
- Altmetric
The strain G184AR is incorrectly reported as G186AR throughout the paper. The authors have provided corrected versions of Table 1, Figs 1 and 2, and Supporting Information files S1, S3–S6, S9, and S10 Figs that use the correct strain name G184AR.

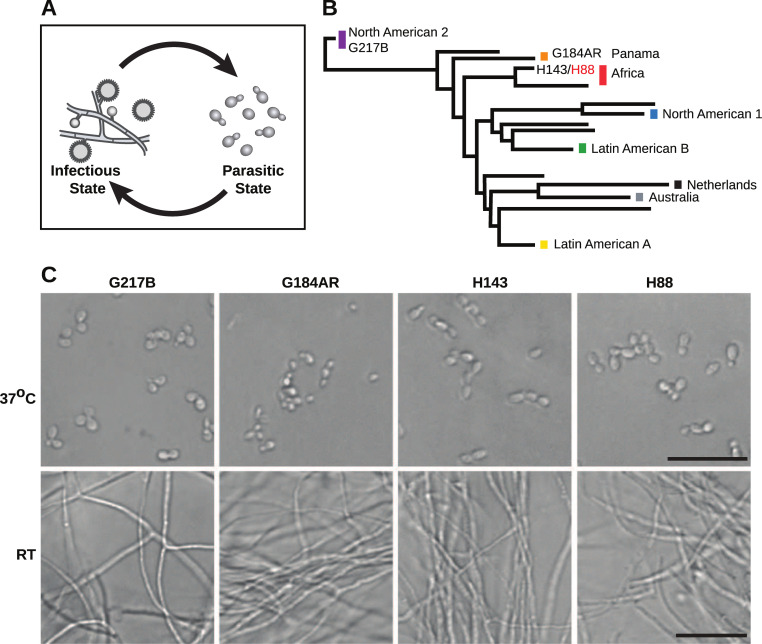
Histoplasma is a global human pathogen with distinct morphological states important for virulence.
(A) Histoplasma hyphae and conidia (infectious state) respond to temperature to differentiate into budding yeast cells (parasitic state) inside of a mammalian host. (B) Phylogeny of Histoplasma illustrating the major lineages of this species, which are geographically and genetically distinct. This neighbor-joining tree was adapted from Katsuga et al. [7] and highlights Histoplasma strains selected for comparative transcriptomics (G217B, G184AR, H88, H143). H88 is highlighted in red as the Hc var. duboisii strain selected for transcriptome analysis. (C) Confocal DIC microscopy showing representative images of yeast (37°C) and hyphal (RT) cell morphologies for the 4 Hc strains used for comparative transcriptomics analyses. Scale bar, 20 μm.

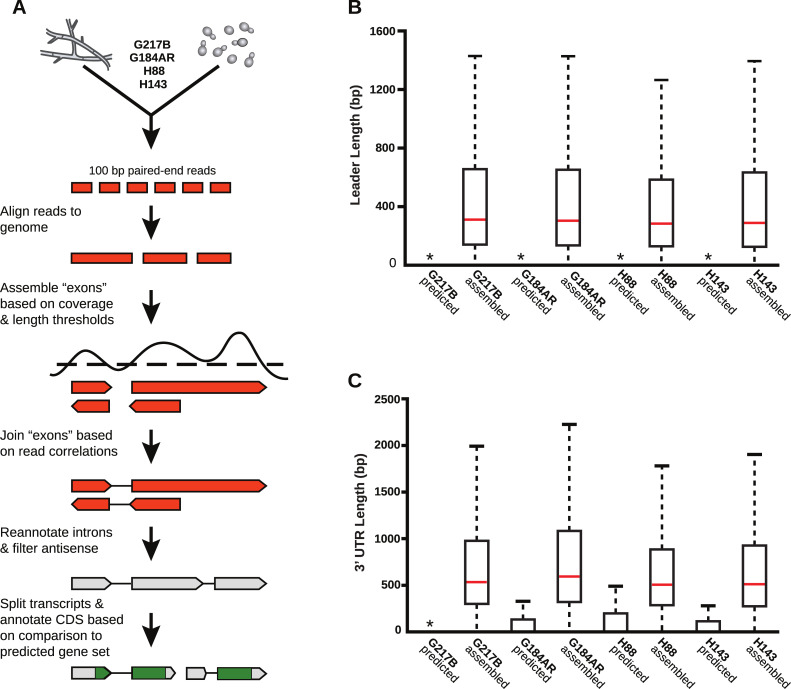
De novo Histoplasma transcriptome reconstruction augments transcript models.
(A) Schematic of transcriptome reconstruction method. (B–C) Boxplots of the length of (B) leader regions that were defined as the distance from the 5’ transcript end to the CDS start codon or (C) 3’ UTR regions that were defined as the distance from the CDS stop codon to the 3’ transcript end were plotted for all assembled and predicted transcripts with CDS regions > 0. Boxes, interquartile range (IQR). Whiskers, 1.5*IQR. For ease of viewing, outliers are not displayed. *, indicates that the predicted transcript leader or 3’ UTR length distributions is not visible on this graph. The predicted transcript leader lengths range from 0–1652 bp and the 3’ UTR lengths range from 0–3112 bp in the 4 Hc strains.

| G217B | G184AR | H88 | H143 | |
|---|---|---|---|---|
| Predicted Transcripts | 11, 330 | 9, 233 | 9, 428 | 9, 532 |
| Predicted Protein Coding Transcripts | 11, 329 | 9, 229 | 9, 424 | 9, 483 |
| Predicted InParanoid Ortholog Pairs (G217B) | 7, 104 | 7, 122 | 6, 831 | |
| Predicted Mercator Orthogroups (per strain) | 7, 485 | 7, 960 | 8, 610 | 8, 325 |
| Assembled Transcripts | 12, 313 | 12, 663 | 12, 175 | 12, 889 |
| Assembled Protein Coding Transcripts | 9, 580 | 9, 844 | 9, 647 | 9, 723 |
| Assembled InParanoid Ortholog Pairs (G217B) | 6, 708 | 6, 670 | 6, 079 | |
| Assembled Mercator Orthogroups (per strain) | 7, 362 | 7, 659 | 8, 288 | 7, 819 |
| 1:1:1:1 Mercator Orthogroups | 6, 791 | 6, 791 | 6, 791 | 6, 791 |
Reference
1
 Correction: Genome-Wide Reprogramming of Transcript Architecture by Temperature Specifies the Developmental States of the Human Pathogen Histoplasma
Correction: Genome-Wide Reprogramming of Transcript Architecture by Temperature Specifies the Developmental States of the Human Pathogen Histoplasma