
- Altmetric
The roles of clock components in salt stress tolerance remain incompletely characterized in rice. Here, we show that, among OsPRR (Oryza sativa Pseudo‐Response Regulator) family members, OsPRR73 specifically confers salt tolerance in rice. Notably, the grain size and yield of osprr73 null mutants were significantly decreased in the presence of salt stress, with accumulated higher level of reactive oxygen species and sodium ions. RNA sequencing and biochemical assays identified OsHKT2;1, encoding a plasma membrane‐localized Na+ transporter, as a transcriptional target of OsPRR73 in mediating salt tolerance. Correspondingly, null mutants of OsHKT2;1 displayed an increased tolerance to salt stress. Immunoprecipitation‐mass spectrometry (IP‐MS) assays further identified HDAC10 as nuclear interactor of OsPRR73 and co‐repressor of OsHKT2;1. Consistently, H3K9ac histone marks at OsHKT2;1 promoter regions were significantly reduced in osprr73 mutant. Together, our findings reveal that salt‐induced OsPRR73 expression confers salt tolerance by recruiting HDAC10 to transcriptionally repress OsHKT2;1, thus reducing cellular Na+ accumulation. This exemplifies a new molecular link between clock components and salt stress tolerance in rice.
A member of the pseudo response regulator (PRR) family in rice is induced upon salt stress to decrease expression of a sodium transporter via chromatin modifications.
Introduction
Salinization of the cultivated land is increasingly imposing severe constraints on plant adaptation and crop yield. Excess sodium (Na+), the most widespread soluble cation in salinized soil, damages plants mainly by resulting in sequential osmotic stress and oxidative stress for glycophyte crops including rice (Munns & Tester, 2008; Ismail et al, 2014; Zhu, 2016; Yang & Guo, 2018a; Yang & Guo, 2018b). On the other hand, plants also need to uptake Na+ as a nutrient, especially in the potassium limited conditions (Horie et al, 2007). Hence, to survive on the salinized soil, plants have evolved intricate mechanisms to maintain Na+ homeostasis, which are mainly gated by ion transporters (Munns & Tester, 2008; Zhu, 2016; Yang & Guo, 2018a). It has been demonstrated that members of the High‐affinity K+ transporters (HKT) are required for maintaining Na+ homeostasis, thus regulating salt tolerance (Oomen et al, 2012; An et al, 2017; Kobayashi et al, 2017).
In plants, there are two subfamilies of HKTs. Subfamily I of HKTs is conserved in both monocotyledonous and dicotyledonous species by acting as Na+‐selective transporter, including rice OsHKT1;1, OsHKT1;4, and OsHKT1;5, which mainly contribute to Na+ exclusion upon salt stress (Wang et al, 2015; Suzuki et al, 2016; Kobayashi et al, 2017). By contrast, the subfamily II of HKTs, only present in monocots, includes the transporters permeable to both Na+ and K+, such as plasma membrane‐localized OsHKT2;1 which mediates Na+ uptake, and its mRNA expression is rapidly repressed under salt stress (Horie et al, 2007). Consistently, oshkt2;1 mutants accumulate less Na+, but not K+ (Horie et al, 2007). The abundance of OsHKT2;1 was intricately regulated at both transcriptional and post‐translational levels. ETHYLENE INSENSITIVE3‐LIKE1 (OsEIL1) and OsEIL2 were shown to directly activate OsHKT2;1 expression, hence negatively regulating salt tolerance in rice seedlings (Yang et al, 2015). Recently, OsSIRH2‐14, a (RING) H2‐type E3 ligase, was found to physically interact with OsHKT2;1 protein at the high salinity condition and regulate its stability through 26S‐proteosome pathway (Park et al, 2019). Notably, OsHKT2;1 overexpressing lines displayed the reduced salt stress tolerance in both rice and Arabidopsis (Ardie et al, 2009), suggesting a potential role of OsHKT2;1 in rice salt stress tolerance. Nonetheless, the molecular mechanism underlying salt‐induced OsHKT2;1 repression that results in reduced Na+ uptake is largely unknown.
