
Edited by Jasper Rine, University of California, Berkeley, CA, and approved March 5, 2021 (received for review November 5, 2020)
Author contributions: K.S. and G.T. designed research; J.F.N., A.K.E., S.J.C., A.M.M., S.C.L.H., and G.T. performed research; A.T. contributed new reagents/analytic tools; J.F.N., A.K.E., S.J.C., A.M.M., S.C.L.H., A.T., K.S., and G.T. analyzed data; and J.F.N., S.J.C., K.S., and G.T. wrote the paper.
1J.F.N., A.K.E., and S.J.C. contributed equally to this work.
- Altmetric
How repressive heterochromatic states propagate along chromosomes and are subsequently maintained for many cell divisions remain important unanswered questions in biology. Combining mathematical modeling and single-cell measurements, we find that heterochromatin does not propagate in a purely linear manner as often assumed. Rather, sudden transitions can affect large domains globally at once. We suggest this is due to long-range interactions that bring distant nucleosomes close to each other to facilitate their modification. This supports the notion that the compartmentalization of chromatin components within the nucleus and the folding of chromatin into loops dynamically control heterochromatin propagation in three dimensions by bringing nucleosomes and their modifiers into close proximity.
Methylation of histone H3K9 is a hallmark of epigenetic silencing in eukaryotes. Nucleosome modifications often rely on positive feedback where enzymes are recruited by modified nucleosomes. A combination of local and global feedbacks has been proposed to account for some dynamic properties of heterochromatin, but the range at which the global feedbacks operate and the exact mode of heterochromatin propagation are not known. We investigated these questions in fission yeast. Guided by mathematical modeling, we incrementally increased the size of the mating-type region and profiled heterochromatin establishment over time. We observed exponential decays in the proportion of cells with active reporters, with rates that decreased with domain size. Establishment periods varied from a few generations in wild type to >200 generations in the longest region examined, and highly correlated silencing of two reporters located outside the nucleation center was observed. On a chromatin level, this indicates that individual regions are silenced in sudden bursts. Mathematical modeling accounts for these bursts if heterochromatic nucleosomes facilitate a deacetylation or methylation reaction at long range, in a distance-independent manner. A likely effector of three-dimensional interactions is the evolutionarily conserved Swi6HP1 H3K9me reader, indicating the bursting behavior might be a general mode of heterochromatin propagation.
Histone-modifying enzymes covalently attach small polypeptides or chemical groups to nucleosomes, thereby controlling gene expression. Some modifications propagate from nucleation sites over large chromosomal distances. They can be maintained clonally upon cell division or overridden to allow for cell differentiation or adaptation to changing environments. Among the many histone modifications that define chromatin structure, the methylation of histone H3K9 maintains large chromosomal regions in a silent heterochromatic state.
Modeling has pointed to key parameters in chromatin formation and maintenance, for instance, the importance of cooperativity in the recruitment of modifying enzymes to chromatin or how various types of positive feedback would contribute differentially to the properties of chromatin (123–4). Nucleosome interconversion models developed with one organism, fission yeast, have been applied to distant species such as Arabidopsis, Drosophila, or mice, transposed to other modifications, for example, from H3K9 to H3K27 methylation (2, 56–7), and merged with other forms of modeling (8). In Arabidopsis, such models have uncovered fundamental aspects of vernalization by showing how epigenetic memory is digitally acquired and stored in cis at the FLC locus for long-term gene silencing and by anticipating the sequence of events taking place at that locus (2, 4, 6). In Drosophila, nucleosome-based models shed new light on the concerted action of PRC complexes (7). Recently, molecular models have been also complemented by an operational model of chromatin-mediated silencing, which supports the view that silencing is a stochastic event that occurs in an all-or-none fashion in individual cells (9).
A central element of nucleosome interconversion models is that they accommodate feedbacks arising through the direct or indirect recruitment of enzymes by modified nucleosomes. In nature, self-recruitment applies to many histone-modifying enzymes and enzymatic complexes (1011–12), some of which might be able to modify nucleosomes brought into proximity by chromatin folding (1314–15), while others would uniquely modify immediate neighbors. Hence, among the fundamental questions that quantitative modeling can address are the impacts of short- and long-range nucleosomal interactions occurring in the nucleus on chromatin dynamics. This is deeply connected to the overall expectation that heterochromatin stability depends on the size of the region in question.
To explore these questions, we used here the mating-type region of the fission yeast Schizosaccharomyces pombe. In this system, heterochromatin formation entails methylation of histone H3K9 by a single Su(var)39-family enzyme, Clr4, in combination with nucleosomal deacetylation by various histone deacetylases (HDACs), but no DNA methylation. The region contains the silent mat2-P and mat3-M loci used in mating-type switching (16) and other elements (Fig. 1), including a central cenH element with homology to centromeric and subtelomeric repeats (17, 18).

