
- Altmetric
Vacuolar protein sorting 41 (VPS41) is as part of the Homotypic fusion and Protein Sorting (HOPS) complex required for lysosomal fusion events and, independent of HOPS, for regulated secretion. Here, we report three patients with compound heterozygous mutations in VPS41 (VPS41S285P and VPS41R662 * ; VPS41 c.1423‐2A>G and VPS41R662 *) displaying neurodegeneration with ataxia and dystonia. Cellular consequences were investigated in patient fibroblasts and VPS41‐depleted HeLa cells. All mutants prevented formation of a functional HOPS complex, causing delayed lysosomal delivery of endocytic and autophagic cargo. By contrast, VPS41S285P enabled regulated secretion. Strikingly, loss of VPS41 function caused a cytosolic redistribution of mTORC1, continuous nuclear localization of Transcription Factor E3 (TFE3), enhanced levels of LC3II, and a reduced autophagic response to nutrient starvation. Phosphorylation of mTORC1 substrates S6K1 and 4EBP1 was not affected. In a C. elegans model of Parkinson’s disease, co‐expression of VPS41S285P/VPS41R662 * abolished the neuroprotective function of VPS41 against α‐synuclein aggregates. We conclude that the VPS41 variants specifically abrogate HOPS function, which interferes with the TFEB/TFE3 axis of mTORC1 signaling, and cause a neurodegenerative disease.
Compound heterozygous mutations in VPS41 were identified in patients with a neurodegenerative phenotype with dystonia and cerebellar atrophy. VPS41 variants obstruct the HOPS complex, leading to decreased endocytic and autophagy cargo transfer to lysosomes and inhibition of mTORC1 signaling.
The paper explained
Problem
VPS41 is, as part of the HOPS complex, required for lysosomal fusion events. Independently of HOPS, VPS41 is required for formation of secretory granules. VPS41 prevents degeneration of dopaminergic neurons overexpressing the Parkinson’s disease‐related protein α‐synuclein. Here, we present three patients bearing compound heterozygote mutations in VPS41 displaying a severe neurological disorder. We address the question how these mutations affect endocytosis, autophagy, secretory pathways, and neuroprotection.
Results
We show that mutations in or depletion of VPS41 causes a delay in HOPS‐dependent endocytic and autophagic cargo delivery to enzymatically active lysosomes and that neuroprotection against α‐synuclein is reduced. Moreover, the disease‐causing mutations specifically impair mTORC1 activity toward TFE3 and TFEB, causing constitutive activation of these transcription factors. As a result, lysosomal biogenesis and autophagosome formation is continuously upregulated independent of nutrient conditions. By contrast, HOPS‐independent function of VPS41 in secretory transport is preserved.
Impact
Our study shows that VPS41 patients represent a new class of lysosomal disorders in which lysosomes are functional, but insufficiently reached by cargo due to a trafficking defect. Besides this, we show that VPS41, as part of the HOPS complex, is involved in the differential regulation of mTORC1 toward TFE3/TFEB, which is of potential importance for treatment of HOPS‐related disorders.
Introduction
While lysosomes are responsible for the degradation and recycling of intra‐ and extracellular substrates (Saftig & Klumperman, 2009), they are increasingly recognized as regulators of cellular homeostasis and key players in nutrient sensing and transcriptional regulation (Luzio et al, 2007; Richardson et al, 2010; Settembre et al, 2015; Mony et al, 2016). Hence, the digestive properties of lysosomes provide them with a major role in the control of cellular metabolism and nutrient homeostasis. To accomplish this, HOPS (Homotypic Fusion and Protein Sorting), a multisubunit tethering complex, regulates fusion of lysosomes with endosomes and autophagosomes (Seals et al, 2000; Wurmser et al, 2000; Caplan et al, 2001; Pols et al, 2013a; Kant et al, 2015; Beek et al, 2019).
Vacuolar Protein Sorting 41 (VPS41) (VPS41, found on Chromosomal location; Chr7p14.1) is a defining component of HOPS, and a 100 kD protein that contains a WD40, TRP‐like (tetratricopeptide repeat), CHCR (Clathrin Heavy Chain Repeat), and RING (Really Interesting New Gene)‐H2 Zinc Finger domain (Radisky et al, 1997; Nickerson et al, 2009). VPS41 knockout mice die early in utero, signifying VPS41 as an essential protein for embryonic development (Aoyama et al, 2012). HOPS‐associated VPS41 is recruited to endosomes by binding to Rab7 and its interactor Rab interacting lysosomal protein, and to lysosomes by interacting with Arf‐like protein 8b (Arl8b; Lin et al, 2014; Khatter et al, 2015). Depletion of VPS41 impairs HOPS‐dependent delivery of endocytic cargo to lysosomes and causes a defect in autophagic flux (Takáts et al, 2009; Pols et al, 2013a). Independent of HOPS, VPS41 is required for transport of lysosomal membrane proteins from the trans‐Golgi‐Network (TGN) to lysosomes (Swetha et al, 2011; Pols et al, 2013b), akin to the Alkaline Phosphatase (ALP) pathway in yeast (Cowles et al, 1997a; Rehling et al, 1999; Darsow et al, 2001; Angers & Merz, 2009; Cabrera et al, 2010). Furthermore, in secretory cells and neurons, VPS41 is required for regulated secretion of neuropeptides (Burgess & Kelly, 1987; Orci et al, 1987; Tooze & Huttner, 1990; Eaton et al, 2000; Asensio et al, 2010; Asensio et al, 2013; Hummer et al, 2017). Together, these data show that VPS41, as part of HOPS, is required for lysosomal fusion events and, independent of HOPS, for transport of lysosomal membrane proteins and regulated secretion.
Interestingly, VPS41 overexpression protects dopaminergic neurons against neurodegeneration. This was shown in a transgenic C. elegans model of Parkinson’s disease, in which α‐synuclein is locally overexpressed (Hamamichi et al, 2008; Ruan et al, 2010; Harrington et al, 2012), and in human neuroglioma cells overexpressing α‐synuclein (Harrington et al, 2012). Neuroprotection against α‐synuclein requires interaction of VPS41 with Rab7 and adaptor protein‐3 (AP‐3) (Griffin et al, 2018). Recent studies in C. elegans showed that overexpression of human VPS41 also mitigates Aβ‐induced neurodegeneration of glutamatergic neurons. This requires the small GTPase Arl8b rather than Rab7 or AP‐3, indicating that VPS41, through different interaction partners, can trigger divergent neuroprotective mechanisms against Parkinson’s disease and Alzheimer’s disease (Griffin et al, 2018). However, how VPS41’s function leads to neuroprotection remains to be elucidated.
Naturally occurring SNPs in VPS41 have been described (T52R, T146P and A187T; Harrington et al, 2012; Ibarrola‐Villava et al, 2015), and very recently a single patient with early onset dystonia and a homozygous canonical splice site variant in VPS41 was identified (Steel et al, 2020). In this patient, cDNA studies demonstrated that the variant leads to in‐frame skipping of exon 7. In our current study, we present three patients with severe neurological features (e.g., ataxia and dystonia accompanied by retinal dystrophy and mental retardation with brain MRI findings of cerebellar atrophy and thin corpus callosum) of unknown etiology. Exome sequencing showed that the patients were compound heterozygous for variants in VPS41 [NM_014396.19: c.853T > C, NP_055211.2: p. Ser285Pro (S285P); NM_014396.6: c.1984C > T, NP_055211.2: p. Arg662Stop (R662*); NM_014396.3: c.1423‐2A > G, r.(spl?)]. At the cellular level, we show that expression of these VPS41 variants prevents the formation of a functional HOPS complex, leading to a kinetic defect in the delivery of endocytosed and autophagic cargo to lysosomes. In addition, we find an inhibition of the mechanistic target of rapamycin complex 1 (mTORC1) toward TFEB/TFE3 and concomitantly a constitutive high level of autophagy with a failure to respond to changing nutrient conditions. Finally, we show that compound expression of VPS41S285P and VPS41R662* abolishes the neuroprotective effect of VPS41 in the C. elegans model of Parkinson’s disease. To the best of our knowledge, this is the first study on the cellular consequences of biallelic VPS41 variants in patients displaying neurological manifestations. Our molecular analysis shows that these mutations result in an unexpected defect in mTORC1 signaling, specifically in the TFEB/TFE3 axis regulating autophagy.
Patients
Clinical presentation of three patients with biallelic variants in VPS41
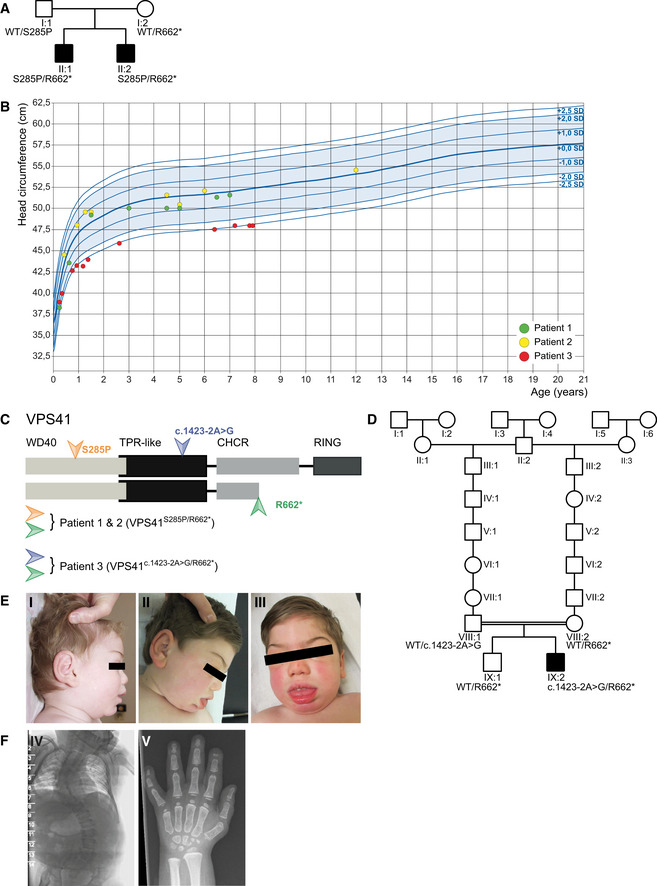
Family 1 (Fig EV1A)—Patients 1 and 2, two brothers, born via spontaneous and vaginal delivery, at term, to healthy and non‐consanguineous parents. Their birth weights were 3.03 kg (10th–50th centile) and 3.34 kg (10th–50th centile), respectively. The birth length and head circumferences were not recorded. No abnormalities were noted after birth, and the babies were discharged on time. Both presented with neonatal hypotonia and poor eye contact and fixation at 2 months of age and both had ophthalmological examination showing hypopigmented and underdeveloped retina which disappeared at 1 year of age. Physical examination at this stage showed a high forehead with frontal bossing, retrognathia, deep set eyes, short and pointed nose, and prominent ears. There was no organomegaly. In infancy, they were noted to have global developmental delay, poor muscle tone, and marked intentional tremor which further impaired their fine and gross motor skills. Both brothers developed progressive spasticity of the lower limbs with some coarsening of the facial features more so in patient 2 with puffy eyelids, heavy eyebrows, and thick lips. Both brothers showed absent deep tendon reflexes (DTRs), and their plantars were extensor. Both developed upper extremity tremor and significant lower limbs’ spasticity and ataxia (Movies [Link], [Link], [Link]). Extensive investigation for metabolic diseases, including lysosomal and mitochondrial disorders, showed no detectable abnormalities. No urinary mucopolysaccharides and oligosaccharides were detected. Cerebrospinal fluid analysis for neurotransmitter levels showed no abnormalities. Brain MRI done in infancy on both brothers showed mild hypomyelination (Fig 1A and B). However, a repeat brain MRI study on the older brother (patient 1) at 4 years 7 months showed thin corpus callosum with a saber‐shape configuration (Fig 1A) and the vermis, although normal in configuration and size initially, demonstrated volume loss on follow‐up examinations. Furthermore, brain MRI done on the younger brother (patient 2) at 10 years of age confirmed the thin corpus callosum, mild progression of the cerebellar atrophy, bilateral hypointensity in the globus pallidus (GP) compatible with early iron deposition, and abnormal hyperintensity in the dentate nucleus (Fig 1B). The overall findings were consistent with neurodegenerative rather than solely malformative brain disease. The growth charts for head circumference were within normal range (Fig EV1B).

Mutations in VPS41 cause a neurodegenerative disease
Pedigrees of family 1 affected by recessive mutations in VPS41. Circle = female, square = male, black‐filled shape = individual phenotypically affected.
Head circumference of patients 1 (green), 2 (yellow), and 3 (red) in cm.
Outline of the VPS41 protein depicting distinct domains. The WD40 domain facilitates protein–protein interactions and the CHCR and RING domains enable homo‐oligomerization and are required for HOPS complex formation and regulated secretion. Mutations in VPS41 were identified using whole exome sequencing. The two siblings (patients 1 and 2) bear compound heterozygous mutations; a missense mutation VPS41S285P in the WD40 domain and a nonsense mutation VPS41R662 * at the C‐terminus resulting in a premature stop codon. Patient 3 bears a canonical splicing variant expected to destroy an acceptor site (VPS41c . 1423‐2A>G) in the TPR‐like domain and shares the nonsense mutation (VPS41R662 *) at the C terminus with the other patients.
Pedigree of family 2 affected by recessive mutations in VPS41. Circle = female, square = male, black‐filled shape = individual phenotypically affected, double line = consanguinity.
Patient 3 developed coarse facial features with heavy eyebrows, gingival hypertrophy, protruding tongue, thick lips, and thick ear lobes (I; age 1 year, 7 months, II and III; age 4 years, 10 months).
A spinal X‐ray showed a kyphosis at C3 and hypoplastic distal phalanges on digits 2, 3, and 5 and a short metacarpal 1, bilaterally (IV; age 5 years, V; age 11 years).

Mutations in VPS41 cause a neurodegenerative disease
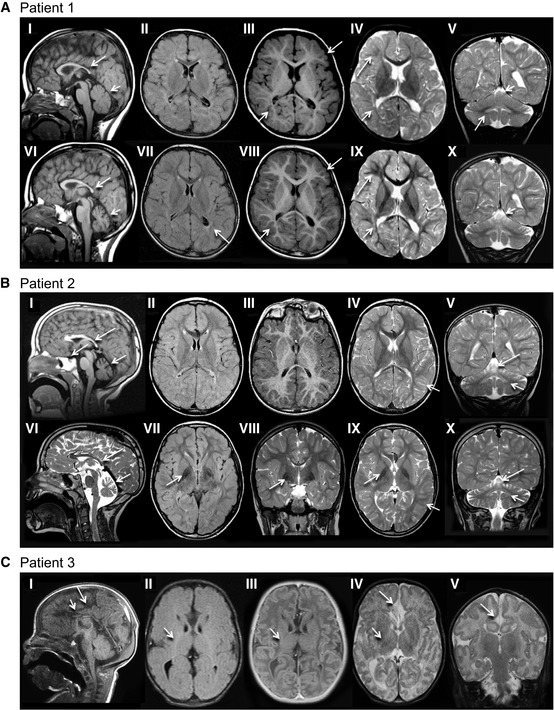
Patient 1 (older sibling) underwent 3 MRI studies, first and last are shown. Top row: 21 months; Bottom row: 4 years 7 months. The corpus callosum is thin on T1‐weighted sagittal images (I, V, VI) with a saber shape (long arrows). The shape remains consistent over time. The vermis is normal in configuration and size initially (I), but demonstrates volume loss on follow‐up (V, VI) examination (short arrows). FLAIR axial image (II) is age appropriate, while (VII) demonstrates periatrial increased signal (arrow). There is delay in myelin maturation present on T1 (III, VIII)‐ and T2 (IV, IX)‐weighted axial images (long arrows) on initial examination, with further slow myelin development over time. There is deficiency of periatrial white matter volume on both T1 (III, VIII)‐ and T2 (IV, IX)‐weighted axial images (short arrows). Coronal T2 image (V) demonstrates abnormally increased signal of the dentate nuclei (arrow), unchanged on follow‐up image (X). Superior vermian atrophy (short arrows) over time.
