
- Altmetric
In many animal species, females undergo physiological and behavioral changes after mating. Some of these changes are driven by male-derived seminal fluid proteins and are critical for fertilization success. Unfortunately, our understanding of the molecular interplay between female and male reproductive proteins remains inadequate. Here, we analyze the postmating response in a Drosophila species that has evolved strong gametic incompatibility with its sister species; Drosophila novamexicana females produce only ∼1% fertilized eggs in crosses with Drosophila americana males, compared to ∼98% produced in within-species crosses. This incompatibility is likely caused by mismatched male and female reproductive molecules. In this study, we use short-read RNA sequencing to examine the evolutionary dynamics of female reproductive genes and the postmating transcriptome response in crosses within and between species. First, we found that most female reproductive tract genes are slow-evolving compared to the genome average. Second, postmating responses in con- and heterospecific matings are largely congruent, but heterospecific matings induce expression of additional stress-response genes. Some of those are immunity genes that are activated by the Imd pathway. We also identify several genes in the JAK/STAT signaling pathway that are induced in heterospecific, but not conspecific mating. While this immune response was most pronounced in the female reproductive tract, we also detect it in the female head and ovaries. These results show that the female’s postmating transcriptome-level response is determined in part by the genotype of the male, and that divergence in male reproductive genes and/or traits can have immunogenic effects on females.
Introduction
In internally fertilizing organisms, gamete fusion is often preceded by a complex array of biochemical interactions within the female reproductive tract that mediate reproductive success (Wolfner 2007, 2009). During copulation, males transfer sperm and a cocktail of seminal fluid proteins (SFPs) that facilitate a variety of postmating effects that are required for successful fertilization (Ravi Ram and Wolfner 2007; Sitnik et al. 2016). In Drosophila melanogaster, several of these SFPs are well-characterized and play important roles in postmating processes within the female reproductive tract (Chapman et al. 2000, 2001; Liu and Kubli 2003; Findlay et al. 2008, 2009; Wong et al. 2008; Ram and Wolfner 2009; Avila et al. 2010, 2011; Rubinstein and Wolfner 2013). These processes include facilitating sperm storage (Neubaum and Wolfner 1999; Tram and Wolfner 1999), inducing ovulation (Herndon and Wolfner 1995), reducing the female’s propensity to remate (Chen et al. 1988; Liu and Kubli 2003), and affecting female longevity and survival (Civetta and Clark 2000; Barnes et al. 2008). In addition, females undergo dramatic physiological and behavioral changes after mating (Carvalho et al. 2006; Mattei et al. 2015; Carmel et al. 2016). However, little is known about the female proteins that facilitate these postmating responses and their role in fertilization success (Prokupek et al. 2008; Yapici et al. 2008; Schnakenberg et al. 2011; Findlay et al. 2014). This shortcoming is due in part to the difficulty in isolating and characterizing the interacting female proteins. One approach that is widely used to understand female postmating reproductive processes is to analyze changes in gene expression after mating (McGraw et al. 2004, 2008; Mack et al. 2006; Kapelnikov et al. 2008; Kocher et al. 2008; Dalton et al. 2010; Bono et al. 2011; Thailayil et al. 2011; Alfonso-Parra et al. 2016; Al-Wathiqui et al. 2016; Hollis et al. 2019).
The majority of studies on the transcriptional dynamics in females after mating have been conducted in D. melanogaster. Overall, these studies show that females typically display a transcriptional response within 3 h after mating (McGraw et al. 2004), and the peak response occurs around 6 h after mating (Mack et al. 2006). These changes in female gene expression are induced by a combination of transferred SFPs, sperm, and nonejaculate components of mating (McGraw et al. 2004), the latter potentially including copulatory and behavioral cues. Furthermore, the functional categories of genes that are typically upregulated in mated females include proteases, protease inhibitors, and immune response genes, which suggests that these classes of proteins play important roles in postcopulatory interactions (Lawniczak and Begun 2004; McGraw et al. 2004; Mack et al. 2006; Kapelnikov et al. 2008; Innocenti and Morrow 2009; Delbare et al. 2017; Hollis et al. 2019). Indeed, these functional categories are typically enriched among postmating response genes in species outside the Drosophila genus—such as Apis mellifera (Kocher et al. 2008), Anopheles gambiae (Thailayil et al. 2011; Shaw et al. 2014), Aedes aegypti (Alfonso-Parra et al. 2016), and Ostrinia nubilalis (Al-Wathiqui et al. 2016)—suggesting broad functional conservation of postmating processes among insects. However, it is not clear if the genes that respond to mating in females across a broad range of insect species are orthologous, even among species in the Drosophila genus.
Reproductive interactions between the sexes are often subject to intense postcopulatory sexual selection (Birkhead and Pizzari 2002; Pitnick et al. 2009). In species where females can simultaneously store sperm from multiple males, the reproductive tract becomes an arena for intense selective forces that act among ejaculates from different males (e.g., sperm competition) and between the sexes (e.g., cryptic female choice) (Parker 1970; Eberhard 1996). Furthermore, opposing fitness interests between the sexes that are caused by interlocus sexual conflict can accelerate coevolutionary arms races (Parker 2006; Perry and Rowe 2015; Rowe et al. 2018). Taken together, these forces can generate rapid evolutionary changes between diverging lineages and can ultimately lead to reproductive isolation between closely related species (Markow 1997). Indeed, male reproductive genes evolve rapidly across a broad range of species (Wyckoff et al. 2000; Swanson and Vacquier 2002; Haerty et al. 2007), but the picture is less clear for female reproductive genes. Several studies suggest that some female reproductive tract genes evolve rapidly (Swanson et al. 2004; Kelleher et al. 2007; Prokupek et al. 2008), as would be expected under a scenario of sexual conflict or cryptic female choice within the female reproductive tract, but the prevalence of this phenomenon has not been explored. We specifically lack direct evidence of the consequences of rapid divergence of reproductive molecules and their consequences for reproductive isolation between species.
Many closely related species are reproductively isolated at the gametic level. Some species pairs show competitive gametic isolation such that conspecific males enjoy a fertilization advantage when simultaneously competing for fertilization with heterospecific males (Robinson et al. 1994; Howard 1999; Price et al. 2001; Chang 2004), while other species show noncompetitive gametic incompatibility where heterospecific sperm has reduced fertilizing ability compared to conspecific sperm (Sweigart 2010; Ahmed-Braimah and McAllister 2012; Marshall and Dirienzo 2012; Ahmed-Braimah et al. 2017). These postmating prezygotic reproductive barriers can evolve rapidly between species owing to the strength of postcopulatory sexual selection within species and can play an important role in species isolation early in the speciation process (Turissini et al. 2018). Although the mechanisms that cause gametic barriers are not yet well-understood, they likely involve defects in postcopulatory processes that can be directly impacted by biochemical mismatches between male and female proteins. Thus, species that are reproductively isolated at the gametic level provide a unique opportunity to identify the molecular mechanisms within females that mediate postcopulatory processes.