The circadian clock, the endogenous time‐keeping system in higher plants, has been demonstrated to function as an important integrator of multiple abiotic stresses signals, including salt stress in Arabidopsis (Greenham & McClung, 2015; Sanchez & Kay, 2016). For instance, the salinity stress responsive genes are expressed rhythmically in constant light condition, suggesting that there are regulated by circadian clock (Covington et al, 2008). In addition, EARLY FLOWERING 3 (ELF3), which mediates light input to circadian clock, is required for salt tolerance by modulating critical regulatory components in salt stress response pathways at both transcriptional and post‐translational levels (Sakuraba et al, 2017). Moreover, salinity stress triggers protein degradation of GIGANTEA (GI), one of the major clock components, resulting in the release of SOS2 (Salt Overly Sensitive 2) sequestration to thus confer salt tolerance by subsequently activating SOS1, the major plant Na+/H+‐antiporter (Kim et al, 2013). In addition, high salinity could alter core clock gene expression, including the repression of PSEUDO‐RESPONSE REGULATOR 7 (PRR7) and TIMING OF CAB EXPRESSION 1 (TOC1), but induction of PRR9 (Kwon et al, 2014). Intriguingly, the triple mutant of prr579 was more tolerant to salt stress (Nakamichi et al, 2009) with unknown mechanism. Together these results indicate that the clock components PRR members may play a key role in mediating salt stress tolerance in Arabidopsis. In rice, it has been implicated that the expression of SOS pathway genes including OsSOS2 and OsSOS3 is affected by diurnal rhythm in seedling stage (Soni et al, 2013). In addition, a number of circadian‐regulated genes were shown to be involved in salinity tolerance in rice, such as cyclophilin 2 (OsCYP2) and Receptor for Activated C Kinase 1 (OsRACK1) (Ruan et al, 2011; Zhang et al, 2018a). Nevertheless, whether rice core clock components participate in salt tolerance and the underlying mechanisms remain largely unclear.
The OsPRR (Oryza sativa Pseudo‐Response Regulator) gene family consists of five members and is expressed with robust oscillation pattern from dawn to dusk, sequentially with approximately 2‐h intervals, in the order OsPRR73 (OsPRR37), OsPRR95 (OsPRR59), and OsPRR1 (Murakami et al, 2003). Similar to AtPRR proteins, all the OsPRR proteins contain a Pseudo‐Response (PR) motif at their N terminus and a CCT (CONSTANS, CONSTANS‐Like, and TOC1) domain at the C terminus, which functions as a DNA‐binding motif. Ectopic expression of OsPRRs in the respective Arabidopsis prr mutants confirmed that they are in part genetically interchangeable with AtPRRs (Murakami et al, 2007). Notably, OsPRR37 was found to be the causal gene for quantitative trait locus (QTL) of HD2 (Heading date 2) and indirectly regulates grain productivity and regional adaptation, which is critical for rice breeding to achieve desired photosensitivity (Murakami et al, 2005; Koo et al, 2013; Yan et al, 2013; Gao et al, 2014). However, the role of OsPRRs in rice abiotic stress tolerance, especially salt stress, and the underlying mechanism remains elusive.
Here, we systematically generated the loss‐of‐function mutants of OsPRRs and found among OsPRR members, OsPRR73 is the unique member required for rice salt tolerance. OsPRR73 was rapidly induced by NaCl treatment and oprr73 null mutants were hypersensitive to sodium ion stress. Transcriptomic and biochemical analyses further identified OsHKT2;1 as a novel transcriptional target of OsPRR73 in mediating salt tolerance. Additionally, OsPRR73 could physically interact with the histone deacetylase 10 (HDAC10) to form a repressive complex to repress OsHKT2;1 transcription by altering its promoter chromatin status. Consistently, null mutant of OsHKT2;1 generated by CRISPR/Cas9 approach exhibits the enhanced salt tolerance phenotype. Together, we conclude that OsPRR73 transcriptionally represses OsHKT2;1 upon salt stress hence to modulate sodium ions homeostasis and confer salt tolerance in rice.