Patient 2 (younger sibling). Top row: 4 years, 11 months; Bottom row: 9 years, 6 months. The corpus callosum is thin with a saber‐shape (long arrows) configuration on T1‐weighted sagittal (I) and T2‐weighted sagittal (V, VI). The shape remains consistent over time, although the volume decreases slightly. The vermis demonstrates volume loss on both sagittal examinations (short arrows). There is iron deposition in the basal ganglia on FLAIR (VII) and T2‐weighted coronal (VIII) and axial (IX) images at 9 years 8 months of age (arrow). This was not present on earlier imaging. Iron deposition was also present in the subthalamic nuclei (not shown). Myelin maturation is age appropriate on the initial T1‐weighted axial image (III) and minimally delayed on T2‐weighted image (IV) at 4 years 11 months of age (short arrow). Myelin is age appropriate (short arrow) on follow‐up T2‐weighted images (VIII, IX). The dentate nuclei (short arrows) are bright on T2 coronal images (V, VI, X) at both ages. There is progressive cerebellar hemisphere volume loss.
Patient 3, 3 weeks old. Sagittal T1‐weighted image (I) demonstrates a very thin severely hypoplastic remnant of corpus callosum (short arrow), a short cingulate sulcus (long arrow), and slender pons. FLAIR image (II) reveals faintly increased signal (short arrow) in the posterior limb of internal capsule (PLIC). There is lack of myelin maturation in the PLIC (short arrows) on T1‐IR (III)‐ and T2 (IV)‐weighted axial images. Focal widening of the interhemispheric CSF space (arrow) on T2‐weighted image (IV) reflects the absence of rostral fiber tracts. Cingulate gyrus is present (long arrow). No traversing callosal fibers are shown at this level (I).
DNA analyses using a gene panel including genes associated with mental retardation and dystonia as well as genes associated with iron deposition in the basal ganglia showed no detectable variants (Appendix Table S1). Microarray analysis (IDT xGen Exome Research Panel v1.0) revealed a de novo duplication of 2.568 Mb at 1q21.1 [GRCh37/hg19 chr1:145,804,790–148,817,029] in patient 1 only. This region partially overlaps with the 1q21.1 microduplication syndrome and is of unknown significance (Brunetti‐pierri et al, 2008). Whole exome sequencing done on the brothers and their parents showed that both patients are compound heterozygote for variants in the VPS41 gene in trans; they carried a missense variant in the WD40 domain [Chr7(GRCh37)[(NM_014396.3:c.853T > C, NP_055211.2:pSer285Pro (S285P)(heterozygote, paternal); hereafter referred to as VPS41S285P] and a nonsense variant in the Clathrin Heavy Chain Repeat (CHCR) resulting in a truncated protein [c.1984C > T,NP_055211.2:pArg662Stop(R662*)(heterozygote, maternal); hereafter referred to as VPS41R662 *] (Fig EV1C). The VPS41S285P variant has mixed in silico predictors and is observed in 2/282128 (0.0007%) alleles in gnomAD. The VPS41R662 * variant is predicted to cause loss of normal protein function either through protein truncation or nonsense‐mediated mRNA decay. It is observed in 98/282180 (0.03%) alleles in gnomAD, and no individuals are reported to be homozygous. The gnomAD frequencies and predictions of pathogenicity by available software programs are shown in Table 1.

| VPS41 (NM_014396.3) | gnomAD global | SIFT | CADD | PolyPhen2 HDIV | PolyPhen2 HVAR | Patient |
|---|---|---|---|---|---|---|
|
c.853 T > C p.S285P | 0.0000071; 2/282128 |
0.048 damaging | 23.5 |
0.956 possibly damaging |
0.361 benign | 1 and 2 |
| c.1984 C > T p.R662* | 0.00035; 98/282180 | (b) | 40 | (b) | (b) | 1, 2 and 3 |
| c.1423‐2A > G p.? (a) | 0.000032; 1/31398 | (b) | 34 | (b) | (b) | 3 |
Frequency of the specific VPS41 variants in the general population based on the gnomAD database ([variants observed]/[total of individuals studied]) (http://gnomad.broadinstitute.org/) and prediction of pathogenicity based on the programs SIFT, (https://sift.bii.a‐star.edu.sg/), CADD (https://cadd.gs.washington.edu/), and PolyPhen (http://genetics.bwh.harvard.edu/pph2/).
(a): Three different splice site analysis programs (MaxEnt, NNSPLICE, and SSF) predict a total loss of wild‐type acceptor splice site (Interactive Biosoftware ‐ Created by Alamut Visual v.2.15.0).
(b): Not determined; SIFT and PolyPhen2 programs can only be used for analysis of missense mutations.
Compound heterozygous missense variants in the ITGB4 gene were also identified; however, ITGB4 is associated with autosomal recessive non‐Herlitz junctional epidermolysis bullosa which does not fit the phenotype observed in this family.
A description of the whole exome sequencing technique used, the filtering strategy, variant analysis and confirmation by Sanger sequencing, as well as a list of rare variants detected, can be found in the Supplements (Appendix Supplementary Methods and Dataset EV1, respectively).
Family 2 (Fig EV1D)—Patient 3, a boy, born to a distantly related parents of Jewish descent (common ancestor in 18th century, Fig EV1D). The pregnancy was uneventful and delivery was at 40wk 1d by Cesarean section for decelerations and meconium‐stained amniotic fluid. His birth weight was 3290 grams (10th–50th centile). On examination, he was noted to have a head circumference at the 3rd centile, retrognathia, short and upturned nose, relatively large ears, clinodactyly of both 5th fingers and hepatosplenomegaly. He was noted to be hypotonic and jittery. He had recurrent infections during infancy and childhood, photophobia, mild sensorineural hearing deficit which required hearing aids, tracheomalacia, and a severe global delay. He developed hypertension and a progressive spasticity of the lower limbs from the age of 5 years onwards. His facial features coarsened over time with heavy eyebrows, gingival hypertrophy, protruding tongue, thick lips, and thick ear lobes (Fig EV1E I–III). He had a pectus carinatum and short stubby hands. The liver and spleen sizes gradually normalized, and by age 5, there was no longer hepatosplenomegaly. At that age, the boy had severe global developmental delay, a convergent strabismus with restricted vertical eye movements, axial hypotonia with peripheral spasticity, brisk DTRs at the upper limbs, and weak DTRs on the lower extremities. He had short Achilles tendons with equinus feet position. There was a dysmetry when reaching for objects, but no tremor. In the following years, the boy developed a scoliosis and severe spastic tetraplegia with contractures, first in the lower extremities and later in the upper extremities (starting in the hands). His skin felt soft and thick, and there was hirsutism on his back and legs. He never developed the ability to sit, walk, or speak. He was able to move in a walking device, but later lost that ability. His ability to cough decreased from the age of 12 years, and he developed swallowing problems, resulting in more upper airway infections. His height and head circumference remained at −2.5 SD during the years (Fig EV1B).
Ophthalmologic investigation was normal apart from retinal hypopigmentation. An ECG and cardiac ultrasound were normal. Extensive metabolic investigation including analysis for peroxisomal and lysosomal disorders, including urinary mucopolysaccharides and oligosaccharides as well as CDG (sialo transferrines), showed no abnormalities. A spinal X‐ray showed a kyphosis at C3 (Fig EV1F IV) and X‐rays of the hands showed hypoplastic distal phalanges on digits 2, 3, and 5 and a short metacarpal 1, bilaterally (Fig EV1F V). A cerebral MRI showed agenesis of the corpus callosum (Fig 1C), bilateral colpocephaly, a dysplastic cerebellum with polymicrogyria of the upper part, and a dysplastic pons.
Microarray analysis (Agilent 180 K custom HD‐DGH microarray; (AMADID‐nr. 27730)) revealed no chromosomal imbalances, targeted genetic analysis for several disease genes, and a gene panel including approximately 800 genes associated with intellectual disability, showed no detectable variants. Trio whole exome sequencing showed compound heterozygous variants in the VPS41 gene; a paternal variant in the TPR‐like domain [NM_014396.3:c.1423‐2A > G p.?; hereafter referred to as VPS41 c.1423‐2A>G] and a maternal nonsense variant in the CHCR domain [c.1984C > T,NP_055211.2:pArg662Stop(R662*)(heterozygote, maternal); hereafter referred to as VPS41R662 *]. The c.1423‐2A > G variant is a splice site variant that destroys the canonical splice acceptor site in intron 17 and is predicted to cause abnormal gene splicing. The VPS41R662 * nonsense variant is shared between all three patients and heterozygously present in the healthy brother of patient 3 (Fig EV1C). Patient 3 also carried a homozygous missense variant of unknown significance in UPF3A [NM_080687.2:c.707G > A, NP_542418.1:pArg236Gln] for which both parents and his healthy sibling were heterozygotes. This variant is located in an evolutionary conserved region, but not within a known functional domain of the protein. At this point, variants in this gene have not been associated with any human disease.
A description of the whole exome sequencing technique used, the filtering strategy, variant analysis, and confirmation by Sanger sequencing, as well as a list of rare variants detected, can be found in the Supplements (Appendix Supplementary Methods and Dataset EV2, respectively). The gnomAD frequencies and prediction of pathogenicity by available software programs are shown in Table 1.
Results
Patient fibroblasts and VPS41KO cells contain small‐sized, acidified lysosomes active for cathepsin B
To study the cellular effects of VPS41 variants in patient cells, we obtained skin biopsies from the youngest brother of the first family (patient 2; VPS41S285P/R662 *), patient 3 (VPS41 c.1423‐2A>G /R662 *), an independent control (VPS41WT/WT), and from the father and mother of patients 1 and 2 (VPS41WT/S285P and VPS41WT/R662 *) and made primary fibroblast cultures. In addition, we mimicked disease conditions in HeLa cells using CRISPR/Cas9 methodology. We chose to study fibroblasts from patient 2, because patient 1 also displayed the de novo duplication at Chr1q21.1, which partially overlaps with the 1q21.1 microduplication syndrome (Brunetti‐pierri et al, 2008). The clinical features of patient 1 are more severe than in individuals suffering from 1q21.1 duplication syndrome; however, we cannot rule out that patient fibroblasts are affected by this duplication. Similarly, patient 3 bears a homozygous mutation in UPF3A of unknown pathogenicity, which possibly could induce alterations in fibroblasts.
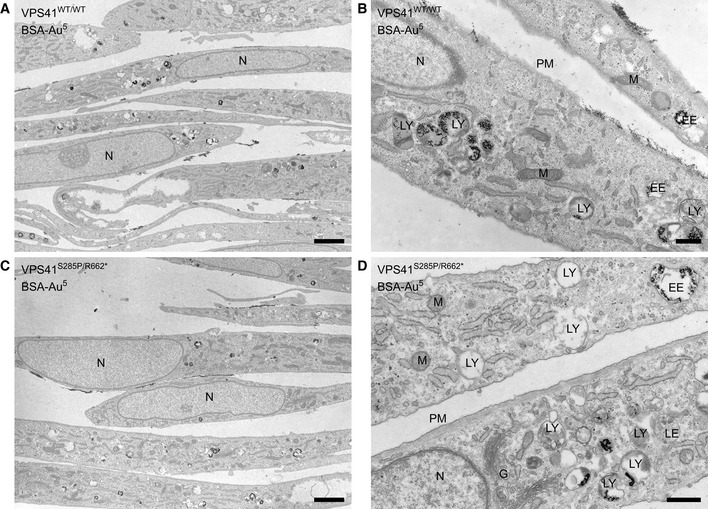
First, we studied the effects of the VPS41 mutations on lysosomal morphology in the VPS41S285P/R662 * fibroblasts. Electron microscopy (EM) showed a high variation in appearance of endolysosomal compartments (Fig EV2A–D), but no inclusion bodies. This indicates that mutations in VPS41 do not cause a classical lysosomal storage phenotype (Papa et al, 2010), which is in agreement with the metabolic studies performed on patient fibroblasts, leucocytes, and urine (see patient descriptions). To study lysosomal acidification and hydrolase activity, we performed fluorescence microscopy using MagicRed cathepsin B. This showed an increase in the number of cathepsin B‐active puncta as compared to control or parental cells (Appendix Fig S1A, quantified in S1A’). Concomitantly, immunofluorescence of the lysosomal membrane protein LAMP‐1 showed a significant increase in both patients (Fig 2A, quantified in 2A’). Similarly, HeLaVPS41KO cells, i.e., HeLa cells knockout for VPS41 using CRISPR/Cas9 methodology (Fig 2B), showed numerous small LAMP‐1 puncta (Fig 2C, quantified in 2C’) and an increase in acidic compartments, investigated using Lysotracker‐Green (Appendix Fig S1B, quantified in S1B’).

Mutations in VPS41 do not cause a lysosomal storage phenotype
A, BElectron micrographs of Epon sections of VPS41WT/WT fibroblasts.
C, DElectron micrographs of Epon sections of VPS41S285P/R662 * fibroblasts.
Data information: Cells were incubated with BSA‐Au5 for 2 h to label endolysosomal compartments. Both primary fibroblast cell lines show a high variation in the appearance of endolysosomal compartments. There is no aberrant swelling of endolysosomal organelles in patient VPS41S285P/R662 * fibroblasts. G = Golgi, EE = Early endosome, LE = Late endosome, LY = Lysosome, M = Mitochondria, N = Nucleus, PM = Plasma membrane. Scale bars low magnification (A, C), 2 µm; High magnification (B, D), 500 nm.

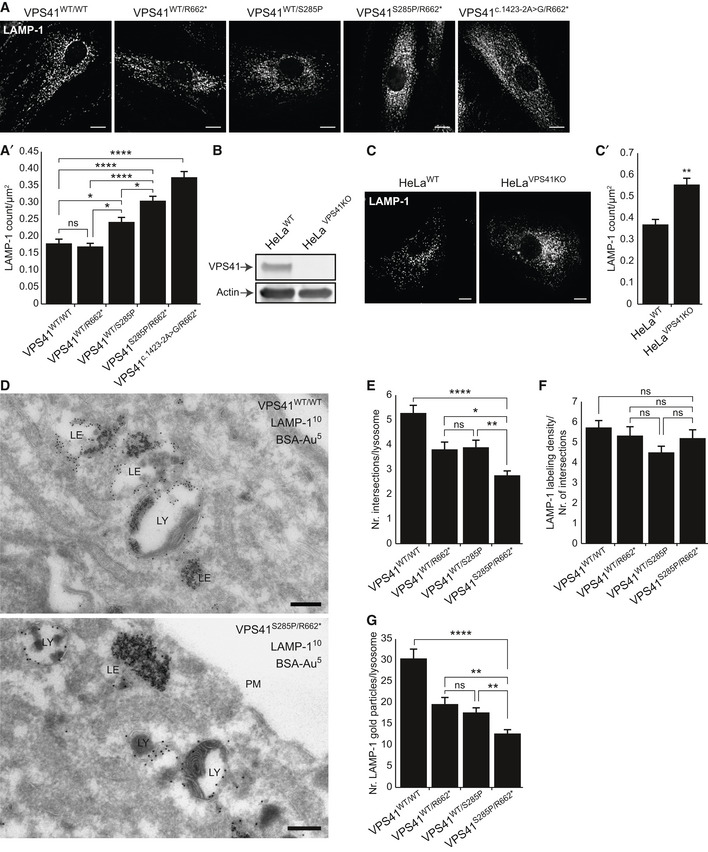
VPS41 patient and HeLa VPS41 knockout cells contain more but smaller lysosomes
Immunofluorescence microscopy of control, parental, and patient fibroblasts labeled for LAMP‐1. Patient fibroblasts (VPS41S285P/R662 * and VPS41c . 1423‐2A>G/R662 *) show more LAMP‐1 puncta, which are distributed throughout the cell. (A’) Quantification shows a significant increase in the number of LAMP‐1‐positive compartments for both patients. > 15 Cells per condition were quantified (n = 3). Scale bars, 10 µm.
HeLa VPS41 knockout (HeLaVPS41KO) cells made using CRISPR/Cas9 methodology. Western blot analysis confirms a full knockout. The same HeLaWT and HeLaVPS41KO samples were analyzed in Appendix Fig S8C, showing the same actin control.