Examining the postmating regulatory response in females after con- or heterospecific mating can provide clues about the processes that disrupt fertilization in heterospecific crosses. For example, Bono et al. (2011) used females from the cactophilic species, D. mojavesis, to analyze the postmating transcriptional changes that take place in the female reproductive tract after mating to conspecific (D. mojavensis) or heterospecific (D. arizonae) males. They found significant perturbations in gene expression in heterospecifically mated females, potentially indicating failed molecular interactions between male and female proteins. However, linking these perturbations with the gametic incompatibility phenotype in heterospecific crosses requires identifying the genetic causes of the incompatibility and the functional consequences of gene expression changes after mating. Bono et al. (2011) also found that males transfer accessory gland-derived mRNAs to females during copulation: heterospecifically mated females received several mRNA transcripts that were derived from males, potentially suggesting a yet unknown mechanism of reproductive communication. [This phenomenon was also observed in the yellow fever mosquito, Aedes aegypti (Alfonso-Parra et al. 2014)]. In addition to being the first of its kind, this study highlighted the utility of using closely related sister species in studying postmating transcriptional responses and additionally suggests that the peculiar phenomenon of paternal mRNA transfer might be of functional significance.
The Drosophila virilis species complex has recently emerged as an ideal system to study the genetic basis of reproductive interactions and their evolutionary consequences (Ahmed-Braimah et al. 2017). This species group exhibits strong postmating prezygotic reproductive isolation between member species (Sweigart 2010; Ahmed-Braimah and McAllister 2012; Ahmed-Braimah 2016; Ahmed-Braimah et al. 2017), in addition to marked gametic incompatibilities between populations of the same species (Jennings et al. 2011, 2014; Ahmed-Braimah and McAllister 2012; Garlovsky and Snook 2018). Between the closely related pair within the virilis subgroup, D. americana and D. novamexicana, fertilization rate is only ∼1% in heterospecific crosses when D. novamexicana is the female in the cross, and ∼35% in the reciprocal cross (Ahmed-Braimah and McAllister 2012). This species pair diverged ∼0.5 million year ago, and maintains allopatric distributions in the continental United States (Caletka and McAllister 2004). Importantly, gametic isolation is the only detectable reproductive barrier between this species pair, which also exhibits no precopulatory isolation, suggesting that postcopulatory sexual selection is a particularly strong divergence force in these species (Spieth 1951; Ahmed-Braimah and McAllister 2012; McNabney 2013; Ahmed-Braimah et al. 2017).
Here, we analyze the postmating transcript abundance changes in D. novamexicana females after mating to D. novamexicana (conspecific) or D. americana (heterospecific) males. While this species pair is characterized by strong interspecific gametic incompatibility, our goal in this study is to understand the transcriptome profile of the female reproductive tract in unmated and mated females. First, we identify candidate female reproductive tract genes based on tissue-biased expression and analyze their functional categories and patterns of molecular evolution. Second, we characterize the transcript abundance landscape in the female reproductive tract, ovaries, and head after con- or heterospecific mating. Third, we compare our results on postmating transcript abundance changes in the female reproductive tract to two other studies on distantly related species (D. melanogaster and D. mojavensis) to identify genes that show a conserved postmating response across the Drosophila genus. Finally, we identify the set of male-derived mRNAs and examine their tissue origin in males. Our results show that female reproductive tract genes with tissue-biased expression are largely slow evolving in this species group compared to their male counterpart and that the female postmating response after heterospecific mating is highly distinct from conspecific mating due to consistent upregulation of stress response genes shortly after mating.
Results
Evolutionary Dynamics of Female Reproductive Tract Genes
In our first analysis, we use unmated female tissue samples to identify female reproductive tract-biased genes to characterize their functional attributes and evolutionary dynamics.
Female Reproductive Tract Genes Are Enriched for Proteolytic Enzymes and Membrane-Bound Receptors
We defined female reproductive tract (fRT) genes in D. novamexicana based on expression bias in the lower female reproductive tract (bursa, spermathecae, seminal receptacle, and oviduct) compared to other female and male tissues. Specifically, we defined fRT genes as those that show >2-fold mRNA abundance (FDR≤0.01) relative to ovaries, head, gonadectomized female carcass, and male tissues. (We used analogous criteria to identify ovary-biased genes). This classification yielded 148 fRT-biased genes and 766 ovary-biased genes (fig. 1A). Of these 148 fRT-biased genes, 118 have orthologs in D. melanogaster. We performed gene ontology (GO) enrichment analysis on the 148 fRT-biased genes and found that they are enriched for serine-type peptidases, suggesting that the fRT plays roles in dictating proteolytic cleavage of male and/or female compounds (Kelleher and Pennington 2009; LaFlamme et al. 2012). All ten proteolytic enzymes are almost exclusively expressed in the fRT, and comprise six trypsins, three serine proteinases (Stubble-like), and one collagenase (supplementary table S1, Supplementary Material online). One of the trypsins is orthologous to the D. melanogaster Send1/Send2 proteins, which are specifically expressed in the secretory cells of the spermatheca (Schnakenberg et al. 2011) (supplementary table S1, Supplementary Material online). We also found a significant enrichment of plasma membrane-bound proteins, some of which are known receptors that might bind male-derived compounds. For example, one of those receptor proteins is the ortholog of a gustatory receptor in D. melanogaster, Gr39a, which is expressed in gustatory sensilla receptor neurons in D. melanogaster, but shows fRT-specific expression in D. novamexicana and has likely been co-opted into a reproductive function in this species (supplementary table S1, Supplementary Material online).

