurance.

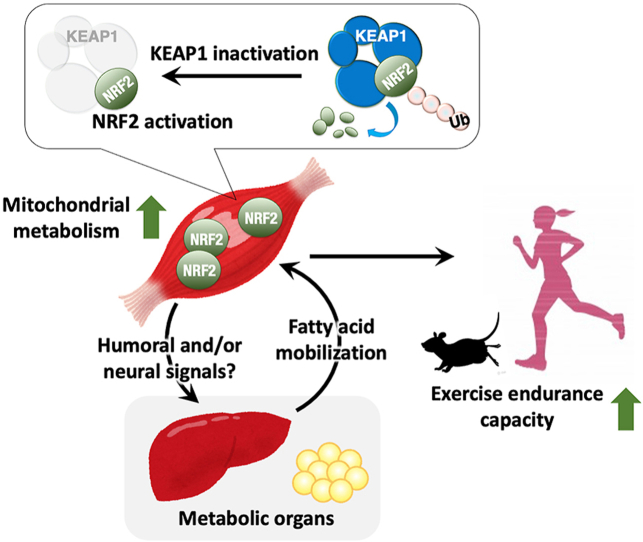
•
Systemic Keap1 knockdown enhances exercise endurance capacity in mice.
•Keap1 deficiency in skeletal muscle activates NRF2 pathway.
•Keap1 deficiency in skeletal muscle enhances endurance capacity in female mice.
•Keap1 deficiency in skeletal muscle promotes exercise-induced fatty acid utilization.
Introduction
Physical exercise produces wide-ranging health benefits. The loss of skeletal muscle mass with aging, which is known as sarcopenia, diminishes muscle strength and exercise capacity, which seriously affects the quality of life of elderly people and is a major sign of age-associated frailty [1,2]. While regular physical exercise is the best and only way to maintain skeletal muscle mass, a recent study reported that exercise intolerance and rapid skeletal muscle energetic decline during exercise are characteristics of age-associated frailty [3], suggesting that when an elderly person begins to suffer from exercise intolerance, a vicious circle is started in which reduction in skeletal muscle mass leads to further deterioration of exercise intolerance. Increasing exercise endurance capacity by improving skeletal muscle energy metabolism is expected to be an effective means of breaking this vicious cycle. However, because of the multilayered regulation of skeletal muscle metabolism by remote organs and tissues, including the liver, pancreas, adipose tissues, neuronal tissues and gut microbiota [4,5], our knowledge on specific target molecules in specific tissues for the modulation of skeletal muscle metabolism remains limited.
The KEAP1-NRF2 system plays a central role in the defense mechanism against oxidative stress and xenobiotic electrophiles [6]. NRF2 is a potent transcription activator for a number of stress-response and cytoprotective genes, and KEAP1 is a negative regulator of NRF2. KEAP1 is a substrate recognition subunit of the CUL3-based ubiquitin E3 ligase and ubiquitinates NRF2 for proteasomal degradation under unstressed conditions, thereby suppressing NRF2 activity at low levels. When cells are exposed to oxidative stress and/or electrophilic chemicals, KEAP1 activity decreases, and stabilized NRF2 translocates into the nucleus and activates the transcription of its target genes. In addition to the thoroughly analyzed antioxidant function of NRF2, this protein has been demonstrated to exert anti-inflammatory functions [7,8], as well as regulating the metabolism and mitochondrial activity [[9], [10], [11], [12]].
Previous studies described the roles played by NRF2 in skeletal muscle regarding regeneration [13], stem cell function [14], mitochondrial biogenesis [15], autophagy [16] and exercise capacity [17] based on analyses of Nrf2 knockout mice. The contribution of NRF2 to skeletal muscle performance and exercise capacity was also investigated by the pharmacological induction of NRF2 with electrophilic chemicals [11,18]. Although these reports suggest that systemic activation of NRF2 increases exercise capacity and is eventually beneficial for the prevention of age-associated sarcopenia and frailty, activation of skeletal muscle NRF2 reduces skeletal muscle mass [11], indicating the complexity of NRF2 involvement in skeletal muscle physiology and differential contributions of NRF2 in each tissue and organ to exercise capacity in the whole body.
Because we consider that maintaining the healthy condition of skeletal muscles is a primary prerequisite for the prevention of sarcopenia and frailty, we attempted to elucidate how NRF2 activation in skeletal muscles impacts local and systemic metabolism and exercise capacity. To this end, we generated skeletal muscle-specific Keap1 knockout mice (Keap1F/F:Mlc1f-Cre mice; MK mice) and investigated their muscle fiber composition, metabolism, gene expression and exercise capacity. These analyses demonstrated that skeletal muscle-specific NRF2 activation by Keap1 disruption enhances the mobilization and oxidation of fatty acids (FAs) and increases exercise capacity.
Materials and methods
Animal studies
Keap1-knockdown (Keap1-KD) mice [19] were provided by Professor Masi Yamamoto at Tohoku University. Keap1F/F mice were provided by Professor Shyam Biswal at Johns Hopkins University [20]. Mlc1f-Cre mice were provided by Professor Steve Burden at New York University [21]. All the mice utilized in this study were in the C57BL/6J genetic background. These mice were bred and housed under specific pathogen-free conditions with standard animal maintenance according to the regulations of The Standards for Human Care and Use of Laboratory Animals of Tohoku University (Tohoku University. 2007. Standards for human care and use of laboratory animals of Tohoku University. Tohoku University, Sendai, Japan.) and The Guidelines for Proper Conduct of Animal Experiments by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Science Council of Japan. 2006. Guidelines for proper conduct of animal experiments. Science Council of Japan, Ministry of Education, Culture, Sports, Science, and Technology of Japan, Tokyo, Japan.). Mice were examined at 2–5 months of age. After mice were sacrificed by cervical dislocation, the soleus (Sol), plantaris (Plant), gastrocnemius (GC) muscles, heart, lung and liver were subsequently dissected for analysis. The tibia length was measured using caliper. Blood was drawn from anesthetized mice using heparinized capillary tubes (Fisher Scientific) into the microtube within 0.5 M EDTA (pH 7.4, 2 μL) and centrifuged (3000 rpm for 15 min at 4 °C) to isolate plasma. Samples were frozen in liquid nitrogen and stored at −80 °C until analysis.
SDH staining
Plant and Sol muscles were dissected and quickly frozen in isopentane (#26405-65, Nacalai Tesque) that was prechilled in liquid nitrogen. The muscles were sectioned into 10-μm slices using cryostat (CM3050S, Leica Biosystems, Hesse, Germany). SDH staining was performed as previously reported with slight modifications [22]. Sections were stained in a solution containing 0.5 mg/mL nitroblue tetrazolium (#N5514, Sigma-Aldrich) and 50 mM sodium succinate in 50 mM phosphate buffer at 37 °C for 20 min. The SDH-positive area was defined by a fixed threshold and automatically quantified at 100x magnification under optical microscopy with BZ II Analyzer software (BZ-9000 series, Keyence, Osaka, Japan).
Immunofluorescence
MHC I immunofluorescence staining was conducted by using frozen sections. The sections were fixed in ice-cold 4% PFA on ice for 10 min. After washing with ice-cold PBS, the sections were permeabilized by ice-cold 3% NP-40 on ice for 10 min. Then, the sections were incubated with 5% normal goat serum (#31872, Invitrogen) in PBS for 1 h at room temperature for blocking non-specific staining, followed by overnight incubation with mouse anti-MHC I antibody (BA-F8) (1:100, DSHB) in blocking solution at 4 °C. After washing with PBS and additional fixing in 4% paraformaldehyde for 2 min followed by PBS wash, sections were incubated with Alexa Fluor 594-labelled anti-mouse IgG antibody (1:200, #A11005, Thermo Fisher Scientific) in blocking solution for 1 h at room temperature. From this procedure, sections were protected from light and mounted in Permafluor mountant (#TA-030-FM, Thermo Fisher Scientific). Histological samples were imaged by a Keyence BZ-9000 fluorescence microscope (BZ-9000 series, Keyence, Osaka, Japan). The area of MHC I positive muscle fiber were quantified by using NIH ImageJ software (http://rsb.info.nih. gov/ij/).
Skeletal muscle performance
Grip force test
Mice were allowed to grasp a horizontal bar with their forelimbs or grid with the four limbs. The bar and grid were connected with a dynamometer (GPM-101B, Melquest, Toyama, Japan). Grip strength was measured by pulling a tail of a mouse slowly backwards until the mouse released its limbs from the instrument. The maximum tension value out of 5 trials was recorded in grams as one measurement, which was repeated 10 times within a week. The mean value of the 10 measurements was calculated and normalized to the body weight.
Hanging inverted grid test
Mice were placed on the grid wire for 1 min to acclimate the grid. Then, the grid was inverted gently with a mouse, and the time to fall was recorded. If a mouse kept hanging more than 1 h, we censored the recording. If the hanging time was less than 1 min, which was considered an accidental fall, the hanging time was reexamined. The measurement trials were limited to 10 times a day. If the record did not reach 1 min, the maximum record out of 10 trials was adopted. The test was conducted twice within a week, and the mean value of the two tests was calculated as a final result. The Hang Impulse Score was calculated as body weight (g) × hanging time (min).
Time to exhaustion
Before an exercise bout, mice were placed in the treadmill for acclimation (MK-680, Muromachi Kikai, Tokyo, Japan). For male Keap1-KD mice, adaptive training was conducted on the first day (starting speed at 15 m/min for 10 min followed by 22 m/min for 15 min). The treadmill test was conducted on the second day. The starting speed was 5 m/min for 3 min, and the speed was increased by 5 m/min every 3 min until the speed reached 28 m/min. The maximum speed of 28 m/min was continued, and the time to exhaustion was measured.
