
- Altmetric
- Background
- Methods
- Results
- Conclusions
- 1 Introduction
- 2 Materials and methods
- 3 Results
- 4 Discussion
- 5 Conclusions section
- Ethics approval and consent to participate
- Consent for publication
- Availability of supporting data
- Funding
- Authors'contributions
- Disclaimers
- Declaration of competing interest
- Abreviations
- Appendix A Supplementary data
Background
The adaptation to hypoxia is mainly controlled by the HIF transcription factors. Increased expression/activity of HIF-1α correlates with poor prognosis in cancer patients. PARP-1 inhibitors are used in the clinic to treat BRCAness breast/ovarian cancer and have been shown to regulate the hypoxic response; therefore, their use could be expanded.
Methods
In this work by integrating molecular/cell biology approaches, genome-wide ChIP-seq, and patient samples, we elucidate the extent to which PARP-1 exerts control over HIF-1-regulated genes.
Results
In human melanoma, PARP-1 and HIF-1α expression are strongly associated. In response to a hypoxic challenge poly(ADP-ribose) (PAR) is synthesized, HIF-1α is post-transcriptionally modified (PTM) and stabilized by PARylation at specific K/R residues located at its C-terminus. Using an unbiased ChIP-seq approach we demonstrate that PARP-1 dictates hypoxia-dependent HIF-recruitment to chromatin in a range of HIF-regulated genes while analysis of HIF-binding motifs (RCGTG) reveals a restriction on the recognition of hypoxia responsive elements in the absence of PARP-1. Consequently, the cells are poorly adapted to hypoxia, showing a reduced fitness during hypoxic induction.
Conclusions
These data characterize the fine-tuning regulation by PARP-1/PARylation of HIF activation and suggest that PARP inhibitors might have therapeutic potential against cancer types displaying HIF-1α over-activation.
Introduction
Hypoxia is a common event during tumor development consequence of accelerated tumor growth. When the mass exceeds a volume of a few mm3, regions of low oxygen concentration occur in the inner parts of the tumor. Under this situation cells must modify their metabolism to cope with this new environmental context. The adaptation to the hypoxic situation involves the expression of hundreds of genes implicated in the maintenance of cellular survival through metabolic adaptation. These adaptations include new vessels formation [1], glycolysis activation [2], cancer stem cells (CSCs) regulation [3] and even tumor exosome production [4]. All these changes facilitate cell survival, tumor growth, migration, and metastasis [[5], [6], [7]]. The hypoxic response is associated with poor overall survival, lower disease-free survival, and diminished loco-regional control [[8], [9], [10]].
This transcriptional induction is mediated by the hypoxia-inducible factors (HIFs). This family of transcription factors are active as an alpha/beta heterodimer which binds to nucleotides sequences, being the most common known as HRE (5′-RCGTG-3’) or hypoxia response element, localized in the promoters of the hypoxic expressed genes, inducing their transcription [11].
The HIF family is composed of 1 beta chain (HIF-1β) and 3 alpha subunits (HIF−1α, HIF-2α and HIF-3α). The fine-tuning of the HIF response depends on the alpha chain, which induces the expression of different genetic patterns, allowing the cell to respond efficiently to different hypoxic intensities and durations. The hypoxic response is regulated under well oxygenated conditions via HIF-α degradation. Prolyl hydroxylase domain proteins (PHDs) hydroxylate proline residues on the alpha subunits specifically during normoxia. This hydroxylation is recognized by the von Hippel-Lindau (VHL) E3 ligase, causing the ubiquitination of the alpha subunits and their subsequent degradation via the proteasome [12]. Although this is the major mechanism controlling HIF stability and activity, many other PTM including phosphorylation, acetylation, SUMOylation and hydroxylation have been described as modulators of HIF induction/activity [13]. The C-terminus domain of HIF-1α has gained increasing attention as a regulatory site of HIF-1α activation. This domain undergoes hydroxylation of an asparagine by FIH (factor inhibiting HIF), reducing the recruitment of co-activators such as the p300 and thus reducing the expression of hypoxic genes in different cancer types [14,15].
The Poly(ADP-ribose) polymerases (PARP) proteins are a family of intracellular enzymes characterized by the presence of a domain referred as “PARP signature”. Substitutions on this domain make only the members PARP-1, 2 and tankyrases able to synthesize a polymer of poly(ADP-Ribose) or PAR, while the rest of the family is inactive or generates mono(ADP-Ribose) or MAR [16]. Both PAR and MAR require NAD+ to be consumed as a substrate in an ATP-dependent reaction [17]. The final product of the PARP enzymatic activity (mono or poly (ADP-ribose)) is then covalently bound onto Glu, Asp, Lys and Ser [18]. PARPs themselves can undergo this modification (auto modification) or they can modify other proteins (hetero modification). These changes are considered a type of reversible PTM and are tightly regulated by enzymes that cleave the linkage between ADP-ribose units and the acceptors (e.g. ADP-ribosyl-acceptor hydrolases (ARH3, MacroD1, MacroD2) or poly(ADP-ribose) glycohydrolases (PARG)).
Through PARylation, PARPs regulate a wide array of cellular processes, including genomic instability and survival [19], chromatin organization [20], protein degradation via proteasome [21], RNA metabolism [22] and cell death [23,24].
PARP-1 was the first described and most active member of the family, generating up to the 90% of the polymer observed in the cell [25]. It is known primarily for its important role during DNA repair. Owing to this property, the inhibition of PARP activity is recognized as a potential therapeutic strategy to enhance the cytotoxic action of anti-cancer drugs or radiotherapy, and for the treatment of cancers with specific DNA repair defects [26,27]. PARP-1 has also been described as a regulator of the hypoxic response [[28], [29], [30]]; however, the signal linking the selective regulation via PARP-1 of HIF-dependent genes is not known. In this study we describe a new pathway demonstrating that early during the response to hypoxia, increased poly(ADP-ribose) activity leads to PARylation of HIF-1α at specific residues located at its C-terminus domain. This PARylation directs HIF-1α to a selective subgroup of genes (mostly excluding glucose metabolism-related genes) suggesting a hierarchy to avoid overlap in the regulation in HIF-dependent gene expression and to protect critical survival functions during hypoxia.
Materials and methods
Cultures and growth medium
HEK 293T cells were cultured using Dulbecco's Modified Eagle medium low glucose supplemented with l-glutamine 4 mM, MEM non-essential amino acids 0.1 mM, 10% heat-inactivated fetal bovine serum (FBS) plus penicillin (50 IU/ml) and streptomycin (50 mg/ml). HepG2 cells were cultured using Dulbecco's Modified Eagle medium high glucose supplemented with 10% heat inactivated FBS plus penicillin (50 IU/ml) and streptomycin (50 mg/ml). COS cells were cultured in Dulbecco's Modified Eagle medium low glucose supplemented with 10% of heat inactivated FBS plus penicillin (50 IU/ml) and streptomycin (50 mg/ml). All cells were grown using a regular incubator at 37 °C in a humidified 21% O2 and 5% CO2 atmosphere. Hela cells were cultured on D-MEM (high glucose), 10% heat inactivated FBS, 0.1 mM MEM Nonessential Amino Acids (NEAA), 2 mM l-glutamine, 1% Pen-Strep. C8161 were cultured on RPM11640 (GIBCO) supplemented with 10% heat inactivated FBS, glutamine (0.8 Mg/ml), and gentamicin (10 ng/ml). MUM2b cells were grown on Eagle's minimum essential medium (EMEM) (BioWhittaker, Walkersville, MD) supplemented with 10% heat-inactivated FBS (Fisher, Ontario, ON, Canada), 1% nonessential-amino acids (NEAA-Mixture, 100; BioWhittaker) 2 mmol/L l-glutamine, and penicillin/streptomycin.
Treatments
The PARP inhibitor PJ34 was purchased from Enzo Life Sciences (San Diego, CA, USA) and was used at a concentration of 10 μM, 90 min before hypoxia. The PARP inhibitor olaparib was used at 5 μM, 90 min before hypoxia and purchased from Selleckchem (Houston, USA). The ROS inhibitor MPG-2, also referred as N-(2-Mercaptopropionyl) glycine, was purchased from Sigma-Aldrich (San Louis. Missouri, USA) and was used at 300 μM 90 min before the exposure to different times of hypoxia. Mitochondrial antioxidant MitoTempo was purchased from Sigma-Aldrich (San Louis. Missouri, USA) and used at a final concentration of 20 μM. Cells were pretreated for 120 min and then exposed to hypoxia.
Hypoxia
Hypoxic incubation was achieved using a sealed hypoxic workstation (Ruskin, Bridgend, UK). Cells were exposed to 1% O2 and 5% CO2 at 37 °C. Hypoxic duration was 4 h if a different duration is not indicated.