LAMP‐1 immunofluorescence of HeLaWT and HeLaVPS41KO cells. Similar to patient‐derived fibroblasts, more LAMP‐1‐positive compartments are seen in HeLaVPS41KO cells (quantified in C’). > 10 Cells per cell line per experiment were quantified (n = 3). Scale bars, 10 µm.
Immuno‐electron microscopy of VPS41WT/WT and VPS41S285P/R662 * fibroblasts incubated for 2 h with BSA conjugated to 5 nm gold (BSA‐Au5) and labeled for LAMP‐1 (10 nm gold particles). Lysosomes are recognized by the presence of degraded, electron‐dense material. LE = late endosome, LY = lysosome, PM = plasma membrane. Scale bar, 200 nm.
Morphometrical analysis showing that lysosomes in patient fibroblasts are significantly smaller than in control and parental cells. > 100 Randomly selected lysosomes per condition were quantified.
Relative labeling density of LAMP‐1. Number of LAMP‐1 gold particles per lysosome was divided by the number of grid intersections, representing lysosomal size. No significant difference was found between VPS41WT/WT, VPS41WT/S285P, VPS41WT/R662 * or VPS41S285P/R662 * fibroblasts. > 53 Lysosomes per condition were quantified.
Quantitation of LAMP‐1 gold particles per lysosome. VPS41S285P/R662 * lysosomes have significantly less LAMP‐1 then control and parental cells. > 53 Lysosomes per condition were quantified. Similar results were obtained for LAMP‐2 (Appendix Fig S2).
Data information: Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ****P < 10−5. One‐way ANOVA with Tukey’s correction (A’, E, F and G) or Unpaired t‐test (C’). Exact P‐values are reported in Appendix Table S3.
Source data are available online for this figure.
LAMP‐1 resides in lysosomes as well as late endosomes, which by EM can be distinguished by the presence or absence of degraded material, respectively (Peden et al, 2004; Huotari & Helenius, 2011; Pols et al, 2013a; Klumperman & Raposo, 2014). By immuno‐EM, we found that VPS41S285P/R662 * fibroblasts contain LAMP‐1‐positive late endosomes and lysosomes (Fig 2D), which were also positively labeled for cathepsin B (Appendix Fig S1C). However, in agreement with the fluorescent microscopy observations, morphologically identified, LAMP‐1‐positive lysosomes in patient‐derived cells were significantly smaller than in VPS41WT/WT, VPS41WT/S285P or VPS41WT/R662 * fibroblasts (Fig 2E). The relative labeling density (an indication for protein concentration per membrane unit) of LAMP‐1 in lysosomes remained equivalent to control cells (Fig 2F), but since lysosomes were smaller, the total number of gold particles (indication for total amount of LAMP‐1) had decreased (Fig 2G). Similar data were obtained for LAMP‐2 (Appendix Fig S2A and B).
These data show that lysosomes in patient cells contain LAMP‐1, LAMP‐2, and cathepsin B, but are considerably smaller in size than in control cells. We conclude that biallelic expression of VPS41R662 * with VPS41S285P or VPS41c . 1423‐2A>G, as well as the absence of VPS41 by gene knockout, induces an increase in small‐sized, enzymatically active lysosomes, which at steady state conditions contain normal LAMP levels.
VPS41R662 * is not detectable in fibroblasts
To establish expression levels of the VPS41 variants, we performed quantitative Western blots of patients and parental fibroblasts. Wild‐type VPS41 was readily detectable in VPS41WT/R662 * fibroblasts from the mother of patients 1 and 2 (Fig 3A). Also in the paternal VPS41WT/S285P fibroblasts there was a strong signal for full‐length VPS41, in this case representing both VPS41WT and VPS41S285P (Fig 3A). VPS41R662*, the variant shared between all 3 patients, encodes for a premature stop codon predicted to result in a truncated protein of lower molecular weight. However, we could not detect a lower VPS41R662* band in either VPS41S285P/R662 * patient or VPS41WT/R662 * maternal fibroblasts (Fig 3A, Appendix Fig S3A). Treatment with proteasome inhibitor MG132, to prevent degradation of VPS41R662*, did not change this outcome. Since the VPS41R662* variant is recognized by the VPS41 antibody, these data indicate that VPS41R662* protein levels in fibroblasts are below detection. RNA bearing a premature stop codon can be prematurely degraded via Nonsense‐Mediated Decay (NMD). To determine whether this is the case for VPS41R662 *, we performed RT–PCR on VPS41WT/WT, VPS41WT/R662 *, VPS41WT/S285P, and VPS41S285P/R662 * fibroblasts. Indeed, maternal and patient fibroblasts showed a significant decrease in VPS41 mRNA levels (Appendix Fig S3B). Together, these data show that VPS41R662 * mRNA levels are below detection due to premature degradation of the mutant mRNA. In contrast to VPS41R662*, the VPS41S285P variant was readily detectable in Western blots of VPS41S285P/R662 * fibroblasts (Fig 3A). Since this is the only allele that is expressed, the total VPS41 expression was lower than in control cells (Fig 3A’). Finally, our Western blot analysis revealed a striking reduction, to circa 10% of wild‐type levels, of VPS41 protein levels in VPS41c . 1423‐2A>G/R662 * fibroblasts (Fig 3A, quantified in 3A’ and Appendix Fig S3A). Since the VPS41R662 * variant is not expressed, these data indicate that the VPS41c . 1423‐2A>G variant is expressed at a very low level.

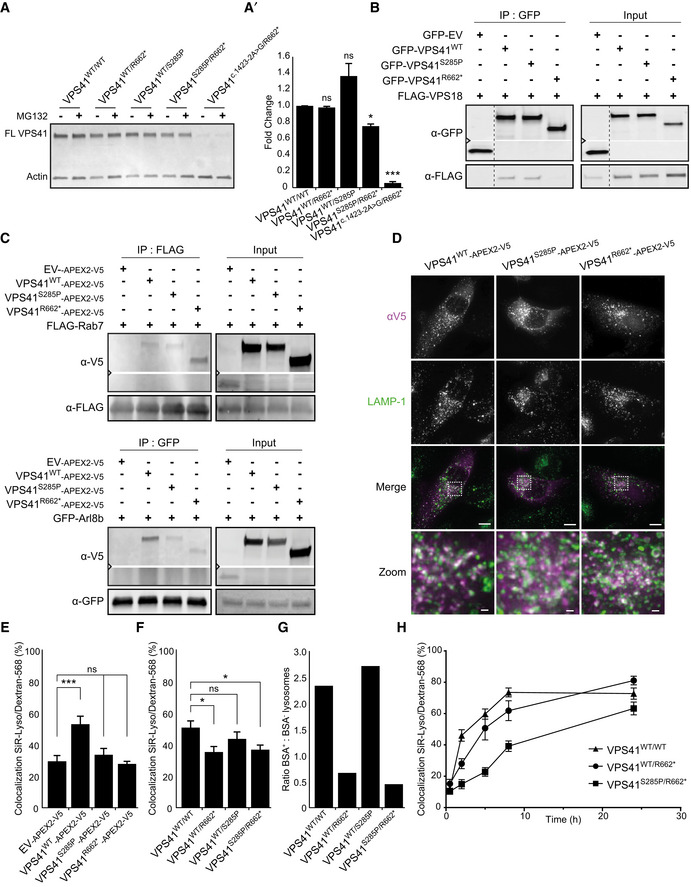
VPS41 variants delay HOPS‐dependent late endosome–lysosome fusion
Western blot of primary fibroblasts derived from patient 2 (VPS41S285P/R662 *), his mother (VPS41WT/R662 *), father (VPS41WT/S285P), patient 3 (VPS41 c.1423‐2A>G /R662 *) and an unrelated, healthy control (VPS41WT/WT) (Western blot of longer exposure time shown in Appendix Fig S3A). Full‐length VPS41 (FL VPS41), representing VPS41WT and VPS41S285P, is observed in all cells. Truncated VPS41R662* in patients and maternal fibroblasts is not detectable. VPS41R662* is also not visible in fibroblasts treated with 50 µM MG132 (4 h) to inhibit proteasomal degradation, indicating that the mRNA encoding for this mutant is degraded. (A’) Quantification of VPS41 protein levels show a reduction in both patients compared with VPS41WT/WT, with only 10% remaining VPS41 levels in VPS41 c.1423‐2A>G /R662 * (n = 2).
Immunoprecipitation (IP) on HeLa cells co‐expressing GFP‐empty vector (EV), GFP‐VPS41WT, GFP‐VPS41S285P, or GFP‐VPS41R662* and FLAG‐VPS18. GFP‐VPS41WT and GFP‐VPS41S285P both interact with FLAG‐VPS18 and as shown in Appendix Fig S4A with HA‐VPS33A. Interaction between GFP‐VPS41R662* and FLAG‐VPS18 is strongly reduced. Note that the stop codon in VPS41R662 * leads to a truncated protein of lower molecular weight (n = 3).
IP on Hela cells co‐expressing VPS41WT‐APEX2‐V5, VPS41S285P‐APEX2‐V5, or VPS41R662*‐APEX2‐V5 and FLAG‐RAB7 or GFP‐Arl8b. The IP was performed on FLAG or GFP, respectively. All VPS41 variants interact with Rab7 and Arl8b (n = 3).
HeLaVPS41KO cells transfected with VPS41WT‐APEX2‐V5, VPS41S285P‐APEX2‐V5, or VPS41R662*‐APEX2‐V5 constructs labeled for LAMP‐1 and V5 immunofluorescence microscopy. All VPS41 variants colocalize with LAMP‐1, indicating that VPS41S285P and VPS41R662* are recruited to late endosomes/lysosomes. Scale bars 10 µm; zoom of squared area, 1 µm.
Quantification of endocytosis‐rescue experiments in HeLa cells. HeLaVPS41KO cells transfected with VPS41WT‐APEX2‐V5 show a significant increase in colocalization of endocytosed Dextran‐568 and SiR‐Lysosome cathepsin D (SiR‐Lyso), indicating rescue of the endocytosis phenotype. Neither VPS41 variant rescues this HOPS complex functionality. >13 Cells per cell line per experiment were quantified (n = 3).
Quantification of lysosomal delivery of endocytosed cargo in fibroblasts, based on fluorescent data. VPS41WT/WT, VPS41WT/S285P, VPS41WT/R662 *, and VPS41S285P/R662 * primary fibroblasts were incubated with Dextran‐568 and SiR‐Lysosome cathepsin D (SiR‐Lyso) for 2 and 3 h, respectively. Colocalization representing delivery of Dextran to enzymatically active lysosomes is reduced in VPS41WT/R662 * and VPS41S285P/R662 * cells. >11 Cells per cell line per experiment were quantified (n = 3).
Quantification of lysosomal delivery of endocytosed cargo in fibroblasts, based on EM data. VPS41WT/WT, VPS41WT/S285P, VPS41WT/R662 *, and VPS41S285P/R662 * fibroblasts were incubated with BSA‐Au5 for 2 h and labeled for LAMP‐1 (10 nm gold particles) immuno‐EM (Fig 2D). LAMP‐1‐positive lysosomes were scored for presence of BSA‐Au5, and the ratio between BSA+ and BSA‐ lysosomes was calculated. > 46 Lysosomes per condition were quantified. Both VPS41WT/R662 * and VPS41S285P/R662 * show a strong decrease in BSA‐positive lysosomes indicating a fusion defect between late endosomes and lysosomes.
VPS41WT/WT, VPS41WT/R662 *, and VPS41S285P/R662 * primary fibroblasts incubated with Dextran‐568 for 0.5, 2, 5, 8, and 24 h. Colocalization of Dextran‐568 and SiR‐Lysosome cathepsin D (SiR‐Lyso) reveals a delay in lysosomal delivery in maternal (VPS41WT/R662 *) and patient (VPS41S285P/R662 *) cells at 2 h of Dextran uptake. After 5 h, maternal cells show similar to control colocalization levels. Patient cells show only after 24 h colocalization levels similar to control, indicating a delay rather than a block in late endosome–lysosome fusion. > 10 Cells per cell line were quantified.
Data information: Data are represented as mean ± SEM. *P < 0.05, ***P < 10−4. Unpaired t‐test (A’), one‐way ANOVA with Bonferroni correction (E) or one‐way ANOVA with Tukey’s correction (F). Exact P‐values are reported in Appendix Table S3.
Source data are available online for this figure.
We conclude from these data that fibroblasts of patients 1 and 2 only contain substantial levels of VPS41S285P, whereas cells of patient 3 only contain circa 10% of the VPS41c.1423‐2A>G variant.
VPS41S285P interacts with other HOPS subunits, Arl8b and Rab7
In control conditions, VPS41 interacts with other HOPS subunits and the small GTPases Rab7 and Arl8b (Lin et al, 2014; Khatter et al, 2015). To study whether VPS41 mutations affect HOPS assembly, we performed co‐immunoprecipitation (co‐IP) studies. We focused on VPS41S285P, being the only variant significantly expressed in patient cells (Fig 3A), and included VPS41R662* as negative control, since VPS41–HOPS interaction requires the presence of the C‐terminal RING domain absent in VPS41R662* (Fig EV1C; Hunter et al, 2017). We performed the co‐IP studies in HeLa cells expressing GFP‐tagged constructs of VPS41WT, VPS41S285P, or VPS41R662* and, respectively, FLAG‐ and HA‐tagged constructs of the HOPS core components VPS18 or VPS33A. As expected, the truncated VPS41R662* mutant clearly showed reduced interactions with the other HOPS subunits (Fig 3B and Appendix Fig S4A). By contrast, the point mutation VPS41S285P did not affect interaction with VPS18 and VPS33A (Fig 3B and Appendix Fig S4A). Additionally, we examined in HeLaVPS41KO cells the interaction between endogenous VPS18 and GFP‐tagged VPS41 constructs. We found that GFP‐VPS41S285P and GFP‐VPS41WT bind endogenous VPS18 with similar affinities, whereas no interaction between endogenous VPS18 and VPS41R662* was found (Appendix Fig S4B). These data indicate that VPS41S285P can correctly bind to the HOPS complex, whereas VPS41R662* lacks this ability. Hence, if VPS41R662* would be expressed at sufficient levels, it would not be incorporated in the HOPS complex.
Next, we performed co‐IPs using transiently transfected HeLaWT cells with APEX‐V5‐tagged VPS41 constructs and FLAG and GFP‐tagged constructs of Rab7 and Arl8b, respectively. This showed that both VPS41S285P and VPS41R662* bind Rab7 and Arl8b to the same extent as VPS41WT (Fig 3C). The subcellular distribution of the distinct VPS41 constructs was explored by immunofluorescence microscopy. VPS41WT was present in the cytoplasm as well as in distinct fluorescent puncta that colocalized with LAMP‐1, consistent with previous studies (Khatter et al, 2015; Jia et al, 2017; Fig 3D). Remarkably, both VPS41S285P and VPS41R662* showed a similar distribution as VPS41WT, despite the lack of the C terminus in VPS41R662*. Since VPS41R662* binds Rab7 and Arl8b (Fig 3C), these interactions could mediate recruitment of VPS41R662* to late endosomes and lysosomes independent of its C‐terminus or the HOPS complex.
Summarizing, these co‐IP experiments show that VPS41S285P has retained the capacity to bind the HOPS components VPS18 and VPS33A, as well as Rab7 and Arl8b. VPS41R662* does not bind other HOPS components, but still interacts with Rab7 and Arl8b. Both VPS41S285P and VPS41R662* are distributed between the cytoplasm and LAMP‐1‐positive endo‐lysosomes.
VPS41S285P causes a defect in HOPS‐dependent late endosome–lysosome fusion
Depletion of VPS41 by RNAi results in a decrease in HOPS‐dependent fusion between late endosomes and lysosomes (Swetha et al, 2011; Pols et al, 2013a). To establish the effect of VPS41S285P on HOPS functionality, we transiently transfected HeLaVPS41KO cells with APEX2‐V5 tagged constructs of VPS41WT, VPS41S285P, or VPS41R662*. Cells were incubated for 2 h with the endocytic marker Dextran‐Alexa Fluor 568 (Dextran‐568) combined with SiR‐Lysosome to mark active cathepsin D compartments. Colocalization between these probes indicates the transfer of endocytosed cargo to enzymatically active lysosomes, which is HOPS dependent. VPS41KO cells transfected with Empty Vector (EV) showed low levels of colocalization (Fig 3E and Appendix Fig S5A), which was increased by transfection with VPS41WT, indicating a restoration of HOPS function (Fig 3E and Appendix Fig S5A). As expected, transfection with VPS41R662* did not rescue the endocytosis defect, since VPS41R662* fails to bind other HOPS components (Fig 3B and Appendix Fig S4). Surprisingly, however, VPS41S285P also failed to rescue the endocytosis defect (Fig 3E and Appendix Fig S5A). Thus, even though VPS41S285P binds VPS18 and VPS33A (Fig 3B and Appendix Fig S4) and localizes to endo‐lysosomes (Fig 3D), it does not form a functional HOPS complex. A plausible explanation for this defect is misfolding of the VPS41S285P protein due to the Arginine to Proline substitution.
