Proceedings of the National Academy of Sciences of the United States of America

Contributed by Wing Hung Wong, March 1, 2021 (sent for review January 7, 2021; reviewed by Faming Liang and Weijie Su)
Author contributions: Q.L., R.J., and W.H.W. designed research; Q.L. and W.H.W. performed research; Q.L. and J.X. contributed new reagents/analytic tools; Q.L. analyzed data; and Q.L., R.J., and W.H.W. wrote the paper.
Reviewers: F.L., Purdue University; and W.S., University of Pennsylvania.
- Altmetric
Density estimation is among the most fundamental problems in statistics. It is notoriously difficult to estimate the density of high-dimensional data due to the “curse of dimensionality.” Here, we introduce a new general-purpose density estimator based on deep generative neural networks. By modeling data normally distributed around a manifold of reduced dimension, we show how the power of bidirectional generative neural networks (e.g., cycleGAN) can be exploited for explicit evaluation of the data density. Simulation and real data experiments suggest that our method is effective in a wide range of problems. This approach should be helpful in many applications where an accurate density estimator is needed.
Density estimation is one of the fundamental problems in both statistics and machine learning. In this study, we propose Roundtrip, a computational framework for general-purpose density estimation based on deep generative neural networks. Roundtrip retains the generative power of deep generative models, such as generative adversarial networks (GANs) while it also provides estimates of density values, thus supporting both data generation and density estimation. Unlike previous neural density estimators that put stringent conditions on the transformation from the latent space to the data space, Roundtrip enables the use of much more general mappings where target density is modeled by learning a manifold induced from a base density (e.g., Gaussian distribution). Roundtrip provides a statistical framework for GAN models where an explicit evaluation of density values is feasible. In numerical experiments, Roundtrip exceeds state-of-the-art performance in a diverse range of density estimation tasks.
Let be a density on a
Traditional density estimators such as histograms (1, 2) and kernel density estimators (KDEs) (3, 4) typically perform well only in low dimension. Recently, neural network-based approaches were proposed for density estimation and yielded promising results in problems with high-dimensional data points such as images. There are mainly two families of such neural density estimators: autoregressive models (56–7) and normalizing flows (8910–11). Autoregression-based neural density estimators decompose the density into the product of conditional densities based on probability chain rule
As suggested in ref. 12, both of these are special cases of the following general framework. Given a differentiable and invertible mapping
To overcome the limitations above, we propose a neural density estimator called Roundtrip. Our approach is motivated by recent advances in deep generative neural networks (15, 17, 18). Roundtrip differs from previous neural density estimators in two ways. 1) It allows the direct use of a deep generative network to model the transformation from the latent variable space to the data space, while previous neural density estimators use neural networks only to learn the parameters in the component functions that are used for building up an invertible transformation. 2) It can efficiently model data densities that are concentrated near learned manifolds, which is difficult to achieve by previous approaches as they require the latent space to have the same dimension as the data space. Importantly, we also provide methods, based on either importance sampling and Laplace approximation, for the pointwise evaluation of the density estimate. We summarize our major contributions in this study as follows: 1) We propose a general-purpose neural density estimator based on deep generative models, which requires less restrictive model assumptions compared to previous neural density estimators. 2) We show that the principle in previous neural density estimators can be regarded as a special case in our Roundtrip framework. 3) We demonstrate state-of-the-art performance of Roundtrip model through a series of experiments, including density estimation tasks in simulations as well as in real data applications ranging from image generation to outlier detection.
Methods
Method Overview.
The key idea of Roundtrip is to approximate the target distribution as a convolution of a Gaussian with a distribution induced on a manifold by transforming a base distribution where the transformation is represented by two GAN models (Fig. 1). After learning the weights of two GAN models by training on data, density estimation is achieved by an offline algorithm.