Western Blot
For Western Blot analysis, cells were plated in six-well plates at a density of 2.5 × 105, lysed using TRE buffer and then the whole cell extract was sonicated, resuspended, and boiled for 5 min in modified Laemli charge buffer (250 mM Tris-HCl (pH 75), glycerol 20%, SDS 10%, 1,4 M of mercaptoethanol and 1% blue bromophenol). Samples were transferred to a nitrocellulose membrane through wet transfer (Amersham Biosciences). Proteins were then visualized using the ECL system (Amersham Biosciences) after using specific antibodies for: HIF-1α (Bethyl), GST antibodies (Bethyl), Tubulin (Sigma-Aldrich), Actin (Sigma-Aldrich), Myc-tag (Cell Signaling Technology), Poly(ADP-ribose) (Trevigen), PARP-1 (Enzo), RPA (Cell Signaling), p-RPA (Bethyl), P53 (Santa Cruz), p-P53 (Millipore), H2AX (Millipore), p-H2AX (Millipore), Histone H1 (Santa Cruz), Laminin B (Abcam).
Transfection assay
Cells were cultured in six-well plates at a density of 2.5 × 105 cells per well. 24 h later they were transfected using the reagent jetPRIME (Polyplus, Illkirch, France) using the concentrations indicated by the manufacturer for the solutions and plasmids of interest. Cells were cultured for 24 h before being exposed to the reactive. Four hours later the medium was refreshed and one day later cells were lysed using TR3 buffer. For pull down and immunoprecipitation assays, cells were plated in p100 plates at 1.5 106 concentration. Then the jetPRIME protocol was completed in the same way following Polyplus instructions.
Pull down assay
Cells were co-transfected with Myc-HIF1α C-ter and different GST-PARP-1 domains. After following the jetPRIME protocol they were harvested in 20 nM Tris-HCl (pH 7.5), 400 mM NaCl, 20% glycerol, 5 mM DTT, 0,5 mM pefabloc and protease inhibitors (Complete Mini; Roche, Mannheim, Germany). Lysates were cleared by centrifugation and incubated for 2 h with glutathione-sepharose 4B (Sigma, St Louis, MO, USA) Beads were washed three times with 20 nM Tris–HCl (pH 7.5), 150–500 nm NaCl, 0.1% NP-40 and protease inhibitors. All samples were resuspended and boiled for 5 min in modified Laemli charge buffer (250 mM Tris-HCl (pH 7.5), glycerol 20%, SDS 10%, 1,4 M mercaptoethanol and 1% blue bromophenol). Then samples were analyzed by western blot. Blots were subsequently incubated with the anti-GST antibody.
Irradiation
Cell cultures were irradiated in an Xstrahl RS225 cabinet using tissue flasks at room temperature. A voltage of 195 kV X‐rays was used at a dose rate of 1.6 Gy/min until a final dose of 5Gy was accumulated on the sample.
Immunoprecipitation
For IP, 2x106 cells per condition were incubated. After undergoing the different treatments, they were collected and exposed to ice for 20 min on a lysis buffer (1% NP-40, 1 mM EGTA (pH 8.0), 100 mM NaCl, 1 mM sodium orthovanadate, 20 mM Tris-Cl (pH 7.6), 10 mM sodium pyrophosphate, 10 mM NaF, 500 mM phenylmethyl sulfonyl fluoride and cocktail of proteases inhibitors (Complete Mini, Roche, Zurich, Switzerland)). Lysates were incubated with Protein A Sepharose to reduce non-specific signaling. Then, the pre-cleared supernatants were isolated and incubated with the antibodies of interest plus Protein A Sepharose overnight at 4 °C. The extracts were washed five times with saline buffer (100 mM NaCl, 20 mM Tris-HCl (pH 8.0), 0,5% NP-40, 1 mM EDTA, and protease inhibitors). The immunoprecipitated complexes were isolated by centrifugation and boiled for 5 min in modified Laemli charge buffer (250 mM Tris-HCl (pH 7.5), glycerol 20%, SDS 10%, 1,4 M mercaptoethanol and 1% blue bromophenol). Then, samples were analyzed by western blot.
Immunostaining
On 6-well plates, sterilized coverslips were placed, then 2.5 × 105 cells were seeded on top of them. After growing overnight cells were exposed to different treatments. They were then washed gently with PBS 1x and fixed using 3% paraformaldehyde during 15 min at room temperature. Then, they were washed using PBS 1x and permeabilized with 0.25% Triton for 10 min. They were washed again with PBS 1x and blocked with BSA 2% during 1 h at room temperature. Samples were incubated with BSA 2% using anti-FLAG (Sigma Aldrich) or anti-53BP1 (Milipore) (1:100) for 45 min at 37 °C. A secondary antibody, anti-rabbit or anti-mouse, was incubated with BSA 2% for 20 min at 37 °C. Cells were washed with PBS 1x and then sealed with Vectashield antifade mounting medium + DAPI. Images were obtained using confocal microscopy (SP5 Confocal Leica Microscope). Detecting the anti-rabbit signal at 488 nm and the anti-mouse at 647 nm.
Real time q-PCR
Cells were harvested and the RNA isolated following the instructions provided for the RNeasy mini kit (Qiagen, Hilden, Germany). Reverse Transcription was performed using the iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad, Hercules, USA). Real time PCR was performed on a CFX96 thermocycler using the iTaq™ Universal SYBR® Green Supermix (Bio-Rad, Hercules, USA). The primers were synthesized by the “PCR Primer Design” service (Sigma-Aldric, Saint Louis, USA). 36B4 was used as housekeeping gene. The list of primers goes as follow:
h HIF1α Fw CTGCAACATGGAAGGTATTGCA
h HIF1α Rv TACCCACACTGAGGTTGGTTACTG
h CAIX Fw TAAGCAGCTCCACACCCTCT
h CAIX Rv TCTCATCTGCACAAGGAACG
h LDHA Fw TGGGAGTTCACCCATTAAGC
h LDHA Rv AGCACTCTCAACCACCTGCT
h ANGPTL4 Fw CGTACCCTTCTCCACTTGGG
h ANGPTL4 Rv GCTCTTGGCGCAGTTCTTG
h GLUT-1 Fw CAGTTTGGCTACAACACTGGAGT
h GLUT-1 Rv ATAGCGGTGGACCCATGTCT
h VEGFA Fw GGGCAGAATCATCACGAAGT
h VEGFA Rv TGGTGATGTTGGACTCCTCA
h DDIT4 Fw: GACAGCAGCAACAGTGGCTTC
h DDIT4 Rv: CCACGCTATGGCAGCTCTTGC
h UBE2M Fw: CCTGCCCAAGACGTGTGATA
h UBE2M Rv: CCCTGGCCCACCTTAAAACT
h GAPDH Fw: ACAGTCAGCCGCATCTTCTT
h GAPDH Rv: ACGACCAAATCCGTTGACTC
ROS determination
To measure ROS, DCFA-Cellular Reactive Oxygen Species Detection Assay Kit (AbCam) was used. Cells were seeded at a concentration of 2.5 × 104 per point of 96 wells, 24 h later they were stained using 25 μM of DCFDA for 45 min. Next, the DCFDA was washed and cells were exposed to the different treatments. ROS were measured by fluorescence at Ex 485 ± 20 nm Em 535 ± 25 nm using a TECAN plate reader Infinite PRO 200.
To confirm the ROS induction observed during hypoxia we used a non DCFDA-related method for ROS detection based on the molecule DHE (Dihydroethidium). Following the instructions provided by the DHE Assay Kit (Abcam) 2.5 × 104 were seeded per point on a 96 well. After 24 h they were washed using PBS 1X and then exposed to DHE 5 μM for 90 min at 37 °C in the dark. The DHE was then removed and the experiments performed. Using the TECAN plate reader Infinite PRO 200, DHE fluorescence was detected at a Ex 550 ± 10 nm and Em 590 ± 10 nm.
32P-PAR synthesis for the in vitro PARylation assay
Purified 32P-PAR was prepared by incubating 20 μg of purified human PARP-1 (Alexis) in a mixture containing 50 mM Tris-HCl (pH 8.0), 4 mM MgCl2, 0.15 M NaCl, 1 mM DTT, 0.5. DNase I treated-DNA, 0.1 mM BSA, 0.4 mM NAD+, 0.5 Ci [32P]-NAD+ (800 Ci/mmol, 5 PerkinElmer) in a total 500 μl reaction. PARP-1 and the mixture were incubated for 1 h at 32 °C. Then, 2 μl of 10 mg/ml DNAse I (Roche) and 2 μl of 1 M CaCl2 were added and incubated for 1 h at 37 °C. Following this step, 250 μl H2O, 5 μl 20% SDS and 20 μl of 10 mg/ml Proteinase K were added to the mix and incubated for 4 h at 37 °C. Proteins were then extracted using a phenol/CHCl3 treatment: 500 μl phenol/CHCl3 were added, vortexed and centrifugated for 5 min at 4 °C. The upper phase was recovered, and the PAR precipitated by adding 100 μl of 3 M potassium acetate and 1 ml isopropyl alcohol. The samples were then left to precipitate at −20 °C overnight, then centrifuged in low binding Eppendorf tubes at 14,000 rpm at 4 °C 1 h. Carefully the liquid was removed, and the precipitate was washed with 1 ml 80% ethanol. The PAR was left then to dry at room temperature for 1 h. Finally, the PAR was resuspended in 400 μl TE buffer.