These data show that, despite its ability to bind other HOPS components, VPS41S285P cannot form a functional HOPS complex.
Patient fibroblasts are compromised in delivery of endocytosed cargo to enzymatically active lysosomes
The data so far indicate that all patient‐derived VPS41 variants prevent formation of a functional HOPS complex. To assess the process of late endosome–lysosome fusion in patient cells, we performed the Dextran‐568 and SiR‐Lysosome cathepsin D colocalization assay in primary fibroblasts of patient 2 and his parents. A similar assay with VPS41 c.1423‐2A>G /R662 cells of patient 3 failed to be successful, since these cells are very vulnerable and died upon incubation with SiR‐Lysosome. We found a significant endocytosis defect in VPS41S285P/R662 * patient fibroblasts (Fig 3F and Appendix Fig S5B) and, unexpectedly, also in the maternal VPS41WT/R662 * cells. By contrast, VPS41WT/S285P (paternal) fibroblasts were not affected in endocytosis (Fig 3F and Appendix Fig S4B).
To pinpoint the block in endocytosis at the EM level, we incubated primary fibroblasts for 2 h with the endocytic marker BSA conjugated to 5 nm gold particles (BSA‐Au5) and processed cells for immuno‐EM. Sections were labeled for LAMP‐1 or LAMP‐2 and randomly screened for LAMP‐positive lysosomes (Klumperman & Raposo, 2014) that were scored positive or negative for BSA‐Au5 (Fig 2D). In agreement with the data from florescence microscopy, this showed a decrease in the delivery of BSA‐Au5 to LAMP‐positive lysosomes in VPS41S285P/R662 * patient and VPS41WT/R662 * maternal fibroblasts (Fig 3G). The paternal VPS41WT/S285P fibroblasts were not affected. Collectively, these fluorescent and EM data indicate that transfer of endocytosed cargo to enzymatically active, LAMP‐positive lysosomes is affected in fibroblasts of patient 2 and of his mother.
Since the mother of the patients does not show a clinical phenotype, we reasoned that the defect in lysosomal delivery could be kinetic rather than a complete block. To test this, we incubated control, maternal, and patient fibroblasts with Dextran‐568 for several time points, after which we determined colocalization with SiR‐Lysosome (Fig 3H). After 2 h, VPS41WT/R662 * as well as VPS41S285P/R662 * fibroblasts again displayed significant lower levels of colocalization than VPS41WT/WT cells. After 5 h, however, the difference between maternal and control fibroblasts was abolished, whereas patient fibroblasts still showed a significant lower level of Dextran in lysosomes. After 24 h of Dextran‐568 uptake, patient fibroblasts showed a similar level of colocalization with SiR‐Lysosome as control and maternal cells (Fig 3H).
Together, these data show that expression of VPS41S285P or absence of VPS41 causes a deficiency in transfer of endocytic cargo to lysosomes, representing a defect in HOPS‐dependent endolysosomal fusion. This defect is a delay rather than a block in fusion, which in patient cells is more severe than in maternal fibroblasts, indicating that transport kinetics is an important determinator of pathogenesis. The data also show that in maternal fibroblasts the consequence of carrying the VPS41R662 * variant is not fully compensated for by the VPS41WT allele, yet this does not result in a pathogenic phenotype.
Patient fibroblasts have a defect in autophagic response to starvation
In addition to late endosome–lysosome fusion, the HOPS complex is required for fusion of autophagosomes with lysosomes (Jiang et al, 2014; McEwan et al, 2015; Nakamura & Yoshimori, 2017). A block in autophagosome‐lysosome fusion results in increased numbers of autophagosomes containing lipidated LC3 (LC3II) and hence an increase in LC3II:LC3I ratio. We quantified this on Western blot and found that under nutrient‐rich conditions, the LC3II:LC3I ratio is increased in VPS41S285P/R662 * (patient 2) fibroblasts compared with control cells, indicating that patient cells have more autophagosomes (Fig 4A, quantified in 4A’). To verify these findings, we performed immunofluorescent labeling of LC3 in patient and control fibroblasts. This confirmed that VPS41S285P/R662 * cells contain significantly more LC3‐positive compartments than VPS41WT/WT cells (Fig 4B, quantified in 4B’). Starvation induced an increase in numbers of autophagosomes in both control and patient fibroblasts (Fig 4B, quantified in 4B’). Intriguingly, however, whereas in control fibroblasts this increase was 13.4‐fold, in patient fibroblasts only a 3.8‐fold increase was attained. These data indicate that VPS41 patient fibroblasts sustain a higher basal level of autophagy and are less responsive to nutrient starvation than control fibroblasts.

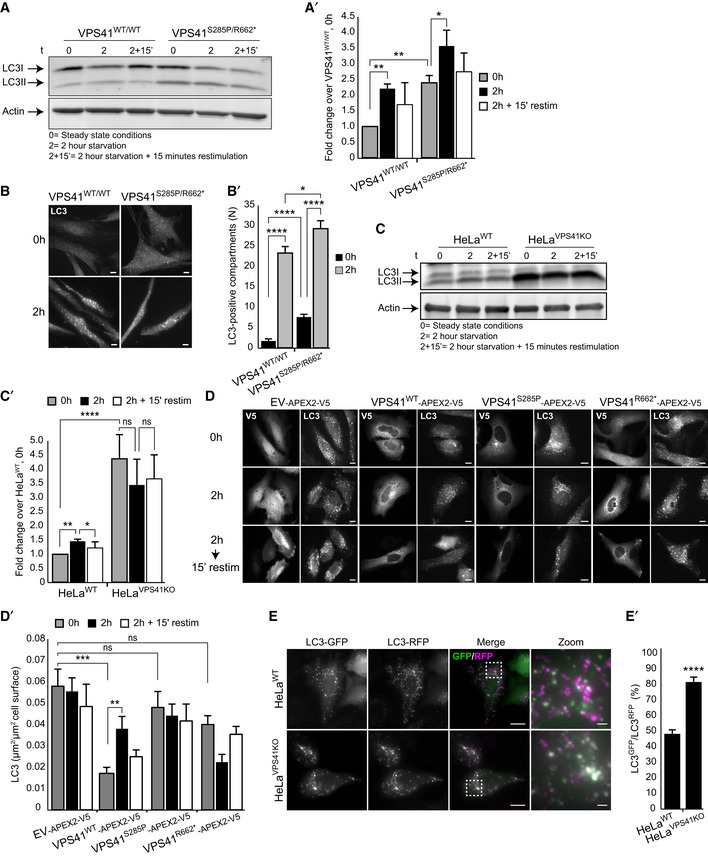
Patient fibroblasts and VPS41KO cells show decreased autophagic flux and response to autophagic stimuli
Western blot of LC3 expression levels in control and patient fibroblasts. In steady state conditions (0 h), patient fibroblasts have a higher ratio of lipidated LC3 (LC3II):LC3I than control cells. Induction of autophagy by nutrient starvation (2 h incubation with minimal EBSS medium) results in a raise in LC3II:LC3I ratio in both control and patient cells, but in VPS41S285P/R662 * fibroblasts this increase is only modest (quantified in A’) (n = 2).
Immunofluorescence of LC3 inVPS41WT/WT and VPS41S285P/R662 * fibroblasts under steady state and starved conditions. At steady state conditions (0 h), patient fibroblasts contain more LC3‐positive compartments. Nutrient starvation (2 h) increases the number of LC3‐positive autophagosomes in control fibroblasts 13.4‐fold, and in VPS41S285P/R662 * fibroblasts only 3.8‐fold, indicating a reduced responsiveness to starvation (quantified in B’). > 54 Cells per cell line were quantified. Scale bars, 10 µm.
Western blot analysis of HeLaVPS41KO cells shows a fourfold increase in LC3II protein levels in steady state conditions (0 h) compared with HeLaWT cells. In contrast to control cells, nutrient starvation (2 h) did not increase LC3II protein levels in HeLaVPS41KO cells, indicating irresponsiveness to nutrient availability (quantified in C’) (n = 3).
Rescue experiments. HeLaVPS41KO cells transfected with EV‐APEX2‐V5, VPS41WT‐APEX2‐V5, VPS41S285P‐APEX2‐V5, or VPS41R662*‐APEX2‐V5 and labeled for V5 and LC3 by immunofluorescence microscopy. Rescue with VPS41WT‐APEX2‐V5 decreases the number of LC3‐positive compartments in steady state conditions (0 h) and restores responsiveness to nutrient starvation (2 h) and replenishment (2‐hour starvation followed by 15 min restimulation). Neither of the mutant VPS41 variants rescues this autophagy phenotype (quantified in D’). >15 Cells per condition were quantified (n = 3). Scale bars, 10 µm.
Immunofluorescence of HeLaWT and HeLaVPS41KO cells transfected with LC3GFP/RFP tandem construct. The increased percentage of GFP/RFP‐positive compartments in HeLaVPS41KO cells indicates a block in autophagic flux (quantified in E’). > 12 Cells per cell line were quantified. Scale bars 10 µm; zoom, 1 µm.
Data information: Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 10−4, ****P < 10−5. Unpaired t‐test (A’ and E’) or one‐way ANOVA with Tukey’s (B’) or Bonferroni correction (C’ and D’). Exact p‐values are reported in Appendix Table S3.
Source data are available online for this figure.
We further studied this phenomenon in HeLaVPS41KO cells. Under nutrient‐rich conditions, LC3II levels in HeLaVPS41KO cells were significantly elevated compared to HeLaWT cells (Fig 4C, quantified in 4C’). A similar upregulation was seen in a previous study in VPS41‐depleted HeLa cells (Ding et al, 2019). Concomitantly, immunofluorescence of HeLaVPS41KO cells revealed an increase in LC3 puncta (Fig 4D, quantified in 4D’). Strikingly, starvation of HeLaVPS41KO cells did not further increase LC3II protein levels or number of LC3 puncta (Fig 4C and D). Rescue of HeLaVPS41KO cells with VPS41WT reduced the number of LC3 puncta and restored the capacity of cells to respond to starvation and restimulation (Fig 4D, quantified in 4D’). By contrast, expression of VPS41S285P or VPS41R662* did not restore the autophagic flux (Fig 4D, quantified in 4D’). Together, these data show that both patient fibroblasts and VPS41KO cells have increased basal autophagy levels. Patient fibroblasts respond to starvation, but not to a similar extent as control fibroblasts. On the contrary, HeLaVPS41KO cells are impaired in their responsiveness to starvation.
To follow the autophagic flux from autophagosome to lysosome, we next transfected HeLaWT and HeLaVPS41KO cells with an LC3GFP/RFP tandem construct. GFP and RFP co‐label autophagosomes, whereas the GFP signal is quenched in the acidic environment after autophagosome–lysosome fusion (Kimura et al, 2007). Immunofluorescence of HeLaVPS41KO cells showed a significant higher level of GFP/RFP colocalization than HeLaWT cells, indicating that in the absence of VPS41, autophagosome–lysosome fusion is impaired (Fig 4E, quantified in 4E’). To confirm these findings, we incubated HeLaWT and HeLaVPS41KO cells with Bafilomycin A1 (BafA1), which increases the lysosomal pH and prevents degradation of LC3. Indeed, Western blots of BafA1‐treated WT cells showed an increase in LC3II protein levels, however, not to a similar level as non‐treated, non‐starved VPS41KO cells (Fig EV3A, quantified in EV3B and C). Strikingly, starvation of WT cells in the presence of Baf1A resulted in similar LC3II levels as starved VPS41KO cells in the absence of Baf1A (Fig EV3C). Overall, BafA1 treatment of VPS41KO cells had little effect on LC3II protein levels (Fig EV3A, quantified in EV3B). These data indicate that LC3II in VPS41KO cells is less well degraded by lysosomes than in control cells, consistent with the HOPS‐dependent role of VPS41 in autophagosome–lysosome fusion.

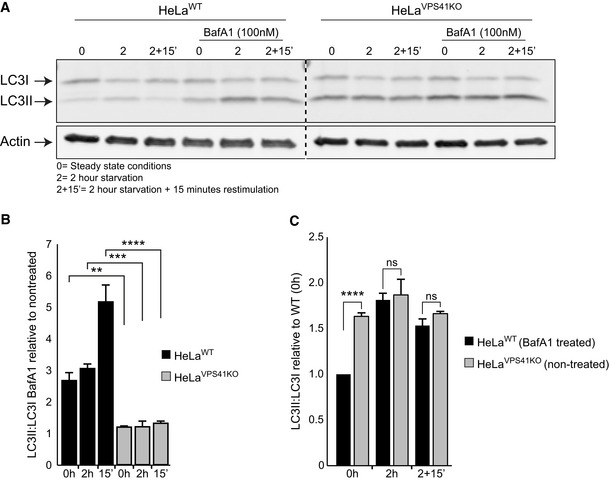
VPS41KO cells are insensitive to Bafilomycin A1 treatment
Western blot of LC3 protein levels in HeLaWT and HeLaVPS41KO cells with and without Bafilomycin A1 (BafA1) treatment. Cells depleted of VPS41 show increased LC3II protein levels compared with WT cells. BafA1 treatment increases LC3II protein levels in HeLaWT cells, whereas HeLaVPS41KO cells are relatively insensitive (n = 2).
Quantification of LC3II:LC3I levels. Per cell line and per condition the increase in LC3II:LC3I after BafA1 treatment are plotted against non‐treated cells. In contrast to HeLaWT cells, the HeLaVPS41KO cells show only a minor increase (n = 2).
Quantification of LC3II:LC3I levels in HeLaWT BafA1 treated compared with non‐treated HeLaVPS41KO cells in steady state (0 h), starved (2 h) and refeeding (2‐h starvation followed by 15‐min restimulation) conditions. Similar LC3II ratios are observed after starvation and BafA1 treatment of HeLaWT cells, indicating that the elevated LC3 levels in HeLaVPS41KO cells are caused by increased autophagic induction and decreased autophagic flux (n = 2).
Data information: Data are represented as mean ± SD. **P < 0.01, ***P < 0.001, ****P < 10‐5. One‐way ANOVA analysis with Tukey’s correction (B) or unpaired t‐test (C). Exact P‐values are reported in Appendix Table S3.
Source data are available online for this figure.
Together, the data indicate that VPS41S285P/R662 * fibroblasts have increased basal autophagic levels and are, to some extent, responsive to autophagic stimuli. VPS41KO cells also have high basal autophagy levels, but are virtually insensitive to starvation. The differences between patient fibroblasts and HeLaVPS41KO cells could be due to differences in autophagic responsiveness in these cell types. Alternatively, the patient cells might have developed epigenetic compensatory mechanisms.
VPS41 deficiency causes a HOPS‐dependent defect on the TFE3 but not S6K1/4EBP1 axis
A complex of v‐ATPase/Ragulator/Rag GTPases present at the lysosomal membrane senses nutrient status in lysosomes and, in nutrient‐rich conditions, recruits the mTORC1 complex. Upon nutrient deprivation, mTORC1 dissociates from the lysosomal membrane and loses its kinase activity toward p70 ribosomal protein S6 kinase 1 (S6K1), 4E‐binding protein 1 (4EBP1), and the transcription factors TFEB and TFE3. Consequently, TFEB and TFE3 translocate to the nucleus where they activate the CLEAR (Coordinated Lysosomal Expression and Regulation) network that induces lysosomal biogenesis and autophagy (Palmieri et al, 2011). LC3 is a target of the CLEAR network, and mTORC1 inhibition results in enhanced LC3 levels (Palmieri et al, 2011). Since patient‐derived fibroblasts and VPS41KO cells show increased LC3 levels under basal conditions (Fig 4A–D), we investigated mTORC1 and TFE3 localization and activity in VPS41 patient fibroblasts and VPS41KO cells.