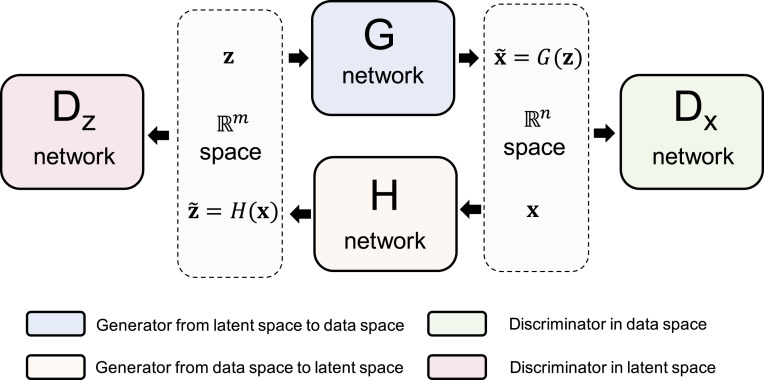
The overview of Roundtrip framework. In the latent space, latent variable
Model for the Data Density.
Consider two random variables
Typically, we set
where
Importance Sampling.
The simplest way to estimate [3] is to use the empirical expectation by
where
Laplace Approximation.
We can also obtain an approximation to the integral in [3] by Laplace’s method. To achieve this goal, we expand
where
In the remaining part of Methods, we discussed how to learn
Adversarial Training Loss.
The Roundtrip model consists of a pair of two GAN models. In the forward GAN model, the network
where
Roundtrip Loss.
During the training, we also aim to minimize the roundtrip loss, which is defined as
where
Full Training Loss.
Combining the adversarial training loss and roundtrip loss together, we get the full training loss for generator networks and discriminator networks as
After an iterative model training process, the learned networks
Model Architecture.
The model architecture of Roundtrip is highly flexible. In most cases, when it is utilized for density estimation tasks with vector-valued data, we used fully connected layers for both generative networks and discriminative networks. Specifically, the
Roundtrip can also handle tensor-valued data such as images. Similar to the model architecture in DCGAN (21), we used transposed convolutional layers for generating the image
Labeled Data and Conditional Density Estimation.
We provide a strategy for conditional density estimation given labeled data. The original Roundtrip model is extended by one-hot encoding the class label
Note that for the image datasets (MNIST and CIFAR-10) used in our study, we regard the marginal distribution of
Results
Experiment Settings.
We test the performance of Roundtrip in a series of experiments, including simulation studies and real data studies. For the density estimation task, we compared our method to the widely used Gaussian KDE as well as several neural density estimators, including MADE (6), RealNVP (10), and MAF (7). For the outlier detection task, comparisons are also made to two commonly used outlier detection methods: one-class SVM (24) and Isolation Forest (25). Note that the default setting of Roundtrip was based on the importance sampling strategy. Results of Roundtrip density estimator based on Laplace approximation are reported in SI Appendix, Fig. S2.
The neural networks in Roundtrip model were implemented with TensorFlow (26). In all experiments, we set
We took Gaussian KDE as a baseline where the bandwidth is selected by Silverman’s “rule of thumb” (28) or Scott’s rule (29). We choose the one with better result to present. The three alternative neural density estimators (MADE, RealNVP, and MAF) were implemented using codes from https://github.com/gpapamak/maf. For outlier detection tasks, we implemented one-class SVM and Isolation Forest using Scikit-learn library (30), where the default parameters were used. To ensure fair model comparison, both simulation and real data were randomly split into a 90% training set and a 10% test set. For neural density estimators including Roundtrip, 10% of the training set was kept as a validation set. The image datasets with training and test set were directly provided which require no further data split.
Evaluation.
For simulation datasets with two dimensions, we directly visualized both true density and estimated density on a two-dimensional (2D) bounded region. For simulation datasets with higher dimensions where the true density can be calculated, we evaluate different density estimators by calculating the Spearman (rank) correlation between true density and estimated density based on the test set. For real data where the ground truth density is not available, the average estimated density (natural log-likelihood) on the test set will be considered as a metric for evaluation.
In the application of outlier detection, we measure performance by calculating the precision at k, which is defined as the proportion of correct results in the top-k ranks. We set k to the number of outliers in the test set.
Simulation Studies.
We first designed three 2D simulation datasets to test the performance of different neural density estimators where the truth density can be calculated.
a)Independent Gaussian mixture.
b)Eight-octagon Gaussian mixture.
c)Involute.