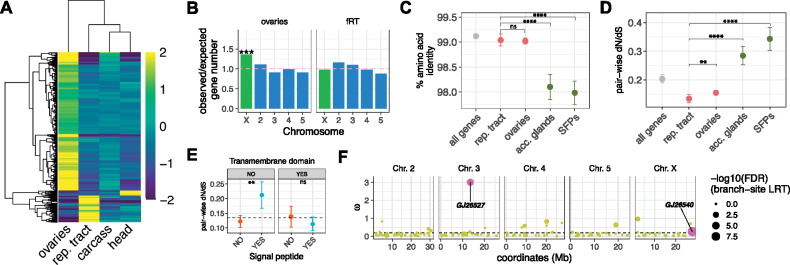
Evolutionary dynamics of female reproductive tract genes. (A) Heatmap of female tissue-biased genes, which shows the transcript abundance as median-centered log2 transcript per million (TPM). The majority of female reproductive genes are expressed in ovaries (n = 766), whereas the fRT contains 148. Only 27 genes show expression bias in the female head. (B) Chromosomal distribution of ovary-biased and fRT-biased genes. The horizontal dashed-line at y = 1 indicates random expectation of observed/expected number of genes on a given chromosome. Significant overrepresentation on a given chromosome is indicated by asterisks (***: P << 0.001). (C) Amino acid conservation between Drosophila americana and Drosophila novamexicana reproductive genes. “SFPs” are the subset of accessory gland-biased genes that contain a predicted signal sequence. Error bars represent standard error. Statistical comparisons are performed between fRT-biased genes and the remaining reproductive genes (ovaries, accessory glands, and SFPs) using a Mann–Whitney U test (****: P < 0.0001; ns: “not significant”). (D) Average pair-wise non-synonymous to synonymous substitution rate (dN/dS) among four classes of reproductive genes (fRT, ovaries, accessory glands, and SFPs) and the genome average. Error bars represent standard error. Statistical comparisons are performed between fRT-biased genes and the remaining reproductive genes (ovaries, accessory glands, and SFPs) using a Mann–Whitney U test (**: P < 0.01; ****: P < 0.0001; ns: “not significant”). (E) Average pair-wise non-synonymous to synonymous substitution rate (dN/dS) among fRT-biased genes, subdivided based on predicted transmembrane domain presence/absence (facet panels) or predicted signal peptide presence/absence (x-axis groups). Error bars represent standard error, and the dotted line represents the average fRT dN/dS ratio prior to subdivision. Statistical comparisons are performed between signal peptide groups within each transmembrane domain classification using a Mann–Whitney U test (**: P < 0.01; ****: P < 0.0001; ns: “not significant”). (F) Gene-wise ω (a proxy for dN/dS) for fRT genes along the five major chromosomes. The dotted line indicates the genome average ω value (0.2). The size of each dot indicates the −log10(FDR) derived from the LRT under a χ2 distribution. The two significant genes (FDR < 0.05) are indicated.
The chromosomal distribution of reproductive genes can often be informative with regards to the evolutionary forces that impact them. For instance, male reproductive proteins—particularly SFPs—tend to be under-represented on the X chromosome (Ravi Ram and Wolfner 2007; Ahmed-Braimah et al. 2017). We found that fRT-biased genes—unlike ovary-biased genes—are not overrepresented on the X chromosome, suggesting that their chromosomal distribution is not restricted by sex-specific selection (fig. 1B).
To examine the repertoire of reproductive genes in both sexes, we performed the expression bias analysis separately using male and female tissues. We were surprised to find that several genes show expression bias in the female reproductive tract relative to other female tissues, and also in male reproductive tissues relative to other, nonreproductive male tissues (fig. 2A). To explore this further, we examined the overlap of reproductive genes that are shared between the sexes. Strikingly, we found that 14, 31, and 20 genes that show higher expression in the female fRT within females also show higher expression in the accessory-glands, ejaculatory bulb, and testes, respectively, within males (fig. 2B). Several of these genes show exclusive expression in the fRT in females and a corresponding male reproductive tissue (examples in fig. 2C).

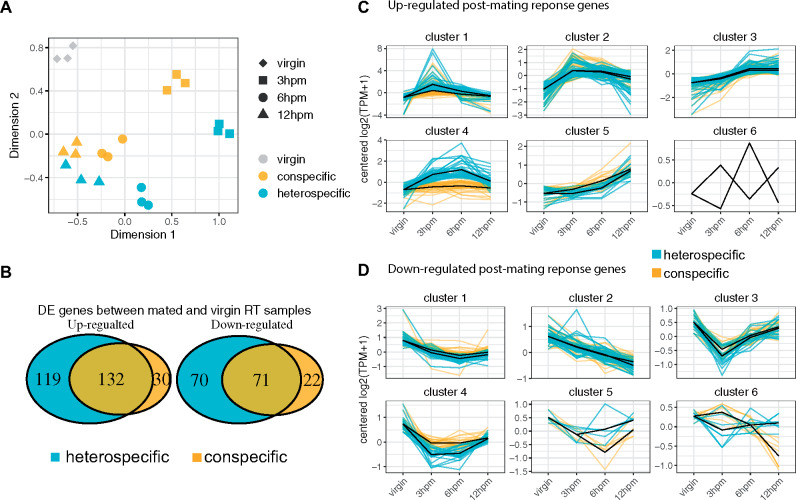
Postmating transcript abundance profiles differ between conspecific and heterospecific crosses. (A) Multidimensional scaling (MDS) plot of unmated and postmating fRT samples, using the first two dimensions. The key on the right indicates the postmating time-point or unmated sample (shape) and the cross type (color). The plot shows distinct grouping of the two cross types, especially at earlier time points. (B) Venn diagram depicting the overlap between all post-mating DE genes, and shows that many more genes respond to heterospecific mating compared to conspecific mating. K-means clustering of (C) up-regulated and (D) down-regulated postmating DE genes. Conspecific and heterospecific transcript abundance responses are plotted together and are distinguished by color (see key). Solid black lines represent the average abundance profile for each of the two cross types. Cluster 4 contain most of the genes that respond uniquely heterospecific mating.
These results show that our classification of female reproductive tract genes using mRNA expression bias can provide robust candidates for analyzing female reproductive genes. These results also show that using sex-bias as a criterion for defining reproductive genes might miss a set of reproductive genes that are shared between the sexes.
Female Reproductive Tract Genes Show a Reduced Rate of Sequence Divergence Compared to Male Accessory Gland-Derived Genes
We analyzed the rate of nucleotide divergence of female reproductive genes (fRT-biased and ovary-biased) by examining 1) conservation of amino acid sequence between D. americana and D. novamexicana, 2) the average pair-wise ratio of nonsynonymous to synonymous substitutions between D. americana and D. novamexicana (dN/dS), and 3) evidence of positive natural selection using the “branch-site” test implemented in PAML (Zhang et al. 2005).
First, we found that fRT- and ovary-biased genes are highly conserved but are slightly less conserved than the genome average (fig. 1C, P = 0.024; Mann–Whitney U test; fRT vs. all genes comparison). In contrast, male accessory gland genes (n = 191), especially those that contain a predicted secretion signal (n = 71), are less conserved compared to fRT genes and the genome average (fig. 1C, P << 0.0001; Mann–Whitney U test). Second, the average dN/dS ratio is lower in fRT genes compared to the genome average, although this difference is not significant (fig. 1D, P = 0.2; Mann–Whitney U test). Ovary-biased genes have a significantly higher dN/dS ratio relative to fRT-biased genes (fig. 1D, P = 0.003; Mann–Whitney U test), but significantly lower than the genome average (fig. 1D, P = 0.01; Mann–Whitney U test). Importantly, fRT-biased genes have a significantly lower dN/dS ratio compared to male accessory gland genes and predicted SFPs (fig. 1D, P << 0.0001; Mann–Whitney U test). We examined dN/dS ratios among fRT-biased genes after subdividing them based on presence/absence of a predicted signal peptide and a transmembrane domain, and found that fRT-biased genes that contain a predicted signal peptide but lack a predicted transmembrane domain have a significantly higher dN/dS ratio compared to the remaining fRT-biased genes (fig. 1E, P = 0.008; Mann–Whitney U test).