PAR binding assay on peptides
Biotinylated peptides were ordered at Genscript. Dried peptides were resuspended in H2O to 3 μg/μl 3 μg of peptides were bound to 15 μl slurry of streptavidin-magnetic beads (Millipore) on ice for 1 h with frequent agitation using 1.5 ml Eppendorf tubes. Beads were washed twice in 1 ml PBS 1x. The beads-peptides complexes were incubated with 50,000 cpm 32P-PAR in 200 μl PBS 1x for 1 h on ice with agitation. The beads were then washed three times with 1 ml PBS 1x, and the tubes were changed after each wash to ensure that no radioactivity was bound non-specifically onto the plastic of the tubes. After the last wash, the supernatant was removed almost to dry the beads and the radioactivity bound to the beads was evaluated by Cerenkov counting using a Packard counter.
CRISPR-Cas9 cells
The production of HEK 293T cells PARP-1 KO was performed thanks to the use of CRISPR-Cas9 technology. Different sgRNAs were selected using the “Zhang Lab Optimized CRISPR design tool” and then cloned into the pL-CRISPR.EFS.GFP obtained from Addgene (#57818). Cells were transfected using JetPrime reagents, then the GeneArt Genomic Cleavage Detection Kit (Invitrogen) was used to validate the sgRNAs following the manufacturer's instructions. The guides number 2 and 4 were selected for, allelic disruption.
Guide 2: GAGTCGAGTACGCCAAGAGC
Guide 4: GCATCCCCAAGGACTCGCTC
These guides were transfected again, the GFP positive cells were isolated and a clone selection was performed after single-cell separation through flow cytometry. Finally, PARP-1 KO status was confirmed by Sanger sequencing and Western Blot.
ChIP-seq
The chromatin immunoprecipitation was performed using the SimpleChIP enzymatic Chromatin IP Kit with magnetic beads #9003 (Cell SignalingTechnology, Danvers, USA). Following the provider instructions, 2 × 107 cells were seeded and 5 μg of HIF−1α antibody ab2185 (AbCam, Cambridge, UK) were used per condition. To test IP efficiency, a quantitative PCR was performed for the hypoxic-induced gen EGLN3. Afterward, the Percent Input was calculated to measure this expression as a percent of the global input chromatin.
ChIP-seq sequencing, data alignment, and peak calling
Immunoprecipitated DNA was processed for sequencing using standard protocols and sequenced on an Illumina Nextseq 500 with 75-bp single end reads. Two biological replicates for each ChIP in hypoxic conditions, and one biological replicate for the remaining normoxic samples were sequenced obtaining 21,064,518 as the average read number. Quality assessment and samples alignment were performed using the miARma-Seq pipeline [31,32]. In detail, fastqc [33] were applied to gather the overall sequence quality and possible adapter accumulation. Then, all samples were aligned to the reference Human GRCH38 genome from the GENCODE portal, version 28. All samples were aligned using Burrows-Wheeler Aligner software [34]. PCR duplicates were removed using samtools markdup with options –s –r [35]. Peak calling was performed using MACS2 [36] with option –f BAM –g hs –s 75 –B –SPMR –nomodel.
All ChIP-Seq raw data are available in the Gene Expression Omnibus (GEO) repository: GEO accession GSE144189.
Immunohistochemistry
Four-micrometer-thick tissue sections from paraffin blocks (melanoma in situ n = 11 and metastasis n = 12) were dewaxed in xylene and rehydrated in a series of graded alcohols. Sections were immersed in 3% H2O2 aqueous solution for 30 min to exhaust endogenous peroxidase activity, then covered with 1% blocking reagent (Roche, Mannheim, Germany) in 0.05% Tween 20-PBS, to block nonspecific binding sites. Antigen retrieval was performed using a pressure cooker and EDTA buffer (pH 8.0). Sections were incubated with primary antibodies during 1 h for anti-PARP1 (#BML-SA250-0050, Enzo Life Sciences, Farmingdale, NY) (1/300) or overnight at 4 °C for anti- HIF-1α (#A300-286A, polyclonal anti–HIF–1α (Bethyl, Montgomery, TX, USA) (1/100). Peroxidase-labelled secondary antibodies and 3,3-diaminobenzidine were applied to develop immunoreactivity, according to manufacturer's protocol (EnVision; Dako, Glostrup, Denmark). Slides were then counterstained with hematoxylin and mounted in DPX (BDH Laboratories, Poole, UK). Sections in which primary antibody was omitted were used as negative controls. Immunostaining was evaluated by an experienced pathologist. The percentage of immunostained tumor cells was scored as follows: 0, negative; <19%, weak: and >20%, positive. Representative images were acquired in a microscope (Olympus BX-61).
Glycolysis assay
The glycolysis assay was performed following the instructions provided for the product “Glycolysis assay” ab197244 (Abcam). This assay measured extracellular acidification due to lactate production during glycolysis. As instructed, cells (8 × 104) were cultured in 96-well plates overnight and then purged from CO2 on a CO2-free incubator at 37 °C for 3 h. Then 150 μl of the Respiration buffer were added in combination with 10 μl of the Glycolysis Assay Reagent, which contained a cell impermeable, pH-sensitive fluorophore. Then cells were exposed to hypoxia and after this step, plates were taken to a TECAN fluorescent spectroscope, where fluorescence (Ex 380 ± 40 nm Em 620 ± 10 nm) was measured every 5 min for 1 h.
O2 consumption assay
The assay was performed following the instructions provided by the supplier for the product “Extracellular Oxygen Consumption Assay” ab197242 (Abcam). This assay gave a real-time kinetic analysis of the oxygen consumption, informing of cellular respiration rate and mitochondrial activity. Cells were cultured at a concentration of 8 × 104 per condition and let to grow overnight. The next day were added 10 μl of the Extracellular oxygen consumption reagent, which contained a fluorescent dye that was quenched in the presence of oxygen. Then cells were exposed to hypoxia. Before oxygen determination, 100 μl of the High sensitivity mineral oil were added on top of the assay medium, limiting oxygen diffusion into the medium. On this context mitochondrial activity reduced oxygen concentration, reducing the dye quenching. O2 consumption was measured by fluorescent microscopy on a TECAN fluorescent spectroscope (Ex 380 ± 20 nm Em 650 ± 20 nm) every 5 min for 1 h.
Results
HIF-1α stability depends on PARP-1 PARylation activity
Previous results from different groups (including ours) [29,30,37] have shown a connection between PARP-1 and HIF-1α activation. However, the mechanism connecting both proteins, the extent to which PARP-1 determines HIF-dependent gene recruitment/transcription, and the relevance in cancer patients, has not been explored yet. In order to assess the clinical relevance of the association between HIF-1α and PARP-1, using the TCGA database we analyzed the correlation in gene expression in metastatic [38], acral [39] and uveal melanoma, where hypoxia plays an important role during tumor development [40]. Using cBioportal [41,42] to query this association we found a statistically significant positive correlation between PARP-1 and HIF-1α gene expression (Fig. 1a). To further support the previous data, we characterized the expression of both proteins in biopsies from in situ melanoma patients using a tissue micro array. PARP-1 and HIF-1α expression was co-incident in consecutive sections and over expressed when compared to the adjacent healthy tissue (Fig. 1b). The reliance of HIF-1α on PARP-1 during early hypoxia was tested in HeLa cells, metastatic (C8161), and uveal (MUM2B) melanoma. PARP inhibition with olaparib led to HIF-1α depletion in all cases (Fig. 1c). To test if this effect was specifically PARP-1-dependent, we confirmed that PARP-1 silencing caused the same depletion on HIF-1α accumulation (Fig. 1d). To discriminate if HIF-1α stability depended on PARP-1 itself or on its PARylation activity, we observed that after PARG silencing HIF-1α was induced in parallel with polymer accumulation (mainly synthesized by PARP-1) (Fig. 1e). These results suggested that HIF-1α stability was affected not only by the presence PARP-1, but also by PARP activity.