For Keap1F/F:Mlc1f-Cre (MK) mice, their control Keap1F/F (Cntl) mice and female Keap1-KD mice, training was performed on four consecutive days: 8 m/min for 10 min twice on the first day, 9 m/min for 10 min twice on the second day, 10 m/min for 10 min twice on the third day and 12 m/min for 10 min twice on the fourth day. On the fifth day, a treadmill test was conducted. The starting speed was 5 m/min for 3 min, and the speed was increased by 5 m/min every 3 min until the speed reached 20 m/min. The maximum speed of 20 m/min was continued, and the time to exhaustion was measured.
Mice were considered to be exhausted when the mice remained on the grid area for more than 10 s. If a mouse ran more than 4 h, we ended the recording except for female Keap1-KD mice and their control WT mice, for which the recording was stopped at 6 h.
Immunoblot analysis
Skeletal muscle samples were homogenized by sonicator in denaturing lysis buffer containing 210 mM Tris-HCl, pH 6.8, 6.2% sodium dodecyl sulfate (SDS), 21.6% glycerol and 0.36 M dithiothreitol on ice. Muscle homogenates were boiled at 100 °C for 5 min and centrifuged at 1000 g for 15 min. Supernatants were used for analysis, and the loading amount of each sample was adjusted according to the intensity of α-tubulin, which was used as a loading control. The proteins were separated by SDS-PAGE gel and transferred to polyvinylidene fluoride (PVDF) membranes (#IPVH00010, Millipore) by using a Bio-Rad transfer apparatus (Mini-PROTEAN® Tetra System, Bio-Rad, Hercules, CA, USA). After the transfer of protein, the membrane was blocked in 5% (w/v) skim milk in Tris-buffered saline (TBS) containing 0.1% (w/v) Tween 20 (TBST) for 1 h. The membranes were incubated with primary antibodies diluted with 5% (w/v) skim milk in TBST overnight at 4 °C. The antibodies used in this study were as follows: rabbit anti-cytochrome c (Cyt c) (1:1250, #11940, Cell Signaling Technology), mouse anti-Complex Ⅳ-subunit (Cox Ⅳ) (1:1250, #A21348, Thermo Fisher Scientific) and mouse anti-α-tubulin (1:3000, #T6199, Sigma-Aldrich). Primary antibodies against MHC I (BA-F8) (1:100, DSHB), MHC IIa (SC-71) (1:100, DSHB), and MHC IIb (BF–F3) (1:100, DSHB) were purchased from Developmental Studies Hybridoma Bank at the University of Iowa. The primary KEAP1 antibody was a generous gift from Professor Masi Yamamoto at Tohoku University [23]. The membranes were washed and incubated with 5% skim milk in TBST containing peroxidase-conjugated secondary antibody for 1 h at room temperature. The bands were detected with ECL prime (GE Healthcare) or Chemi-Lumi One L (Nacalai Tesque) using ImageQuant LAS 4000 mini (GE Healthcare, Chicago, IL, USA). Band intensities were quantified using NIH ImageJ software (http://rsb.info.nih. gov/ij/).
RNA extraction and real-time PCR
Total RNA was isolated from skeletal muscle by using Sepazol-RNA I Super G (#09379-55, Nacalai Tesque) according to the manufacturer's protocol, and RNA quantity was measured using a Nano Drop (ND-2000, Thermo Fisher Scientific, Waltham, MA, USA). cDNA was synthesized from total RNA using ReverTra Ace® qPCR RT Master Mix with gDNA Remover (#FSQ-301, TOYOBO CO., LTD., Osaka, Japan). Real-time PCRs were conducted on an Applied Biosystems 7300 Real-Time PCR System (7300 M, Thermo Fisher Scientific, Waltham, MA, USA) using THUNDERBIRD® SYBR qPCR Mix (#QPS-201, TOYOBO Co., Ltd., Osaka, Japan) or THUNDERBIRD® Probe qPCR Mix (#QPS-101, TOYOBO Co., Ltd., Osaka, Japan). β-Actin was employed for normalization. All primers used for real-time PCR are described in Table 2.

| Gene symbol | Gene description | log2 [fold change] | FDR |
|---|---|---|---|
| exercise vs. sedentary in Cntl mice | |||
| Atf3 | Activating transcription factor 3 | 3.656 | 5.660E-02 |
| Ppargc1a | PPARG coactivator 1 alpha | 3.248 | 6.400E-04 |
| Nr4a3 | Nuclear receptor subfamily 4 group a member 3 | 2.483 | 7.250E-02 |
| Otud1 | OTU deubiquitinase 1 | 2.232 | 7.250E-02 |
| Trim36 | Tripartite motif containing 36 | −2.215 | 7.640E-02 |
| Arhgap8 | Rho GTPase activating protein 8 | 2.195 | 5.660E-02 |
| Impact | Impact RWD domain protein | 2.095 | 8.270E-03 |
| Tctex1d2 | Tctex1 domain containing 2 | −1.862 | 7.250E-02 |
| Irs2 | Insulin receptor substrate 2 | 1.783 | 7.640E-02 |
| Slc19a2 | Solute carrier family 19 member 2 | −1.543 | 7.280E-02 |
| Tnfsf9 | TNF superfamily member 9 | 1.528 | 8.640E-02 |
| Slc20a1 | Solute carrier family 20 member 1 | 1.469 | 7.250E-02 |
| Sik1 | Salt inducible kinase 1 | 1.462 | 8.640E-02 |
| exercise vs. sedentary in MK mice | |||
| Ppargc1a | PPARG coactivator 1 alpha | 2.608 | 6.530E-03 |
| Il12rb1 | Interleukin 12 receptor subunit beta 1 | 2.215 | 7.980E-02 |
| Impact | Impact RWD domain protein | 1.760 | 3.170E-02 |
| Clec2e | C-type lectin domain family 1 member e | −1.723 | 3.170E-02 |
| Slc20a1 | Solute carrier family 20 member 1 | 1.552 | 7.980E-02 |
| Dscam | DS cell adhesion molecule | 1.195 | 9.590E-02 |

| Gene | Primer/Probe | Sequence (5’- -3′) |
|---|---|---|
| Mouse Acox2 | Forward primer | CAATGACTTCCATCAAGTGGTG |
| Reverse primer | GTCTATGTTTTCGAAGCCCATC | |
| Mouse β-Actin | Forward primer | CGGTTCCGATGCCCTGAGGCTCTT |
| Reverse primer | CGTCACACTTCATGATGGAATTGA | |
| Mouse Cytochrome b | Forward primer | GGCTACGTCCTTCCATGAGGAC |
| Reverse primer | GAAGCCCCCTCAAATTCATTCGAC | |
| Mouse Gbe1 | Forward primer | CAAGAGCTATACGGACTACCGAGT |
| Reverse primer | CCGCTGCGTCAGAATCTAGT | |
| Mouse Gclc | Forward primer | ATCTGCAAAGGCGGCAAC |
| Reverse primer | ACTCCTCTGCAGCTGGCTC | |
| Probe | FAM-ACGGGTGCAGCAAGGCCCA-TAMRA | |
| Mouse Gclm | Forward primer | TGGAGCAGCTGTATCAGTGG |
| Reverse primer | AAATCTGGTGGCATCACACA | |
| Mouse Keap1 | Forward primer | CCCATGAGGCATCACCGTAG |
| Reverse primer | CATAGCCTCCGAGGACGTAG | |
| Probe | FAM-GGATTACTGTGCACCAGGGCAAG-TAMRA | |
| Mouse Nfe2l2 | Forward primer | CAAGACTTGGGCCACTTAAAAGAC |
| Reverse primer | AGTAAGGCTTTCCATCCTCATCAC | |
| Probe | FAM-AGGCGGCTCAGCACCTTGTATCTTGA-TAMRA | |
| Mouse Nqo1 | Forward primer | AGCTGGAAGCTGCAGACCTG |
| Reverse primer | CCTTTCAGAATGGCTGGCA | |
| Probe | FAM-ATTTCAGTTCCCATTGCAGTGGTTTGGG-TAMRA | |
| Mouse Phka1 | Forward primer | TGGCAACAGAACTAGCACACTC |
| Reverse primer | GATTCATCAGGCCCTCTGTG | |
| Mouse Srxn1 | Forward primer | AGCCTGGTGGACACGATC |
| Reverse primer | AGGAATAGTAGTAGTCGCCA | |
| Mouse Txnrd1 | Forward primer | AGAAAGTGCTGGTCTTGGATTTTG |
| Reverse primer | ACACGTTCCTCCGAGACCC | |
| Probe | FAM-TCTGGTCCCAAGAGGAGTCGGTGTG-TAMRA |
RNA purification and RNA-sequence analysis
Total RNA was isolated as described above and further purified using the RNeasy® MinElute® Cleanup Kit (#74204, QIAGEN, Hilden, Germany) according to the manufacturer's instructions. Subsequently, 0.8 μg of total RNA was subjected to rRNA removal using the NEB Next® rRNA Depletion Kit (#E6310S, New England BioLabs, Ipswich, MA, USA). cDNA sequencing libraries were subsequently prepared using the SureSelect Strand-Specific RNA library preparation kit (#G9691B, Agilent Technologies, Santa Clara, CA, USA) with a modified protocol omitting the polyA selection step. The libraries were quantified by the qMiSeq method [24] and sequenced on a HiSeq2500 System (Illumina, San Diego, CA, USA) to generate 76-bp single-end reads. Approximately 30 million reads per sample were obtained. Raw data in fastq sequencing files were mapped and counted using the Galaxy platform [[25], [26], [27]]. Expression level estimations were reported for each sample as fragments per kilobase of transcript sequence per million mapped fragments (FPKM). iDEP (integrated Differential Expression and Pathway analysis, version 0.91) was used for data analysis and identification of DEGs. A heat map of DEGs was created using GraphPad Prism 8 (version 8. 4. 3, GraphPad Software, San Diego, CA, USA).