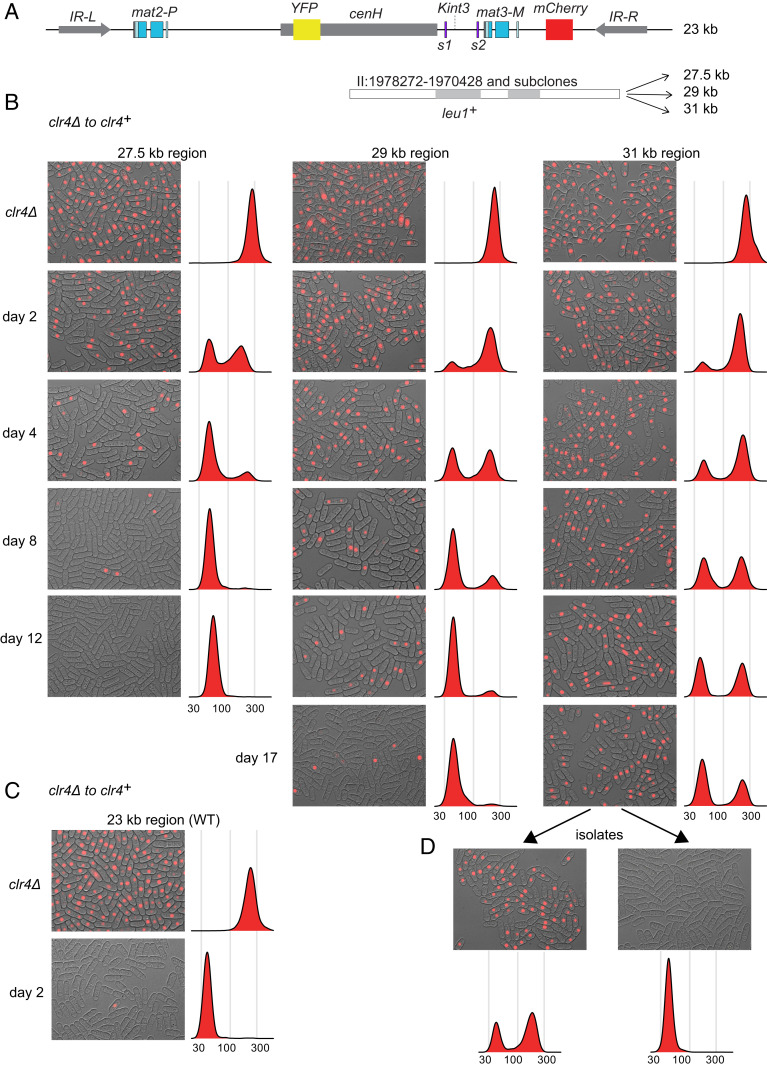
Domain-size-dependent establishment of heterochromatin. (A) Mating-type region of S. pombe showing two reporters used to monitor the establishment of heterochromatic silencing, YFP at the Kint2 site within the nucleation center cenH and mCherry at the (EcoRV) site. The heterochromatic domain is delimited by the IR-L and IR-R boundary elements and contains two Atf1 binding sites, s1 and s2. The region was extended by inserting, at the indicated Kint3 site, DNA originating from the leu1+ chromosomal region, respectively, a 4.5-kb, two distinct 5.9-kb, and a 7.8-kb fragment (see also SI Appendix, Fig. S1) generating mating-type regions of, respectively, 27.5 kb, 29 kb (SH4), and 31 kb. (B and C) The clr4+ gene was reintroduced into clr4 cells with extended mating-type regions (B) or original length (C) through genetic crosses, and the establishment of (EcoRV)::mCherry silencing was monitored in cultures maintained in exponential growth for 19 d, taking one time point per day. Overlays of brightfield and red fluorescence micrographs are presented for days 2, 4, 8, 12, and 17 as indicated. Histograms of cell fluorescence intensities are shown for the same cultures, each compiling >15,000 cells. Individual channels including YFP are shown in SI Appendix, Figs. S2–S4. (D) Two examples of clonal populations derived from the strain with the 31-kb mating-type region at day 17 and propagated for approximately 40 generations. Each micrograph shows a field 86.5 × 66
Time course experiments have found that the nucleation of heterochromatin occurs at cenH, stochastically and at a high rate (3). It is driven by RNA interference (RNAi) (19, 20) as at centromeres (2122–23). Outward expansion relies on positive feedback by several chromodomain proteins that bind H3K9me. Thus, in Clr4 chromodomain mutants catalytically proficient but incapable of binding H3K9me (20, 24), or in mutants lacking the HP1 homolog Swi6 (19, 24, 25), H3K9me heterochromatin is restricted to cenH. At centromeres or when heterochromatin is induced by artificially tethering Clr4, the Clr4 chromodomain is also critical to the epigenetic heterochromatic state (23, 26, 27). For Swi6, the positive feedback might be exerted by interacting HDAC complexes (2829–30) that foster H3K9 methylation (25, 31), and the Swi6 paralog Chp2 is likely to function in a similar way (28, 29, 323334–35). These H3K9me/chromodomain feedbacks would be missing in organisms where heterochromatin relies exclusively on other modifications (36, 37).
Following stochastic nucleation at cenH, it is not known whether heterochromatin propagates outward gradually as a front of modified nucleosomes or whether the propagation follows intermittent dynamics. The experiments monitoring the de novo establishment of heterochromatin in the wild-type mating-type region could not distinguish between the two possibilities (3). We reasoned that increasing system size would exacerbate the differences in outcome of various modes of propagation, allowing distinguishing between them. We thus expanded the size of the mating-type region incrementally to a total length of 31 kb and determined the effects on the profiles of establishment of heterochromatic silencing. The experiment revealed a stunning mode of heterochromatin propagation exhibiting an all-or-none dynamics even across large regions. The discovery of slower establishment for larger regions favors a scenario where heterochromatin formation is facilitated by distance-independent recruitment from the silenced state.
Results
Construction of Fission Yeast Strains with Extended Mating-Type Regions.
We created a set of S. pombe strains with two fluorescent reporters, YFP and mCherry, to monitor the nucleation and propagation of heterochromatin in mating-type regions of varied length. The gene encoding YFP was embedded in the cenH nucleation center (Kint2::YFP), while mCherry was expressed from a peripheral location, (EcoRV)::mCherry. The expression of both genes was driven by the weak ura4 promoter, and the proteins were targeted to the nucleus with the SV40 nuclear localization signal to take advantage of the very low autofluorescence in the nucleus. The setup is identical to refs. 3 and 38 except that intervening DNA was introduced between cenH and (EcoRV)::mCherry to extend the heterochromatic region.
We chose to extend the region by adding S. pombe genomic DNA, less likely to introduce nucleotide biases or changes in nucleosome density than DNA from other organisms. Compared to other organisms, S. pombe has a relatively high nucleosome density [6.5 nucleosomes per kilobase, on average (39)]. The DNA selected here contains the S. pombe leu1+ gene and flanking DNA amounting to, respectively, 4.5, 5.9, and 7.8 kb free of highly expressed genes (40) (Fig. 1A and SI Appendix, Fig. S1). This DNA was integrated at a location designated Kint3. Two distinct 5.9-kb inserts were generated by removing different parts of the 7.8-kb insert in strains SH4 and SH5 (SI Appendix, Fig. S1). Altogether, mating-type regions of 23, 27.5, 29 (SH4 and SH5), and 31 kb were obtained.
Heterochromatin Establishment Is Domain Size Dependent and Occurs in Two Consecutive Stochastic Switches.
Strains with the engineered mating-type regions in a clr4
Silencing followed very different kinetics at cenH and outside cenH (Fig. 1 and SI Appendix, Figs. S2–S4), as seen previously for the wild-type mating-type region (3, 38). Silencing within cenH occurred fast in all cases; the Kint2::YFP reporter was silenced at the population level within a day. In contrast, establishment of silencing at the (EcoRV)::mCherry reporter was slower and differed greatly between strains. Bimodal distributions of fluorescence intensities were observed throughout the establishment periods (Fig. 1 and SI Appendix, Figs. S3 and S4), indicating that, during heterochromatin establishment, (EcoRV)::mCherry adopts one of two expression states, ON or OFF, rather than numerous intermediate states.