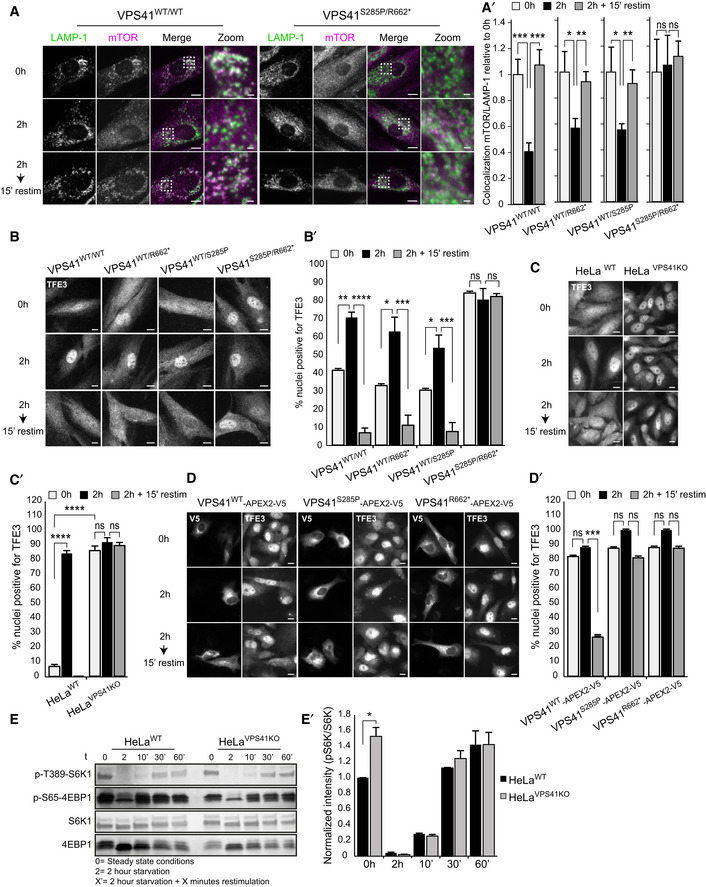
We first studied recruitment of mTORC1 to lysosomes by immunofluorescence microscopy, monitoring colocalization with LAMP‐1. As expected, we found significant overlap between mTORC1 and LAMP‐1 puncta in control VPS41WT/WT fibroblasts grown in nutrient‐rich conditions. After 2‐h starvation, mTORC1 had redistributed to the cytoplasm, and after 15‐min restimulation, the colocalization with LAMP‐1‐positive lysosomes was partially restored (Fig 5A). The parental derived fibroblasts VPS41WT/S285P and VPS41WT/R662 * followed this same pattern (Appendix Fig S6). However, in patient‐derived VPS41S285P/R662 * fibroblasts, mTORC1–LAMP‐1 colocalization was strikingly less in all conditions and did not change upon starvation or restimulation. Quantitation of mTORC1/LAMP‐1 colocalization, for each cell type relative to the control condition (0 h), clearly showed that VPS41S285P/R662 fibroblasts do not alter mTORC1 localization in response to starvation (Fig 5A, quantified in 5A’). To monitor the effect of mTORC1 dissociation on TFEB/TFE3 localization, we labeled fibroblasts for endogenous TFE3 (Fig 5B). In VPS41WT/WT, VPS41WT/S285P, and VPS41WT/R662 * fibroblasts, TFE3 showed a normal localization pattern, and translocated to the nucleus in response to starvation (Fig 5B, quantified in 5B’). However, in VPS41S285P/R662 * fibroblasts, TFE3 was constitutively found in the nucleus, regardless of nutrient conditions. A similar constitutive nuclear localization of TFE3 was observed in fibroblasts obtained from patient 3 (VPS41 c.1423‐2A>G /R662 *, Fig EV4A). These data show that TFE3 is continuously present in the nucleus of patient cells, regardless of nutrient status.

Patient fibroblasts and VPS41KO cells exhibit mTORC1 inhibition toward TFE3
Immunofluorescence of control, parental, and patient fibroblasts labeled for LAMP‐1 and mTOR. In steady state conditions (0 h), VPS41S285P/R662 * fibroblasts show less colocalization between mTOR and LAMP‐1. VPS41WT/WT, VPS41WT/S285P, and VPS41WT/R662 * show an appropriate mTOR response upon nutrient deprivation (2 h) or restimulation (2‐h starvation followed by 15‐min restimulation) (Appendix Fig S5A). > 10 Cells per cell line were quantified (A’). Quantifications are performed relative to colocalization under steady state conditions per cell line (n = 3). Scale bars, 10 µm; zoom, 1 µm.
Immunofluorescence of VPS41WT/WT, VPS41WT/S285P, VPS41WT/R662 *, and VPS41S285P/R662 * fibroblasts labeled for TFE3. In VPS41S285P/R662 * fibroblasts, TFE3 is constitutively localized in the nucleus regardless of nutrient state (quantified in B’). > 25 Cells per condition were quantified (n = 3). Scale bars, 10 µm.
TFE3 immunofluorescence in HeLaWT and HeLaVPS41KO cells. In HeLaVPS41KO cells, TFE3 constitutively localizes in the nucleus (quantified in C’). > 83 Cells per condition were quantified (n = 3). Scale bars, 10 µm.
Rescue experiments of HeLaVPS41KO cells. Expression of VPS41WT‐APEX2‐V5, VPS41S285P‐APEX2‐V5, or VPS41R662*‐APEX2‐V5. Reintroduction of VPS41WT rescues TFE3 localization, whereas expression of mutant VPS41 has no effect (quantified in D’). > 83 Cells per condition were quantified (n = 3). Scale bars, 10 µm.
Western blot of phosphorylated mTORC1 substrates S6K and 4EBP1 after starvation (2 h) and restimulation (10, 30 or 60 min). HeLaVPS41KO cells show comparable levels of phospho‐S6K and phospho‐4EBP1, with no difference in recovery after restimulation (quantified in E’) (n = 3).
Data information: Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 10−4, ****P < 10−5. One‐way ANOVA with Bonferroni correction (A’–D’) or unpaired t‐test (E’). Exact P‐values are reported in Appendix Table S3.
Source data are available online for this figure.

VPS41 deficiency causes decreased lysosomal localization of mTORC1 and defects in TFE3/TFEB signaling
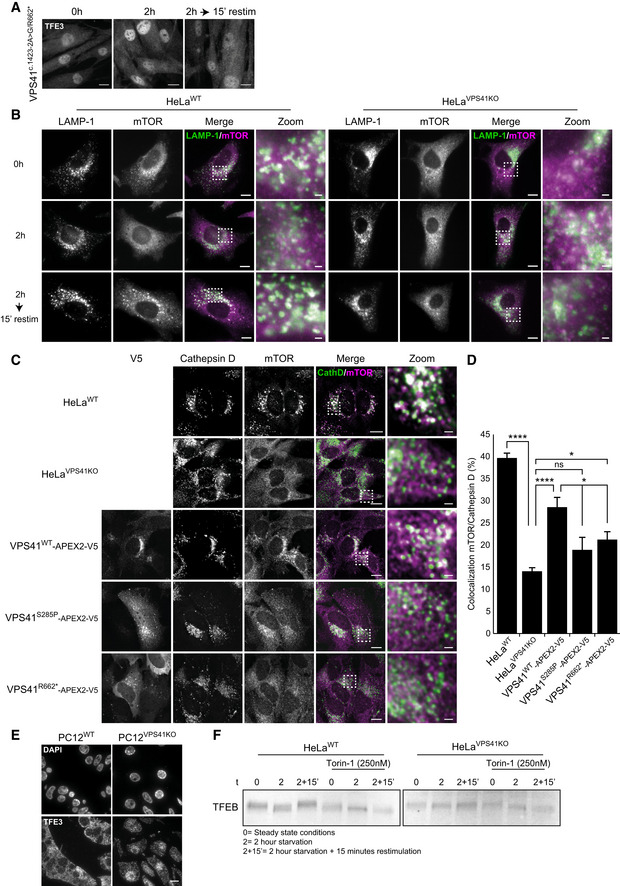
VPS41c . 1423‐2A>G/R662 * fibroblasts show constitutive nuclear localization of TFE3. Scale bars, 10 µm.
HeLaWT and HeLaVPS41KO cells labeled for LAMP‐1 and mTOR immunofluorescence. In HeLaVPS41KO cells, mTOR is dissociated from LAMP‐1‐positive compartments regardless of nutrient state. In HeLaWT cells, mTOR colocalizes with LAMP‐1 in steady state (0 h) and after nutrient restimulation (2‐h starvation followed by 15‐min restimulation). Scale bars, 10 µm; zoom 1 µm.
Rescue experiment of mTOR localization on cathepsin D‐positive lysosomes in HeLaVPS41KO cells transfected with VPS41WT‐APEX2‐V5, VPS41S285P‐APEX2‐V5, or VPS41R662*‐APEX2‐V5. Cells were starved for 2 h and restimulated with full medium for 15 min. Scale bars, 10 µm; zoom, 1 µm.
Quantification of mTOR colocalization with cathepsin D in rescued VPS41KO cells. Reintroducing VPS41WT results in increased colocalization of mTOR with lysosomes. > 15 cells per condition were quantified in this assay (n = 2).
Immunofluorescence of 300 nm thick cryosections of PC12WT and PC12VPS41KO cells labeled for TFE3. PC12VPS41KO cells show constitutive nuclear localization of TFE3. Scale bar, 10 µm.
Western blots of TFEB. In HeLaWT cells, molecular weight shifts are indicative for (de)phosphorylation events dependent on nutrient availability. mTORC1 inhibitor Torin‐1 impairs TFEB phosphorylation and no shift in molecular weight is seen. In VPS41KO cells, no molecular weight shift of TFEB is seen either, indicating impaired phosphorylation of TFEB.
Data information: Data are represented as mean ± SEM. *P < 0.05, ****P < 10−5. One‐way ANOVA analysis with Bonferroni correction. Exact P‐values are reported in Appendix Table S3.
Source data are available online for this figure.
We then performed similar experiments in HeLa VPS41KO cells. Like patient fibroblasts, these cells showed impaired lysosomal recruitment of mTORC1 (Fig EV4B). Reintroducing VPS41WT into HeLaVPS41KO cells increased the colocalization of mTORC1 with the lysosomal marker cathepsin D from 15 to 30%. Expression of VPS41S285P or VPS41R662* slightly increased lysosomal recruitment of mTORC1, but to a much lesser extent than VPS41WT (Fig EV4C and D). Concomitantly, HeLaVPS41KO cells showed the “TFE3 translocation to the nucleus phenotype” (Fig 5C, quantified in 5C’), and the same was observed in PC12VPS41KO cells (Fig EV4E, Appendix Fig S7A and B). Expression of VPS41WT in HeLaVPS41KO cells rescued the TFE3 phenotype after starvation and restimulation, whereas expression of VPS41S285P or VPS41R662* had no effect; TFE3 was present in the nucleus in all conditions (Fig 5D, quantified in 5D’). Notably, transfection of cells affects membrane integrity causing lysosomal stress. This likely explains the nuclear localization of TFE3 in HeLaVPS41KO cells transfected with VPS41WT when cultured under steady state conditions (Fig 5D). Similar to TFE3, TFEB is also phosphorylated by mTORC1 to prevent nuclear translocation. Phosphorylation induces a molecular weight shift visible on Western blot. To address whether TFEB phosphorylation is affected in VPS41KO cells, we analyzed this in control conditions and upon starvation. We used the mTORC1 inhibitor Torin‐1 as positive control to show the molecular weight shift of TFEB. Indeed, starvation of HeLaWT cells resulted in a similar molecular weight shift as observed after Torin‐1 treatment (Fig EV4F). By contrast, HeLaVPS41KO cells did not respond to starvation or Torin‐1 treatment, indicating that TFEB phosphorylation is impaired (Fig EV4F). The continuous nuclear localization of TFE3 in patient cells predicts an increase in expression of lysosomal and autophagy proteins (Settembre et al, 2011; Martina et al, 2014). Western blot analysis indeed showed that protein levels of LAMP‐1 and the lysosomal hydrolase cathepsin B, both CLEAR targets, are increased in patient fibroblasts (Appendix Fig S8A and B). Similar results were obtained for LAMP‐1 and cathepsin D in VPS41KO cells (Appendix Fig S8C). Collectively, these data show that in cells lacking VPS41 or expressing VPS41S285P and/or VPS41R662*, regulation of the mTORC1/TFE3 axis is perturbed, resulting in higher levels of autophagy and lysosome proteins and a continuous activation of autophagy, independent of nutrient status.
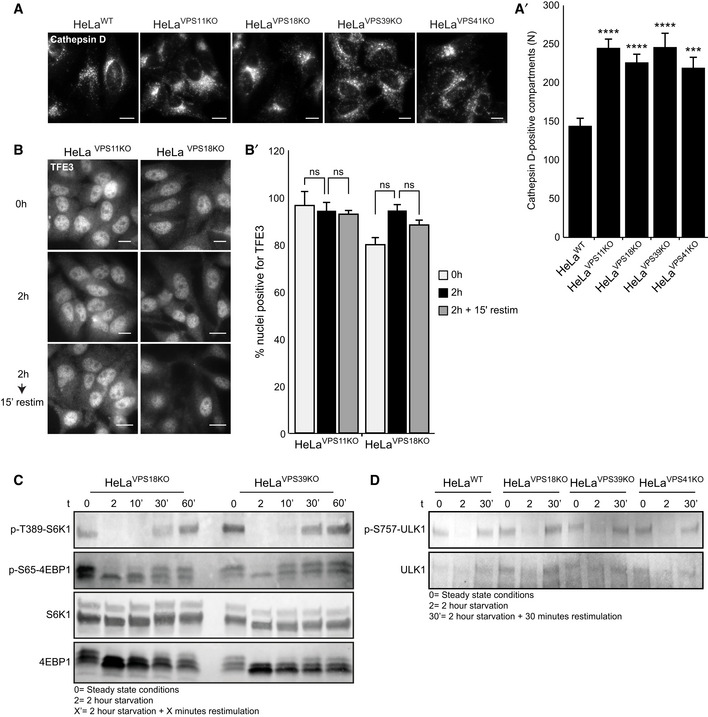
A possible explanation for the mTORC1/TFE3 phenotype is that the defect in HOPS function results in insufficient delivery of nutrients to lysosomes and subsequent mTORC1 dissociation. If so, any block in HOPS function should give this phenotype. To test this, we made HeLaKO cells for VPS11, VPS18, and VPS39, which are part of HOPS but not required for the ALP/LAMP pathway (Pols et al, 2013b; Appendix Fig S9A and B). Similar to VPS41KO cells, an increase in endolysosomal compartments was observed by immunofluorescent labeling of cathepsin D (Fig EV5A, quantified in EV5A’). VPS11KO and VPS18KO cell lines showed increased LC3II protein levels in basal conditions and a constitutive nuclear localization of TFE3, independent of nutrient status (Fig EV5B, quantified in EV5B’ and Appendix Fig S9C). These data show that the increase in endolysosomal compartments and the mTORC1/TFE3 phenotype is caused by a dysfunctional HOPS complex. We then studied the effect of VPS41 or HOPS depletion on the canonical mTORC1 substrates S6K1 and 4EBP1, required for cellular growth. Intriguingly, phosphorylation of these substrates was not affected by the absence of VPS41 or other HOPS subunits (Fig 5E, quantified in 5E’ and Fig EV5C). Likewise, phosphorylation of ULK1, another substrate of mTORC1 involved in autophagy initiation, was not affected in HOPS knockout cells (Fig EV5D). These data imply that the HOPS complex selectively regulates the mTORC1‐dependent control of the MiT/TFE family of transcription factors.

The HOPS complex is required for mTORC1‐dependent TFE3 regulation
Immunofluorescence microscopy of HeLaWT, HeLaVPS11KO, HeLaVPS18KO, HeLaVPS39KO, and HeLaVPS41KO cells labeled for cathepsin D. All KO cells lines show more cathepsin D‐positive compartments (quantified in A’). > 56 Cells per cell line were quantified (n = 3). Scale bars, 10 µm.