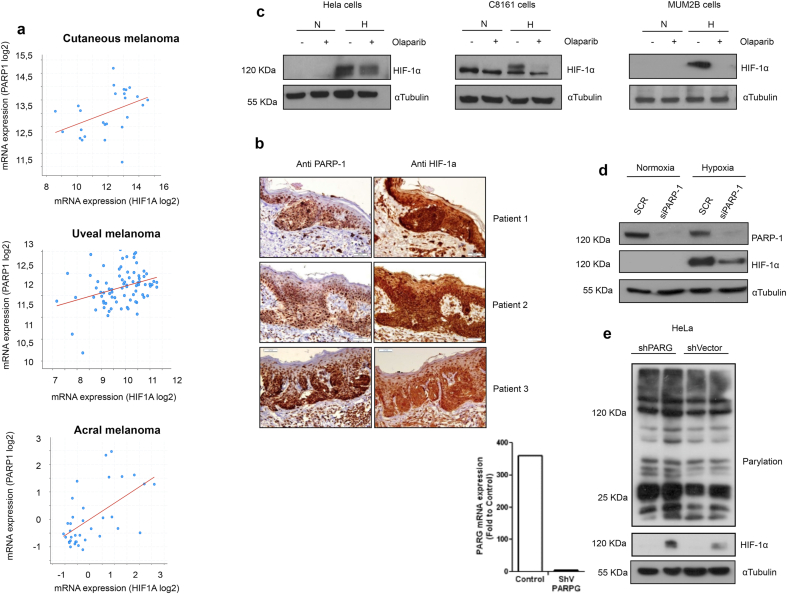
a Correlation of gene In vivo study of PARP1 and HIF-1α expression in melanoma patients using the publicly available database cBioportal; top panel, metastatic melanoma (CM Spearman: 0.50 (p = 8.722e-3)Pearson:0.42 (p = 0.0309)); middle panel, uveal melanoma (UM Spearman: 0.30 (p = 6.295e-3), Pearson: 0.33 (p = 2.655e-3)); lower panel, acral melanoma AM Spearman: 0.55 (p = 4.636e-4)Pearson: 0.59 (p = 1.516e-4)). b Tissue array on 3 melanoma patients, immunochemistry analysis is performed to observe PARP-1 and HIF1α expression. Consecutive sections are shown. c Three different cell lines (MUM2B, C8161 and Hela) show a reduction on HIF-1α after PARP inhibition using Olaparib 5 μM during hypoxia 4 h. d Transient silencing of PARP-1 on HEK 293T cells shows an impairment on HIF-1α accumulation during early (4 h) hypoxia. e In Hela cells polymer accumulation is induced on PARG silenced cell. After 4 h of hypoxia HIF-1α is more stable on the polymer enhanced context.
Hypoxia-induced ROS leads to PARP activation and HIF-1α accumulation
To understand HIF-1α and PARP-1 interaction, we focused in the signals arising during early hypoxia (4 h, 1% O2). This is the case for reactive oxygen species (ROS), that have been described to increase very early after acute hypoxia [43]; ROS are a family of molecules known to trigger the poly (ADP-ribose) activity [44]. After different minutes of hypoxia ROS, were measured using two different probes: DCFA (Fig. 2a) and DHE (Figure S1b). In both cases, ROS peaked during the first hour and their induction was prevented using the general ROS scavenger MPG-2 (N-(2-mercaptopropionyl) glycine) (Fig. 2a and S1a). During early hypoxia PAR levels were rapidly up-regulated in parallel with HIF-1α accumulation. The inhibition of PAR synthesis completely prevented PAR accumulation, consequently reducing HIF-1α stabilization (Fig. 2b and S1c). Cells treated with the ROS scavenger also displayed decreased PAR synthesis and reduced HIF-1α accumulation (Fig. 2c and S1d), unveiling the importance of ROS induction for PARP-1 activation, and subsequently, for HIF-1α stabilization. Using the mitochondrial ROS scavenger MitoTempo we were unable to reduce the ROS induction during hypoxia, implying that maybe other ROS sources (non-mitochondrial) were involved (Figure S1e).

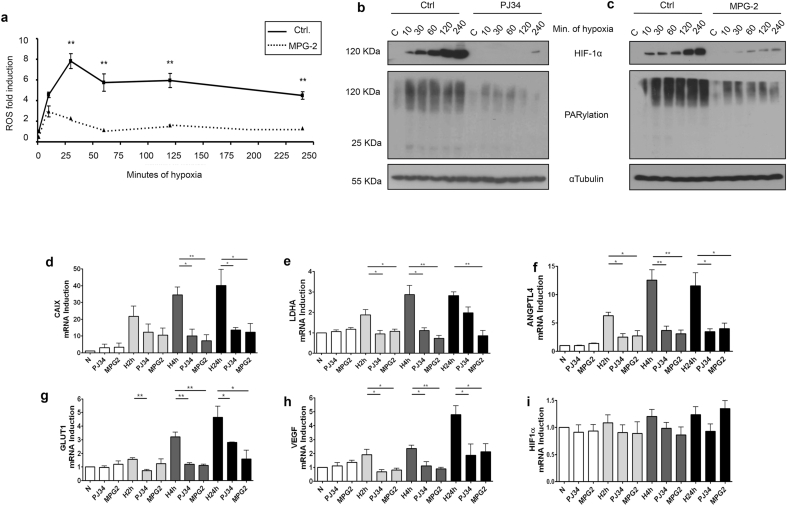
Hypoxia induces ROS and PAR accumulation, both events necessary for HIF-1α stability and activity. a ROS measurement during increasing times of hypoxia (from 10 to 240 min) on HEK 293T cells with and without pretreating with the ROS scavenger MPG2 at 300 μM b,c Western blot showing PARylation levels and HIF-1α accumulation on HEK 293T during the same increasing times of hypoxia, HIF-1α accumulation is impaired when the cells were pretreated with PJ34 at 10 μM or MPG2 300 μM respectively. d-i Gene expression of different hypoxic-related genes. During normoxia and hypoxia (2, 4 and 24 h). Cells were pretreated with PJ34 10 μM and MPG-2 at 300 μM. In both cases HIF-1α transcriptional activity is impaired.
As total HIF-1α levels were decreased after PARP inhibition, we asked if HIF-1α transcriptional activity was also down-regulated. This was indeed the case, as reflected by the expression of five different genes known to be HIF-1α targets: CAIX, ANGPTLA4, GLUT1, VEGF and LDH (Fig. 2d–h). A significant down-regulation was found for all tested genes after treating with either ROS scavenging or PARP inhibition, with exception of LDHA after 24 h of hypoxia following PJ34 treatment. Non-significant alterations on HIF-1α mRNA levels were described (Fig. 2i). These data strengthen the idea of a tight link between hypoxia, ROS production, PARylation induction, and HIF-1α stabilization and activation.
PARP-1 interacts with the C-terminus domain of HIF-1α regulating the stability of the protein
It has been previously shown that PARP-1 and HIF-1α form a complex after ciclopirox olamine treatment (hypoxia mimetic) and in response to Epstein Barr virus (EBV) infection [28,29,37]. However, this interaction and the domains mediating its formation have not been addressed in the hypoxic context, where HIF-1α performs its major activity. To further dissect the influence of PARP-1 in HIF−1α stability during normoxia and hypoxia, we generated a double mutant HIF-1α for the PHD sites P402 and P564. These prolines were changed into alanines using site-directed mutagenesis on two constructs spanning the HIF-1α full length (DML: 1–826), and a truncated HIF-1α lacking the C-terminus domain (DMS: 1–657). In addition, we generated a C-terminus fragment of HIF-1α (C-ter: 630–826) that lacks sites for PHD regulation (Fig. 3a). pVHL overexpression affected the stability of the wild type full length and wild type short HIF-1α, but not to the C-ter domain (Fig. 3b, c and d). As expected, when the PHD sites were mutated, the DML and DMS became insensitive to pVHL over-expression. Therefore, both constructs were stable during normoxia (Fig. 3e and f). We then exposed cells transfected with the different constructs to PARP inhibition, this prevented the accumulation of the DML and C-ter but did not affect the stability of the DMS construct (Fig. 3g). These results showed that the C-terminus (present in both the C-ter and the DML domain) was responsible for this regulation. Moreover, GST-pull down assay using different PARP-1 fragments (Fig. 3h) demonstrated that the C-ter of HIF-1α interacted with the PARP-1 domain D, which is known to be responsible for protein-protein interactions (Fig. 3i).