The proportions of ON and OFF cells were estimated from histograms at each time point (Fig. 2 and more below). For all strains examined, the proportion of cells expressing mCherry decayed exponentially over time. The rates of silencing establishment proved considerably slower in strains with added DNA compared to the wild-type region, most dramatically so in the strain with the 31-kb mating-type region. Compared with the mean lifetime of the mCherry ON state of 5.4 generations in the 23-kb region, the mean lifetime was 15 generations in the 27.5-kb region, 45 and 48 generations, respectively, in the two 29-kb regions, and 110 generations, more than 3 wk, in the 31-kb region.

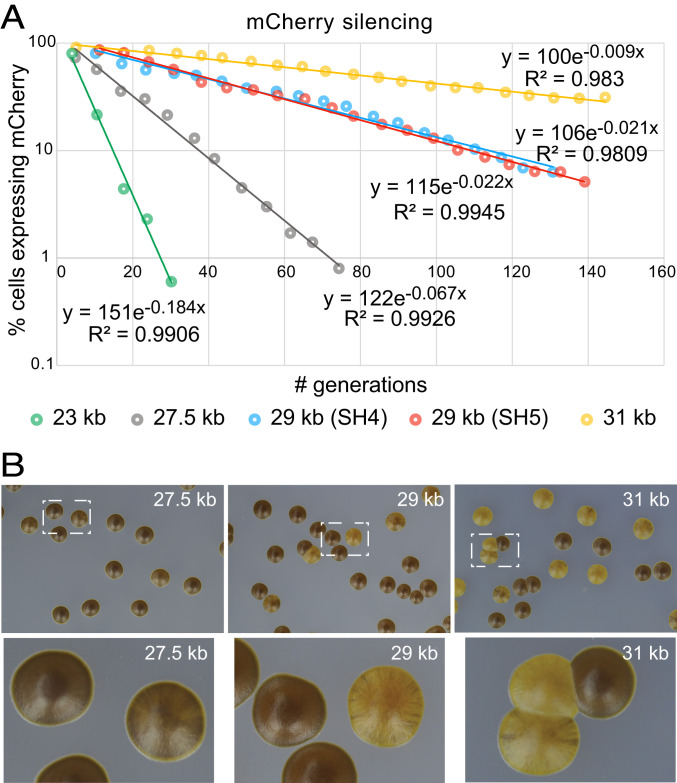
Stochastic switches to a stable heterochromatic state. (A) Exponential decay of (EcoRV)::mCherry ON state following reintroduction of clr4+. The proportion of mCherry-positive cells in populations undergoing heterochromatin formation in mating-type regions of various lengths was measured over time as described in Fig. 1 and in the text, by sampling liquid cultures maintained in exponential phase for long periods of time. Fits with exponential functions are shown. The histograms used to generate the plots are displayed in SI Appendix, Fig. S4. (B) Heterochromatin establishment monitored by iodine staining of colonies. Cells from the same cultures as shown in A were plated on MSA sporulation plates supplemented with leucine, incubated at
The profiles of silencing establishment were nearly identical for the two strains with a 29-kb mating-type region despite their different sequence contents (Fig. 2A). One 29-kb region (SH4) contains the apc10+ gene, whereas the other 29-kb region (SH5) does not (SI Appendix, Fig. S1), indicating that this transcription unit at least does not affect the kinetics. Moreover, the 29-kb region with the apc10+ gene has the same gene content as the 31-kb region (SI Appendix, Fig. S1) that showed much slower kinetics (Fig. 2 and SI Appendix, Figs. S3 and S4). Thus, altogether, the differences in establishment profiles seemed due to region length, predominantly, rather than content.
We wondered whether establishment might appear slow in the strain with the 31-kb region, due to instability of the OFF state. To test the stability of the ON and OFF states, clonal populations were derived from single cells during the establishment period, and they were examined by fluorescence microscopy. Two types of cultures were obtained. The first type displayed just the silent state, and the second type displayed bimodal mCherry expression (examples shown in Fig. 1D). This showed that a stable heterochromatic state could be attained for the 31-kb region as for the shorter regions and that the relatively high proportion of ON cells during the establishment period was not due to great instability of the OFF state. Indeed, cells in the OFF state could be propagated stably in liquid cultures for >200 generations (SI Appendix, Fig. S5).
The properties revealed by fluorescence microscopy, system-size-dependent silencing kinetics, bimodal cell distributions during establishment periods, and stability of the heterochromatic state, once established, were corroborated by independent tests for mating-type switching and for leu1+ expression.
Heterochromatin, Once Established, Is Stable in Both Short and Long Domains.
As an independent way of monitoring heterochromatin formation, we examined the ability of cells to switch mating type following reintroduction of the clr4+ gene, by iodine staining of colonies. This assay relies on a biological function of heterochromatin, required for efficient mating-type switching and homothallic sporulation. As spores but not vegetative cells are stained by iodine, colonies of cells with fully established heterochromatin display a uniform dark-brown staining, while cells lacking heterochromatin form lighter colonies. We thus plated cells on sporulation plates at various time points during establishment experiments, let isolated cells develop into colonies for approximately 20 generations, and stained the plates with iodine vapors (Fig. 2B and SI Appendix, Fig. S5).
The short mating-type region produced exclusively dark-staining colonies starting at early time points, but longer regions showed a delay correlated with the slow silencing of (EcoRV)::mCherry in them, and distinct patterns of staining were observed (Fig. 2D). The 27.5-kb region produced more heavily streaked colonies than the longer regions, reflecting more frequent successful establishment events during colony growth. For the longest region examined, the 31-kb region, colonies with wild-type iodine staining coexisted side by side with very light colonies for the entire length of the experiment. In this assay, uniformly brown staining shows stability of the heterochromatic state at the timescale of colony formation. The brown colony phenotype was also stable upon growth in culture and replating, and cells in the brown colonies had silenced both leu1+ and (EcoRV)::mCherry, while cells in the light colonies showed unstable leu1+ and (EcoRV)::mCherry expression (SI Appendix, Fig. S5). These phenotypes reflect a long-lasting epigenetic inheritance of alternate chromatin states over the mating-type region and denote the stability of the heterochromatic state once it is established. They also show that (EcoRV)::mCherry, the mating-type cassettes, and leu1+ all tend to be euchromatic or heterochromatic in individual cells in a correlated manner.