Results
OsPRR73 positively regulates rice salinity stress tolerance
To examine whether circadian clock components contribute to salt tolerance in rice, we investigated the roles of rice orthologs of the Arabidopsis Pseudo‐Response Regulator (PRR) gene family members in salinity response. The transcript profiles of OsPRR1, OsPRR37, OsPRR59, OsPRR73, and OsPRR95 were first determined under salt stress. Evidently, among OsPRR members, OsPRR73 showed most pronounced induction in response to 180 mM NaCl, while the transcript levels of OsPRR1 and OsPRR95 showed no discernible changes (Appendix Fig S1). The expression of OsPRR73 in the roots was also induced by salt (Appendix Fig S1F) as seen in shoots (Appendix Fig S1D).
By searching a rice T‐DNA insertion mutant database (Jeon et al, 2000), we identified an OsPRR73 mutant 3A‐12296 in Dongjin (DJ) background, in which the T‐DNA was inserted into the fifth intron of OsPRR73 (Fig 1A). Genomic PCR and RT–qPCR analyses confirmed that 3A‐12296 (hereafter referred as osprr73) is a null mutant allele of OsPRR73 (Fig EV1). To eliminate the unrelated traits potentially caused by additional T‐DNA insertions elsewhere, we backcrossed the osprr73 mutant with DJ and conducted subsequent physiological analysis using homozygous F3 progenies. We also generated null mutants osprr37, osprr59, and osprr95 by using CRISPR/Cas9 technology as previously described (Ma et al, 2015). By using TILLING approach, we also identified an osprr1 null mutant with a premature stop codon in OsPRR1 which resulted in 147 aa truncated protein (Fig EV2). The salt stress tolerance phenotypes were determined by transferring 4‐week‐old seedlings into media containing 180 mM NaCl for 21 days, followed by recovery in regular hydroponic culture for additional 7 days unless otherwise noted. Intriguingly, only the null mutant of OsPRR73 displayed hypersensitivity to salinity stress (Fig 1B and C) with a significant reduction of survival rate, while other osprr null mutants showed similar salinity sensitivity to their respective wild‐type control (Fig EV3). Importantly, the genetic complementation lines of OsPRR73 could reverse the hypersensitive phenotypes of osprr73 mutant to salinity stress.


OsPRR73 positively regulates salinity tolerance in rice
ASchematic diagram showing the T‐DNA insertion sites in rice osprr73T‐DNA mutant (hereafter named as osprr73).
B OsPRR73 confers salinity stress tolerance in rice. The seedlings of Dongjin (DJ, the wild‐type control), osprr73, and the complementation line of osprr73 (abbreviated as Com‐L1) grown under 12‐h light/12‐h dark conditions for 28 days (left panel), transferred to 180 mM NaCl for 21 days (middle panel) and recovered for 12 and 24 days (the two right panels), respectively. Scale bar, 5 cm.
CStatistical analysis of the survival rate of DJ, osprr73, and Com‐L1 plants in (B) after recovery 12 days. Data are presented as mean ± SD. (n from 4 biological replicates, and 24 plants were tested in each of biological replicates). (***) P ≤ 0.001 were generated by Student’s t‐test.
DDiagram showing the target and mutated sites of OsPPR73 by CRISPR/Cas9‐based genome editing. The PR and CCT are the abbreviations of Pseudo‐Response and COSTANS, COSTANS‐LIKE, and TOC1 domains, respectively. The target sequences within the first exon of OsPRR73 were shown with the green boxes. PAM sequences were labeled with underline within the green boxes.