DNA purification and mitochondrial DNA quantification
Skeletal muscle samples were digested by proteinase K in lysis buffer containing 0.1 M Tris-HCl (pH 8.0), 20 mM EDTA, pH 8.0, 0.2 M NaCl and 0.2% sodium dodecyl sulfate (SDS) at 55 °C overnight. Genomic DNA was isolated from skeletal muscle lysate by phenol-chloroform extraction and ethanol precipitation. DNA quantity was measured by a Nano Drop (ND-2000, Thermo Fisher Scientific, Waltham, MA, USA). Real-time PCRs were performed by an Applied Biosystems 7300 Real-Time PCR System (7300 M, Thermo Fisher Scientific, Waltham, MA, USA) using THUNDERBIRD® SYBR qPCR Mix (#QPS-201, TOYOBO CO., LTD., Osaka, Japan). The gene dose of Cytochrome b encoded in mitochondrial DNA was quantified and normalized against the gene dose of β-Actin encoded in nuclear DNA.
Sample preparation for metabolome and lipidome analyses
Total metabolites and lipids were extracted from plasma and Sol muscle according to the Bligh and Dyer method with minor modifications [28]. Briefly, each 50 μL of plasma was mixed with 910 μL of methanol, 15 μL of internal standard (IS) solution A (chloroform) containing DG 15:0–18:1 (d7) (0.23 nmol) and TG 15:0–18:1 (d7)−15:0 (0.53 nmol), 15 μL of IS solution B (methanol/chloroform, 1/1, v/v) containing FA 16:0 (13C16) (0.15 nmol) and MG 18:1 (d7) (1.5 nmol), and 10 μL of IS solution C (water) containing 10-camphorsulfonic acid (2.0 nmol) and piperazine-1,4-bis(2-ethanesulfonic acid) (2.0 nmol). The samples were centrifuged at 16,000 g at 4 °C for 5 min, and the supernatant (400 μL) was collected in clean tubes. After mixing with 400 μL of chloroform and 320 μL of water, phase separation of aqueous and organic layers was performed via centrifugation (16,000 g, 4 °C, 5 min). The aqueous (upper) layer (500 μL) was transferred into a clean tube. After the aqueous layer extracts were evaporated under vacuum, the dried extracts were stored at −80 °C until the analysis of hydrophilic metabolites. Prior to analysis, the dried aqueous layer was reconstituted in 50 μL of water. The organic (lower) layer (250 μL) was placed in another tube and diluted twice with methanol and stored at −80 °C until hydrophobic metabolite analysis.
Dissected Sol muscles were snap frozen in liquid nitrogen. Frozen muscles were plunged into 1 mL of cold methanol (−30 °C) containing 10-camphorsulfonic acid (1.5 nmol) and piperazine-1,4-bis(2-ethanesulfonic acid) (1.5 nmol) as ISs and homogenized with zirconia beads at 20 Hz for 1 min using a tissue homogenizer (ball mill mixer MM301, Retsch, Haan, Germany). Subsequently, the homogenate sample was centrifuged at 16,000 g and 4 °C for 5 min. Then, 600 μL supernatants were transferred to another tube, and 600 μL of chloroform and 480 μL of water were added. After centrifugation at 16,000 g and 4 °C for 5 min, 800 μL of the upper layer was isolated and used for hydrophilic metabolite analysis.
Metabolome and lipidome analyses
Anionic polar metabolites (e.g., organic acids, nucleotides, and 3-hydroxybutyric acid) were analyzed via ion chromatography (Dionex ICS-5000+ HPIC system, Thermo Fisher Scientific) with a Dionex IonPac AG11-HC-4 μm guard column (2 mm i.d. × 50 mm, 4 μm particle size, Thermo Fisher Scientific) and a Dionex IonPac AS11-HC-4 μm column (2 mm i.d. × 250 mm, 4 μm particle size, Thermo Fisher Scientific) coupled with a Q Exactive, high-performance benchtop quadrupole Orbitrap high-resolution tandem mass spectrometer (Thermo Fisher Scientific) (IC/HRMS/MS) [29]. Cationic polar metabolites (e.g., amino acids, bases, and nucleosides) were analyzed via liquid chromatography (Nexera X2 UHPLC system, Shimadzu Co., Kyoto, Japan) with a Discovery HS F5 column (2.1 mm i.d. × 150 mm, 3 μm particle size, Merck) coupled with a Q Exactive instrument (PFPP-LC/HRMS/MS) [57]. MGs, DGs, and TGs were analyzed using supercritical fluid chromatography (SFC) (Nexera UC system, Shimadzu Co.) equipped with an ACQUITY UPC2 Torus diethylamine (DEA) (3.0 mm i.d. × 100 mm, 1.7 μm particle size, Waters, Milford, MA) and triple quadrupole mass spectrometry (TQMS, LCMS-8060, Shimadzu Co.) (DEA-SFC/MS/MS) in multiple reaction monitoring (MRM) mode [30]. FAs were analyzed using an SFC (Shimadzu Co.) with an ACQUITY UPC2 HSS C18 SB column (3.0 mm i.d. × 100 mm, 1.8 μm particle size, Waters) coupled with a TQMS (Shimadzu Co.) (C18-SFC/MS/MS) in MRM mode [31]. Xcalibur 4.2.47 (Thermo Fisher Scientific), LabSolutions, version 5.9 (Shimadzu Co.), MRMPROBS 2.86 [32], and Microsoft Excel 2010 were used for data processing. The details of the analytical conditions for the analyses of hydrophilic and hydrophobic metabolites are described in the Supporting Information.
Statistical analysis
All data are presented as the mean ± SD. The sample size (n) of each experiment is described in each corresponding figure legend. Statistical analyses were generated using Student's t-test (two-sided unpaired), one-way analysis of variance (ANOVA) followed by Tukey's multiple comparison test and log-rank (Mantel-Cox) test with GraphPad Prism 8 (GraphPad Software). P-values less than 0.05 (*), 0.01 (**), 0.001 (***) and 0.0001 (****) were considered to be significant.
Results
Systemic inhibition of the Keap1 gene in mice increases skeletal muscle mass and enhances exercise capacity
Previous studies reported that the administration of NRF2-inducing electrophiles enhances exercise capacity in mice [11,18]. To determine whether genetic activation of NRF2 recapitulates this effect, we used Keap1-knockdown (Keap1-KD) mice [33]. We first verified reduced expression of KEAP1 protein and mRNA in the skeletal muscle of Keap1-KD mice (Fig. 1A and B). Nqo1, one of the typical target genes of NRF2, was determined to be upregulated in Keap1-KD skeletal muscle (Fig. 1B), suggesting that the NRF2 pathway is activated in skeletal muscles of Keap1-KD mice. The body weight of Keap1-KD mice was observed to be smaller than that of wild-type (WT) mice (Fig. 1C), which was due to less fat mass according to the previous study [33]. Skeletal muscle wet weight normalized by tibia length was not different for gastrocnemius (GC) and plantaris (Plant) muscles, which are known as fast glycolytic muscles, between Keap1-KD and WT mice (Fig. 1D). In contrast, soleus (Sol) muscle, known as slow oxidative muscle, in Keap1-KD mice was significantly heavier than in WT mice (Fig. 1D). To evaluate exercise capacity, we conducted a treadmill test using the protocol shown in Fig. 1E. Male Keap1-KD mice exhibited significantly longer running times than male WT mice did (Fig. 1F), suggesting that the NRF2 pathway activation due to systemic KEAP1 inhibition increased exercise capacity.

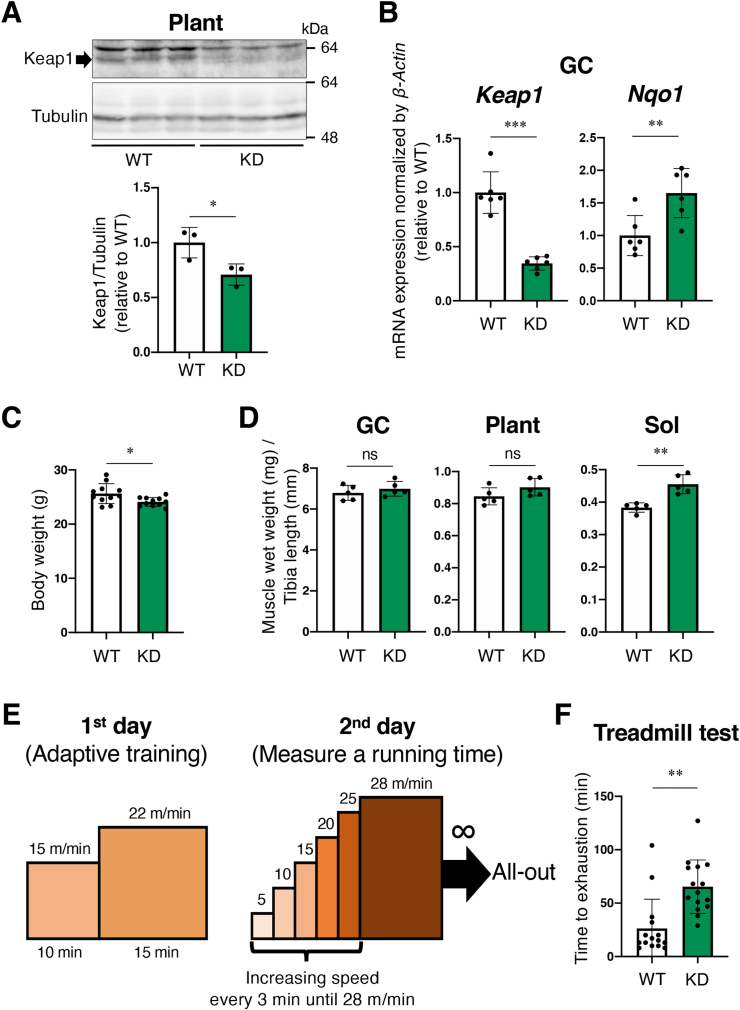
Systemic KEAP1 inhibition increases skeletal muscle mass and enhances endurance exercise capacity.