Temporally Correlated Silencing of Genes outside cenH.
To assess the relative timing of silencing of reporters at a finer scale, we performed a clr4+ reintroduction experiment in a strain where the YFP gene, instead of being at cenH, was at a location between cenH and (EcoRV)::mCherry, approximately 5 kb from (EcoRV)::mCherry (Fig. 3). In that setup, YFP and mCherry were silenced in a highly correlated fashion. Throughout the experiment, with few exceptions, cells either expressed or had silenced both reporters (Fig. 3). Hence, despite being 5 kb apart, the two reporters were silenced at the same time, or nearly. This was confirmed by time-lapse microscopy of dividing cells during the establishment period (examples shown in Movies S1–S6). In cases where a short delay separated silencing of the two reporters, either mCherry (Movies S3 and S4) or YFP (Movies S5 and S6) could be silenced first. The fact that mCherry could be silenced before YFP is interesting, as it argues against a linear spreading of heterochromatin from cenH. It does not exclude that linear spreading might occur from elements located between the YFP and mCherry genes, such as Atf1 binding sites (Fig. 1). However, even in this case, spreading would be catalyzed at a distance by cenH, as, in the absence of cenH, heterochromatic silencing at EcoRV occurs once in 2,000 cell divisions, on average, whereas, here, in the presence of cenH, it occurred two orders of magnitude more efficiently. The possibility of linear spreading from Atf1 binding sites facilitated by physical contacts with cenH through looping was investigated as part of our modeling.

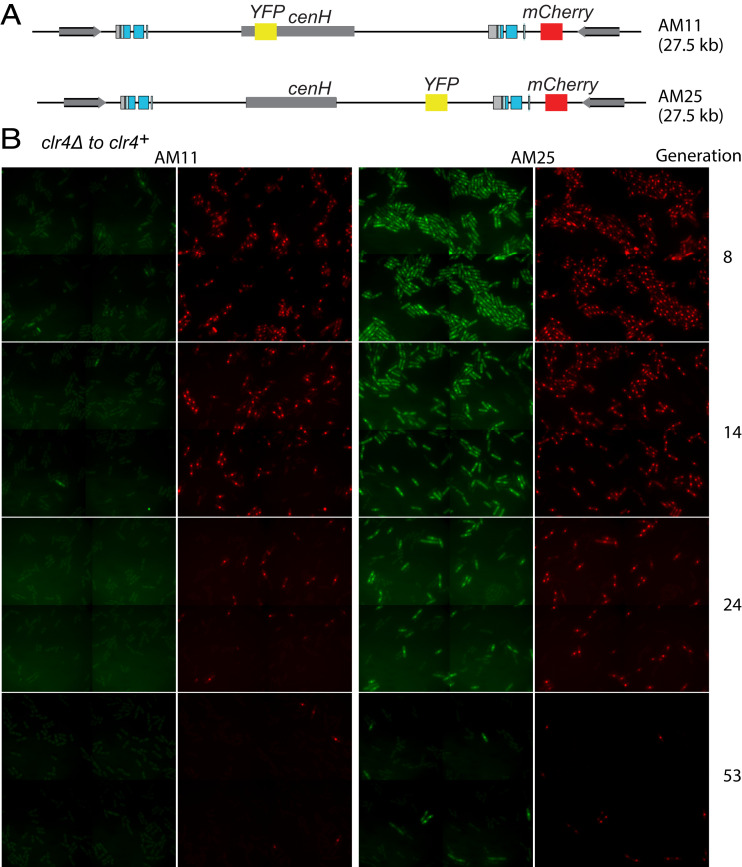
Temporally correlated silencing of two fluorescent reporters placed outside cenH. (A) Mating-type region of strains AM11 and AM25. (B) Micrographs of clr4+ cells from exponentially growing cultures in the process of establishing heterochromatin in the AM11 (Left) or AM25 (Right) mating-type region at four time points, showing fast repression of YFP for AM11 and slow and temporally correlated repression of the YFP and mCherry reporters for AM25. Four contiguous fields are shown for each sample, each field 110
Modeling Heterochromatin Formation in the Mating-Type Region.
To model heterochromatin establishment, the wild-type and three expanded mating-type regions were represented by 153, 182, 191, and 203 nucleosomes, respectively, each containing a special 31-nucleosome region to mimic the ability of the cenH element to recruit the Clr4 methyltransferase through RNAi. In addition to explaining the observed heterochromatin establishment kinetics of the differently sized mating-type regions, we demanded that the model reproduce bistability of a 76-nucleosome system lacking the special region, to mimic the phenomenon observed with
The model considers two types of reactions, recruited conversions, where an enzyme is recruited by a nucleosome and modifies another nucleosome in the system, and direct conversions that do not result from nucleosome-based recruitment processes. Direct conversions encompass nucleosome state changes due to nucleosome turnover, to the binding of enzymes to DNA or transcription factors, or to chromatin contacts from outside the mating-type region, among others. The reactions were simulated using an event-based Gillespie algorithm in which each discrete event with an assigned rate parameter represents one modification event (presented in SI Appendix). The algorithm was executed by first selecting an event, either a direct conversion or a recruitment. When a direct conversion attempt was selected, one nucleosome in the system was chosen randomly, and its state was changed accordingly. In case of a recruited conversion attempt being chosen, a recruiting nucleosome was chosen first. If this nucleosome was either in an A state or an S state, a second nucleosome in the system was chosen, and, if this second nucleosome was in a state that permitted a conversion, the state of this second nucleosome was changed according to the action of the recruited enzyme.
In ref. 3, the necessity of a combination of nonlocal (global) and local interactions was highlighted, and several possible models were analyzed and discussed. Here, the consideration of different system sizes restrained the possible model versions further. Fig. 4 shows simulation results of a model version where the S-mediated feedback on A- to U-state conversions is global, and the remaining three feedbacks act only locally. Other models, including looping models, are discussed farther down and presented in SI Appendix, Figs. S7–S19.