Immunofluorescence microscopy of HeLaVPS11KO and HeLaVPS18KO cells labeled for TFE3 at steady state (0 h), starved (2 h), or restimulated (2‐h starvation followed by 15‐min restimulation) conditions. In both HeLaVPS11KO and HeLaVPS18KO cells, TFE3 is present in the nucleus regardless of nutrient availability, indicating that mTORC1‐dependent regulation of TFE3 is impaired in cells depleted of HOPS subunits (quantified in B’) (n = 3). Scale bars, 10 µm.
Western blot of phosphorylated mTORC1 substrates S6K1 and 4EBP1 after starvation (2 h) and restimulation with full medium for 10, 30, or 60 min in HeLaVPS18KO and HeLaVPS39KO cells. After 30‐min restimulation, both substrates are phosphorylated in both cell lines. These data show that phosphorylation of S6K1 and 4EBP1 is independent of the HOPS complex.
Western blot showing ULK1, a substrate of mTORC1 involved in autophagy initiation, and phosphorylated upon nutrient availability. HeLaWT, HeLaVPS18KO, HeLaVPS39KO, and HeLaVPS41KO cell lines show an appropriate response in (de)phosphorylation of ULK1 upon starvation (2 h) and restimulation (2‐h starvation followed by 30‐min restimulation).
Data information: Data are represented as mean ± SEM. ***P < 0.001, ****P < 10−5. One‐way ANOVA analysis with Bonferroni correction. Exact P‐values are reported in Appendix Table S3.
Source data are available online for this figure.
We conclude from these data that VPS41 patient cells show a decreased association of mTORC1 with lysosomes, a continuous nuclear localization of TFE3, and increased expression of LC3II and other CLEAR proteins. Notably, the autophagy defect is specific for the patient‐derived fibroblasts and not observed in any of the parental cell lines. VPS41KO cells show a similar lysosomal dissociation of mTORC1 and constitutive nuclear localization of TFE3 whereas phosphorylation of S6K1 and 4EBP1 is unaffected. This mTORC1/TFE3 phenotype is HOPS dependent, since depletion of other HOPS subunits results in a similar phenotype. The phenotype cannot be rescued by expression of VPS41S285P or VPS41R662*, which is in line with our other findings showing that these variants cannot restore HOPS function.
VPS41S285P allows for normal regulated secretion in PC12 cells
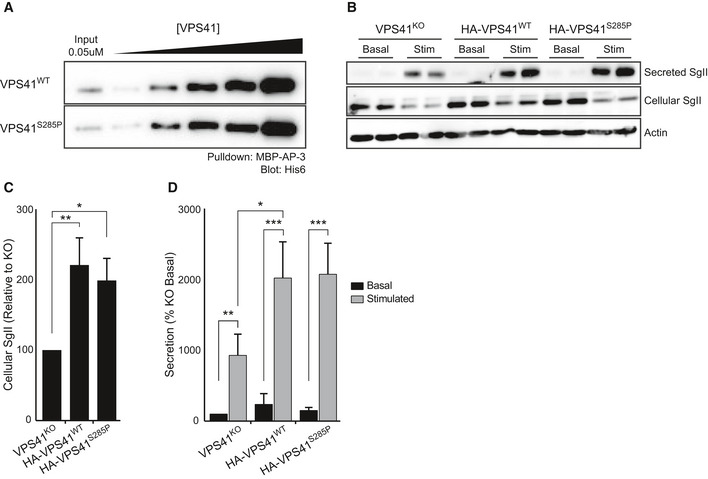
In secretory cells, VPS41 is required for secretory protein sorting and secretory granule biogenesis in a pathway that is independent of the HOPS complex (Asensio et al, 2013; Burns et al, 2020). This VPS41 function requires the N‐terminal residues 1–36 for interaction with AP‐3 and the presence of the C‐terminal located CHCR domain (Asensio et al, 2013; Margarita Cabrera et al, 2010). It was suggested that VPS41 might form a coat on AP‐3 containing membranes that exit the TGN (Rehling et al, 1999; Darsow et al, 2001; Asensio et al, 2013). Since VPS41R662* lacks both the RING domain and part of the CHCR domain, a function of this variant in regulated secretion is prohibited (Asensio et al, 2013). However, VPS41S285P could theoretically still exert this HOPS‐independent function. To test this, we first investigated whether VPS41S285P can interact with AP‐3. Pulldowns of recombinant VPS41 constructs with the hinge‐ear domain of AP‐3D1 showed that VPS41S285P binds AP‐3 with equivalent affinity as VPS41WT (Fig 6A and Appendix Fig S10).

VPS41S285P rescues regulated secretion in PC12 VPS41KO cells
AWestern blot of pulldown of AP‐3 incubated with HIS‐6 VPS41WT or VPS41S285P. There is no difference in affinity between VPS41WT and VPS41S285P (See Appendix Fig S10 for MBP‐AP‐3 pulldown input).
BWestern blot on cellular or secreted secretogranin II (SgII) of PC12 cells VPS41KO, or VPS41KO transduced with HA‐VPS41WT or HA‐VPS41S285P lentivirus. Cells were washed and incubated for 30 min in Tyrode’s solution containing 2.5 mM K+ (basal) or 90 mM K+ (stimulated). No difference in secreted SgII levels was observed between VPS41WT and VPS41S285P.
C, DCellular (C) and secreted (D) secretogranin II (SgII) were measured by quantitative fluorescence immunoblotting (n = 3). VPS41S285P rescues intracellular SgII levels and regulated secretion of SgII to the same extent as VPS41WT.
Data information: Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 10−4. One‐way ANOVA (C and D). Exact P‐values are reported in Appendix Table S3.
To directly test the effect of VPS41S285P on regulated secretion, we made use of PC12VPS41KO cells. By using EGF‐ALEXA647 degradation as readout, we established that these cells show a similar endocytosis defect as patient fibroblasts and VPS41KO HeLa cells (Appendix Fig S11A and B) and also display the HOPS‐dependent mTORC1/TFE3 phenotype (Fig EV4E). Regulated protein secretion was measured as previously described (Asensio et al, 2010). Briefly, cells were incubated in Tyrode’s buffer containing 2.5 mM (basal) or 90 mM (stimulated) K+ and the supernatant and cell lysates were analyzed by quantitative fluorescent immunoblotting (Fig 6B). As previously shown, depletion of VPS41 from PC12 cells resulted in decreased cellular SgII protein levels, as well as in a reduction in the regulated secretion of the proteins that are present (Asensio et al, 2013). Reintroducing VPS41WT in PC12 VPS41KO cells rescued cellular SgII levels and recovered regulated secretion (Fig 6C and D). Interestingly, similar results were obtained by expression of the VPS41S285P variant (Fig 6C and D), indicating that its role in regulated secretion has been preserved.
Together, these data show that expression of VPS41S285P rescues the regulated secretory pathway in VPS41KO PC12 cells. Thus, the VPS41S285P variant is defective in HOPS‐dependent endocytosis and autophagy pathways, but retains its HOPS‐independent role in regulated secretion.
Co‐expression of VPS41S285 and VPS41R662 * abolishes neuroprotection in a C. elegans model of Parkinson’s disease
The clinical symptoms of the VPS41 patients (Fig 1) overlap with Parkinson’s disease (Jankovic, 2008; Reich and Savitt, 2016). Previously, a screen for neuroprotective factors in a transgenic C. elegans model for Parkinson’s disease showed that overexpression of human VPS41 protects against α‐synuclein‐induced neurodegeneration (Hamamichi et al, 2008; Ruan et al, 2010; Harrington et al, 2012). The neuroprotective effect of VPS41 depends on the presence of the WD40 and CHCR domains and ability to interact with Rab7 and AP‐3 (Harrington et al, 2012; Griffin et al, 2018). To explore the impact of patient mutations in this Parkinson’s disease model, we made transgenic nematodes, co‐expressing the human VPS41 (hVPS41) variants found in patients 1 and 2. We crossed these with isogenic strains overexpressing either GFP alone or human α‐synuclein‐GFP to mimic Parkinson’s disease.
First, we combined VPS41WT with VPS41S285P or VPS41R662* expression, reflecting paternal and maternal conditions of patients 1 and 2, and studied if expression of these mutants induces neurodegeneration in the absence of α‐synuclein. At 7 or 10 days post‐hatching, there was no statistically significant change in neurodegeneration in any of these strains (Fig 7A and B). Likewise, when we co‐expressed VPS41S285P with VPS41R662* there was no discernable increase in neurodegeneration. This demonstrates that co‐expression of the patient‐specific VPS41 variants does not increase neurodegeneration in (aging) animals.

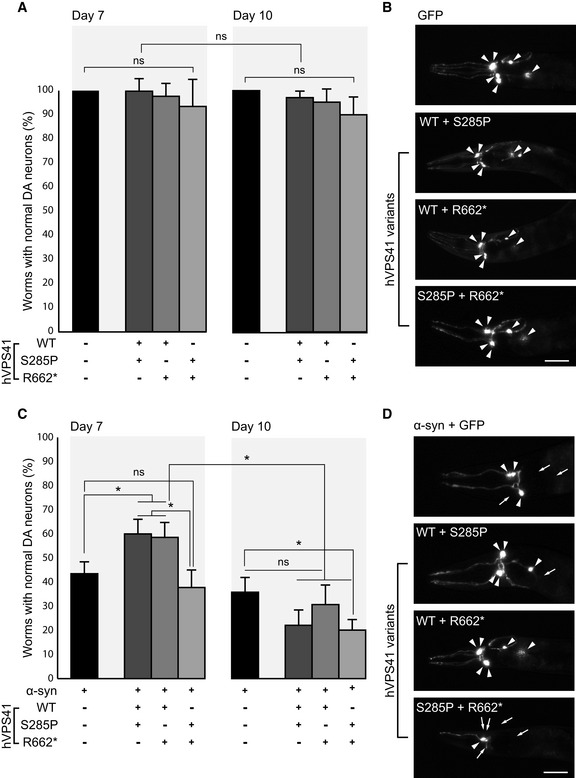
Compound heterozygous expression of hVps41 variants fails to rescue α‐synuclein‐induced neurodegeneration in C. elegans
Graph representing the percentage of adult C. elegans animals with 6 normal dopaminergic neurons. Heterozygous expression with hVPS41WT (strains UA386 or UA387) or compound heterozygous expression of hVPS41 variants (strain UA388) does not yield significant changes in neurodegeneration compared with the Pdat‐1::GFP (strain BY250) control at either day 7 or day 10 post‐hatching. n = 30 Adult worms for each of 3 independent experiments for GFP (total of 90 worms) and n = 90 for each independent transgenic strain (30 worms/trial × 3 independent transgenic lines = 270 worms), for each of 3 independent experiments.
Representative images of the neuroprotection assay described in (A), where worms express GFP specifically in the anterior 6 DA neurons. Intact DA neurons are indicated with arrowheads. hVPS41 variant backgrounds are expressed as indicated. Scale bar 20 µm.
Graph representing the percentage of animals with 6 normal dopaminergic neurons in the anterior region of Pdat‐1::GFP; Pdat‐1:: α‐syn (strain UA44) animals with heterozygous expression of hVPS41 variants. Heterozygous expression of hVPS41WT with either variant (strains UA389 or UA390) significantly rescues neurons from α‐synuclein‐induced degeneration at day 7 whereas compound heterozygous expression (strain UA391) fails to rescue neurodegeneration. At day 10, none of the heterozygous backgrounds significantly rescues α‐synuclein‐induced neurodegeneration. n = 30 Adult worms for each of 3 independent experiments for the α‐synuclein (α‐syn) strain (total of 90 worms) and n = 90 for each independent transgenic strain (30 worms/trial × 3 independent transgenic lines = 270 worms) for each of 3 independent experiments.
Representative images of DA neurons from C. elegans expressing Pdat‐1::GFP; Pdat‐1:: α‐syn, with or without hVPS41 variants, as described in (C). Intact DA neurons are indicated with arrowheads and missing neurons are indicated with arrows. Scale bar 20 µm.
Data information: Data are represented as mean ± SD. *P < 0.05. One‐way ANOVA with Tukey’s correction (A and C). Exact P‐values are reported in Appendix Table S3.
We then analyzed VPS41 neuroprotective function in strains overexpressing α‐synuclein. At 7 days post‐hatching, only ~43% of the animal population overexpressing α‐synuclein retains the full complement of dopaminergic neurons and at 10 days only ~35%. Previous studies show that expression of VPS41WT rescues neurodegeneration at day 7 and day 10, albeit it with reduced efficiency at day 10 (Ruan et al, 2010; Harrington et al, 2012). Here, we found that co‐expression of VPS41WT with either VPS41S285P or VPS41R662* significantly enhanced survival of dopaminergic neurons at day 7, but at day 10 this protective effect was lost (Fig 7C and D). In animals co‐expressing VPS41S285P and VPS41R662*, reflecting patient conditions, we found no protective effect at all at either day 7 or 10 (Fig 7C and D). Moreover, neurodegeneration at day 10 was significantly increased indicating that co‐expression of the clinical VPS41 variants adds additional stress besides α‐synuclein overexpressing and aging. Collectively, these data indicate that the capacity of VPS41 to protect dopaminergic neurons against α‐synuclein overexpression is lost upon co‐expression of the patient variants. This implies that VPS41‐mediated neuroprotection depends on the HOPS complex.
Discussion
The vacuolar protein sorting‐associated protein 41 (VPS41) has a major contribution in vesicle‐mediated trafficking to lysosomal compartments including endocytic transport and the autophagic pathway. VPS41 is part of the HOPS complex, a multisubunit tethering complex which mediates fusion of lysosomes with late endosomes and autophagosomes (Radisky et al, 1997; Nickerson et al, 2009). Independently of the HOPS complex, VPS41 is involved in the transport of lysosomal membrane proteins from the trans‐Golgi network (Pols et al, 2013b) and in regulated secretion of neuropeptides (Asensio et al, 2013). Intriguingly, VPS41 was previously identified as neuroprotector in C. elegans and mammalian models of Parkinson’s disease (Ruan et al, 2010; Harrington et al, 2012; Griffin et al, 2018). Here, we present three patients with a neuronal disorder, bearing compound heterozygous variants in VPS41 and show the consequences of this on the molecular and cellular level. We demonstrate that all VPS41 variants give rise to a non‐functional HOPS complex and a kinetic defect in the delivery of endocytic and autophagic cargo to enzymatically active lysosomes. Most strikingly, we find that lack of HOPS function causes increased dissociation of mTORC1 from lysosomes and the transfer of TFE3 to the nucleus, resulting in continuously increased autophagy levels. By contrast, phosphorylation of mTORC1 substrates S6K and 4EBP1 is not affected. Our studies thereby link HOPS complex functionality to mTORC1 signaling, specifically for the TFEB/TFE3 axis.
Patients 1 and 2, two brothers, were diagnosed with global developmental delay, hypotonia, ataxia, and dystonia. By exome sequencing, we identified a missense variant in the WD40 domain (VPS41S285P) and a nonsense variant in the CHCR domain resulting in a premature stopcodon (VPS41R662 *). The third patient, presenting with severe developmental delay, axial hypotonia, and spastic quadriplegia, was identified with a splice site variant in the TPR‐like domain (VPS41 c.1423‐2A>G) and the same nonsense variant in the CHCR domain as observed in the siblings (VPS41R662 *). By Western blot, we found that only the VPS41S285P variant was expressed at significant levels in patient fibroblasts. The truncated VPS41R662* form could not be detected at all, whereas expression levels of VPS41 c.1423‐2A>G were strikingly low. Hence, cells from patients 1 and 2 only contained the VPS41S285P variant, whereas patient 3 only contained VPS41c.1423‐2A>G at about 10% of wild‐type VPS41 levels which could explain the more severe phenotype observed in patient 3. The three patients described here share features with a recently reported patient bearing a homozygous VPS41 splice site variant (NM_014396.3c.450 + 1G>T) (Steel et al, 2020). All patients show the autosomal recessive inheritance pattern, early onset generalized dystonia with developmental delay, and brain MRI characterized by thinning of the corpus callosum and atrophy of the cerebellar vermis. One of our patients (Patient 1) presents a slightly different phenotype, characterized by a less severe intellectual disability and ataxia without overt dystonia, clearly indicating that the phenotypic expression of VPS41‐related disorders is variable.