Two-sided Student's t-test was conducted for statistical significance. *P < 0.05, **P < 0.01, ***P < 0.001, ns; not significant.
A. Immunoblot analysis of KEAP1 protein in plantaris muscle (Plant). (n = 3 mice per group). Tubulin was detected as a loading control. Intensities of the immunoblot bands (upper panel) were quantified using image J software (lower panel). Mean and SD are indicated. Mean values of WT mice are set as 1. WT; wild-type mice, KD; Keap1-knockdown mice.
B. RT-PCR for measuring mRNA expression of Keap1 and Nqo1 in gastrocnemius muscle (GC) (n = 6 mice per group). β-Actin was employed for normalization. Mean and SD are indicated. Mean values of WT mice are set as 1.
C. Body weight. (n = 11 mice per group). Mean and SD are indicated.
D. Skeletal muscle wet weights normalized by the length of the tibia. GC, Plant and soleus (Sol) muscles were examined (n = 5 mice per group). Mean and SD are indicated.
E. Protocol for the treadmill test. On the first day, adaptive training was undertaken using a condition of 15 m/min for 10 min followed by 22 m/min for 15 min. On the second day, a treadmill test was undertaken. A starting speed was 5 m/min, and the speed was increased by 5 m/min every 3 min until the speed reached 28 m/min. The maximum speed of 28 m/min was continued, and the time to exhaustion was measured.
F. Time to exhaustion at the speed of 28 m/min (n = 15 mice per group). Mean and SD are indicated.
Generation of skeletal muscle-specific Keap1 knockout mice
To further investigate the effects of NRF2 activation specifically in skeletal muscles, we generated skeletal muscle-specific Keap1 knockout mice through the Cre-loxP recombination system. Mlc1f-Cre mice [21] were crossed with Keap1 floxed mice [20] to generate Keap1F/F:Mlc1f-Cre mice (MK mice). Keap1F/F mice were employed as a control (Cntl mice). We verified the reduction of KEAP1 protein and mRNA in skeletal muscle in MK mice (Fig. 2A and B). Meanwhile, the mRNA expression of the NRF2 target genes Nqo1 and Txnrd1 was significantly upregulated in MK mice (Fig. 2B). No apparent differences were observed in the expression levels of Keap1 or Nqo1 in other organs (Fig. 2C and D), indicating that the Keap1 inhibition and consequent NRF2 pathway activation were induced specifically in the skeletal muscle.

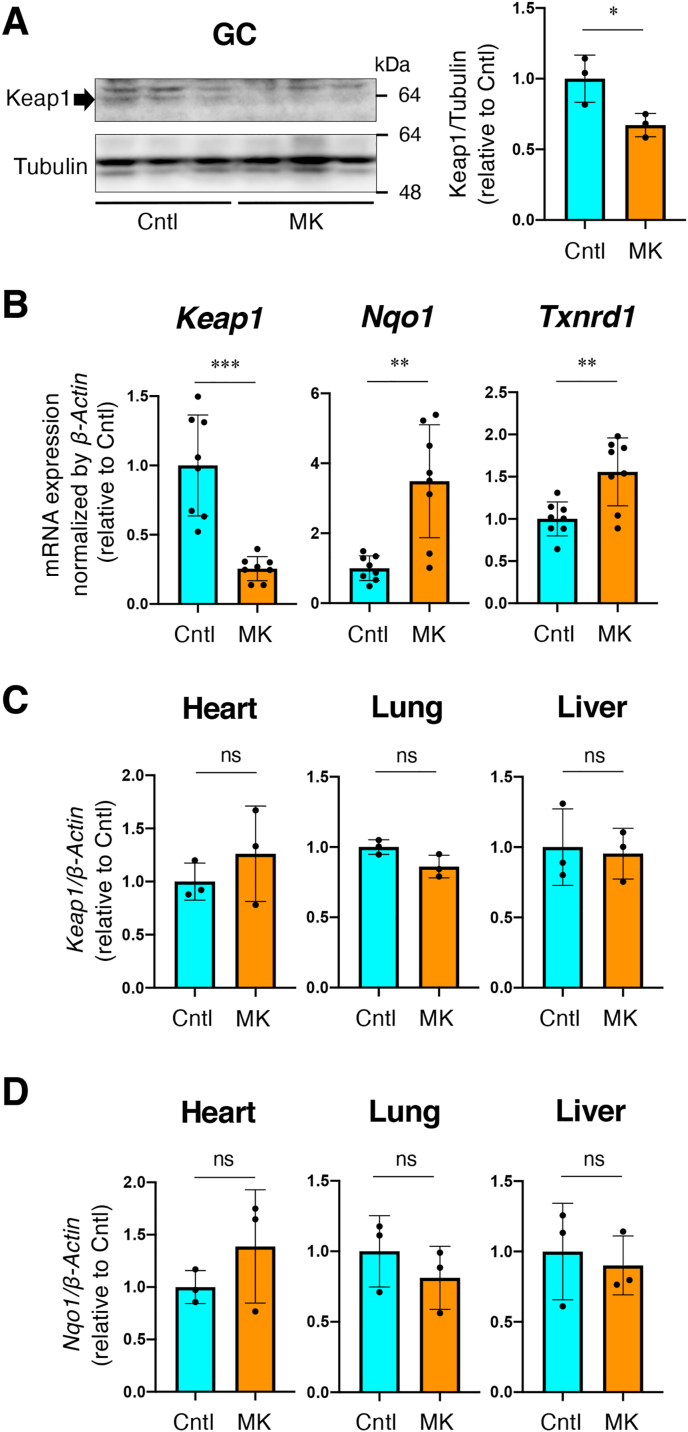
Generation of skeletal muscle-specific Keap1 knockout mice.
Cntl; Keap1F/F mice, MK; Keap1F/F:Mlc1f-Cre mice. Two-sided Student's t-test was conducted for statistical significance. *P < 0.05, **P < 0.01, ***P < 0.001, ns; not significant.
A. Immunoblot analysis of KEAP1 protein in GC muscle. (n = 3 mice per group). Tubulin was detected as a loading control. Intensities of the immunoblot bands (left panel) were quantified using image J software (right panel). Mean and SD are indicated. Mean values of Cntl mice are set as 1.
B. RT-PCR for measuring mRNA expression of Keap1, Nqo1 and Txnrd1 in gastrocnemius muscle (n = 8 mice per group). β-Actin was employed for normalization. Mean and SD are indicated. Mean values of Cntl mice are set as 1.
C, D. RT-PCR for measuring mRNA expression of Keap1 (C) and Nqo1 (D) in representative tissues (n = 3 mice per group). β-Actin was employedfor normalization. Mean and SD are indicated. Mean values of Cntl mice are set as 1.
Skeletal muscle-specific Keap1 disruption alters the content of skeletal muscle fiber MHC subtypes on soleus muscle in female mice
Skeletal muscle-specific Keap1 disruption did not alter body weight or skeletal muscle wet weight normalized by tibia length (Fig. 3A–C), which was different from the results obtained with Keap1-KD mice. To determine whether there were any changes in the contents of muscle fiber subtypes in MK mice, we detected each fiber subtype by immunoblot analysis. In male mice, there were no apparent changes in the content of muscle fiber subtypes except for a modest decrease in MHC IIa, known as a fast-oxidative muscle fiber subtype, in GC muscle (Fig. 3D and E). In female mice, the MHC Ⅱa fiber was significantly decreased in Sol muscle, although there were no changes in GC. These results indicated that the contents of muscle fiber subtypes were substantially altered in female MK mice, and we utilized female mice for further investigation.

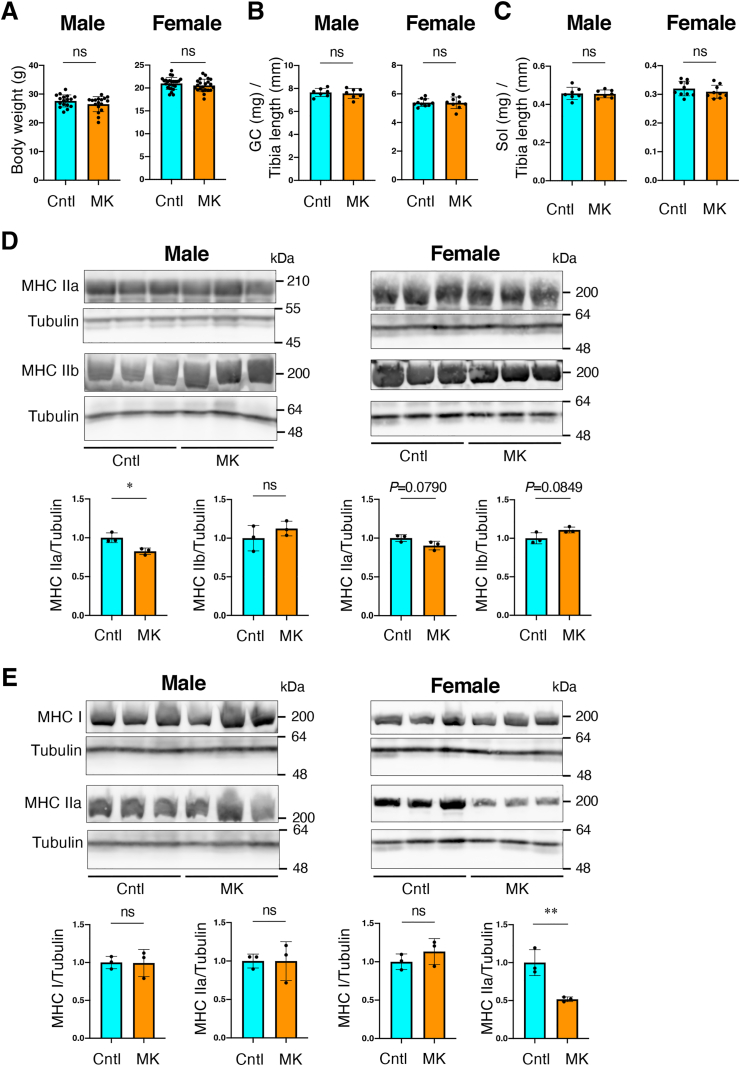
Muscle weight and fiber types of skeletal muscle-specific Keap1knockout mice
Cntl; Keap1F/F mice, MK; Keap1F/F:Mlc1f-Cre mice. Two-sided Student's t-test was conducted for statistical significance. *P < 0.05, **P < 0.01, ns; not significant.