ENull mutant of osprr73 in Nipponbare (NIP) background by genome editing displayed hypersensitivity to NaCl treatment. Scale bar, 5 cm.
FSurvival rate of NIP (wild type of CRISPR lines) and osprr73‐C (C stands for CRISPR) plants in (E) after recovery 9 days. Data are presented as mean ± SD. n = 4 biological replicates, 24 plants for each biological replicate. (*) P ≤ 0.05 and (***) P ≤ 0.001 were generated by Student’s t‐test.
G, HQuantification of chlorophyll content and electrolyte leakage of WT and osprr73 mutants with or without 180 mM NaCl treatment for 7 days. Data are presented as mean ± SD. n = 3, biological replicates, and asterisks represent significant difference among means by Student’s t‐test with (*) P ≤ 0.05, (**) P ≤ 0.01, (***) P ≤ 0.001.
Source data are available online for this figure.


Molecular identification of OsPRR73 T‐DNA mutant
Insertion site of T‐DNA (osprr73 3A‐12296) in the fifth intron of OsPRR73 genome. Green arrowheads (LP and RP) represent positions of gene‐specific primers and primers used for detection of the mutant allele. Red and pink arrowheads (UF and UR, DF and DR) represent positions of RT–PCR primers used for OsPRR73 expression analysis in the upstream and downstream of the T‐DNA insertion position.
Testing of the osprr73 T‐DNA knockout mutant, LB primer was on the T‐DNA fragment and RP was on genome. Dongjin (DJ) as the wild‐type control.
OsPRR73 expression level in the osprr73 mutant of the upstream and downstream of the T‐DNA insertion position. OsUbiquitin (OsUBI) was used as an internal control.
RT–PCR to test the OsPRR73 expression level in the osprr73 mutant and the upstream and downstream genes of OsPRR73. OsUbiquitin (OsUBI) was used as an internal control.
OsPRR73 expression level in the osprr73 by RT–qPCR. Data represent means ± SD. n = 3 technical replicates. Data are representative from three independent biological replicates with similar result. The asterisks represent significant difference among means by Student’s t‐test with (***) P ≤ 0.001.
Source data are available online for this figure.


Characterization of osprrs mutants generated by TILLING approach or genome editing
ASanger sequencing showing the point mutation of OsPRR1 screened by TILLING approach that caused a premature stop codon with encoding a 147 aa truncated peptide.
B–GSanger sequencing showing the respective OsPRR mutants generated by genome editing, which harbors one bp insertion in OsPRR73 (B) that results in a premature stop codon and only encodes a 86 aa truncated peptide, 5 bp deletion in the first exon of OsPRR59 with premature stop codon which only encodes 180 aa truncated protein (C), a 3 bp deletion in OsPRR95 (D) which caused one aa deletion in its C‐terminal CCT motif, and a 33‐bp deletion in OsPRR73 (E) that results a deletion of 11 amino acids in its PR domain, a 5‐bp deletion in OsPRR73 causing a premature stop (F), and one bp insertion in OsPRR73 that causes a premature stop codon only with 17 aa truncated protein (G).

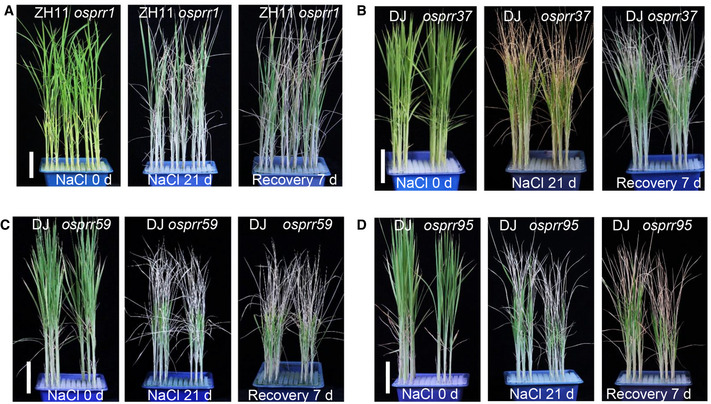
Phenotypes of osprr1, osprr37, osprr59, and osprr95 mutants upon NaCl stress treatment
AFour‐week‐old seedlings of ZH11 and osprr1 were treated with 180 mM NaCl for 21 days (middle panel) and recovered for additional 7 days (right panel). Scale bar, 5 cm.