Since VPS41S285P is the only variant expressed at substantial levels, we investigated whether it could form a functional HOPS complex. By co‐IPs using tagged constructs, we found that VPS41S285P interacts with other HOPS subunits and by immunofluorescence that this variant is recruited to endolysosomal membranes. However, expression of VPS41S285P in VPS41KO HeLa cells did not rescue the HOPS‐dependent fusion between lysosomes and late endosomes or autophagosomes. Since a serine to proline substitution is predicted to affect protein folding, the inability of VPS41S285P to form a functional HOPS complex may be due to a folding defect. Importantly, these data demonstrate that all patient variants affect HOPS function, either because of lack of expression and/or an inability to form a functional complex. In accordance, HOPS‐dependent transport of endocytosed Dextran to enzymatically active lysosomes is significantly decreased in patient cells as compared to the healthy controls. Intriguingly, the VPS41WT/R662 * cells of the mother of the two siblings also showed decreased delivery to lysosomes. This effect was apparent after 2 h Dextran incubation but disappeared after 5 h. By contrast, lysosomal delivery in patient cells reached control levels only after 24 h uptake. This shows that expression of the VPS41 variants causes a delay rather than a block in HOPS‐dependent transport to lysosomes and implies that the kinetics of cargo delivery to lysosomes is a factor of importance in the development of VPS41‐associated pathology.
Independent of HOPS, VPS41 is also required for formation of secretory granules and transport of lysosomal membrane proteins via the ALP/LAMP‐carrier pathway (Cowles et al, 1997b; Darsow et al, 2001; Swetha et al, 2011; Pols et al, 2013b). Both functions were reported to depend on the interaction of VPS41 with AP‐3 (Cowles et al, 1997a; Rehling et al, 1999; Darsow et al, 2001; Angers & Merz, 2009; Cabrera et al, 2010). By co‐IP, we found that VPS41S285P still binds AP‐3 with an affinity comparable to VPS41WT. By immuno‐EM of patient‐derived fibroblasts, we found no effect on the concentration of LAMP‐1 or LAMP‐2 in lysosomal membranes. Also, cathepsin B and cathepsin D were readily transported to and activated in lysosomes. Though these steady state assays do not give information on transport kinetics, they do indicate that lysosomes in patient cells contain normal levels of lysosomal proteins. Interestingly, we found that expression of VPS41S285P fully rescued the secretion defect in VPS41KO PC12 cells. Likewise, in a parallel study we found that expression of VPS41S285P in VPS41KO INS‐1 cells restored insulin secretion (Burns et al, 2020). This shows that expression of VPS41S285P fails to restore HOPS function, but does rescue the regulated secretion pathway in PC12 and INS cells. It is conceivable that this activity of VPS41S285P in secretion is important for disease development, but how exactly VPS41 regulates secretion—direct or indirect?—remains to be elucidated. In future studies, we can use the VPS41S285P variant to distinguish between these HOPS‐dependent and HOPS‐independent functions of VPS41.
A striking finding in our studies is that expression of the VPS41 variants leads to a decreased lysosomal localization of mTORC1, a constitutive nuclear localization of TFE3, and impaired TFEB phosphorylation. This was seen in patient‐derived fibroblasts and recapitulated in HeLa and PC12 cells knockout for VPS41, indicating that this phenotype occurs across different cell types. TFE3 and TFEB are part of the MiTF/TFE family of transcription factors (Slade & Pulinilkunnil, 2017). Both TFE3 and TFEB translocate to the nucleus upon nutrient starvation and bind CLEAR elements in promotors of autophagic and lysosomal genes (Sardiello et al, 2009; Palmieri et al, 2011). In VPS41 disease cells, the continuous nuclear localization of TFE3 results in elevated levels of target genes like LAMP‐1 and cathepsin B, which is in agreement with the observed increase in number of small‐sized, LAMP‐1‐positive compartments. Even though lysosomal size is controlled by a diversity of pathways (de Araujo et al, 2020), we speculate that due to HOPS complex dysfunctionality and subsequent impaired fusion between endolysosomal compartments, these newly formed organelles remain small because of decreased membrane input, as is observed in both patient and VPS41KO cells.
It is tempting to speculate that impaired endocytic cargo delivery deprives lysosomes of nutrients, thereby causing a state of starvation. However, phosphorylation of S6K and 4EBP1, canonical substrates of mTORC1, was not affected in cells depleted of a functional HOPS complex. Indeed, also VPS18KO and VPS39KO cells showed constitutive nuclear localization of TFE3, but normal S6K and 4EBP1 phosphorylation in response to nutrient starvation and replenishment. Also, phosphorylation of ULK1 was not affected in any of the knockout cells. These findings suggest that HOPS‐deficient cells are not continuously starved or impaired in nutrient sensing, but that the HOPS complex specifically regulates mTORC1‐dependent control of the TFEB/TFE3 axis. Interestingly, cells depleted of Folliculin (FLCN), the GTPase activating protein (GAP) for the lysosome‐associated RagC/D GTPases necessary for membrane recruitment of mTORC1, are also selectively affected in the TFEB/TFE3 axis, with no effect on S6K and 4EBP1 phosphorylation (Lawrence et al, 2019). As explanation for a differential regulation of mTORC1 substrates, it was recently proposed that amino acids, and not growth factors, are required for FLCN‐mediated regulation of TFEB/TFE3 (Napolitano et al, 2020). Possibly the HOPS complex, by facilitating late endosome–lysosome fusion, is required for the delivery of degradable material for the supply of amino acids. This endolysosomal fusion might not be necessary for the growth factor‐dependent regulation of S6K/4EBP1. Indeed, cells treated with chloroquine, which recently was shown to block late endosome–lysosome fusion, also show translocation of TFE3 to the nucleus (Roczniak‐ferguson et al, 2012; Mauthe et al, 2018). Additionally, HOPS could be involved in the association of mTORC1 and/or TFEB/TFE3 with lysosomes. TFEB/TFE3 and mTORC1 both bind to active Rags on the lysosomal membrane, thereby enabling the phosphorylation of TFEB/TFE3 (Martina & Puertollano, 2013). By contrast, S6K and 4EBP1 do not associate with the lysosomal membrane and, therefore, lysosomal association of mTORC1 is less crucial for phosphorylation of these latter substrates (Napolitano et al, 2020). Since patient and VPS41KO cells show decreased association of mTORC1 with the lysosomal membrane, an interesting hypothesis is that the HOPS complex enables lysosomal association and retention of mTORC1 and TFE3/TFEB, possibly by cooperating with the Rags.
Currently, patients with mutations in HOPS core components VPS11, VPS16, VPS33A, and VPS41 have been reported in literature, which all show a neurodegenerative phenotype (Chintala et al, 2009; Edvardson et al, 2015; Zhen & Li, 2015; Cai et al, 2016; Hörtnagel et al, 2016; Kondo et al, 2016; Zhang et al, 2016; Dursun et al, 2017; Beek et al, 2019; Pavlova et al, 2019; Steel et al, 2020). During our studies, an additional patient with a homozygous splice site variant (NM_014396.3c.450 + 1G>T) in VPS41 was identified, displaying a neurological phenotype similar to the patients discussed here (Steel et al, 2020). For future studies, it will be interesting to obtain cells from this patient and screen for the mTORC1/TFE3 phenotype.
VPS41 was previously reported to confer neuroprotection in both C. elegans and mammalian models for Parkinson’s disease (Hamamichi et al, 2008; Ruan et al, 2010; Harrington et al, 2012; Griffin et al, 2018). Overexpression of α‐synuclein results in degeneration of dopaminergic neurons, which is prevented by overexpression of human VPS41. Both the WD40 and CHCR domain of VPS41 are necessary for this neuroprotection (Ruan et al, 2010; Harrington et al, 2012), which are mutated in VPS41S285P and VPS41R662 *, respectively. We found that transgenic nematodes co‐expressing hVPS41WT with either VPS41S285P or VPS41R662*retained the neuroprotective effect, resulting in a decrease in neurodegeneration. However, co‐expressing VPS41S285P and VPS41R662* did not result in neuroprotection and even exacerbated cell death in aging worms. Since we found that the VPS41 patient cells completely lack HOPS function, the neurodegenerative phenotype in the nematodes is likely HOPS related. Moreover, since HOPS‐dependent autophagosome–lysosome fusion is important for clearance of aggregated material, which particularly tends to accumulate in neurons over the course of aging (Takáts et al, 2009; Jiang et al, 2014; McEwan et al, 2015; Jia et al, 2017), overexpression of VPS41 in the C. elegans model possibly induces protein clearance through autophagy. The intersection of lysosomal dysfunction and neurodegeneration has become a critical junction in the mechanistic underpinnings of Parkinson’s disease (Mazzulli et al, 2011). This is best exemplified by mutations in the gene encoding the lysosomal protein GBA1, which causes Gaucher’s disease and has become the most prevalent genetic risk factor for Parkinson’s disease (Do et al, 2019). In the context of these results, it is tempting to speculate that an increased understanding of autolysosomal trafficking defects in VPS41 patient cells may serve to inform therapeutic strategies for neurodegenerative disorders (Griffin et al, 2018).
In summary, our data imply that VPS41 patients may represent a novel class of lysosomal disorders in which lysosomes are enzymatically active, but due to delayed trafficking kinetics inefficiently reached by endocytic and autophagic cargo. Moreover, patient cells are selectively inhibited in mTORC1‐mediated TFEB/TFE3 regulation. Interestingly, many other neurodegenerative diseases show an opposite phenotype, i.e., with hyperactivity of mTORC1 (An et al, 2003; Wong, 2013; Lam et al, 2017). These observations are of particular importance with regard to the design of a possible treatment of VPS41 and other HOPS‐related disorders (Chintala et al, 2009; Laplante & Sabatini, 2012; Wong, 2013; Dursun et al, 2019; Pavlova et al, 2019).
Materials and Methods
Patient material and identification of VPS41 variants
Informed consent was obtained from all subjects and the experiments conformed to the principles set out in the WMA Declaration of Helsinki and the Department of Health and Human Services Belmont Report. Parents provided written informed consent for publication of clinical pictures. Information about whole exome sequencing, filtering, and variant analysis is presented in Appendix Supplementary Methods.
Antibodies, reagents, and lysosomal functionality assays
Antibodies and reagents used in this study and their specific dilutions or used concentrations are specified in Appendix Table S2. To study endocytosis, cells were incubated for 2 h with 10,000 MW Dextran‐Alexa Fluor 568 (Invitrogen), washed five times using warm PBS, fixed with 4% PFA, embedded in Prolong DAPI (Invitrogen), and imaged. Lysosomal acidity was determined using Lysotracker™ Red (Molecular Probes) for 30 min after which cells were washed five times using 37°C PBS, fixed with 4% PFA, embedded in Prolong DAPI (Invitrogen), and imaged. To visualize active lysosomes, we used the SiR‐Lysosome (Spirochrome) probe. Cells were incubated for 3 h, washed with PBS, fixed with 4% PFA, embedded in Prolong DAPI (Invitrogen), and imaged. For the visualization of cathepsin B‐active compartments, cells were incubated with MagicRed cathepsin B substrate (Molecular Probes) for 30 min and imaged at RT using live‐cell imaging. To assess EGF degradation, WT and VPS41KO PC12 cells were washed twice with PBS and starved of serum for 2 h in DMEM with 0.1% BSA (GoldBio). During starvation, EGF‐biotin (GoldBio) streptavidin‐647 conjugate was prepared. EGF‐biotin (5 μg/ml) was incubated for 30 min at 4°C at a 5:1 ratio to streptavidin‐Alexa647 (Life Technologies). Following starvation, cells were washed twice with ice‐cold PBS on ice and incubated with EGF‐A647 conjugate at a final concentration of 100 ng/ml for 1 h on ice. Excess unbound EGF was removed by washing with ice‐cold PBS with 0.5% BSA. Cells were chased for indicated times at 37°C before fixation with 4% PFA in PBS for 20 min at room temperature. Cells were analyzed by flow cytometry (CyAn ADP Analyzer, Beckman Coulter, USA) or scanning confocal microscopy.
For the RT–PCR, mRNA was isolated from cells using RNeasy kit (Qiagen) and converted to cDNA using TaqMan reverse transcriptase reagents (Applied Biosystems). Real‐time PCR was performed using PCR mastermix with SYBR green (Applied Biosystems) with primers that were ordered from realtimeprimers.com.
Cell culture and light and electron microscopy
Patient‐derived fibroblasts and HeLa cells were cultured in High Glucose Dulbecco's Modified Eagle’s Medium (Invitrogen) supplemented with 10% FCS in a 5% CO2‐humidified incubator at 37°C. Cells were transfected using X‐tremeGENE™ HP (Roche) according to manufacturer’s instructions. To induce autophagy, cells were starved for 2 h at 37°C using EBSS (Thermo Fisher) and restimulated for 15 min with complete medium. Cells destined for immunofluorescence were washed with PBS once and fixed using 4% wt/vol paraformaldehyde (PFA, Polysciences Inc.) for 20 min. Cells were washed three times with PBS and permeabilized with 0.1% Triton X‐100 (Sigma) for 5 min. Samples were blocked for 15 min using a 1% BSA solution. Samples were labeled, embedded in Prolong DAPI (Invitrogen), and imaged. When imaging transfected cells, cells with similar transfection levels were selected. Images were taken using a Deltavision wide‐field microscope (Applied Precision) with a 100x/1.4A oil immersion objective. Image analysis was performed using Volocity software (Perkin Elmer) and macros written in Fiji (Schindelin et al, 2012). The ComDet plugin was used for dot detection for quantifications of immunofluorescent images (Eugene Katrukha, Utrecht University). For TFE3 localization experiments, cells were scored positive when a defined nuclear outline was visible due to nuclear labeling of TFE3.
For resin EM, cells grown in 6cm dishes were fixed in 2% wt/vol PFA, 2.5% wt/vol GA (Electron Microscopy Sciences) in Na‐cacodylate buffer (Karnovsky fixative) for 2 h at RT. Subsequently, the fixative was replaced for 0.1 M Na‐cacodylate buffer, pH 7.4. Post‐fixation was performed using 1% wt/vol OsO4, 1.5% wt/vol K3Fe(III)(CN)6 in 0.065 M Na‐cacodylate buffer for 2h at 4°C. Next, cells were stained with 0.5% uranyl acetate for 1 h at 4°C, dehydrated with ethanol, and embedded in Epon (Electron Microscopy Sciences). Ultrathin sections were stained with uranyl acetate and lead citrate using AC20 (Leica). Images were taken on a TECNAI T12 electron microscope (FEI Thermo Fisher Scientific).
For immuno‐EM, cells were grown in 6cm dishes were incubated with BSA‐Au5 (Cell Microscopy Core, UMC Utrecht) for 2h in culture medium at 37°C, washed with medium, and fixed by adding freshly made 2% FA, 0.2% GA in 0.1 M phosphate buffer (pH 7.4) to an equal volume of medium for 15 min. After 15 min, the fixative was refreshed to continue fixation for 2h at RT. Cells were stored in 1% formaldehyde at 4°C until further processing. Then, cells were washed with PBS/0.05 M glycine, scraped in 1% gelatin/PBS, pelleted in 12% gelatin/PBS, solidified on ice, and cut into small blocks. The blocks were infiltrated in 2.3 M sucrose overnight at 4ᵒC, mounted on aluminum pins, and frozen in liquid nitrogen (Slot & Geuze, 2007). Ultrathin sections were prepared using an ultra‐cryomicrotome (Leica). To pick up ultrathin sections, a 1:1 mixture of 2.3 M and 1.8% wt/vol methylcellulose was used. Ultrathin sections were immuno‐gold labeled and stained as described (Slot & Geuze, 2007). Protein A‐conjugated colloidal gold particles were made in house (Cell Microscopy Core, UMC Utrecht).
CRISPR/Cas9 knockout cells
HeLaVPS41KO
Hela cells were transiently transfected with pSpCas9(BB)‐2A‐GFP (PX458) (Ran et al, 2013) encoding sgRNAs targeting VPS41 using X‐tremeGENE (Merck) according to the manufacturers recommendation. sgRNAs were designed with http://crispor.tefor.net/ (Haeussler et al, 2016) and target coding sequences in the N‐terminal region encoded by the third coding exon. The sgRNA used is sgRNA2 AAGTATTTCAGTTACCCCAT. GFP‐positive cells were sorted using a FACSAria II flow cytometer (BD) and plated in 10‐cm dishes. Colonies were picked from these plates after 1 week and expanded. To confirm VPS41 absence, total cell lysates were analyzed by Western blotting using mouse anti‐VPS41 (SC‐377271, Santa Cruz).