Finally, we performed the “branch-site” test in PAML to examine evidence of positive selection at any of the 148 fRT-biased genes. We performed the test using the core D. virilis species subgroup (D. americana, D. lummei, D. novamexicana, and D. virilis) and found that only two genes show evidence of a significant signature of positive selection on the branches leading to D. novamexicana or its sister species—this is in contrast to three accessory gland-biased genes that show evidence of positive selection using the branch-site test (Ahmed-Braimah et al. 2017). One gene (GJ26540; LRT = 52.3, FDR = 6.2 × 10−10) resides on the X chromosome, has a substitution ratio that is equal to the genome average, and has no known ortholog or function. The other gene (GJ26527; LRT = 26.1, FDR = 0.0001) resides on chromosome 3, has the highest ω ratio among fRT-biased genes, and is orthologous to Titin, which is a muscle-associated protein. Thus, we conclude that fRT genes tend to be conserved relative to male reproductive genes in this species group, but a small subset of genes could experience rapid bouts of selection.
Postmating Transcript Abundance Changes in Females after Con- or Heterospecific Matings
Next, we analyzed the postmating transcript abundance changes that occur in D. novamexicana females after mating with a conspecific male or a heterospecific male. This allows us to identify the impact of divergent reproductive male molecules—or other divergent male traits—on postmating transcript abundance in females.
Heterospecifically Mated Females Show a Distinct Transcript Abundance Profile in the fRT Compared to Conspecifically Mated Females
We collected mRNA abundance data from the fRT after two distinct mating conditions (conspecific or heterospecific), at three different time points after mating (3 h, 6 h, and 12 h), and from unmated females (virgin) as a control. First, after filtering transcripts with low counts, we examined the grouping of replicates within each sample using a Pearson correlation matrix to assess the suitability of our downstream analyses and found that replicates do indeed cluster as expected (supplementary fig. S3B, Supplementary Material online). Next, we examined replicate and sample groupings by constructing a multidimensional scaling (MDS) plot of fRT data using the filtered counts, and observe distinct clustering of postmating samples based on the cross type and postmating time point (fig. 2A). Specifically, early postmating time points (3 hpm and 6 hpm) were clearly distinct from the virgin control sample and showed the strongest difference between the two cross types. At 12 hpm the two cross types were somewhat congruent and clustered closer to each other than the two preceding time points. These observations suggest that, shortly after mating, the transcript abundance profile between females mated to a conspecific or a heterospecific male are highly distinct.
Next, we performed pair-wise differential abundance analysis between each postmating time point and the virgin control sample for both cross types separately. We define significantly differentially expressed (DE) genes as those with >2-fold change in abundance (FDR < 0.05) in the postmating sample relative to the virgin control. We found many more genes that are up-regulated after mating (281) than down-regulated (163) (fig. 2B). Furthermore, the number of DE genes in the heterospecific mating samples is greater than the conspecific samples: 251 upregulated and 141 downregulated in heterospecific matings, 162 upregulated, and 93 downregulated in conspecific matings, and 132 shared upregulated and 72 shared downregulated in both cross types (fig. 2B).
We performed GO analysis for all up-regulated genes and found several enriched GO categories (supplementary fig. S4A, Supplementary Material online; FDR < 0.05). Prominent among these were terms associated with the proteasome complex, which is involved in the degradation of poly-ubiquitinated proteins in the nucleus and the cytosol (Tsakiri et al. 2013). Seventeen genes that code for various subunits of the proteasome core complex gradually increase in abundance after mating to either a con- or heterospecific male; however, the magnitude of the increase is markedly higher among heterospecifically mated females than conspecifically mated ones (supplementary fig. S4B, Supplementary Material online). The amplified response in the heterospecific cross is also observed for other genes that significantly respond to mating in both con- and heterospecific crosses (supplementary fig. S5, Supplementary Material online). Other enriched GO terms represent additional proteolytic processes and the immune response (see below). Only two GO terms are significant among down-regulated genes: oxidoreductase and alanine-glyoxylate transaminase activity.
To further explore the postmating expression profiles across the three time-points, we performed separate k-means clustering of the up- and down-regulated DE genes, with k = 6 as estimated by the “elbow” method (Syakur et al. 2018) (fig. 2C and D, respectively). This analysis revealed that the expression profiles between the con- and heterospecific cross types are largely congruent, but two clusters in the upregulated set (cluster 1 and cluster 4) contained several genes that show distinct expression profiles between the con- and heterospecific cross types (fig. 2C). We performed GO enrichment analysis on the up-regulated clusters and found that clusters 2, 3, and 4 were enriched for several GO categories. Cluster 2 contained several terms related to stress response mechanisms (e.g., response to hypoxia, topologically incorrect proteins, and acid chemicals), which are primarily driven by several heat shock proteins that are up-regulated at 3 hpm in both con- and heterospecific crosses (supplementary table S3, Supplementary Material online). Cluster 3 was enriched for terms related to proteolytic activity and the proteasome complex, which reflect the overall composition of all up-regulated transcripts. Notably, cluster 4 primarily contained an enrichment of immune response genes, and these appear to show the most distinct abundance response between con- and heterospecific cross types.
To probe the abundance differences between the con- and heterospecific cross types directly, we performed DE analysis between the two postmating samples at each time-point. We identified 65 genes that show significantly higher abundance in the heterospecific samples across all time-points, and only 15 that show significantly higher abundance in the conspecific sample (supplementary fig. S4C, Supplementary Material online). Of the 65 genes that show higher abundance in the heterospecific cross, 47 are significant at the earliest time-point (3 hpm). We performed GO enrichment analysis on these 65 genes and recovered significant enrichment of immune response terms (supplementary fig. S4D, Supplementary Material online). These immune genes include the NF-κB signaling protein, Relish, which is a transcription factor that is a master regulator of immunity through the Imd pathway (Hetru and Hoffmann 2009) (fig. 3A). Several immune effectors that act as antimicrobial peptides (Attacin, Cecropin, Defensin, and Diptericin) are upregulated distinctly in the heterospecific cross, and show little or no response after conspecific mating (fig. 3A). Furthermore, other classes of immune-related genes show a distinct response in the heterospecific cross, including a recognition protein with beta-glucan binding activity [Gram-negative bacteria binding protein (GNBP)] and a coagulation effector protein (Tiggrin) (supplementary table S3, Supplementary Material online).