A. Body weight of Cntl and MK mice. (male: n = 17 mice per group, female: n = 25 for Cntl mice and n = 22 for MK mice). Mean and SD are indicated.
B, C. Skeletal muscle wet weights normalized by the length of the tibia. GC (B) and Sol (C) are shown. (male: n = 7 mice per group, female: n = 10 for Cntl mice and n = 9 for MK mice). Mean and SD are indicated.
D. Immunoblot analysis of MHC IIa and IIb in GC muscle. (n = 3 mice per group). Tubulin was detected as a loading control. Intensities of the immunoblot bands (upper panel) were quantified using image J software (lower panel). Mean and SD are indicated. Mean values of Cntl mice are set as 1.
E. Immunoblot analysis of MHC I and IIa in Sol muscle. (n = 3 mice per group). Tubulin was detected as a loading control. Intensities of the immunoblot bands (upper panel) were quantified using image J software (lower panel). Mean and SD are indicated. Mean values of Cntl mice are set as 1.
Skeletal muscle-specific Keap1 disruption enhances mitochondrial activity
The reduction in MHC Ⅱa suggested a relative increase in MHC I, a slow-oxidative muscle fiber subtype, in the Sol muscle of female MK mice. Indeed, % distribution of MHC I was higher in MK mice than Cntl mice by immunofluorescence using anti-MHC I antibody in Sol muscle (Fig. 4A and B). Because anti-MHC IIa antibody did not work for us, MHC I-negative fibers in Sol muscle were regarded as MHC IIa-positive based on an observation that MHC IIb was not detected in Sol muscle. We then examined mitochondrial proteins and DNA and conducted histological assessment of succinate dehydrogenase (SDH) activity to evaluate mitochondrial biogenesis and activity, respectively. The expression levels of mitochondrial proteins, Cytochrome c (Cyt c) and COX4, were examined using immunoblot analysis. Cyt c protein levels were higher in GC and Sol muscles of MK mice than in those of Cntl mice (Fig. 4C). Mitochondrial DNA, examined as the Cytochrome b (Mt-cyb) gene dose, in GC and Sol muscles were comparable between Cntl and MK mice (Fig. 4D). SDH activity in Plant muscle was higher in MK mice than in Cntl mice (Fig. 4E and F). Sol exhibited a similar tendency, although the difference was not significant. These results suggest that skeletal muscle-specific Keap1 disruption promotes mitochondrial metabolic activity, rather than biogenesis, which is consistent with a previous finding that NRF2 pathway activation in skeletal muscle increased oxygen consumption but did not alter mitochondrial DNA levels [11].

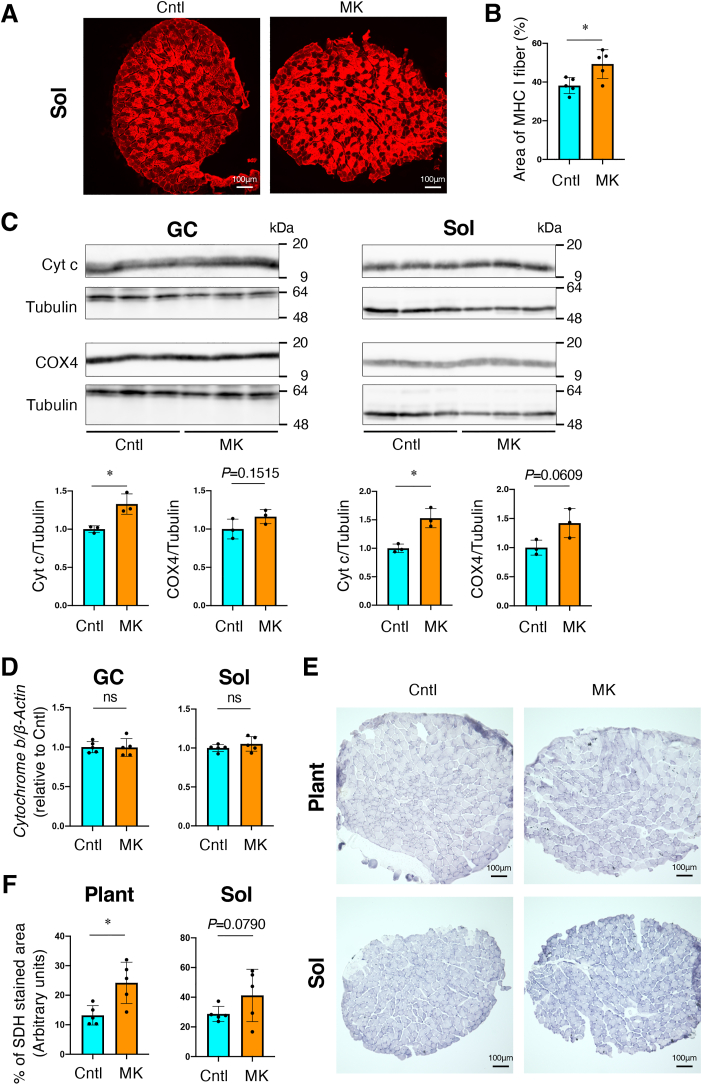
Mitochondrial biogenesis and activity in skeletal muscle-specific Keap1 knockout mice.
Cntl; Keap1F/F mice, MK; Keap1F/F:Mlc1f-Cre mice. Two-sided Student t-test was conducted for statistical significance. *P < 0.05, ns; not significant.
A, B. Immunofluorescence with MHC I antibody of Sol muscle. Representative results from 5 mice per group are shown (A). Area of MHC I fiber were quantified by using image J software (F). Mean and SD are indicated
C. Immunoblot analysis of Cytochrome c (Cyt c) and Cytochrome c oxidase subunit 4 (Cox4) in GC and Sol muscles (n = 3 mice per group). Tubulin was detected as a loading control. Intensities of the immunoblot bands (upper panel) were quantified using image J software (lower panel). Mean and SD are indicated. Mean and SD are indicated. Mean values of Cntl mice are set as 1.
D. Quantification of mitochondrial DNA normalized to genomic DNA in GC and Sol muscles (n = 5 mice per group). Cytochrome b and β-Actin were amplified as representatives of mitochondrial and genomic DNA, respectively (n = 5 mice per group). Mean and SD are indicated. Mean values of Cntl mice were set as 1.
E, F. Succinate dehydrogenase (SDH) staining of Plant and Sol muscles. Representative results from 5 mice per group are shown (E). Staining intensities were quantified (F). Mean and SD are indicated.
Metabolomic profiling of the soleus muscle in skeletal muscle-specific Keap1 knockout female mice.
Because increased mitochondrial activity was suggested in skeletal muscles of female MK mice, we investigated the metabolic alterations in MK mice by conducting metabolome analysis using Sol muscle. Although NRF2 is a well-known activator of glutathione synthesis, reduced and oxidized glutathione levels did not exhibit significant differences (Fig. 5A). The uric acid level tended to be higher in MK female Sol muscle (Fig. 5B), which was consistent with a previous finding obtained in forestomach epithelia of Keap1-KD mice [9]. Other metabolites of central metabolism did not show any significant differences between MK mice and Cntl mice (data not shown), except for nucleoside triphosphates. The nucleoside triphosphates ATP, GTP and CTP were unexpectedly all reduced in the Sol muscles of MK mice, whereas nucleoside di- and monophosphate levels were mostly comparable between the two groups (Fig. 5C). Considering the increased SDH activity (see Fig. 4E and F) and increased oxygen consumption [11] in Keap1-deficient muscles, we surmise that decreased ATP production due to mitochondrial uncoupling and/or increased ATP consumption account for the decreased levels of nucleoside triphosphates.

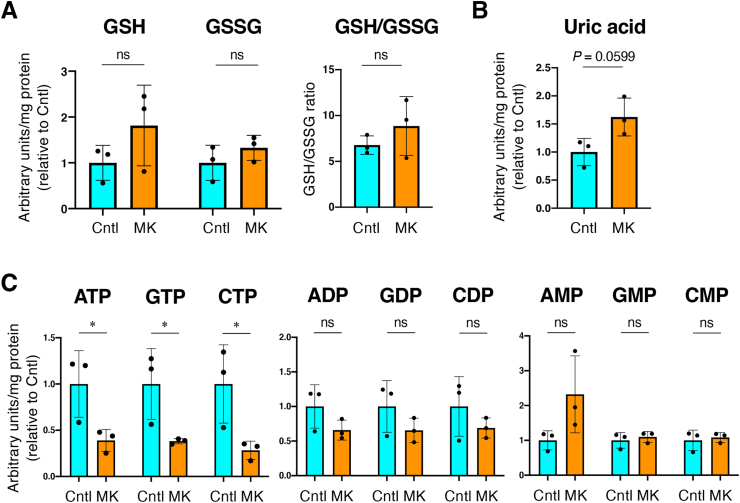
Metabolome analysis using Sol muscle of skeletal muscle-specific Keap1 knockout mice.
Cntl; Keap1F/F mice, MK; KeapF/F:Mlc1f-Cre mice. Two-sided Student's t-test was conducted for statistical significance. *P < 0.05, ns; not significant.