The 20,000 i.i.d. points were sampled from each of the above true data distribution. After model training, we directly estimated the density in a 2D bounded region (

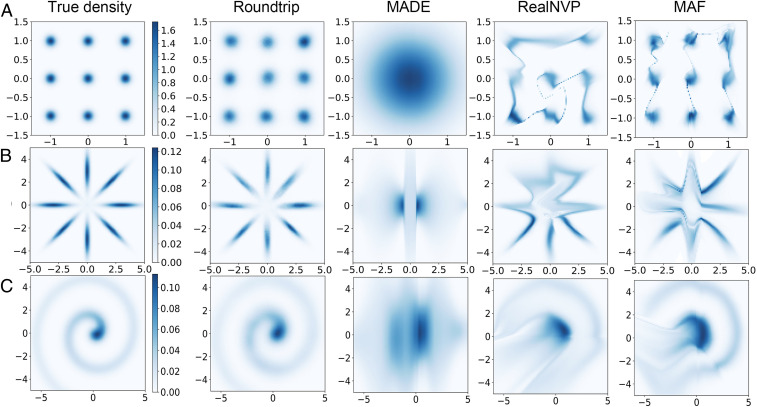
True density and estimated density by different neural density estimators (Roundtrip, MADE, RealNVP, and MAF) with three simulation datasets. Density plots were shown on a 100 × 100 grid 2D bounded region. True densities were shown in the first column. Each row gives results of a dataset: (A) independent Gaussian mixture; (B) eight-octagon Gaussian mixture; (C) Involute.
Real Data Studies.
University of California, Irvine, datasets.
We collected five datasets (AReM, CASP, HEPMASS, BANK, and YPMSD) from the University of California, Irvine (UCI) machine learning repository (31) with dimensions ranging from 6 to 90 and sample size from 42,240 to 515,345 (see more details about data description and data preprocessing in SI Appendix, Text S3). Unlike simulation data, these real datasets have no ground truth for the density. Hence, we evaluated different methods by calculating the average log-likelihood on the test set as suggested by previous work (7). Table 1 illustrates the performance of Roundtrip and the other neural density estimators. A Gaussian KDE fitted to the training data is also reported as a baseline. The results show that Roundtrip significantly outperforms other neural density estimators by achieving the highest average log-likelihood on the test set of each dataset.

| AReM | CASP | HEPMASS | BANK | YPMSD | |
| KDE | 6.26 ± 0.07 | 20.47 ± 0.10 | −25.46 ± 0.03 | 15.84 ± 0.12 | 247.03 ± 0.61 |
| MADE | 6.00 ± 0.11 | 21.82 ± 0.23 | −15.15 ± 0.02 | 14.97 ± 0.53 | 273.20 ± 0.35 |
| RealNVP | 9.52 ± 0.18 | 26.81 ± 0.15 | −18.71 ± 0.02 | 26.33 ± 0.22 | 287.74 ± 0.34 |
| MAF | 9.49 ± 0.17 | 27.61 ± 0.13 | −17.39 ± 0.02 | 20.09 ± 0.20 | 290.76 ± 0.33 |
| Roundtrip | 11.74 ± 0.04 | 28.38 ± 0.08 | −4.18 ± 0.02 | 35.16 ± 0.14 | 297.98 ± 0.52 |
The average log likelihood and 2 SDs are shown. The best performance among five methods is shown in bold.
Image datasets.
Deep generative models have demonstrated their power in generating synthetic images. However, a conventional deep generative model alone cannot provide quality scores for generated images. Here, we propose to use our Roundtrip method to generate both the image and its quality score based on the density of the image. We test this approach on two commonly used image datasets, MNIST (32) and CIFAR-10 (33), where in each of these datasets, the image comes from 10 distinct classes. Roundtrip model was modified by introducing an additional class label


True images and generated images from Roundtrip and MAF models trained on MNIST and CIFAR-10 databases. Each row denotes the true or generated images of a specific class. (A) True and generated images of MNIST. (B) True and generated images of CIFAR-10. Images generated by Roundtrip and MAF are sorted by decreased likelihood for each class.