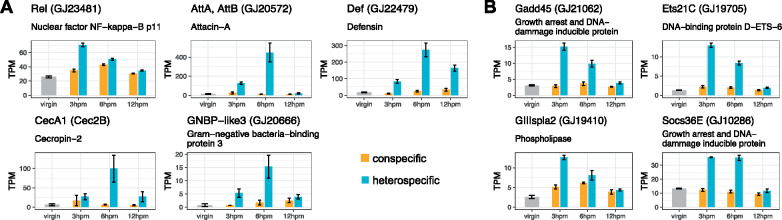
Misregulated postmating genes in the fRT. (A) The known immune response genes and (B) stress response genes that are up-regulated in the heterospecific cross but remain largely unchanged in the conspecific cross. The Drosophila melanogaster ortholog gene name is shown, followed by the Drosophila virilis gene name and a gene description derived from a SwissProt database search. Error bars represent standard error, and expression is in TPM units.
We also found that several genes that are involved in some stress response pathways in Drosophila are up-regulated in the heterospecific cross but mRNA levels remain largely unchanged in the conspecific cross. These include a negative regulator of the Janus Kinase/Signal Transduction and Activator of Transcription (JAK/STAT) signaling pathway, Socs36E, which shows a ∼3-fold increase in abundance at 3 hpm and 6 hpm only after heterospecific mating (fig. 3B). Another stress response gene is a phospholipase, GIIIspla2, which acts as a downstream target of various stress response pathways and metabolizes phospholipids.
These results show that the two cross types largely induce congruent changes in transcript abundance that are likely required for normal postmating processing, but that the heterospecific cross induces additional abnormal changes in transcript abundance that indicate a heightened stress response.
Several Genes Are Mis-Regulated in Ovaries after Heterospecific Mating
We analyzed transcript abundance changes in ovaries and female heads after con- or heterospecific mating at 6 hpm. In D. melanogaster, mating increases oogenesis and stimulates ovulation in part through the action of transferred male SFPs (e.g., Sex Peptide and ovulin; Soller et al. 1997; Heifetz et al. 2000; Rubinstein and Wolfner 2013). We therefore reasoned that heterospecific mating could cause mis-regulation in ovaries—that is, transcript abundance changes that are induced in the conspecific cross might fail induction in the heterospecific cross.
Our analysis of the ovaries revealed that, indeed, transcript abundance profiles of mated female ovaries at 6 hpm are distinct between the two cross types: conspecifically mated female samples cluster separately from heterospecifically mated females (supplementary fig. S6A and B, Supplementary Material online). Differential expression tests revealed that more genes are differentially abundant after conspecific mating relative to virgin (96 up-regulated and 25 down-regulated; supplementary table S4, Supplementary Material online), compared to heterospecific mating relative to virgin (23 up-regulated and 4 down-regulated; supplementary table S4, Supplementary Material online). However, when we compared the two postmating samples against each other (conspecific vs. heterospecific), we found that only 11 genes show higher abundance in the conspecific sample. Of the 11 genes with higher abundance in the conspecific sample, three are orthologous to Cytochrome C oxidase subunit 1 or 2 (fig. 4). In addition, one of the 11 genes is orthologous to the D. melanogaster actin gene, Act79B, which localizes to the actin cytoskeleton and is often expressed in muscle tissue (Dohn and Cripps 2018). Conversely, only one gene—an fRT-biased, textilinin-like protease inhibitor—shows higher abundance in the heterospecific sample compared to the conspecific sample. The D. melanogaster ortholog of this gene has not been characterized, but is a predicted kunitz-type serine protease inhibitor that is predicted to localize to the extracellular matrix (FlyBase.org).

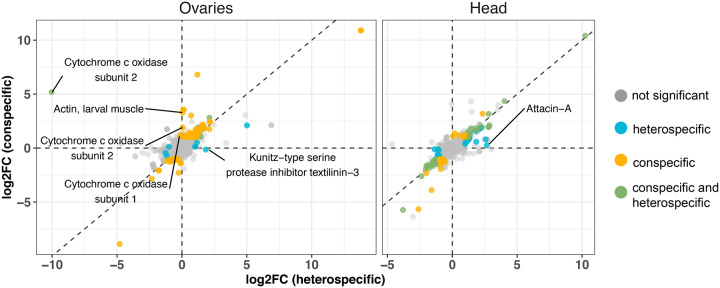
Log-fold change in the ovaries and female head at 6 hpm. The log2 fold-change of heterospecific crosses is represented on the x-axis, and the log2 fold-change of conspecific crosses is represented on the y-axis. Each point represents a single gene, and the DE status is indicated by color (see legend). “Normal” gene data points are defined as those that are DE in both the conspecific and heterospecific cross when compared to the unmated control, and show a log2 fold-change in the same direction. “Conspecific” and “heterospecific” gene data points refer to those that are significantly DE between the conspecific postmating sample—and not the heterospecific sample—and the unmated control, or between the heterospecific sample—and not the conspecific sample—and the unmated control, respectively. Genes that are significantly differentially expressed between the conspecific and the heterospecific sample are indicated by text.
We performed GO enrichment analysis of up-regulated genes in the ovaries and, as expected, recovered significant terms associated with developmental processes involved with reproduction and vitelline membrane formation (supplementary table S6, Supplementary Material online). We also recovered terms associated with the immune response, and this enrichment was driven by ten genes—for example, two peptidoglycan-recognition proteins, thioester-containing protein, and Cathepsin—that were similarly up-regulated in the con- and heterospecific cross. Interestingly, one antimicrobial peptide, Cecropin 2B, is up-regulated in the conspecific sample after mating (∼2k-fold) but massively up-regulated after the heterospecific cross (∼17k-fold; supplementary table S4, Supplementary Material online).
These findings suggest that heterospecific mating also induces distinct postmating transcriptional changes in ovaries, and particularly fails to induce some “normal” changes that are observed in the conspecific cross.
A Single Antimicrobial Peptide is Detected as Mis-Regulated in the Female Head after Heterospecific Mating
Insect females undergo various behavioral changes after mating that might reflect underlying transcriptional changes in the head. Indeed, female D. novamexicana up-regulate 51 genes and down-regulate 56 genes at 6 h postmating in head tissue (supplementary table S7, Supplementary Material online). Notably, the postmating transcript abundance changes in the female head are almost identical after con- or heterospecific mating and replicates do not cluster by cross (supplementary table fig. S6B, Supplementary Material online). We sought to examine whether the two cross types induce different transcript abundance patterns at 6 hpm, but found that only one gene shows a significant increase in the heterospecific cross but remains unchanged in the conspecific cross: the antimicrobial peptide Attacin-A. This gene is also up-regulated in the fRT after heterospecific mating but not conspecific mating, suggesting that the mechanism of induction could be organism-wide.