A. Quantities of reduced and oxidized glutathione (GSH and GSSG, respectively) and their ratios (n = 3 mice per group). Mean and SD are indicated. Mean values of Cntl mice are set as 1 for GSH and GSSG quantities.
B. Uric acid quantity (n = 3 mice per group). Mean and SD are indicated. Mean values of Cntl mice are set as 1.
C. Quantities of nucleoside tri-, di- and monophosphates (n = 3 mice per group). Mean and SD are indicated. Mean values of Cntl mice are set as 1.
Skeletal muscle-specific Keap1 disruption enhances exercise endurance capacity but not muscle strength
We asked how Keap1 disruption in skeletal muscles, which was accompanied by a significant reduction in nucleoside triphosphates, including ATP, affects exercise capacity. MK mice and their control mice were subjected to physical performance tests, i.e., the grip force test, inverted grid hanging test and treadmill running test. The grip force test was employed for measuring muscle strength, while the inverted grid hanging test and treadmill running test were used for measuring muscle endurance capacity. The grip force test did not indicate any differences between MK and Cntl mice (Fig. 6A). In contrast, MK mice exhibited dramatic gain of hanging capacity in the inverted grid hanging test (Fig. 6B and C) and significant extension of running time in the treadmill running test (Fig. 6D and E). These results suggest that skeletal muscle-specific Keap1 disruption enhances exercise endurance capacity in female mice. As for male mice, physical performance tests did not give any conclusive results, implying that impacts of skeletal muscle-specific disruption of Keap1 are smaller than those of unidentified fluctuating factors in individual male mice.

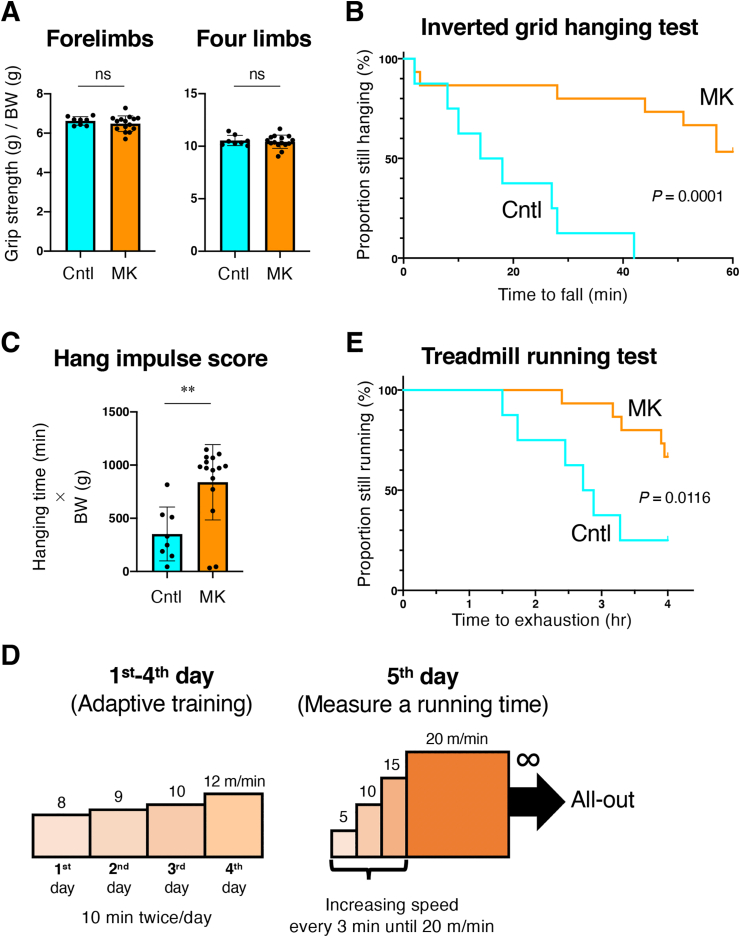
Skeletal muscle-specific Keap1 disruption enhances endurance exercise capacity.
Cntl; Keap1F/F mice, MK; Keap1F/F:Mlc1f-Cre mice. Two-sided Student t-test (A, C) and Log-rank (Mantel-Cox) test (B, D) were conducted for statistical significance. **P < 0.01, ns; not significant.
A. Grip strength of forelimbs and four limbs normalized by body weight (BW) (n = 8 for Cntl mice and n = 15 for MK mice). Mean and SD are indicated.
B. Inverted grid hanging test (n = 8 for Cntl mice and n = 15 for MK mice).
C. Hang impulse score calculated as hanging time (min) × BW (g) (n = 8 for Cntl mice and n = 15 for MK mice). Mean and SD are indicated.
D. Protocol for the treadmill test. The adaptive training was undertaken for four consecutive days: 8 m/min for 10 min twice on the first day, 9 m/min for 10 min twice on the second day, 10 m/min for 10 min twice on the third day and 12 m/min for 10 min twice on the fourth day. On the fifth day, a treadmill test was undertaken. The starting speed was 5 m/min, and the speed was increased by 5 m/min every 3 min until the speed reached 20 m/min. The maximum speed of 20 m/min was continued, and the time to exhaustion was measured.
E. Treadmill running test (n = 8 for Cntl mice and n = 15 for MK mice).
To elucidate cell-autonomous and non-cell-autonomous function of NRF2 for the improvement of physical performance, we examined skeletal muscle mass and conducted physical performance test using female Keap1-KD mice for comparison. Treadmill running test was conducted according to the protocol shown in Fig. 6D. The body weight was almost comparable between Keap1-KD and WT mice (Fig. 7A). Slight increases in Sol muscle mass and in grip strength were observed in Keap1-KD mice (Fig. 7B and C) but not in MK mice (see Fig. 3, Fig. 6A), whereas improvements in hanging capacity and treadmill running capacity were similarly observed in Keap1-KD (Fig. 7D–F) and MK mice (see Fig. 6B, C and 6E). Thus, muscle NRF2 activity accounts for the improvement of muscle endurance capacity, and extra-muscle NRF2 activity accounts for the increase of muscle mass and strength although the effect size was small.

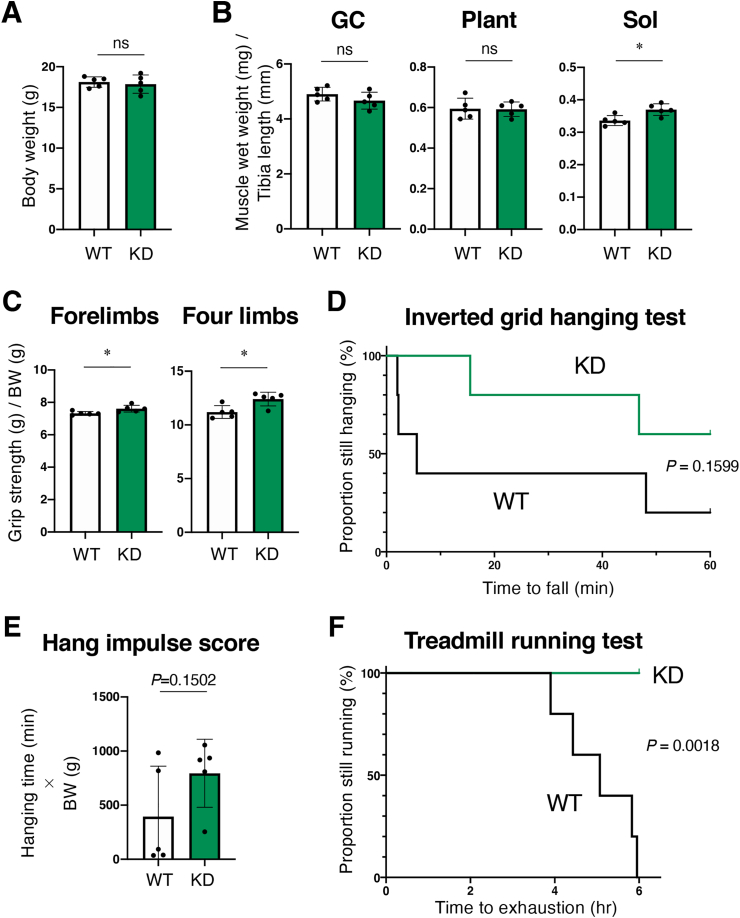
Systemic KEAP1 inhibition increases skeletal muscle mass and enhances endurance exercise capacity in female mice.
WT; wild-type mice, KD; Keap1-knockdown mice. Two-sided Student t-test (A, C) and Log-rank (Mantel-Cox) test (B, D) were conducted for statistical significance. *P < 0.05, ns; not significant.
A. Body weight. (n = 5 mice per group). Mean and SD are indicated.
B. Skeletal muscle wet weights normalized by the length of the tibia. Gastrocnemius (GC), plantaris (Plant) and soleus (Sol) muscles were examined (n = 5 mice per group). Mean and SD are indicated.
C. Grip strength of forelimbs and four limbs normalized by body weight (BW) (n = 5 mice per group). Mean and SD are indicated.
D. Inverted grid hanging test (n = 5 mice per group).
E. Hang impulse score calculated as hanging time (min) × BW (g) (n = 5 mice per group). Mean and SD are indicated.
F. Treadmill running test (n = 5 mice per group).
Transcriptomic profiling of soleus muscle in skeletal muscle-specific Keap1 knockout female mice with or without acute exercise
Because exercise endurance was markedly increased in MK mice despite the reduced ATP levels in resting skeletal muscle, we expected that the exercise-induced response was augmented in skeletal muscles of MK mice and conferred elevated endurance on MK mice. We examined the changes in gene expression induced by acute exercise in the Sol muscles of MK and Cntl mice. MK and Cntl mice were randomly allocated into two groups, the sedentary and acute exercise groups, and the Sol muscles of these mice were utilized for RNA-seq analysis (Fig. 8A).