HeLaVPS11KO/VPS18KO/VPS39KO
To generate VPS11, VPS18, and VPS39 knockout HeLa cells, a WT HeLa parental stain was transfected with pSpCas9(BB)‐2A‐Puro (PX459) plasmids containing gsRNAs for VPS11, VPS18, or VPS39 using Effectene transfection reagent (QIAGEN). gsRNAs were designed using against the first exons of VPS11, VPS18, and VPS39 using an optimized CRISPR design tool. sgRNA sequences that generated the knockout cells used in this study were GTGTGTCACCCGTAGTTTGT for VPS11, CGAGAACTCGCTGTCCCGCT for VPS18, and GCCTCTGCAAATCGACTGTC for VPS39. We selected for transfected cells using 1 µg/ml Puromycin. Surviving cells were recovered for 2 weeks and split into 96‐well plates at single cell/well dilution. Single cell colonies were selected and expanded. For VPS18 and VPS11 clones, knockout was confirmed by the absence of VPS18 or VPS11 protein on Western blot. VPS39 knockout clones were confirmed via genomic DNA isolation followed by PCR of VPS39 exon 1, sequencing, and analysis using the TIDE sequence analysis tool (https://tide.nki.nl/). The selected VPS39 knockout clone had two deletions of 5 and 8 nucleotides in the first exon, resulting in a frameshift.
PC12VPS41KO
The human codon‐optimized Cas9 and chimeric guide RNA expression plasmid (pX459v2) developed by the Zhang laboratory were obtained from Addgene (Ran et al, 2013). To generate gRNA plasmids, a pair of annealed oligos (20 base pairs) were ligated into the single guide RNA scaffold of pX459v2. The following gRNAs sequences were used: Forward (rat): 5′‐CACCGACTCTCAGACTGAGCTATGG‐3′; Reverse (rat): 5′‐AAACCCATAGCTCAGTCTGAGAGTC‐3′ to generate the PC12VPS41KO line. Rat VPS41 lentiviral plasmid was generated by amplifying VPS41 from rat cDNA using the following primers: WT Forward: 5′‐CCTCCATAGAAGACACCGACTCTAGACACCATGGCGGAAGCAGAGGAG‐3′; WT Reverse: 5′‐TATGGGTAACCCCCAGATCCACCGGTCTTCTTCATCTCCAGGATGGCA‐3′. The PCR products were then subcloned by Gibson ligation into pLenti‐CMV‐GFP‐Puro. pLenti‐CMV‐GFP‐Puro was a gift from Paul Odgren (Addgene plasmid #73582). To test for the presence of indels, the resulting PCR products were ligated into pBlueScript II KS. Isolated plasmids from 10 random colonies were analyzed for the presence of indels by Sanger sequencing. Insertions in the initiator codon of the VPS41 gene were observed in all clones of either a single or a triple T.
Co‐immunoprecipitation and Western blotting
Cells were seeded in 15‐cm dishes and transfected with the appropriate constructs as indicated in the respective figures and according to abovementioned transfection protocols. Cells were washed three times with ice‐cold PBS and lysed using a CHAPS lysis buffer (50 mM Tris pH = 7.5, 150 mM NaCl, 5 mM MgCl2, 1mM DTT, 1% (w/v) CHAPS) complemented with protease and phosphatase inhibitor (Roche). Cells were scraped, collected, and spun down at 16,000 rcf/g for 15 min at 4°C. Protein levels were equalized between samples using a Bradford Protein Assay (Bio‐Rad). 10% of the sample was saved as input control. The remainder of the samples was incubated for 1 h with uncoated protein G beads (Millipore) to remove a‐specifically bound proteins. Beads were spun down and the supernatant was incubated with beads together with 2 µg antibody against the prey. Samples were incubated overnight, extensively washed, and eluted using SDS sample buffer and run on precast gradient (4–20%) gels (Bio‐Rad). Gels were transferred using Trans‐Blot® Turbo™ RTA Mini PVDF Transfer Kit and the Trans‐Blot® Turbo™ Transfer system (Bio‐Rad). Membranes were blocked with Odyssey® Blocking Buffer (LI‐COR) in PBS for 1h at RT and incubated with primary antibody diluted in Odyssey® Blocking Buffer (LI‐COR) in 0.1% TBST overnight at 4°C, rocking. Membranes were washed extensively with 0.1% TBST and incubated with secondary antibodies, diluted in Odyssey® Blocking Buffer (LI‐COR) in 0.1% TBST at RT for 1 h, rocking. The membranes were again washed extensively with 0.1% TBST, followed by 2 washing steps with PBS and 1 washing step with MiliQ. Membranes were scanned using the Amersham™ Typhoon™ Laser Scanner (GE Healthcare Life Sciences).
Plasmids
The GFP‐VPS41 constructs were cloned from hVPS41 cDNA (Origene) into a pDonor201 vector (Invitrogen) using PCR. Next, using the Gateway system and GFP‐pcDNA‐Dest53 (kindly provided by Prof. R. Roepman, Radboud University Medical Center Nijmegen), a recombination reaction was performed. The VPS41 mutant variants were made using the QuickChange Site‐Directed Mutagenesis kit (Agilent) using the primers 5’‐CAGCTTGTTGTACTTCCGTATGTAAAGGAGA and 5’‐TCTCCTTTACATACGGAAGTACAACAAGCTG to generate GFP‐VPS41S285P and 5’‐GTTTATCTTCTGAGCTGAATGGGTTAATAGCC and 5’‐GGCTATTACCCATTCAGCTCAGAAGATAAAC to generate GFP‐VPS41R662*. The pcDNA3‐APEX2‐V5 construct (kindly provided by Dr. S. de Poot) was used as empty vector in pulldown experiments. VPS41‐APEX2‐V5 was generated via a BamHI insert using pEGFP‐C1‐mCherry‐VPS41 (kindly provided by Prof. J.J.C. Neefjes, University Medical Center Leiden) using the primers 5’‐AGAGGGATCCATGGCGGAAGCAGAGGAGCAG and 5’‐AGAGTCTAGACTATTTTTTCATCTCCAAAATTG. VPS41S285P‐APEX2‐V5 and VPS41R662*‐APEX2‐V5 were generated via an identical approach using pEGFP‐C1‐mCherry‐VPS41S285P and pEGFP‐C1‐mCherry‐VPS41R662*. These VPS41 variants were made using the abovementioned Mutagenesis kit (Agilent) and the same primer pairs.
For the C. elegans experiments, the wild‐type (wt) Pdat‐1::hVPS41 construct was assembled as previously described (Harrington et al, 2012). Briefly, human VPS41 cDNA was obtained from Open Biosystems (Huntsville, AL). Plasmid entry vectors were generated using Gateway Technology (Invitrogen) to clone PCR amplified constructs into pDONR221 to generate a hVPS41 entry clone. The hVPS41 entry clone was used to clone hVPS41 into the Gateway expression vector, pDEST‐Pdat‐1. The mutant hVPS41 alleles were generated by site‐directed mutagenesis of the Pdat‐1::hVPS41 construct using the primers 5’‐ATTCTACATCAGTGGACTTGCACCTCTCTGTGATCAGCTTGTTGTACTTCCGTATGTAAAGGAGATTTCAGAAAAAACGGAAAGAGAATACTGTGCCAG and 5’‐CTGGCACAGTATTCTCTTTCCGTTTTTTCTGAAATCTCCTTTACA TACGGAAGTACAACAAGCTGATCACAGAGAGGTGCAAGTCCACTGATGTAGAAT to generate the Pdat‐1::hVPS41S285P construct and 5’‐GATCTGTCAACAGAGAAACTTTGTAGAAGAGACAGTTTATCTTCTGA GCTGAATGGGTAATAGCCGAAGTGCCCTGAAGATGATTATGGAGGAATTACA and 5’‐TGTAATTCCTCCATA ATCATCTTCAGGGCACTTCGGCTATTACCCATTCAGCTCAGAAATAAACTGTCTCTTCTACAAAGTTTCTCTGTTACAGATC to generate the Pdat‐1::hVPS41R662* construct. The identities of the Gateway entry and expression constructs were validated by DNA sequencing.
Caenorhabditis elegans strains
Transgenic C. elegans strains were generated by directly injecting a Punc‐54::tdTomato co‐injection marker (50 ng/µl) and Pdat‐1::hVPS41 wt (25 ng/µl) with Pdat‐1::hVPS41S285P (25 ng/µl) or Pdat‐1::hVPS41R662* (25 ng/µl) constructs or Pdat‐1::hVPS41S285P (25 ng/µl) with Pdat‐1::hVPS41R662* (25 ng/µl) into the gonads of strain N2 Bristol hermaphrodites. Progeny of injected animals were screened for expression of the hVPS41 expression constructs by evidence of fluorescence in the body‐wall muscles from the Punc‐54::tdTomato co‐expression marker. Several stable lines of each were isolated and crossed with isogenic UA44 (baIn11[Pdat‐1::α‐syn, Pdat‐1::GFP]) and BY250 (vtIs7[Pdat‐1::GFP]) males. Heterozygous hermaphroditic progeny expressing both tdTomato and GFP were transferred and allowed to self‐fertilize. Several lines of each Pdat‐1::hVPS41wt + Pdat‐1::hVPS41S285P [baEx215] in UA44 (UA389) and BY250 (UA386) were analyzed for neurodegeneration at days 7 and 10. Likewise, Pdat‐1::hVPS41wt + Pdat‐1::hVPS41R662 *[baEx216] in UA44 (UA390) and BY250 (UA387) was analyzed. Additionally, Pdat‐1::hVPS41S285P + Pdat‐1::hVPS41R662 * (baEx217] in the UA44 (UA391) or BY250 (UA388) backgrounds was analyzed. For all strains, three representative lines of each were selected for further analysis.
Dopaminergic neurodegeneration analysis in Caenorhabditis elegans
Caenorhabditis elegans dopaminergic neurons were analyzed for degeneration as previously described (Hamamichi et al, 2008; Harrington et al, 2012). Briefly, synchronized progeny from the BY250, UA44, and experimental hVPS41 variant strains were produced from a 3‐h egg‐lay and grown at 20°C. Animals were analyzed at days 7 and 10 post‐hatching (4‐ and 7‐day‐old adults). On the day of analysis, the 6 anterior dopaminergic neurons [4 CEP (cephalic) and 2 ADE (anterior deirid)] were examined in 30 adult hermaphrodite worms, which were immobilized on glass cover slips using 3 mM levamisole and transferred onto 2% agarose pads on microscope slides. In quantifying neurodegeneration, animals are scored as “normal” when a full complement of all 6 anterior dopaminergic neurons are present and the neuronal processes are fully intact, as previously reported (Cao et al, 2005; Hamamichi et al, 2008; Harrington et al, 2010). In total, at least 90 adult worms (30 worms/trial × 3 trials = 90 total animals) were analyzed for each independent strain and at least 270 adult worms were analyzed for each independent transgenic strain (30 worms/trial × 3 independent transgenic lines × 3 trials = 270 total animals/construct). Values of each independent experiment were normalized to the BY250 control, and representative analyses were averaged and statistically analyzed by Prism and one‐way ANOVA.
Protein purification
The appendage domain of rat AP‐3 delta was amplified by PCR and cloned in‐frame into pET15b‐MBP‐MCS. The recombinant proteins were induced overnight at 20°C in Escherichia coli BL21 (DE3) using 0.1 mM IPTG. The bacteria were lysed by sonication in in 50 mM Tris, 300 mM NaCl, pH 8.0, and MBP fusions purified by incubation with amylose resin beads (NEB) and eluted with 50 mM maltose in 50 mM Tris, 300 mM NaCl, pH 8.0. Two days after infection with baculoviruses produced according to the manufacturer’s instructions (Invitrogen), Hi5 cells were harvested by centrifugation and resuspended in binding buffer (50 mM Tris, 300 mM NaCl, pH 8.0) with 1 mM PMSF and protease inhibitors (Complete, Roche). The cells were lysed by sonication and pelleted for 20 min at 21,000 g using a 4°C Eppendorf 5424 centrifuge. Supernatant was passed through a 0.2 μm filter and incubated with Co‐Nta agarose (GoldBio) for 1 h at 4°C. The agarose was then washed five times in binding buffer with 10–40 mM imidazole and His‐hVPS41 eluted with 500 mM imidazole. The eluates were immediately desalted on a Secadex‐5 column (Prometheus) into lysis buffer. 50 µg of MBP or MBP‐AP‐3 were loaded onto amylose resin for 30 min at 4°C. Unbound protein was removed by several washing steps, and varying amounts of VPS41 were incubated with the MBP resin to assess interaction. Bound protein was eluted and evaluated by quantitative fluorescence immunoblotting.
Secretion assays
PC12 cells were plated on poly‐L‐lysine, washed, and incubated in Tyrode’s buffer containing 2.5 mM K+ (basal) or 90 mM K+ (stimulated) for 30 min at 37°C. The supernatant was collected, cell lysates prepared as previously described (Asensio et al, 2010), and samples were analyzed by quantitative fluorescence immunoblotting.
Statistics
Quantification of all Western blots was performed using Fiji software (Schindelin et al, 2012). Two‐tailed Students t‐tests were performed to compare two samples whereas ANOVA was performed for comparison of multiple samples to analyze statistical significance. Data distribution was assumed to be normal, but this was not formally tested. All error bars represent the Standard Error of the Mean (SEM) or Standard Deviation (SD) as indicated in the specific figure legends.
Author contributions
Experiments: RENW, CB, PS, CB, LC, FJZ, SZ, EFG, JAB, and TV; Clinical investigation on the patients: RJ, AF, CMAR‐A, HHL, SB, CS, AML, GY, and DC; Sequencing analysis and variant identification: HHL, RP, and TS‐S; Study design: RENW, CB, PS, JAB, NL, GAC, CSA, KAC, DC, and JK; Manuscript reviewing and revision: AF, FJZ, JAB, NL, CMAR‐A, SB, CSA, GAC, KAC, and DC; Study design and supervision: CA, GAC, KAC, DC, and JK; Funding: JK; Manuscript writing: RENW, CMAR‐A, SB, DC, and JK.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
We are indebted to our colleagues of the Center for Molecular Medicine for fruitful discussions. We thank MCLS master student Bas van Zuijlen for the generation of the VPS11 and VPS18 knockout cell lines, Dr. Stephanie de Poot (Cell biology, UMC Utrecht) for kindly providing the APEX2 constructs and Prof. R. Roepman (Radboud UMC, Nijmegen, Netherlands) and Prof. J.J.C. Neefjes (Leiden UMC, Netherlands) for providing the GFP‐pcDNA‐Dest53 and pEGFP‐C1‐mCherry‐VPS41 constructs, respectively. Prof. Catherine Rabouille (Hubrecht, The Netherlands) is acknowledged for her critical input throughout our studies. We thank Dr. I. Stolte‐Dijkstra (UMC Groningen, Netherlands) for providing information and patient fibroblasts from the Dutch VPS41 patient and Dr. H.E. Westerlaan (UMC Groningen, Netherlands) for providing the MRI data of this patient. Support for the C. elegans studies came from a grant from the National Institutes of Health [R15 NS075684‐01 to G.A.C.]. N.L. is supported by a ZonMW TOP grant [40‐00812‐98‐16006 to J.K]. P.S and RvdW are supported by a DFG grant [FOR2625 to J.K. as part of the Research Consortium]. The electron microscopy within this work is part of the research program National Roadmap for Large‐Scale Research Infrastructure (NEMI) 2017 – 2018 with project number 184.034.014, which is (partly) financed by the Dutch Research Council (NWO). We are very grateful for the patients and their parents for their help and support.
Data availability
This study includes no data deposited in external repositories.
 Neurodegenerative VPS41 variants inhibit HOPS function and mTORC1‐dependent TFEB/TFE3 regulation
Neurodegenerative VPS41 variants inhibit HOPS function and mTORC1‐dependent TFEB/TFE3 regulation