As a more systematic and quantitative comparison of the different methods for image data, we use the estimated conditional densities of a test image (conditional on label values) to obtain Bayesian predictions of the label (Methods). Roundtrip achieves the highest test accuracy on both MNIST and CIFAR-10 datasets (SI Appendix, Fig. S3). For example, on the MNIST dataset, Roundtrip gives a test accuracy of 0.983, compared to 0.911 of MADE, 0.744 of RealNVP, and 0.926 of MAF, which again demonstrates the superiority of Roundtrip.
Outlier detection.
Finally, we applied Roundtrip to an outlier detection task, where a data point with a very low density value is regarded as likely to be an outlier. We tested this method on three outlier detection datasets (Shuttle, Mammography, and ForestCover) from ODDS database (34).
Each dataset is split into training, validation, and test set (details of data description can be found in SI Appendix, Text S4). Besides the neural density estimators, we also provide comparisons with two baseline methods, namely one-class SVM (24) and Isolation Forest (25). The results shown in Table 2 were based on the average precision of three independent runs of each algorithm. The performance of Roundtrip is either the best or tied for the best for each dataset. For the ForestCover dataset, where the outlier percentage is very low (0.9%), Roundtrip achieves a precision of 17.7% while the precision of other neural density estimators is less than 6%. Roundtrip’s performance is robust to the choice of the sample size used in importance sampling (SI Appendix, Fig. S4A). As for computational efficiency, Roundtrip is more efficient in training but less efficient in testing, compared to the other neural density estimators (SI Appendix, Fig. S4B).

| OC-SVM | I-Forest | RealNVP | MAF | Roundtrip | |
| Shuttle | 0.953 | 0.956 | 0.784 | 0.929 | 0.959 |
| Mammography | 0.037 | 0.482 | 0.474 | 0.407 | 0.482 |
| ForestCover | 0.127 | 0.058 | 0.054 | 0.046 | 0.177 |
The best performance among five methods is shown in bold.
Discussion
We propose Roundtrip as a general-purpose neural density estimator based on deep generative models. Unlike prior studies that focus on modeling the invertible transformation between a base density and the target density, where the parameters of the component functions are learned by neural networks, Roundtrip allows the direct use of a deep generative network to model the transformation from the latent variable space to the data space. In contrast to the change of variable rule used by previous methods, which requires equal dimension in the base density and the target density, Roundtrip provides a more flexible transformation between the base density and target density. In numerical experiments, Roundtrip outperforms previous neural density estimators in a variety of density estimation tasks, including simulation/real data studies and an outlier detection application.
Given the observed data, density estimation aims to recover the underlying density while deep generative modeling aims to generate new data similar to the observed ones. Our work provides a way to leverage the power of deep generative models for an accurate evaluation of density values.
Acknowledgements
The work of W.H.W. was supported by the NSF Grants (DMS1811920 and DMS195238). The work of R.J. was supported by the National Key Research and Development Program of China Grant (2018YFC0910404) and the National Natural Science Foundation of China Grants (61873141, 61721003, and 61573207). Q.L. and J.X. were supported by a China Scholarship Council scholarship.
Data Availability
Source code and data for Roundtrip are freely available on Zenodo repository (DOI: 10.5281/zenodo.3747161) (35) and (DOI: 10.5281/zenodo.3747144) (36). All data in this study are included in the article and/or supporting information. Previously published data were used for this work (https://archive.ics.uci.edu/ml/datasets/Activity+Recognition+system+based+on+Multisensor+data+fusion+%28AReM%29, https://archive.ics.uci.edu/ml/datasets/Physicochemical+Properties+of+Protein+Tertiary+Structure, https://archive.ics.uci.edu/ml/datasets/HEPMASS, https://archive.ics.uci.edu/ml/datasets/Bank+Marketing, https://archive.ics.uci.edu/ml/datasets/YearPredictionMSD, odds.cs.stonybrook.edu/, http://yann.lecun.com/exdb/mnist/, and https://www.cs.toronto.edu/∼kriz/cifar.html).
References
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
 Density estimation using deep generative neural networks
Density estimation using deep generative neural networks