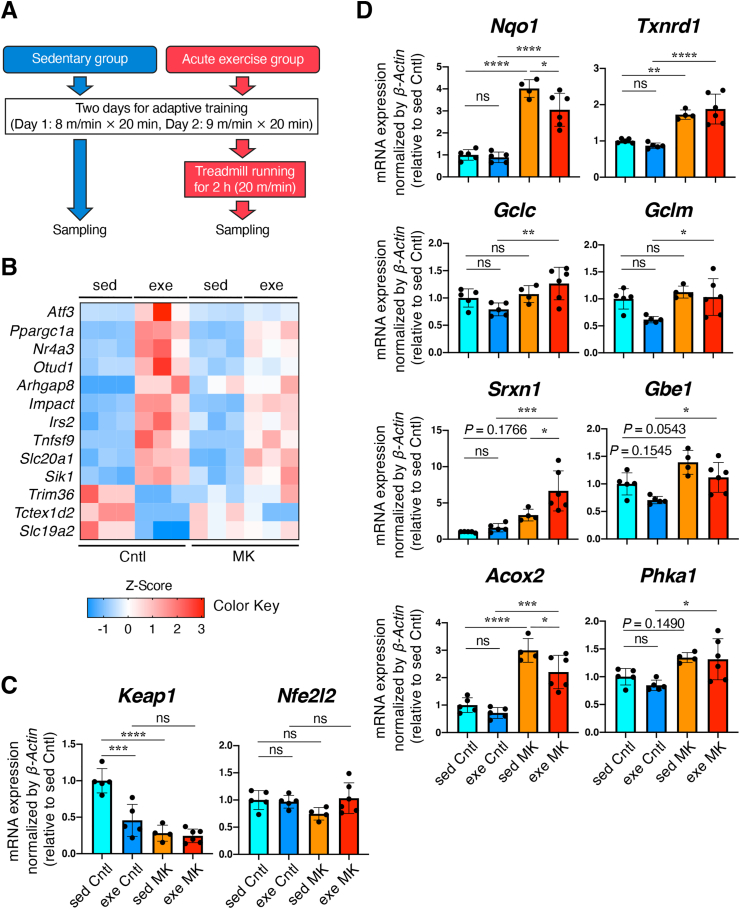
RNA-seq analysis using Sol muscle of skeletal muscle-specific Keap1 knockout mice.
A. Experimental design. Both sedentary and exercise group mice underwent 2-day adaptive training. The exercise group mice were run for 2 h at 20 m/min and were sacrificed for sampling just after running on day 3. Sedentary group mice were killed and sampled without running on day 3. B. Heat map of differentially expressed genes in Sol muscles of Cntl mice from sedentary vs acute exercise groups. Cntl; Keap1F/F mice, MK; KeapF/F:Mlc1f-Cre mice, sed; sedentary group, exe; acute exercise group.
C, D. RT-PCR for measuring mRNA expression of Keap1 and Nef2l2 (Nrf2) (C) and NRF2 target genes (D). β-Actin was employed for normalization. Mean and SD are indicated (Cntl: n = 5 for sed and exe mice, MK: n = 4 for sed mice and n = 6 for exe mice). Mean values of sed Cntl mice are set as 1. One-way analysis of variance (ANOVA) followed by Tukey's multiple comparison test was conducted for statistical significance. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns; not significant.
Following the criteria of FDR >0.1, log2FC > 1 or < −1 and protein-encoding genes, the transcriptome was compared between sedentary and acute exercise groups in each genotype. Thirteen genes and 6 genes were identified as differentially expressed genes (DEGs) in Cntl and MK mice, respectively (Table 1). Contrary to our expectation, the exercise-induced changes in gene expression were attenuated in Sol muscles of MK mice (Fig. 8B), suggesting that reactive oxygen species (ROS), which are regarded as mediators of exercise-induced signaling that induces trophic effects on skeletal muscles, are quenched in skeletal muscles in MK mice due to the antioxidant function of NRF2.
RT-PCR was conducted using mouse Sol muscles obtained in the second independent experiment shown in Fig. 8A. Keap1 expression in Cntl mice was decreased after the acute exercise to the similar level of MK mice whereas Nfe2l2 expression was not changed (Fig. 8C). In the sedentary groups, NRF2 target genes in Sol muscles were mostly upregulated in MK mice compared with those in Cntl mice, except for Gclc and Gclm (Fig. 8D). Gclc and Gclm encode catalytic and modifier subunits of a rate-limiting enzyme of glutathione synthesis, γ-glutamylcysteine synthetase, respectively. Comparable expression of Gclc and Gclm between Cntl and MK mice, implying that NRF2 contribution to Gclc and Gclm expression in Sol muscle is not big enough in the sedentary condition, may explain why glutathione levels were not different in the Sol muscles of Cntl and MK mice in the sedentary condition (see Fig. 5A). Gbe1 and Phka1, described as NRF2 target genes in skeletal muscles, were expected to facilitate glycogen utilization [11]. In the sedentary groups, Gbe1 and Phka1 tended to be upregulated in MK mice, but statistical significance was not reached (Fig. 8D). Srxn1, another NRF2 target gene, was also marginally upregulated in MK mice. One of the enzymes catalyzing β-oxidation of FAs, Acox2, was found to be remarkably increased in MK mice compared with Cntl mice, which is consistent with the findings of a recent report describing that Acox2 is a target gene of NRF2 [34].
In the acute exercise groups, MK mice exhibited elevated expression of these NRF2 target genes, including Gclc and Gclm, compared with Cntl mice (Fig. 8D). Upregulation of the NRF2 target genes in Cntl mice in the acute exercise group compared with those in the sedentary group was not apparent in this experimental setting (Fig. 8D) although previous studies described that muscle contraction and exercise increase NRF2 activity in skeletal muscle [[35], [36], [37], [38]].
However, these transcriptomic changes induced by acute exercise could not explain the increased endurance capacity of MK mice. Considering that NRF2 pathway activation in skeletal muscles has systemic impacts, serving to improve glucose tolerance and increase energy expenditure [11], soluble factors and/or neural signals were likely to be emitted from NRF2-activated skeletal muscles. We suspected that such signals derived from NRF2-activated skeletal muscles acted on liver and/or adipose tissues to supply nutrients to skeletal muscles. Namely, systemic metabolism appeared to be a key to underlying the increase in the exercise capacity of MK mice.
Skeletal muscle-specific Keap1 disruption facilitates the exercise-induced β-oxidation of FAs
We examined changes in systemic metabolism with or without exercise by measuring the plasma metabolites of MK and Cntl mice (see Fig. 8A). Neither genotype nor exercise had significant effects on the plasma levels of glucose and its intermediary metabolites (Fig. 9A and B). However, when we calculated fold changes of metabolite levels with vs. without acute exercise, we observed a significantly lower fold change of lactate in MK mice than in Cntl mice (Fig. 9C). Specifically, in this exercise protocol, the plasma lactate levels were not changed in Cntl mice but decreased in MK mice, suggesting that the skeletal muscles of MK mice utilize the glycolytic pathway less during exercise than do those of Cntl mice. Alternatively or simultaneously, the conversion of lactate to glucose in the liver may be facilitated in MK mice.

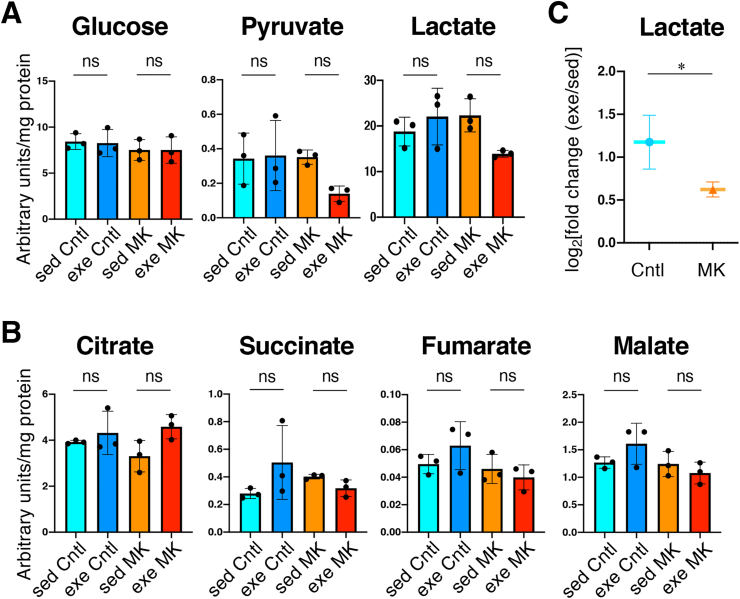
Metabolome analysis using plasma from skeletal muscle-specific Keap1 knockout mice with or without acute exercise.A, B. Glucose and its intermediary metabolites of glycolysis (A) and tricarboxylic acid cycle (B) in plasma (n = 3 mice per group). Mean and SD are indicated. One-way analysis of variance (ANOVA) followed by Tukey's multiple comparison test was conducted for statistical significance. C. Fold change in plasma lactate in the exercise group vs. the sedentary group. The mean and SD of the fold change were calculated from those of lactate shown in panel A.
Cntl; Keap1F/F mice, MK; KeapF/F:Mlc1f-Cre mice, sed; sedentary group, exe; acute exercise group. *P < 0.05, ns; not significant.
Considering the enhanced mitochondrial activity in skeletal muscles of MK mice, we suspected that exercise-induced enhancement of fatty acid utilization can explain the increased exercise endurance of MK mice. To support this hypothesis, we conducted lipidome analysis of the plasma samples. The total concentration of FAs was increased by exercise in both genotypes (Fig. 10A). Exercise did not alter the plasma levels of other lipid species, including monoacylglycerols (MGs), diacylglycerols (DGs) or triacylglycerols (TGs), regardless of genotype (Fig. 10A). Measurement of individual FAs indicated that MK mice had a greater response to exercise than Cntl mice; among 8 FAs quantified in the lipidome analysis, six FAs were significantly elevated in MK plasma, whereas 4 were elevated in Cntl plasma (Fig. 10B). Moreover, after acute exercise, MK mice showed a robust increase in the plasma level of 3-hydroxybutyric acid, which was generated from the β-oxidation of FAs (Fig. 10C). These results strongly suggest that MK mice are more efficient in utilizing FAs as an energy source during exercise via enhanced mobilization of FAs and their subsequent catabolism by β-oxidation.