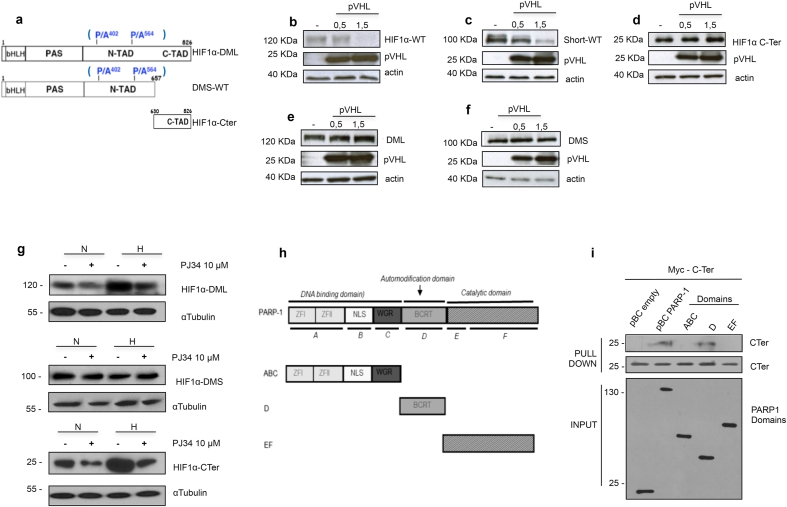
HIF-1α interacts with PARP-1 at its C-terminus domain regulating its stability. a HIF-1α constructs presenting the wild type sequences for the HIF-1α wt, short WT and C-ter. The PHD residues mutated are presented in blue, obtaining the DML and DMS. b-f On HEK 293T western blots showing the downregulation during PHD overexpression of the HIF-1α and short wt. This reduction is impaired on the PHD insensitive mutants. g Western blot study on HEK 293T of the three PHD-insensitive domains of HIF-1α, their stability is compared during normoxia and hypoxia 4 h with PARP activated or inhibited using PJ34 10 μM h PARP-1 domains presenting the full protein, and separately, the DNA-binding, the automodification and the catalytic domain. i pull-Down assay exposing the different domains of PARP-1 to the C-ter domain of HIF-1α. The C-ter binds to PARP-1 full protein and the auto modification domain.
HIF-1α is modified by poly(ADP-ribose) at its C-terminus
Having demonstrated that HIF-1α and PARP-1 do interact, we asked if HIF-1α is a substrate for PARP-1-mediated PARylation during hypoxia. Using a co-IP approach with anti HIF-1α as bate, we found that HIF-1α is PARylated in normoxia and that this modification increased during hypoxia (Fig. 4a). Co-IP with anti-PARP-1 and HIF-1α confirmed the presence of both proteins forming a complex that increased during hypoxia and that was destabilized in the presence of PARP inhibitors (Fig. 4b). To further deepen in the characteristics of this interaction, co-IP assays were performed against poly(ADP-ribose) in cells overexpressing the HIF-1α domains C-ter and DMS. PARylation was observed in the endogenous HIF-1α protein and the C-terminus domain, and it was reduced after PARP inhibition. No modification was observed on the DMS domain (Fig. 4c).

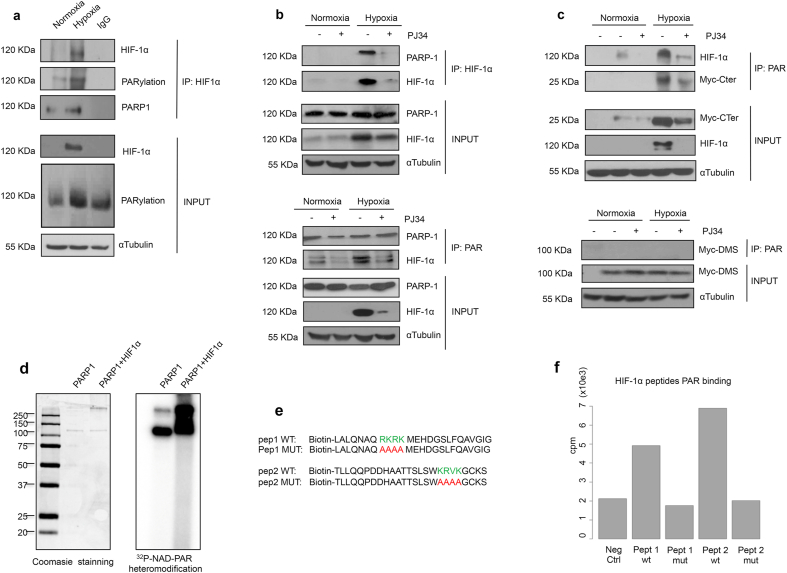
PARP-1 physically interacts and PARylates HIF-1α at its C-Ter domain. a Immunoprecipitation of HIF-1α on HEK 293T cells undergoing normoxia and hypoxia 4 h. A complex is form leading to PARylation. b Immunoprecipitation of HIF-1α and PARP-1 on HEK 293T cells exposed to normoxia or hypoxia 4 h. During PARP-1 inhibition using PJ34 10 μM the complex is destabilized. c Immunoprecipitation of PAR polymer on HEK 293T cells transfected with the C-ter or DMS domain of HIF-1α during normoxia and early hypoxia 4 h. During PARP inhibition with PJ34 10 μM the complex observed between PARP-1 and the endogenous HIF-1α or the C-ter is lost. d In vitro PARylation assay for PARP-1 and HIF-1α. Coomasie staining and 32P-NAD-PAR autoradiography is presented. e Synthesis of four peptides located on the HIF-1α C-ter. Two WT and their correspondent non PARylable analogs are presented. fin vitro PARylation assay performed in the four peptides. The results are presented in counts per minute.
To identify the specific amino acids undergoing this modification, we interrogated the C-ter sequence for putative PARylable sites. It has been described that amino acid sequences presenting repeated lysine and arginine motifs can be acceptors of poly(ADP-ribose) [45]. We identified two sequences with similar characteristics on the C-ter domain. Site directed mutagenesis was performed to change these lysins and arginines residues into non-PARylable alanines (Figure S2a). Cells were then transfected with these HIF-1α mutants. Unexpectedly, the mutations directed HIF-1α to the cytosol (Figure S2b). Further research revealed that those potential PARylable sites were located in a secondary bipartite NLS described for HIF-1α [46]. We decided then to perform an in vitro approach where we observed how purified HIF-1α was PARylated by PARP-1 (Fig. 4d). To study the C-ter domain, four different biotin-modified peptides containing the previously described sequences, were synthesized on their wild type and mutant form (Fig. 4e). Again, an in vitro PARylation assay was performed, showing that both wild type peptides were modified with PAR; after the alanine mutation, this PTM was reduced to the same level than the negative control (Fig. 4f).
PARP-1 conditioned HIF-1α recruitment to target promoters during hypoxia
Once studied the nature of the interactions between PARP-1 and HIF-1α on the hypoxic context, we evaluated the impact that PARP-1 had on the recruitment of HIF-1α to the chromatin. Firstly, we generated a cell line of HEK 293T PARP-1 knockout cells using CRISPR/Cas9. We confirmed that PARP-1 was correctly removed and tested the capacity of the cells to produce poly(ADP-ribose) during peroxide-induced stress (Figure S2c). We also determined that, similarly to PARPi, the absence of PARP-1 diminished the amount of HIF-1α accumulated during hypoxia (Figure S2d).
Then, a ChIP-Seq analysis was performed during early hypoxia (4 hours) on the HEK 293T WT and PARP-1 KO cells. The ChIP-Seq revealed that most of the HIF-1α binding peaks detected were located within the first kilobase upstream of translation start sites: 48,16% in wild type and 43,9% in PARP-1 KO (Fig. 5, Fig. 6) which includes the 5′-UTR and the promoter region of the genes. Statistical analyses were completed to eliminate non-significant peaks and to compare them with the input and the normoxic control signals. 123 peaks were detected on the hypoxic wild type cells and 68 in the PARP-1 KO cells. Comparing with a previous study by Schödel et al. [47] 400 high stringency HIF-1-binding sites were found. The differences with our study could be ascribed to the different cell type used in their study (MCF7), and the fact that they were analysing the hypoxic response after 16 h at 0,5% O2. In our case we focused on the early response (4 h) using a less limiting oxygen concentration (1% O2).

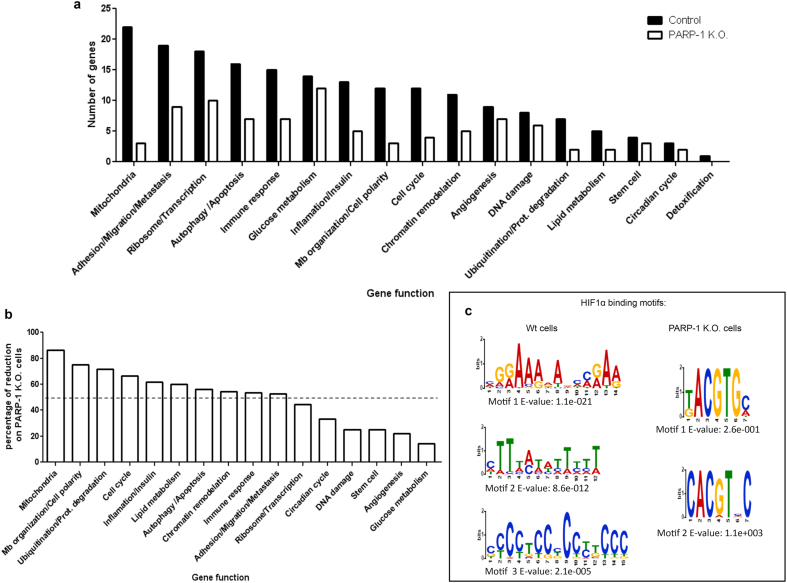
Chip-seq analysis of HIF-1α capacity to bind the promoters of its target genes on WT vs. PARP-1 KO cells. a ChIP-Seq performed for HIF-1α after 4 h of hypoxia on HEK 293T and PARP-1 K.O cells. The HIF-1α binding sites are presented considering the function of the closest gene. b Representation of the percentage of loss on the different functions on the PARP-1 KO cells. c MEME analysis showing the most common sequences where HIF-1α binds and their E-value.