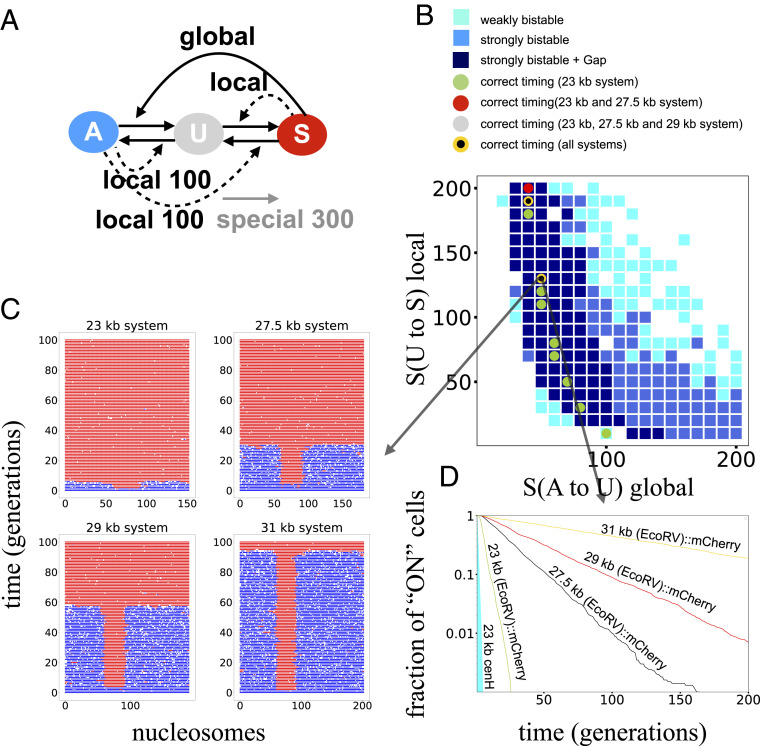
A combination of three local and one distance-independent silencing recruitment reaction can explain the domain-size-dependent kinetics of de novo heterochromatin establishment. (A) The model considers four types of recruited conversions based on positive feedback, where enzymes are recruited by the same histone modifications that they deposit. Only the S-mediated A- to U-nucleosome conversions are global, whereas the remaining three reactions act locally on the nearest-neighbor nucleosomes. Spontaneous conversions between states with a higher rate of U-state to S-state nucleosomes within the cenH region are also included in the model. (B) Scanning revealed that this model can recapitulate the observed bistability of the
A Combination of Three Local and One Distance-Independent Silencing Recruitment Reactions Can Explain the Profiles of de Novo Heterochromatin Formation.
For the model presented in Fig. 4, the rates of A-mediated conversions of U to A nucleosomes and S to U nucleosomes were assigned a fixed rate of 100 conversion attempts per nucleosome per cell generation, whereas the remaining two recruitment reactions were varied as shown. In addition to the spontaneous conversions indicated by the connecting straight black arrows (each having a rate of one conversion attempt per nucleosome per generation), the special cenH region was assigned a higher rate of direct U- to S-nucleosome conversions (300 conversion attempts per nucleosome per generation). To ensure that the ability of the model to reproduce the observed establishment kinetics is not restricted to the relatively low direct conversion rates, we also performed simulations with higher direct conversion rates and were still able to obtain good fits (SI Appendix, Fig. S8). In order to examine whether the model can recapitulate the experimentally observed kinetics, the S-mediated U-to-S and A-to-U nucleosome conversion rate constants were varied as shown in Fig. 4B. A total of 400 pairs of rate constants were tested, 1,000 simulations were performed for each pair, and the operation was repeated for each size of the mating-type region. Bistability is illustrated by cyan and blue squares, where simulations of the 76-nucleosome system are started with either only A- or S-state nucleosomes, and the system remains in each state for more than 100 (cyan) or 500 (blue) generations, respectively. From experimental kinetic measurements performed in five independent experiments plotted in Fig. 5, we could estimate, for each system, the average lifetime (b) of (EcoRV)::mCherry expression following reintroduction of the clr4+ gene (Figs. 2A and 5). The average lifetime (b) for the simulated systems was determined by fitting exponential functions to the simulation results. The measure

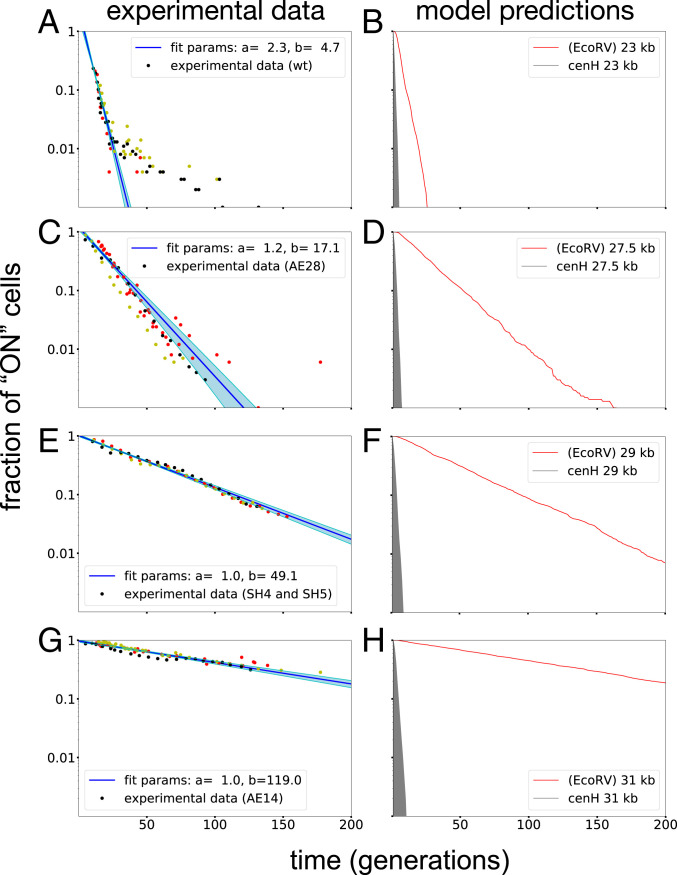
Concordance of data fitting to exponential function and model predictions. (A, C, E, and G) Experimental data obtained in five independent experiments with various strain combinations was fitted to an exponential function of the form
All in all, the model can recapitulate bistability of the 76-nucleosome system and the observed establishment dynamics of all system sizes. Slight deviations of the lifetime between simulations and experiments might be explained by simplifications in the model that could influence the establishment dynamics in the experimental setup such as DNA composition or small temperature fluctuations. Some variation also occurred in experimentally determined average lifetimes, even though this did not change the overall observation that strains with larger mating-type regions showed drastically slower establishment dynamics of heterochromatin. We also observed that the model can recapitulate the observed establishment kinetics with higher noise rates (SI Appendix, Fig. S8). Moreover, in simulations, we could see that small changes of the global rate had substantial effects on the speed of heterochromatin formation. Interestingly, this effect was stronger in larger system, whereas the wild-type 23-kb system was more robust to these changes (SI Appendix, Fig. S8).