B–DFour‐week‐old seedlings of DJ and osprr37 (B), osprr59 (C), and osprr95 (D) mutants, were treated with 180 mM NaCl for 21 days (middle panel) and recovered for additional 7 days (right panel). Scale bar, 5 cm. Data are representative from three independent biological replicates with similar result.
To further validate the role of OsPRR73 in salinity stress tolerance, we generated three osprr73 CRISPR/Cas9 alleles with a 33 bp deletion 184 bp downstream of ATG (osprr73‐C1), a premature stop codon (osprr73‐C2) caused by 5 bp deletion, and a premature stop codon (osprr73‐C3) caused by 1 bp insertion (Fig EV2). The Cas9‐free homozygous lines were subsequently used for examining salt stress tolerance phenotype. Notably, the osprr73‐C2 and osprr73‐C3 plants were more sensitive to salinity stress, while the osprr73‐C1 displayed modest phenotype, indicating that osprr73‐C1 might be a weak allele, while osprr73‐C2 and osprr73‐C3 were likely null alleles (Fig 1E and F). Moreover, both osprr73 allele in DJ background and osprr73‐Cs alleles in NIP background contained significantly lower levels of chlorophyll (Fig 1G) and much higher levels of membrane ion leakage (Fig 1H) in the presence of NaCl. In addition, the grain yield and fertility of osprr73 mutant were dramatically reduced upon irrigation with 75 mM NaCl solution (Appendix Fig S2). Collectively, these results suggest that OsPRR73 specifically regulates rice salinity stress tolerance by acting as a positive regulator.
Next to investigate whether OsPRR73 functions as a clock component in rice, we performed time course RT–qPCR analysis to examine the temporal expression patterns of circadian clock‐related genes (Izawa et al, 2011; Nagano et al, 2012; Bendix et al, 2015; Matsuzaki et al, 2015) including OsPRRs, OsGI, OsFKF1, OsCCA1, OsLHY, OsLUX, OsELF3.1, and OsLKP2, in the free‐running condition which is usually used for assaying circadian phenotypes (Millar et al, 1995; Wang & Tobin, 1998). We found that their oscillation profiles were unanimously altered in osprr73 mutant with evident delayed peak or trough phase (Fig EV4). Moreover, we conducted time course RT–qPCR of osprr73 plants grown in the natural photoperiodic conditions, with 2‐h interval over a day–night cycle. We found the transcriptional peaks of OsPRR1, OsPRR37, and OsLUX genes were dramatically reduced (Appendix Fig S3A, B and J), whereas the transcriptional peaks of OsGI and OsFKF1 were markedly increased in osprr73 mutant, in relation to wild‐type DJ (Appendix Fig S3F and G). Additionally, there were marginal discrepancies of the expression patterns of OsCCA1, OsLHY, OsELF3.1, and OsLKP2 between osprr73 and DJ (Appendix Fig S3H, I, K and L), suggesting that OsPRR73 may not directly regulate these genes under this natural diurnal condition. Noticeably, the transcriptional peak of OsGI, previously proposed as a core clock component (Izawa et al, 2011), was elevated in osprr73 plants under both free‐running and natural diurnal conditions (Fig EV4F and Appendix Fig S3F). Together, these data support a notion that circadian clock was altered in osprr73 plants, and OsPRR73 likely acts as a clock component in rice, as its ortholog in Arabidopsis.