Few Genes Have a Conserved Response to Mating in D. novamexicana, D. mojavensis, and D. melanogaster
We have identified several orthologs of D. novamexicana genes in D. mojavensis and D. melanogaster that show a significant postmating response in at least two out of the three species. First, we compared the list of significant genes in Bono et al. (2011) to significant genes in the current study and found that only 4/18 significant genes in D. mojavensis are also significant in D. novamexicana and change in abundance in the same direction (FBgn0197613, FBgn0209253, FBgn0209991, and FBgn0211115). Unfortunately, none of these genes have a known function in D. melanogaster (CG7685, CG6770, CG6972, and CG6770).
Second, we identified orthologs of D. novamexicana genes in D. melanogaster in the only study thus far that examined postmating transcript abundance changes in the fRT (Mack et al. 2006). The comparison with the Mack et al. (2006) data set revealed six genes that respond similarly to mating in the D. melanogaster and D. novamexicana fRT: Hsp26, CG16721, CG2918, CG7840, CG2471, and Phk-3. Phk-3, or Pherokine 3, increases in abundance in D. melanogaster in response to viral and bacterial infection (Sabatier et al. 2003). CG2471—also known as Sclp—is thought to be involved in muscle system processes (Montana and Littleton 2006), which is consistent with a role in fRT remodeling after mating (Mattei et al. 2015).
Males Transfer Testes-Biased mRNAs to Females during Copulation
We identified several genes that were “up-regulated” after mating and that exhibited high expression in male reproductive tissues. This prompted us to examine whether these transcripts are transferred from the male during copulation. First, we identified all mRNAs that were up-regulated in the fRT of mated females at 3 hpm and gradually decreased in abundance in subsequent time points, which is an abundance pattern that may indicate transfer from males and subsequent decay or usage (see cluster 1 and 2 in fig. 2C). We confirmed—using species-specific single nucleotide polymorphisms (SNPs)—that 13 of these mRNAs are male-derived (supplementary table S7, Supplementary Material online). Notably, three of these encode heat shock proteins (Hsp23, Hsp68, and Hsp70Ab) that show testis-biased expression but have a relatively low normalized abundance (transcripts per million, or “TPM” ∼7–60, supplementary table S4, Supplementary Material online). All but two of the remaining genes also show strong testis-biased expression but high relative normalized abundance (TPM ∼280–61,000, supplementary table S7, Supplementary Material online).
These results suggest that paternally derived mRNA transcripts that are found in the mated fRT may originate from testes, which shows that testes can contribute nonsperm components to the ejaculate.
Discussion
Here, we investigated the mRNA abundance patterns within the reproductive tract of unmated D. novamexicana females and females mated to either conspecific or heterospecific (D. americana) males. This species pair is well-suited for genetic investigations of postcopulatory interactions as they have diverged recently (∼0.5 mya) and have evolved strong gametic incompatibilities (Caletka and McAllister 2004; Ahmed-Braimah and McAllister 2012). These incompatibilities manifest as reduced fertilization and premature loss of sperm from storage after heterospecific mating (Ahmed-Braimah and McAllister 2012). Because these incompatibilities are likely caused by a mismatch between the male ejaculate and the female reproductive tract, here we sought to identify incongruent transcript abundance changes in females after mating with conspecific or heterospecific males to identify potential molecular mechanisms within the female that mediate reproductive success. We also used the mRNA abundance data to identify fRT-biased genes, as these are likely to have specialized reproductive functions.
One of the most pervasive patterns in molecular evolution is the rapid divergence of reproductive genes in a wide variety of organisms (Swanson et al. 2001; Swanson and Vacquier 2002; Haerty et al. 2007). Male reproductive genes in Drosophila, particularly SFPs, are a classic example of this phenomenon. However, little is known about the evolutionary dynamics of female reproductive tract genes because they are not well-characterized across a broad range of species. Studies in other Drosophila species show that some fRT genes, particularly those that code for proteases, show elevated rates of amino acid substitution (Swanson et al. 2004; Kelleher et al. 2007; Prokupek et al. 2008, 2010). Thus, these rapidly evolving fRT genes might directly interact with male seminal proteins. Here, we do not observe a strong signature of rapid divergence in fRT genes between D. americana and D. novamexicana: most fRT genes have ω values that are below the genome average and below the 0.5 threshold used by studies in D. melanogaster (Swanson et al. 2004; Prokupek et al. 2008, 2010). These observations might be due to differences in divergence patterns between these disparate species or they might be due to methodological differences in identifying fRT genes: we used a strict expression bias criterion to identify fRT genes, whereas other studies in Drosophila used expressed sequence tags (ESTs) from fRT tissues. In addition, we found that categorizing fRT genes based on predicted secretion signal and transmembrane domains results in significantly different dN/dS ratios: fRT-biased genes whose predicted proteins contain a signal peptide but lack a transmembrane domain have an elevated dN/dS ratio compared to those without a signal peptide. Still, these elevated dN/dS ratios are close to the genome average ratios and well below those observed for male accessory gland genes. The overall pattern of reduced divergence in fRT-biased genes could be explained by selective constraints due to pleiotropy, that is, fRT-biased genes might be playing other roles in the female that limit their sequence variation. We have attempted to minimize this possibility by applying a strict expression bias criteria, but still some fRT-biased gene transcripts show appreciable abundance in other female tissues. A better understanding of these fRT-biased genes would allow us to disentangle the possible explanations of this observed sequence divergence pattern.
Our analysis of the postmating transcript abundance changes shows that some of the same functional categories of genes that are up-regulated in other insect taxa are also up-regulated in D. novamexicana females after mating (McGraw et al. 2004, 2008; Mack et al. 2006; Kapelnikov et al. 2008; Kocher et al. 2008; Dalton et al. 2010; Al-Wathiqui et al. 2014; Alfonso-Parra et al. 2016; Delbare et al. 2017). A commonly enriched functional class is proteolytic enzymes, which are thought to process peptides and activate enzymatic reactions within the female reproductive tract (Kelleher and Penningto 2009). However, we did not identify an appreciable number of orthologous genes that respond to mating in D. novamexicana females and also respond to mating in D. mojavensis (Bono et al. 2011) or D. melanogaster (Mack et al. 2006) females. This lack of congruence in gene identity suggests that divergent reproductive mechanisms might underlie the female postmating response, even though the overall functional categories of postmating response genes might be conserved. This might also reflect a difference in methodology and approach between these studies and a refined comparison is warranted to compare postmating responses across disparate species.