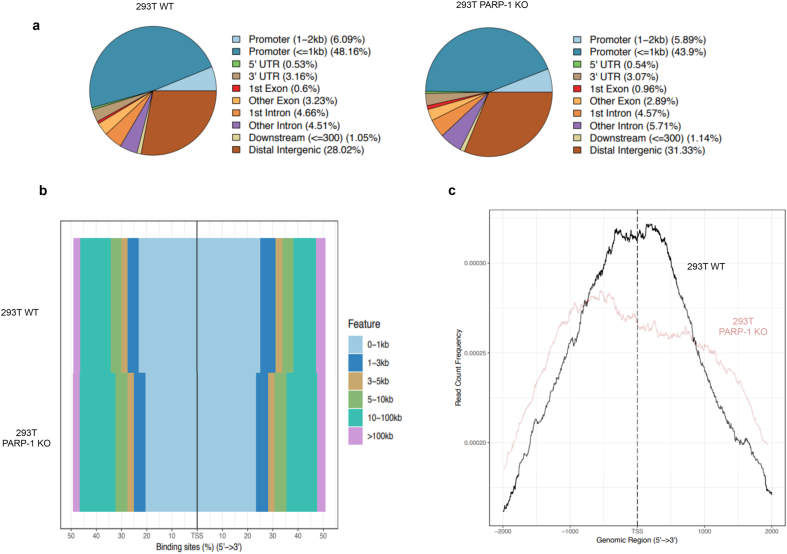
a. On 293T WT and PARP-1 KO, pie charts presenting the porcentaje of HIF-1α binding sites detected on different areas of the gene structure. b Percentaje of HIF-1α accumulation on the genome regarding the distance (measured as kilobases) from the TSS. c Study of the promoter area (showing 2 kb around the TSS), presenting the frequency of HIF-1α detection on 293T WT and PARP-1 KO cells during hypoxia (1% O2, 4 h).
During hypoxia, PARP-1 KO cells presented 44,72% less HIF-1α binding sites and not a single new peak gained with respect to the wild type cells. After grouping these genes according to their functions (Fig. 5a), we found that the effect of PARP-1 on HIF-1α association to the chromatin was not randomly distributed, but there was a range of hierarchical reduction depending on the gene ontology. While some cellular activities remained almost unchanged (this was the case of glucose metabolism, angiogenesis, or DNA repair), other functions almost disappeared (mitochondrial activity, membrane organization and cell cycle regulation) (Fig. 5b). We then compared the common peaks found among the top 30 binding sites reported in pervious ChIP-seq and our results [47] and we found that 7 out of 10 common genes (ALDOA, ANKRD37, EGLN3, ENO1, GPI, PDK1, PFKFB3, PFKFB4, PKM, RSBN1) are involved in glycolysis, confirming that the regulation of glucose metabolism it is likely to be a key parameter to be regulated during cell's adaptation to hypoxia.
Intrigued by this differential loss we completed a MEME analysis [48] on the sequences to which HIF-1α bound during hypoxia in the wild type and PARP-1 KO context (Fig. 5c). Interestingly, HIF-1α presented high flexibility binding to different sequences during the normal hypoxic induction, being HIF-1α peaks located on long and variable sequences. In contrast, in PARP-1 KO cells HIF-1α capacity to bind to the DNA became more rigid and limited, locating in shorter and more HRE-similar sequences.
In order to understand the reduced binding of HIF-1α to its target genes in the absence of PARP-1, we performed a more extensive analysis of the ChIP-Seq to observe in detail this new binding pattern. When studying the recruitment of HIF-1α considering the gene arquitecture we observed how HIF-1α located primarily in the promoter regions of its target genes (Fig. 6a). Being this area the one that presented the stronger reduction in HIF-1α association in PARP-1 KO cells.
We analyzed the data considering the distance between HIF-1α binding sites and the closest TSS (measured in kilobases, or Kb) (Fig. 6b). We observed how the most frequent accumulation of HIF-1α took place within the first 1 Kb around the TSS. To study in detail these promoter regions, we focused on the frequency of HIF-1α detection only 2 Kb around the TSS (Fig. 6c). HIF-1α presented a predominant concentration on the first 1 Kb circling the TSS. In PARP-1 KO cells however, HIF-1α was reduced mainly on that region, where as a transcription factor its expected to perform its main activity.
Beyond HIF-1α recruitment, it was intriguing the effect that PARP-1 ablation had over hypoxic gene expression when measured via mRNA levels. Not only those genes that presented less HIF-1α recruitment on their promoters showed reduced mRNA levels, but even those genes that presented similar or even stronger HIF-1α accumulation on their promoters, displayed a reduced mRNA expression (Figure S3) showing that HIF-1α recruitment per se was not enough to guarantee the expression of its target genes during PARP-1 ablation. The possible mechanisms underlaying this regulation will be addressed at the discussion section.
PARP-1 inhibition/ablation impacts cellular fitness during hypoxia
Once studied in depth the role of PARP-1 over different aspects of HIF-1α biology, and how it altered the expression of genes involved in hypoxic adaptation. We evaluated how PARP-1 inhibition or ablation could be affecting cellular fitness during hypoxia.
We evaluated the status of stress routes like the DNA damage response (DDR) and the replication stress, as well as the cells capacity to consume O2 and the glycolysis status. To finish we observed the impact of these metabolic alterations on cellular proliferation and migration capacity.
The induction of the DDR in hypoxia has been previously reported in conditions close to anoxia (<0.1% O2) and it has been shown to include replication stress via p-RPA accumulation [[49], [50], [51]]. We measured DNA damage using the accumulation of 53BP1 foci in immunofluorescence and analyzed other DDR and replication stress markers via western blot. Interestingly, hypoxia treatment alone did not induce the activation of the DDR. However, an increase on 53BP1 foci was observed after olaparib treatment (Figure S4a and b) which was reported previously in different settings [52,53]. On the same direction, western blot analysis of different proteins involved in the DDR showed no clear difference in DDR or replicative stress induction (Fig S4c). Indicating that the ROS induced during hypoxia were activating PARP-1 through a mechanism independent of DNA damage.
Then we evaluated the status of the cells regarding oxygen consumption and glycolysis, key processes during hypoxic adaptation. The study of oxygen consumption (Fig. 7a) revealed that during normoxia both HEK 293T WT and PARP-1 KO cells had elevated and similar oxygen consumption levels. However, during hypoxia the HEK 293T PARP-1 KO cells presented only a partial reduction in oxygen consumption compared with the observed on the HEK 293T WT cells. Given the crucial significance of the inhibition of the oxidative phosphorylation during hypoxia, this difference in the rate of oxygen consumption could result in increased oxidative stress in the absence of PARP-1.

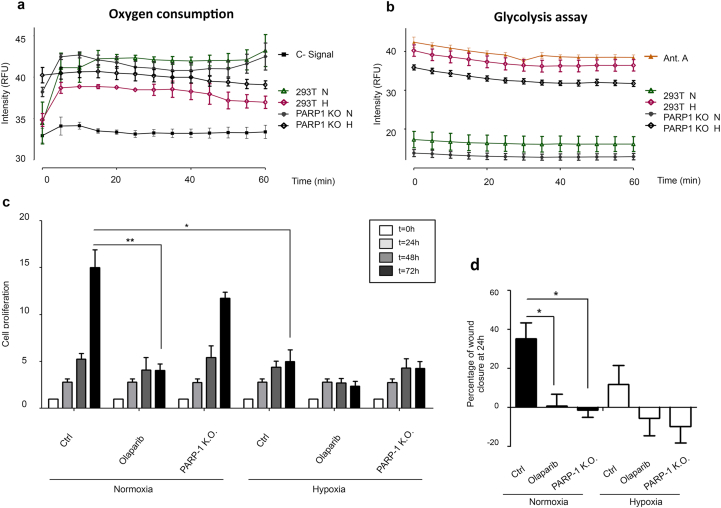
Impact of PARP-1 inhibition/knockout on cellular metabolism and fitness. a Oxygen consumption analysis. HEK 293T WT cells and HEK 293T PARP-1 KO were exposed to normoxia or 4 h of hypoxia. Then fluorescent emission was measured and compared during 60 min via fluorescence spectroscopy. b Glycolysis assay measured as cytoplasmic acidification. HEK 293T WT cells and HEK 293T PARP-1 KO were exposed to 4 h of normoxia and hypoxia, following glycolysis measurement for 60 min via fluorescent emission. Antimycin A was used as a positive control. c Cell growth during different times of normoxia and hypoxia. HEK 293T cells undergoing PARP inhibition (olaparib 5 μM) or PARP-1 KO cells. 24 h after culture, cells were treated and exposed to 1% hypoxia during crescent times. d Wound healing assay performed on HEK 293T cells, being PARP inhibited (olaparib 5 μM) or PARP-1 KO, wound healing measurement was performed 24 h after wound opening.