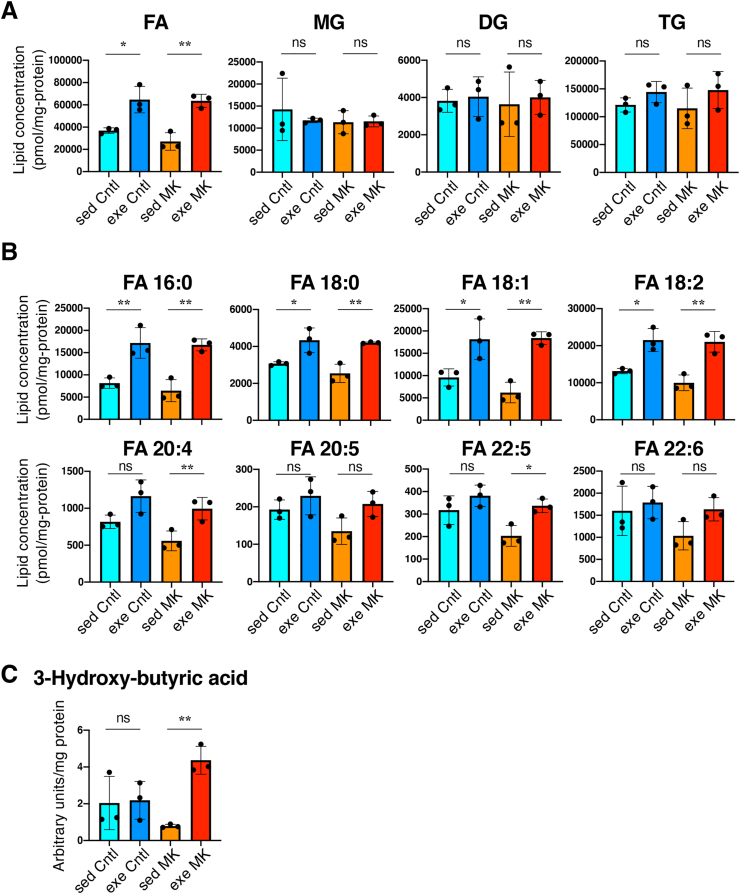
Lipidome analysis using plasma from skeletal muscle-specific Keap1 knockout mice with or without acute exercise.
Cntl; Keap1F/F mice, MK; KeapF/F:Mlc1f-Cre mice, sed; sedentary group, exe; acute exercise group. Mean and SD are indicated. One-way analysis of variance (ANOVA) followed by Tukey's multiple comparison test was conducted for statistical significance. *P < 0.05, **P < 0.01, ns; not significant.
A. Total amount of fatty acids (FAs), monoglycerides (MGs), diglycerides (DGs) and triglycerides (TGs) in plasma (n = 3 mice per group).
B. Individual fatty acids in plasma (n = 3 mice per group).
C. 3-Hydroxybutyric acid quantity in plasma (n = 3 mice per group).
Discussion
This study demonstrated a beneficial role of KEAP1 inhibition in skeletal muscle for endurance capacity, which was accompanied by increased FA mobilization and β-oxidation. To the best of our knowledge, this study is the first to investigate the impact of skeletal muscle-specific KEAP1 inhibition on exercise capacity. Considering a highly strict relationship between KEAP1 and NRF2, we consider that the beneficial effects of KEAP1 inhibition are mostly attributable to NRF2 activation because the currently reported in vivo phenotypes caused by KEAP1 inhibition are all abolished by simultaneous inhibition of NRF2 [[39], [40], [41]] and because Nrf2-deficient mice indeed exhibit impaired exercise endurance [18]. NRF2 activation in skeletal muscles is likely to facilitate interorgan communications between skeletal muscles and adipose tissues via humoral factors and/or neuronal signaling, resulting in increased fatty acid mobilization from adipose tissues. NRF2 activation in skeletal muscles is also likely to exert a cell-autonomous effect, i.e., increasing mitochondrial activity, which probably accounts for the enhanced β-oxidation of FAs. With these results, this study demonstrates multimodal metabolic regulation by NRF2, which enables increased exercise endurance.
It is well established that muscle contractions during exercise produce ROS [42]. Although the precise mechanisms governing ROS production have not been elucidated, mitochondria are regarded as a predominant source of ROS, especially through complexes I and III of the electron transport chain [[43], [44], [45]]. While excessive ROS production by prolonged or high-intensity exercise results in oxidative damage to skeletal muscle [46], recent studies indicate that exercise-induced ROS play a physiological role and act as an important mediator for skeletal muscle adaptations [47] and that muscle contraction and exercise increase NRF2 protein levels and activity via ROS production [35,37]. In our experimental setting, however, we could not detect such tendency of NRF2 pathway activation after acute exercise in Cntl mice (see Fig. 7D). Because forced activation of NRF2 by Keap1 disruption in MK mice suppressed the exercise-induced transcriptional response (see Fig. 7B and C), we believe that optimal activation of NRF2 balances maintaining beneficial ROS levels required for physiological signaling and limiting harmful ROS levels for the protection of tissues from oxidative damage.
NRF2 has been shown to activate mitochondrial activity by controlling substrate availability [48], which supports our current results that the Sol and Plant muscles of MK mice exhibited higher mitochondrial activity, as measured by SDH staining, but did not show a significant increase in mitochondrial DNA. Moreover, facilitation of fatty acid oxidation has been suggested as a major cause of NRF2-mediated enhancement of mitochondrial activity [49], which also appears to be consistent with our results. We found that 3-hydroxybutyric acid was significantly increased in the plasma of MK mice after acute exercise (see Fig. 10C) and that skeletal muscle of MK mice exhibited upregulation of Acox2 (see Fig. 8D), which encodes the branched-chain acyl-CoA oxidase catalyzing degradation of long branched FAs as the first main enzymatic reaction controlling the β-oxidation flux [50]. Although Acox1, which encodes another acyl-CoA oxidase, is abundantly expressed in skeletal muscles and is unlikely to be regulated by NRF2, Acox2 may contribute to the facilitation of β-oxidation in skeletal muscles.
An important remaining question is how Nrf2 activation in skeletal muscles leads to the increased mobilization of FAs in plasma. Exercise-induced FA elevation is derived from both intramuscular lipids and adipose tissue [51]. The relative contribution of the energy source between intramuscular lipolysis and peripheral adipocyte lipolysis depends on the exercise intensity and duration. For low-intensity exercise, which is likely to correspond to the training protocol in our experimental setting, peripheral lipolysis accounts for most of the fuel source. We surmise that NRF2 activation in skeletal muscles facilitates lipolysis in adipose tissues catalyzed by hormone-sensitive lipase via humoral or neuronal signals during exercise. We could not find any clues to the interorgan communication in our RNA-seq data concerning Sol muscles with or without acute exercise.
Another remaining question is the sex difference of NRF2 contributions in skeletal muscles. After observing that fast-oxidative fiber subtype MHC IIa was notably decreased in MK female mice compared with Cntl female mice but not in males, we focused on females. We do not know the reason why skeletal muscle-specific Keap1 disruption generates clear differences in fiber subtypes in females but not in males (see Fig. 3E). Many aspects of skeletal muscles, including overall muscle mass, size of cross-sectional area, lipid content and utilization, relative expression of different myosin isoforms, fatigability and gene expression, differ between males and females [[52], [53], [54], [55], [56], [57]]. In particular, the mean proportion of slow oxidative fiber is higher than that of fast glycolytic fiber in females, and the mean proportion of fast glycolytic fiber is higher than that of slow oxidative fiber in males. Furthermore, intramuscular triacylglycerol content and the fatty acid transporter expression levels are higher in women than in men, and women use more lipids during exercise than men [58,59]. These differences might have augmented the impacts of NRF2 activation in the skeletal muscles of female MK mice.
This study has highlighted the roles played by NRF2 in the regulation of fatty acid metabolism via interorgan communication, which is advantageous for endurance exercise. The function of slow oxidative muscle fibers responsible for endurance exercise is especially critical for preventing sarcopenia and frailty in elderly people [60]. We propose that appropriate NRF2 activation is useful for anti-frailty interventions.
Author contributions
T.O. designed the study, conducted the experiments, analyzed the data and wrote an early draft of the paper. S.M., D.M., N.O., S.M.W. and N.H. conducted the experiments. F.K., Y.I., M.T. and T.B. provided critical experimental systems, conducted the experiments, and wrote the paper. M.O., Y.Y. and M.T. conducted the experiments and analyzed the data. Y.H., M.K. and E.I. analyzed the data, supervised the research and wrote the paper. H.M. designed the study, supervised the research, analyzed the data and wrote an early draft of the paper.
Declaration of competing interest
The authors declare no competing financial or non-financial interests.
References
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
Acknowledgments
We thank Professor Masi Yamamoto, Professor Shyam Biswal and Professor Steve Burden for providing Keap1 knockdown mice, Keap1 floxed mice and Mlc1f-Cre mice, respectively. We also thank the Biomedical Research Cores of the Tohoku University Graduate School of Medicine and Institute of Development, Aging and Cancer for providing technical support. This work was supported by
 Skeletal muscle-specific Keap1 disruption modulates fatty acid utilization and enhances exercise capacity in female mice
Skeletal muscle-specific Keap1 disruption modulates fatty acid utilization and enhances exercise capacity in female mice