Another class of genes that is often up-regulated after mating is immune response genes (Lawniczak and Begun2004; McGraw et al. 2004; Mack et al. 2006; Kapelnikov et al. 2008; Alfonso-Parra et al. 2016; Delbare et al. 2017). Our results show that postmating induction of immune effector genes in the female reproductive tract is largely determined by male genotype: heterospecific males induce a heightened immune response in the female reproductive tract that peaks at 6 hpm, whereas conspecific males induce a mild immune response. The downstream activation of effector immune genes such as antimicrobial peptides (AMPs) is triggered by the transcription factor, Relish, which is an NF-κB Imd pathway activator of immune defenses, typically against Gram-positive bacteria and fungi (Hetru and Hoffmann 2009). Relish mRNA levels significantly increase at 3 hpm after heterospecific, but not conspecific mating, suggesting that AMP activation after heterospecific mating is likely triggered through the Imd pathway.
The initiation of immune defenses after mating in insects has been widely regarded as a mechanism to thwart potential infection during copulation and can potentially trigger an investment trade-off between immune defense and reproduction (Siva-Jothy et al. 1998; McKean and Nunney 2001; Rolff and Siva-Jothy 2002; Schwenke and Lazzaro 2017). However, recent evidence suggests that the magnitude and direction of postmating immune responses varies across study systems and may have additional explanations (Fedorka et al. 2007; Lawniczak et al. 2007; Innocenti and Morrow 2009; Morrow and Innocenti 2012; Hollis et al. 2019; Oku et al. 2019). In particular, several studies suggest that male ejaculate components can be “immunogenic” such that they are directly triggering immune responses in females (Morrow and Innocenti 2012). Our results support this hypothesis, and specifically show that postmating immune responses are elevated as a consequence of heterospecific mating compared to the immune response observed in conspecific mating, where male seminal proteins have diverged from their conspecific counterparts. Furthermore, our results indicate that additional stress responses through alternative pathways (e.g., JAK/STAT) are exacerbated in the heterospecific cross and may be a consequence of divergent ejaculate components. In addition to ejaculate components, however, copulation itself can induce a variety of responses in females (McGraw et al. 2004), and copulation between species can conceivably cause a distinct response if copulatory structures have diverged between species (Masly and Kamimura 2014). Furthermore, postmating immune responses could also be caused by ejaculate-derived microbes: D. americana and D. novamexicana might differ in their ejaculate microbiome composition and thus elicit distinct responses after mating. This hypothesis is difficult to assess at this time as ejaculate microbiomes have not been characterized in Drosophila. However, a recent study shows that some immune genes in D. melanogaster are up-regulated after mating even if the male is germ-free, indicating that copulation and/or ejaculate proteins are sufficient to initiate a postmating immune response (Delbare et al. 2020). Additional premating and copulatory cues may have diverged between species and we cannot rule out the possibility that these divergent cues induce distinct postmating responses in females mated to con- or heterospecific males. Finally, because we use a single strain from each species to examine the postmating female response, we may have only captured a subset of the possible modalities of postmating transcript abundance changes within and between species. Overall the D. novamexicana system provides a unique opportunity to investigate the consequences of postmating immune activation and can provide a tractable experimental system to uncouple hypotheses of immune activation as a reproductive process or pathogen defense mechanism (Lawniczak et al. 2007; Oku et al. 2019).
During copulation, males are thought to transfer a cocktail of sperm and SFPs, but recent work has shown that males can also transfer mRNAs that can be detected in the female’s postmating transcriptome (Bono et al. 2011; Alfonso-Parra et al. 2014). In both of the reported cases in insects, the mRNAs appear to originate from the male accessory glands as the mRNAs are derived from known SFP genes or show strong expression bias in the accessory glands. We examined transcripts that were identified as up-regulated after mating in D. novamexicana and confirmed that these were transferred during copulation. Surprisingly, we found that these transcripts often show strong testis-biased expression. Previous work in mammals has identified spermatozoal associated mRNAs (Zhao et al. 2006; Dadoune 2009), and these mRNAs share functional properties with mRNAs that were identified in D. melanogaster sperm, such as ribosomal proteins (Fischer et al. 2012). Our results show that ejaculate mRNAs may originate from testes, such as heat shock proteins, and suggest that nonsperm components of the ejaculate can originate in the testes. However, some of these transcripts are also expressed in the accessory glands and may have originated there instead, despite the strong testes expression. It is not clear if the transfer of mRNAs in the male ejaculate has functional significance, and more work is needed to rule out that this mRNA transfer simply reflects the presence of cellular debris from the testes or accessory glands.
The heterospecific gametic incompatibility between D. novamexicana and D. americana manifests as an inability of sperm to fertilize eggs, even though copulation and sperm storage proceed normally (Ahmed-Braimah and McAllister 2012). The results that we present here show that heterospecific mating 1) induces a stress/immune response in the fRT, 2) amplifies the abundance of a set of genes that are also up-regulated in the conspecific cross, and 3) fails to up-regulate a set of genes in ovaries. Any of these mechanisms can contribute to the observed failure of fertilization between species. For example, the heightened immune defenses may render sperm incapacitated and unable to fertilize, or some genes that are normally expressed in ovaries after mating and are required for normal fertilization are not activated in heterospecific crosses and thus fertilization fails. While a direct link between the results presented here and the gametic incompatibility remains unclear, we have made significant progress in understanding the fRT transcriptional landscape and can formulate several hypotheses about the nature of gametic isolation in this species group. Overall, this study lays the groundwork for future investigations into the genetic basis of gamete interactions in Drosophila.
Materials and Methods
Single-Pair Matings and Dissections
The D. novamexicana (15010-1031.04) and D. americana (SB02.06) strains were maintained at a constant temperature (22°C) in a 12-h day/night cycle on cornmeal/sucrose/yeast media. Virgin males (D. americana and D. novamexicana) and females (D. novamexicana only) were collected under CO2 anesthesia within a day after eclosion and housed in single-sex groups of ∼20. On day 12 postecolsion, individual males and females were paired without anesthesia and mating was observed. D. novamexicana females were either mated to a D. novamexicana male (conspecific), a D. americana male (heterospecific), or were unmated (virgin). All matings were performed in the morning between 8:00 a.m. and 11:00 a.m., and males were removed immediately after mating. Females were subsequently allocated to one of three postmating dissection time points: 3, 6, and 12 h postmating (hereafter 3 hpm, 6 hpm, and 12 hpm). Virgin females were also allocated to the three dissection time points to control for dissection time. At each postmating time point, conspecific/heterospecific/virgin females were individually anesthetized and immediately dissected in 1×PBS. The lower reproductive tract (bursa, seminal receptacle, spermathecae, and lower oviduct) was extracted, then the ovaries were removed, and finally, the head was severed from the thorax; the three tissues were placed in separate tubes containing ice-cold TRIzol. In addition to those three tissues, the gonadectomized carcass was also preserved for the virgin sample (supplementary fig. S1, Supplementary Material online). Each virgin and postmating reproductive tract sample contained three replicates, and ≥100 reproductive tracts were pooled for each replicate of each treatment. The head, ovaries and carcass samples had two replicates and contained pooled tissues from ∼50 individuals.