Variations on the Model Show the Need for a Global S-Mediated Feedback to Account for the Observed Kinetics.
Fig. 6 shows simulation results of the best fits of other model versions with one global recruitment reaction and three local recruitment moves. Fig. 6A shows the simulated dynamics of the model from Fig. 4 with an altered global reaction, where nucleosomes to be modified are now chosen with a probability inversely proportional to their distance to the recruiting nucleosome. The simulations illustrate that, in addition to an extensive weakening of the model’s ability to reproduce the bistable behavior of the

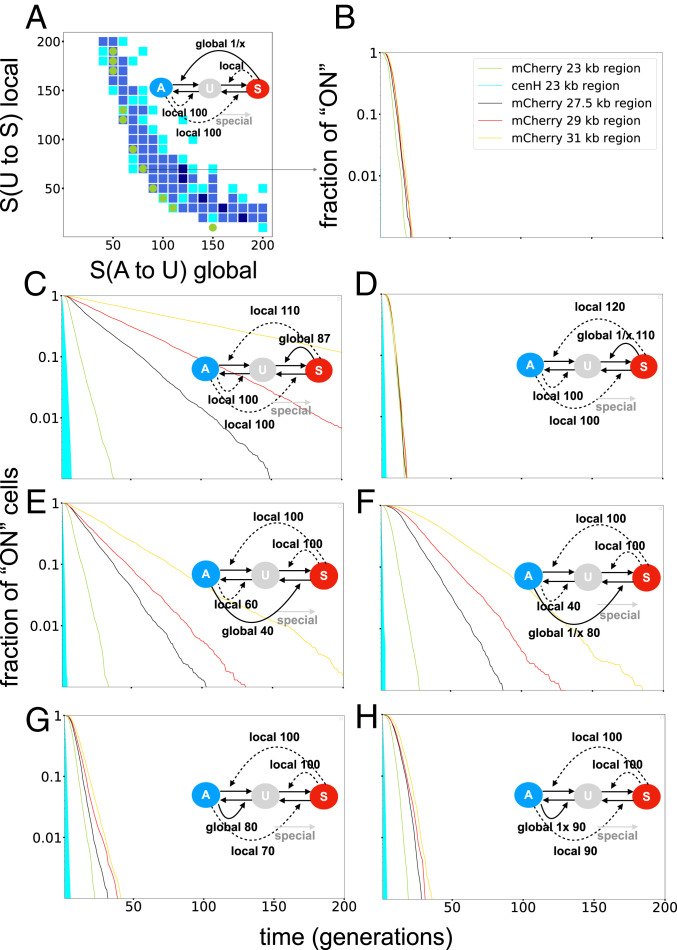
Simulation results show the need for a global, distance-independent feedback for models with one global and three local feedbacks. (A) A scan of the model shown in Fig. 4 and (B) an example plot of the heterochromatin formation over time when the global reaction is changed to a distance-dependent global reaction (conversion of one nucleosome at distance x from a recruiting nucleosome happens with probability 1/x), illustrating the failure of the 1/x global recruitment to produce system-size dependency for systems larger than the wild-type system. C, E, and G show simulation results of all other possible models with one global and three local recruitment reactions, and D, F, and H show simulation results of the same models as shown in C, E, and G but with distance-dependent global reactions of the form 1/x. The model versions with the distance-independent global S-mediated A-to-U and global distance-independent A-mediated S-to-U nucleosome conversions show system-size-dependent kinetics. However, bistability produced by the global A-mediated S-to-U model version is fragile (see SI Appendix, Fig. S11), and the establishment dynamics of larger systems is too fast. Thus, only the model shown in Fig. 4 and the model version with global S-mediated U-to-S conversions can recapitulate the experimental results. All model versions with distance-dependent global reactions fail to reproduce the reporter switch-off time dynamics. The values for the special rate constants are 90 (B), 45 (C), 45 (D), 20 (E), 20 (F), 50 (G), and 50 (H).
In SI Appendix, Figs. S7–S19, a total of seven model motifs (four motifs with each one global and three local feedbacks, and three motifs with each two global and two local feedbacks) have been investigated. Each motif was examined in two different versions, with or without internal looping between cenH and two Atf1 binding sites (s1 and s2 in Fig. 1). Each standard model version, without specific internal looping, was simulated with four different modes of the global feedback reactions (three distance-dependent modes and one distance-independent mode), and the model with an interaction between cenH and Atf1 was simulated with a distance-dependent global feedback reaction (1/x contact probability). Additionally, an Atf1 looping-model version with only local feedbacks was simulated. Distance-dependent feedback reactions followed a power law-distributed contact probability between substrate and recruiting nucleosome as a function of distance between the two nucleosomes. In total, 36 model versions were examined (described in SI Appendix, SI Text and Figs. S7–S19). Of the 36 models, 7 were able to fit the experimentally observed silencing kinetics and fulfill the bistability requirement of the smaller 76-nucleosome system with the same set of parameter values (SI Appendix, Fig. S19). Six of the “working” models have in common that they all have one or two global feedback reactions that are distance independent or very weakly distance dependent (power law exponent
Only one model version, with an S-mediated A-to-U and an A-mediated S-to-U feedback reaction, could recapitulate the experimental observations with distance-dependent global feedback reactions following a 1/x power law contact probability. Although intriguing, we consider this model version to be problematic, because biological evidence is lacking for a global A-mediated S-to-U feedback reaction, in contrast to the feedbacks used by other models that are well documented experimentally, as discussed farther down. Furthermore, any global feedback toward active nucleosomes (either A(S-to-U) or A(U-to-A)) might lead to repeated attacks by A nucleosomes surrounding the mating-type region and destabilize heterochromatin. Similarly a global S(U-to-S) feedback would allow a silenced mating-type region to destabilize the active surroundings of the mating-type region by converting U nucleosomes to S nucleosomes. A global S(A-to-U) feedback would potentially have less destabilizing effects on active states, since it only slightly increases the proportion of U nucleosomes and thus has an effect similar to DNA replication or higher noise. SI Appendix, Fig. S19 presents an overview and discussion of all working models.