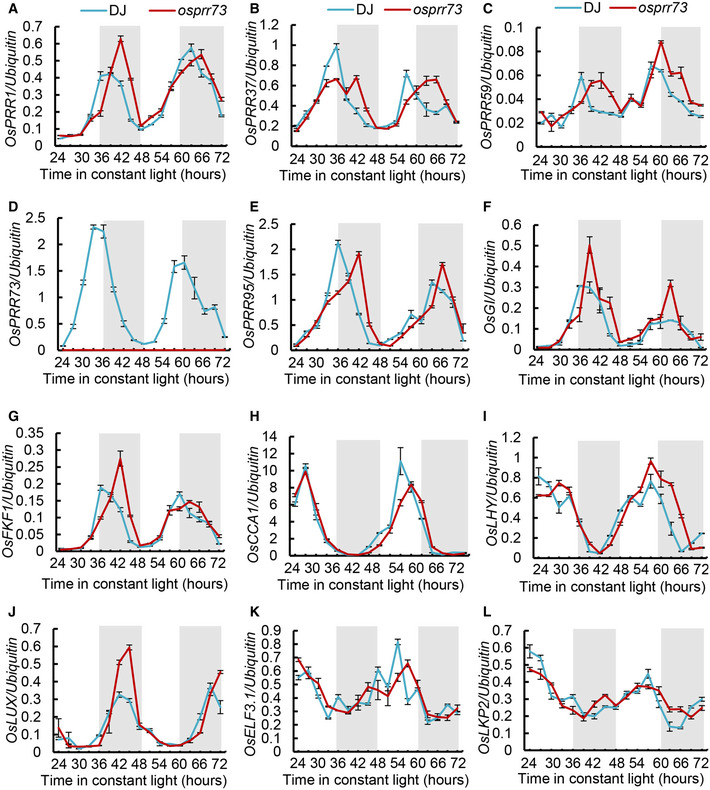
Transcriptional profiles of clock‐related genes in osprr73 mutant under free‐running condition
A–LTime course RT–qPCR were conducted to detect the expression patterns of OsPRR1 (A), OsPRR37 (B), OsPRR59 (C), OsPRR73 (D), OsPRR95 (E), OsGI (F), OsFKF1 (G), OsCCA1 (H), OsLHY (I), OsLUX (J), OsELF3.1 (K), and OsLKP2 (L), with the penultimate leaf blades samples of WT and osprr73 plants harvested under constant light as indicated time points, after entraining in 12‐h light/12‐h dark photocycles for 21 days. Gray rectangles indicate subjective nights in constant light. Data represent means ± SE. n = 3 technical replicates. Data are representative from two independent biological replicates with similar result.
Source data are available online for this figure.
Transcriptomic analysis of the osprr73 mutant
To determine the subcellular localization of OsPRR73, we transiently expressed the GFP‐OsPRR73 fusion gene in leaves of Nicotiana benthamiana and found it was predominantly localized in the nuclei of epidermal cells (Appendix Fig S4A), consistent with its annotated function as a transcriptional regulator. We then reasoned that the transcriptomic analysis might provide additional insights into the role of OsPRR73 in salinity stress response. Hence, RNA‐seq analysis was performed to reveal the transcriptomic profiles of osprr73 mutant and DJ in the shoots of the 2‐week‐old plants treated with NaCl 4 and 28 h, respectively, as OsPRR73 were highly expressed in shoots (Appendix Fig S4B). We set P < 0.05 and the value of fold change (log2) over 1.5 relative to DJ as the cutoff threshold. In total, we identified 690, 627, and 1,419 up‐regulated genes and 742, 795, and 996 down‐regulated genes in osprr73 mutant compared with the DJ control of mock, 4‐h, and 28‐h NaCl treatment, using three biological replicates (Fig 2A). RT–qPCR results confirmed the transcript profiles of eight randomly selected differentially expressed genes (DEGs) in osprr73 mutants (Appendix Fig S5). Venn diagram analysis revealed 1,030, 999, and 1,663 unique DEGs in osprr73 mutant treated with mock, NaCl 4 h, and NaCl 28 h, respectively (Fig 2B). Gene Ontology (GO) analysis further revealed that the top enriched GO terms in the short‐term NaCl (4 h) treatment were involved in response to stimulus (GO:0050896), response to stress (GO:0006950), and oxidation reduction (GO:0055114) (Fig 2C). By contrast, the most enriched GO terms in the long‐term NaCl treatment (28 h) were the regulation of cellular metabolic process (GO:0031323), regulation of metabolic process (GO:0019222), and regulation of primary metabolic process (GO:0080090) (Appendix Fig S6). Under the mock treatment, the DEGs in osprr73 mutant were mainly involved in response to stress and stimulus (Fig 2D). However, the most affected biological processes upon NaCl treatment were the DEGs in response to oxidative stress and oxidation reduction (Fig 2D).