The study of the glycolysis pathway on both cell lines showed low levels of glucose consumption in normoxia. However, during hypoxia a reduced glycolysis induction was observed in the PARP-1 KO cells compared to the WT cells (Fig. 7b). This result could indicate the lack of a proper switch from the oxidative to the glycolytic pathway. However, this observation is hard to conclude because glycolysis was already lower on PARP-1 KO cells during normoxia.
We determined the impact of the alterations observed in these bioenergetic/metabolic pathways in the cellular fitness. For this purpose, we performed cell proliferation and wound healing assays (as a readout of cell migration). During normoxia, PARP inhibition had a stronger effect reducing cell proliferation respect to the effect of PARP-1 absence, probably because the inhibitor affected other PARP members. However, during hypoxia, the elimination of PARP-1 became as effective as global PARP inhibition reducing proliferation, showing the important role of PARP-1 during the hypoxic adaptation (Fig. 7c). In the same way, cell migration measured via wound healing assay manifested how the cellular mobility was reduced during normoxia after both PARP-1 inhibition/knockout. During hypoxia however, these effects became more drastic leading to a widening of the wound (Fig. 7d). All these results reinforced the idea of a cellular dependence on PARP-1 activity for the optimal adaptation and survival to hypoxia.
Discussion
Our data demonstrate that during early hypoxia ROS production and PARP-1 activation are concomitantly produced and are needed to fine-tune signaling through HIF-1α. PARP-1 activation leads to HIF-1α PARylation at its C-ter, being this PTM necessary for its optimal stability and activity. In the absence of PARP-1, HIF-1α downregulation reduces the binding of HIF-1α to its target genes, being preserved only those with a classic HRE sequence. This causes a reduction of gene expression, impairing hypoxic adaptation and having a negative impact on the cellular fitness (Fig. 8).

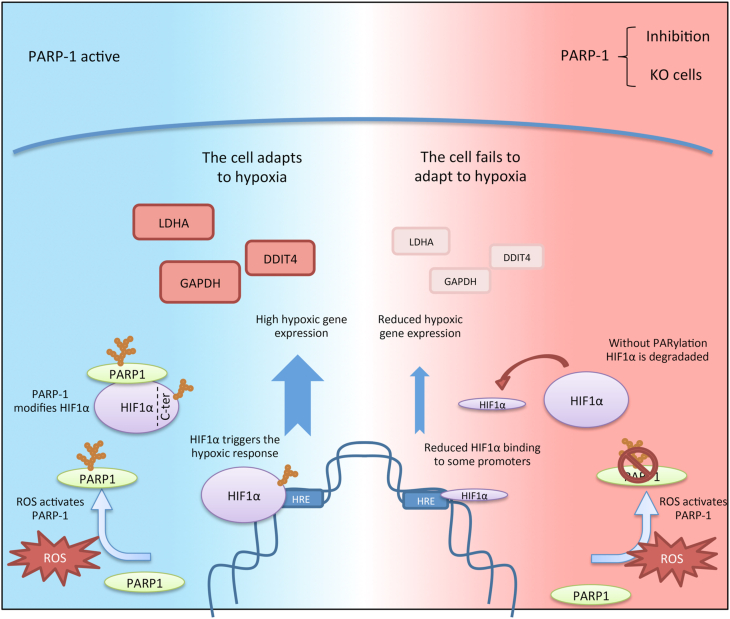
During hypoxia, ROS production leads to PARP-1 activation. This protein interacts with HIF-1α at its C-terminus domain, causing its PTM with poly(ADP-ribose). This modification leads to HIF-1α accumulation. HIF-1α then binds to the promoters of its target genes, causing the expression of hypoxic genes and allowing the cell to adapt to hypoxia. On the other hand, when PARP-1 is inhibited or knocked out, it causes HIF-1α downregulation. This reduces HIF-1α binding to its promoters causing a reduced hypoxic gene expression. These changes lead to a poorer adaptation to hypoxia.
Reactive oxygen species (ROS) such as superoxide anions, hydrogen peroxide and free radicals are a family of molecules that have been shown to be pivotal modulators of the hypoxic response. Owing to the reduced activity of the electron transport chain (ETC) in hypoxia, ROS production escalates [54]. Mitochondrial ROS have been described not just as a by-product of the hypoxic response but as a key regulator of this biological response. ROS production threatens cell integrity and survival. Consequently, during hypoxia mitochondrial protective inhibition is induced, causing a decrease in ATP levels which are compensated by the shift to glycolytic metabolism via the Warburg effect [55,56]. In addition to the impact over the global metabolism, ROS have been directly implicated in HIF-1α stability and activity not only during hypoxia [57] but also during normoxia [58,59]. However, there is controversy regarding how ROS stabilize HIF-1α and alternative mechanisms have been proposed including ROS inhibition of PHD activity [60,61] or through H2O2 [62]. Our work identified a novel mechanism by which hypoxia-induced ROS activate PARP-1, leading to HIF−1α PARylation at its C-terminus domain; PARP-1 activity is required for HIF−1α accumulation and binding to a diversity of promoters, allowing the expression of genes required for the cell adaptation to hypoxia.
Hypoxia-induced DDR is distinct from classical pathways induced by damaging agents due to the repression of DNA repair elements in hypoxic conditions [63,64]. It has been shown that hypoxic conditions induce a rapid compaction of the chromatin, which is associated with a general inhibition of transcription levels [65]. Previous studies have shown that tumor cells display defective DNA repair pathways, increasing mutation rates, and chromosomal instability [50,66]. The link between hypoxia and the DDR is not fully understood and seems to be oxygen dependent: severe hypoxia (<0.2% O2) has been described to cause S-phase arrest, accumulation of γ-H2AX, P53 and ATR activation. However, moderate hypoxia (1–3% O2) can lead to replication stress and activation of the DDR pathway even when no DNA damage is detected [67]. In this context hypoxia‐induced ROS could lead to the activation of PARP-1 on a DNA damage-independent manner. PARP-1 is known to undergo different PTM modifications [68] being some of them known to alter its activity in contexts independent of DNA damage accumulation [[69], [70], [71]] we consider this to be a plausible hypothesis that deserves future exploration.
To decipher the results observed on the ChIP-seq assay, we consider essential to understand the chromatin regulation processes induced in the conditions of our study: On the one hand, the changes produced during the hypoxic induction, on the other hand, those observed during PARP-1 ablation. Finding-out the processes regulating the DNA architecture in both scenarios, will help us to understand the results obtained on the ChIP-seq when we combine both conditions.
As we have point out, the hypoxic situation is characterized by a decrease on the global transcriptional activity [72]. This seeks a reduction on ATP consumption and promotes chromatin protection. However, during this repressive scenario, the response to hypoxia must be triggered to guarantee cellular adaptation and survival. How this hypoxic response is triggered on a transcriptionally repressive context, requires the combination of different mechanisms that go far beyond the simple stabilization of the HIF family.
Respect to the role that PARP-1 can play in this context, we need to point out the increasing number of studies that implicate PARP-1 in other cellular activities beyond DNA repair. It is especially remarkable the implication of PARP-1 as a facilitator of transcription, locating around the TSS of active genes [73], being a mediator of heterochromatin to euchromatin transition [74], participating on gene expression by inducing promoter activation [75,76], chromatin remodelling and relaxation [77] and RNA metabolism [78].
Bearing in mind the roles describing PARP-1 as a transcriptional facilitator, we now analyse the impact of PARP-1 absence during hypoxia. On a chromatin repressive context, as it is the case during hypoxia, PARP inhibition/absence is expected to represent a “double trouble” for the hypoxic response. On the one hand, it reduces HIF-1α stability limiting the number of molecules available to perform their transcriptional activity. On the other hand, PARP-1 absence is expected to promote an even more repressive state of the chromatin, increasing heterochromatin condensation. As described above, PARP-1 induces promoter activation by binding mainly 1 Kb around the TSS. Therefore, it is not surprising that during hypoxia, the absence of PARP-1 causes the loss of HIF-1α location mainly 1 Kb around its target TSS and not on distant regions. Furthermore, in the absence of PARP-1, the MEME study has shown that HIF-1α capacity to bind to the DNA is reduced to its most appealing sequences (HREs). Concerning the MEME analysis, it is important to note that although the motifs on the wild type cells give a significant E-value (E-value ≤0.01) it is not the same situation for PARP-1 knockout cells. This situation is not uncommon and MEME E-values are known to be overly conservative, especially when the input dataset is reduced (which is the case for the PARP-1 KO cells due to HIF-1α down-regulation). Different studies have indicated that this situation can lead to the rejection of real functional motifs that are biologically significant [79,80]. We consider that to be our case. It is difficult to ignore that the sequences obtained on the PARP-1 KO cells are the best known and best described HIF-1α binding sites.