Mimicking Chromosome Looping between cenH and Atf1 Binding Sites near mat3-M Does Not Produce Comprehensively Effective Models.
To test how a loop domain might contribute to heterochromatin establishment, we simulated looping between internal Atf1 binding sites (42, 43) (Fig. 1) and cenH. The interaction between cenH and the Atf1 binding sites was simulated as an S(U-to-S) recruited conversion. Whenever a substrate nucleosome was chosen at either position 101 or 42 nucleosomes upstream of the last nucleosome in the region (Atf1 binding sites) and a recruiting nucleosome was chosen within cenH, an attempt to convert the substrate nucleosome to an S nucleosome was made. Conversely, if a substrate nucleosome was chosen inside cenH combined with a recruiting nucleosome at either of the two Atf1 binding sites, an attempt to convert the substrate nucleosome to an S nucleosome was carried out (see SI Appendix for a more detailed description). The modeling results suggest that, although internal looping helps with the timing of the observed system-size-dependent silencing dynamics in some model versions, it always fails to simultaneously meet our primary modeling requirement of conferring bistability to the small 76-nucleosome system and system-size-dependent silencing. All in all, based on our model simulations, we find that, although we cannot rule out the possibility of an interaction between cenH and Atf1 binding sites, looping does not change the need for distance-independent or weakly distance-dependent global feedback reactions.
Discussion
Overall, kinetic measurements performed at the single-cell level and stochastic modeling show how the mating-type region undergoes two stochastic switches during heterochromatin formation where the cenH region always switches to a heterochromatic state before the rest of the region. Moreover, the second stochastic switch occurs at a size-dependent rate. At the molecular level, this means that heterochromatin does not spread gradually from the nucleating region at cenH to the remaining region; rather, heterochromatin propagates in an all-or-none fashion, where whole regions collapse suddenly. This mode of propagation differs radically from the linear propagation proposed in other cases (44).
Our modeling shows that a previously suggested three-state model with a distance-dependent global S(A-to-U) feedback (3) falls short of explaining the size dependence observed here, unless the global reaction is changed to a distance-independent feedback. In general, models with one global feedback and three local feedbacks can only explain the newly observed silencing dynamics and the bistability of
A key aspect captured by modeling is the role played by the cenH element. Many experiments combined have determined that RNAi acting at cenH facilitates heterochromatin formation over the entire mating-type region (3, 19, 41, 45, 46), but the mechanism has remained obscure. cenH also potentiates the effect of silencing elements at ectopic locations (19, 47, 48). Providing mechanistic insight, modeling shows how the buildup of an H3K9me nucleosome reservoir at cenH could drive the conversion of a larger region to the heterochromatic state via a global positive feedback. A first RNAi-mediated switch would happen within cenH, and a cross-talk would develop between cenH and the rest of the domain, such that only a limited number of nucleosomes would have to be converted to S nucleosomes outside cenH for an irreversible switch to heterochromatin to happen in the entire domain. This property would allow DNA-binding proteins to change the chromatin state of a large region when placed in reach of a block of heterochromatin, even when the DNA-binding proteins themselves are only capable of inducing modifications in a more local manner. This could apply to protosilencers (49) and to cases where heterochromatin silences genes naturally present, or transposed or integrated in its vicinity in Drosophila (50) or human cells (51). Our observations and modeling also show how long delays might separate transgene integration at a locus from the stable repression of the transgene by nearby heterochromatin.
Among the multiple feedbacks that originate from heterochromatic nucleosomes, the best documented and most direct are mediated by chromodomain proteins, in S. pombe, the HP1-like proteins Swi6 and Chp2, the RNAi-effector protein Chp1, and the Clr4 histone H3K9 methyltransferase, that bind dimethylated and trimethylated histone H3K9 (20, 25, 52, 53) (SI Appendix, Fig. S1). In vitro, the binding of Clr4 to an H3Kc9me3 nucleosome mimicking the trimethyl state of histone H3K9 stimulates methylation by Clr4 locally in cis on an adjacent nucleosome (54). In vivo, this local reaction might operate in concert with global deacetylation driven by the association of HDACs with Swi6 and Chp2 (28, 29). HP1 proteins display several properties that, alone or in combination, are susceptible to favor global reactions. Human HP1alpha (55) and Drosophila HP1 (56) can form liquid–liquid phase-separated condensates that, in the case of Drosophila, have been suggested to facilitate early stages of heterochromatin formation in vivo and later underlie important biophysical properties of mature heterochromatin (56). In S. pombe, Swi6 compacts chromatin through oligomerization (57, 58), and deformation of the nucleosome octamer upon Swi6 binding results in phase separation of Swi6 chromatin in vitro (59). Thus, during heterochromatin establishment, global reactions might take place in the context of biomolecular condensates. This would account for the effectiveness of distance-independent reactions in models (Fig. 6 and SI Appendix, Figs. S7–10 and S13), indicating that the global effects emerge from forms of chromatin compaction or subnuclear organization of chromosomes that do not merely reflect nucleosomal distances on the linear chromatin fiber (Fig. 7).

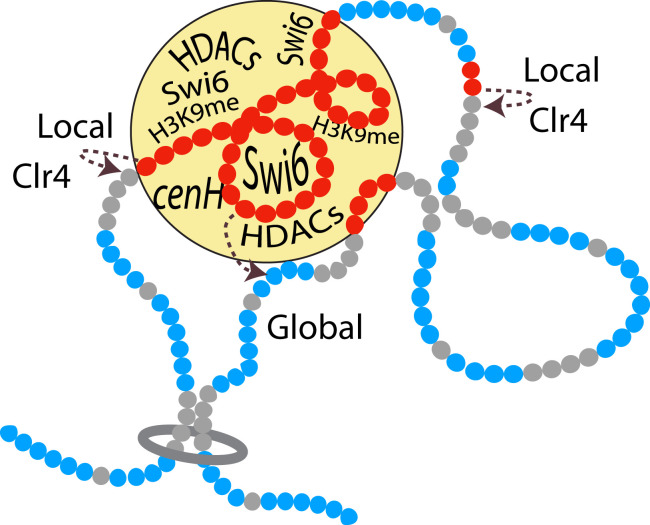
Modes of heterochromatin propagation suggested by experiments and modeling combined. Positive feedbacks exerted by heterochromatic nucleosomes during heterochromatin formation are represented. A global positive feedback exerted by S nucleosomes on A-to-U deacetylation might be imposed by phase separation of Swi6-associated chromatin with bound HDACs. In contrast, the local positive feedback exerted by S nucleosomes on U-to-S methylation reactions could be due to guided catalysis by nucleosome-bound Clr4. This particular feedback combination was suggested by mathematical modeling and literature to account for the kinetics of heterochromatin formation over the fission yeast mating-type region measured here. During establishment, a first stochastic switch at the nucleation center cenH leads to the fast buildup of a patch of heterochromatin from which heterochromatin expands to the entire mating-type region in a second stochastic switch with size-dependent delays.