RNA Isolation, Library Preparation, Sequencing, and Mapping
Total RNA was extracted from each sample using TRIzol reagent following manufacturer guidelines (Invitrogen, Carlsbad, CA). Single-end, strand-specific mRNA libraries were prepared using the Illumina TruSeq Stranded mRNA library kit (cat. no. 20020594) following manufacturer guidelines. Libraries were sequenced to 100 bp read lengths on an Illumina HiSeq 2500 at the Cornell Biotechnology Resource Center. Raw reads were first processed by filtering clusters from low-quality tiles on the flow cell. Subsequently, the first 10 bases of each read were clipped, followed by quality-trimming at both ends to a minimum PHRED quality score of 20. Processed reads were mapped to the D. virilis transcriptome (FlyBase r1.06) using Bowtie2 v2.2.2 (Langmead and Salzberg 2012) with the following parameters: “-no-mixed –no-discordant –gbar 1000 –end-to-end -k 200 –score-min L,-0.6,-1.6.” Finally, a genome-guided de novo transcriptome was generated using Trinity r20140717, using reads generated from the virgin and conspecific samples, in combination with male D. novamexicana reads from a previous study (SRP100565; Ahmed-Braimah et al. 2017).
Differential Expression Analysis
Tissue-Biased Genes
To identify genes in D. novamexicana with female tissue-biased expression, we used a filtered count matrix (CPM > 5) of male and virgin female tissue samples. The differential expression tests used a design matrix where the focal tissue is compared against all other male and female tissues. Gene-wise differential expression tests were performed by fitting a quasilikelihood negative binomial generalized log-linear model to the filtered count data and subsequent gene-wise F-test, implemented in edgeR (glmQLFit and glmQLFTest; Robinson et al. 2010). Female tissue-biased genes are defined as genes that have >2-fold mRNA abundance and FDR <0.01 when compared to other female tissues in pair-wise comparisons. We performed tissue-bias tests using female samples and separately using tissues from the two sexes. We also include an analysis of tissue-biased genes from male reproductive organs that were obtained from a previous study (Ahmed-Braimah et al. 2017).
Postmating Transcript Abundance
To analyze the postmating transcript abundance landscape in females, we analyzed six female reproductive tract samples (three conspecifics and three heterospecifics), four ovary samples (two conspecifics, two heterospecifics), and four head samples (two conspecifics, two heterospecifics). We compared these postmating transcript abundance states in each tissue to their respective unmated control samples, in addition to comparisons between the conspecific and heterospecific postmating samples. For each tissue type, we created a subsetted matrix that only included the tissue to be analyzed and filtered the genes to only include rows where the CPM value was >5 in at least 3 (female RT) or 2 (head and ovary) replicate samples. For the differential expression analysis, we implemented the removal of erroneous variation between samples using the RUVseq package (Risso et al. 2014) with k = 2 (supplementary fig. S3A, Supplementary Material online). We included the residuals from this model in the design matrix that was used to fit a quasilikelihood negative binomial generalized log-linear model to the count data (edgeR). We then performed differential expression (DE) contrasts between the con- and heterospecific postmating samples at each time point or between postmating samples and the virgin sample and classified significantly differentially expressed genes as those that were up- or down-regulated by >2-fold and FDR <0.05.
Transcript Annotations and Gene Ontology (GO) Analysis
Custom annotations of the D. virilis genome (FlyBase version 1.06) were produced in a previous study (Ahmed-Braimah et al. 2017). These annotations included UniProt orthologies and orthologs against the D. melanogaster genome that were checked against the FlyBase orthology calls. These annotations also included gene ontology (GO) associations for protein sequences that contain an orthologous hit in the UniProt database. GO ontology enrichment analyses were performed using the GOseq R package (Young et al. 2010).
Molecular Evolution Analysis
The rate of nucleotide and amino acid substitution between species in the D. virilis group was calculated using custom Perl scripts and BioPerl libraries (“SAPA.pl”. See GitHub repository below). Whole-genome DNA-seq data from the four group members (D. virilis, D. lummei, D. americana, and D. novamexicana) was aligned to the D. virilis genome and known coding gene sequences (CDS) were extracted. We then generated multiple sequence alignment for each CDS and performed pairwise dN/dS between D. americana and D. novamexicana, and calulcated ω (a close proxy for dN/dS; Yang 2007) across the subgroup phylogeny using PAML (Yang 2007). We also used the multiple sequence alignments to calculate the pair-wise percent conservation of amino acids from cDNA sequences using our custom Perl script. Finally, we performed the “branch-site” test along each branch of the phylogeny to test for the impact of natural selection on each gene (Zhang et al. 2005). To identify genes with a significant signature of positive selection, we performed a likelihood ratio test (LRT) between a model with ω = 1 and a model where ω is estimated from the data, and derived P-values using the χ2 distribution.
Analysis of Transferred Paternal mRNAs
To identify paternally transferred mRNAs we first identified genes that show higher abundance in the fRT at 3 hpm compared to virgin, and only analyzed genes that show a decline in abundance at subsequent time points, which would indicate that the transcript is either being degraded or processed. We then analyzed single nucleotide polymorphisms (SNPs) that are D. americana-specific and identified whether mapped reads from the heterospecific 3 hpm sample contained these SNPs; if the sequence reads in the heterospecific 3 hpm sample contains D. americana-specific SNPs, then we infer that these reads—and thus, the mRNA that produced them—are paternally derived.
Acknowledgments
We thank Amanda Manfredo for technical assistance, Sofie Delbare for discussion and comments on the manuscript, the Cornell Biotechnology Resource Center for sequencing and computational resources, and members of the Clark and Wolfner labs for comments and suggestions. We especially thank four anonymous reviewers for helpful comments and suggestions that significantly improved the manuscript. This work was supported by NIH grant R01-HD059060 to A.G.C and M.F.W.
Data and Script Availability
The Illumina sequence reads are available through the Sequence Read Archive (SRA) under project accession PRJNA611072. The processed data files and analysis scripts are available through a GitHub repository: github.com/YazBraimah/DnovPmRNAseq.
 Differences in Postmating Transcriptional Responses between Conspecific and Heterospecific Matings in Drosophila
Differences in Postmating Transcriptional Responses between Conspecific and Heterospecific Matings in Drosophila