One important question to address is how, even when HIF-1α locates to its promoters on the PARP-1 KO cells (according to the ChIP-seq analysis), gene expression is still enabled and mRNA is not accumulated. This shows that HIF-1α binding to its target genes is not enough on its own to induce gene expression. This is not surprising knowing that the absence of PARP-1 impacts chromatin relaxation at the TSS, co-activators recruitment, or even limit RNA maturation. All these events prove that PARP-1 role is pivotal for the hypoxic response, altering HIF-1α stability, inducing repressive configurations on the DNA, and affecting the transcription process.
Clinically, activation of hypoxia is a negative prognostic factor which influences multiple aspects of the tumor biology. It is central in the control of tumor progression and one of the most well validated targets yet to be controlled in oncology. During hypoxia there is an induction of angiogenesis, invasiveness and metastasis associated to a reduced patient response to chemo and radiotherapy. In the same direction, overactive DDR rescues cells from treatment-induced death. In the present work we connect both pathways through HIF-1α C-terminal PARylation via PARP-1. On this context, PARP inhibitors could be doubly beneficial, reducing cell capacity to repair DNA damage whilst disabling the hypoxic adaptation. As PARP inhibitors have already been approved for their use in the clinic, the identification of tumors expressing elevated HIF-1α, could expand the use of PARP inhibitors to target hypoxic tumors.
Conclusions section
Hypoxia plays a central role as driving force of tumor development and the search of strategies to avoid tumor adaptation to hypoxia represent a major translational challenge. In the present study we show that PARP1 and HIF-1α are over-expressed and co-expressed in human melanoma samples. We have analyzed the molecular insights on this interaction with the following main conclusions (see also graphic abstract in Fig. 8):
1.During hypoxia, there is an early ROS induction that leads to PARP-1 activation. This activation is necessary for HIF-1α stability and activity. During hypoxia PARP-1 and HIF-1α physically interact, and HIF-1α is PARylated at specific residues of two regions of its C-ter.
2.ChIP-seq reveals that in the absence of PARP1, HIF1α binding to its target genes is reduced to 44,72% in a gene function-dependent manner: while some functions are preserved (like glucose metabolism), others are severely reduced (like mitochondrial regulation and cell polarity).
3.ChIP-seq analysis shows that PARP-1 determines HIF1α location close to the promoters (1 kb from Transcription Star Site, TSS) but not in distant regions of TSS. Moreover, in the absence of PARP-1, recognition of the hypoxia responsive elements (HRE) by HIF1α is restricted to the consensus sequence.
4.HIF1-dependent gene expression in the absence of PARP-1 is impaired in all cases irrespective whether HIF-1α recruitment to chromatin is lost, or maintained, indicating that PARP-1 plays a role in HIF-1-dependent transcriptional regulation beyond HIF-1α recruitment.
5.As a consequence of these previous results, we observed that PARP-1 loss or inhibition has an effect in reducing cellular fitness, being this effect especially severe during hypoxia.
6.Globally, as PARP inhibitors have already been approved for their use in the clinic, the identification of tumors expressing elevated HIF1α might expand the use PARP inhibitors to target the tumor adaptation to hypoxia.
Ethics approval and consent to participate
These studies were approved and performed in strict accordance with the recommendations of Ethic and Research Committee of Virgen del Rocio University Hospital, Seville.
Consent for publication
Not applicable.
Availability of supporting data
The datasets for ChIP-seq analysis presented in the current study and otherdatasets analyzed are available from thecorresponding author on reasonable request. ChIP-seq sequences have been uploaded in GEO database GSE144189.
Funding
This work was supported by Junta de Andalucía, project of Excellence from Junta de Andalucía P10-CTS-0662, P12-CTS-383 to FJO,
Authors'contributions
JMM, performed in vitro experiments, analyzed data, analyzed ChIP-seq data, performed bioinformatic interpretation. and wrote manuscript. AGD, DDB, OC, JMRV, JCA, AGF, KD performed experiments and analyzed data. EdA, FOV and ATA performed and analyzed immunohistochemistry. FD, GLK, EB, EH and FJO were responsible for the coordination of the project and contributed to study design. All authors read and approved the final manuscript.
Disclaimers
The authors declare no conflicts of interest.
Declaration of competing interest
The authors declare that they have no competing interests.
References
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
Abreviations
| 53BP1 | p53 binding protein 1 |
| ANGPT4 | Angiopoietin Like 4 |
| ATP | Adenosine triphosphate |
| ATR | Ataxia telangiectasia and Rad3-related protein |
| BRCA1&2 | BReast CAncer gene 1 & 2 |
| CAIX | Carbonic Anhydrase 9 |
| Ch-IP Seq | Chromatin immunoprecipitation |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| CSC | Cancer stem cells |
| C-TER | C-terminus domain |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DCFDA | 2ʹ,7ʹ-Dichlorofluorescin Diacetate |
| DDIT4 | DNA Damage Inducible Transcript 4 |
| DDR | DNA damage response |
| DHE | Dihydroethidium |
| DML | HIF-1a long domain |
| DMS | HIF-1a short domain |
| EBV | Epstein-Barr Virus |
| FIH | Factor inhibiting HIF |
| GAPDH | Gliceraldehído-3-fosfato deshidrogenasa |
| GFP | Green fluorescent protein |
| GLUT | Glucose Transporter protein |
| GST | Glutathione S-transferase |
| Gy | Gray unit |
| H2AX | H2A.X Variant Histone |
| H3K27me3 | Tri-methylation at the 27th lysine residue to the Histone H3 |
| H3K4me3 | Tri-methylation at the 4th lysine residue to the Histone H3 |
| H3K9me3 | Tri-methylation at the 9th lysine residue to the Histone H3 |
| HIF | Hypoxia-inducible factor |
| HRE | Hypoxic response element |
| IP | Immunoprecipitation |
| JMJ demethilase | Jumonji demethilase |
| Kb | Kilobase |
| Kda | Kilodalton |
| KDM5B | Lysine Demethylase 5B |
| KLF4 | Kruppel Like Factor 4 |
| LDHA | Lactate dehydrogenase A |
| MEME | Multiple em for motif elicitation |
| MPG-2 | N-(2-mercaptopropionil |
| NAD+ | Nicotinamide adenine dinucleotide |
| NFKB | Nuclear factor kappa B |
| NLS | Nuclear location sequence |
| P300 | Histone acetyltransferase protein 300 |
| P53 | Tumor protein p53 |
| PAR | poly ADP-ribose |
| PARG | poli(ADP-ribosa) glycohydrolase |
| PARilation | poly (ADP-ribosilation) |
| PARP | Poly (ADP-ribose) polimerase |
| PCAF | p300/CREB-binding protein-associated factor |
| PCR | Polymerase chain reaction |
| PFK | Phosphofructokinase |
| PHD | Prolyl hydroxylase domain protein |
| PI3K | Phosphoinositide 3-kinase |
| PIC | Preinitiation complex |
| POL2 | RNA polymerase II |
| PolyA | Poliadenilation |
| PTM | Posttranslational modification |
| PVHL | Von Hippel-Lindau protein |
| RNS | Reactive nitrogen species |
| ROS | Reactive oxigen species |
| RPA | Replication protein A |
| TCGA | The Cancer Genome Atlas |
| TET1 | Ten-eleven translocation methylcytosine dioxygenase 1 |
| TIP60 | Tat-interactive protein 60 |
| TNFa | Tumor necrosis factor alpha |
| TSS | Transcription start site |
| TWIST | Twist family bHLH transcription factor 1 |
| UBE2M | Ubiquitin Conjugating Enzyme E2 M |
| VEGF | Vascular endothelial growth factor |
Acknowledgements
We would like to acknowledge Laura López for technical assistance; Eduardo Andrés and Laura Terrón (Bioinformatic core IPBLN, CSIC) and Pan Hui (Bioinformatic Core, Joslin Diabetes center, Harvard Medical School).
 Selective modulation by PARP-1 of HIF-1α-recruitment to chromatin during hypoxia is required for tumor adaptation to hypoxic conditions
Selective modulation by PARP-1 of HIF-1α-recruitment to chromatin during hypoxia is required for tumor adaptation to hypoxic conditions