Mimicking looping between cenH and Atf1 binding sites did not suffice to account for the kinetics by linear spreading. This does not exclude that looping between these or other elements could contribute to long-range interactions with weak distance dependency, for example, by permitting supercoiling of the domain. Additional global effects might result from the localization of the mating-type region at the nuclear periphery driven by heterochromatin as it forms (60).
Other than deacetylation, the chromatin structures and condensates encountered in vivo might globally facilitate methylation by Clr4, as these structures would differ from the dinucleosomes used in vitro to assay the catalytic activity of Clr4 (54). Thus, the possibility of a global feedback by heterochromatic nucleosomes on the methylation reaction will also have to be considered in the future. Moreover, in vivo Clr4 is part of the multisubunit complex ClrC containing, for example, a ubiquitin ligase whose ubiquitylation of histone H3K14 is required for the methylation of H3K9 by Clr4 (61). Potential feedbacks through the whole complex remain to be studied. RNAi reaching outside cenH might also operate over long distances to recruit Clr4 (24, 62). Compared to a global feedback on A-to-U transitions which might be easily counteracted by histone acetyl transferases in euchromatin, the effects of a global feedback on U-to-S transitions could be more difficult to control genome wide. In all cases, the compartmentalization of chromatin components by phase separation and the organization of chromosomes into loop domains would be important factors in limiting the range of global reactions.
In the future, feedbacks can be investigated genetically using chromodomain mutants (e.g., ref. 53) to alter the affinity for H3K9me of the Clr4 chromodomain affecting the S-mediated (U-to-S) feedback, or the Swi6 chromodomain affecting the S-mediated (A-to-U) feedback. In the other direction, the A-mediated (U-to-A) feedback could possibly be perturbed by targeting a positive feedback loop acting on the Mst2 acetyl transferase (63) and the A-mediated (S to U) feedback by targeting the bromodomain protein Bdf2, an interactor of Epe1 (64). Moreover, in an independent approach, varying system size in the absence of the cenH region would allow us to determine the relative effects of cenH with size, and the relative increase in the contribution of global recruitment when considering larger systems.
Materials and Methods
Strain Constructions and Growth Conditions.
Standard procedures were used to propagate S. pombe cells and to construct new strains by genetic crosses (65) or by transformation of existing strains with recombinant DNA (66) as detailed in SI Appendix. Strain genotypes are shown in SI Appendix, Table S1, and the oligonucleotides used for strain construction are shown in SI Appendix, Table S2. Yeast extract with supplements (YES) was used as rich medium; minimal sporulation agar (MSA) (with 2 g/L arginine as nitrogen source) supplemented with 100 mg/L uracil, 100 mg/L adenine, and 200 mg/L leucine) was used for crosses; MSA (with 2 g/L glutamate as nitrogen source) supplemented with 200 mg/L leucine, when indicated, was used to plate cells during establishment experiments prior to iodine staining; and EMM2 supplemented with 200 mg/L leucine was used as growth medium for the heterochromatin establishment experiments. Cultures were propagated at
Reintroduction of clr4+ Gene into clr4Δ
The S. pombe strains PG3713 or PG3716 were used to reintroduce clr4+ into clr4
Fluorescence Microscopy and Image Analyses.
Cells were imaged with a Nikon Ti Eclipse epifluorescence microscope equipped with a 100× objective, an Andor Luca camera, and a temperature-controlled chamber which was set to
Data Acquisition with Xcyto-10 Quantitative Cell Imager.
As an alternative to the Nikon Ti Eclipse microscope, fluorescence images were acquired and processed with an Xcyto-10 Quantitative Cell Imager (ChemoMetec) to generate the data presented in Fig. 1 and SI Appendix, Fig. S3. The LED535 lamp and filter 582 to 636 were used for mCherry, LED488 lamp and filter 513 to 555 for YFP, with exposure times of 1 s in both cases. The acquired images were visually inspected and analyzed with the XcytoView 1.0.109.0 software (ChemoMetec) to measure the intensity of fluorescence within individual cells for YFP and mCherry. Files (.fcs) were retrieved to plot histograms in R, and these were used to determine the proportions of OFF and ON cells by setting intensities thresholds of 150 a.u. for YFP and 100 a.u. for mCherry based on clr4
Curve Fitting and Confidence Bands of the Fit.
The simulation results and the data shown in Figs. 4–6 were fitted with the curve_fit function of the Scipy.optimize Python package which uses the method of nonlinear least squares to fit the function
Code Availability.
Python code files are available at GitHub (https://github.com/njm226/Heterochromatin-establishment) (67).
Acknowledgements
We thank members of our groups for discussions and granting agencies for financial support: the Novo Nordisk foundation (Grant NNF19OC0058686 to G.T.), the Carlsberg Foundation (Grants CF15-0853 and CF19-0200 to G.T.), the Danish National Research Foundation (Grant DNRF116 to A.T.), and the European Union’s Horizon 2020 Research and Innovation Program (Marie Skłodowska-Curie Grant 813282 to K.S.). S.J.C. received a Novo Nordisk scholarship. We also acknowledge Søren Kjærulff and Rune Troelsgaard Pedersen at ChemoMetec for kindly providing access to an Xcyto 10 Quantitative Cell Imager.
Data Availability
All data generated or analyzed during this study are included in this article and SI Appendix.
References
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
 Establishment of heterochromatin in domain-size-dependent bursts
Establishment of heterochromatin in domain-size-dependent bursts