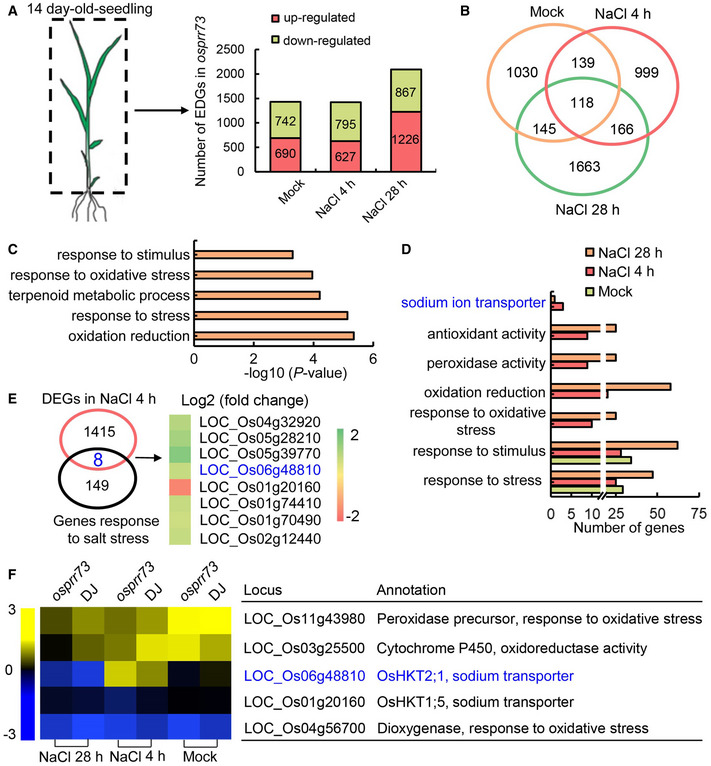
Differential expressed genes in osprr73 mutant upon salinity stress as revealed by RNA‐seq
Schematic diagram for tissues (left panel) and the number of differentially expressed genes (right panel) in osprr73 mutant compared with their respective wild type (DJ).
Venn diagram showing the number of differentially expressed genes in osprr73 mutant with or without salinity stress.
GO (Gene ontology) classification of differentially expressed genes in osprr73 mutant treated by NaCl for 4 h, based on biological process (BP). P‐values were generated by Fisher’s test.
The number of differentially expressed genes, which belongs to BP cluster in GO analysis, in osprr73 mutant.
Venn diagram showing the overlapped DEGs in osprr73 mutants (pink circle) with previously identified salt‐responsive genes which derived from China rice data center (black circle). Heatmap showing 6 common genes, the colored bar represents log2 (fold change).
Heat map of OsHKT2;1, OsHKT1;5, and the genes related to the oxidative stress in GO analysis, showing their transcriptional abundance in osprr73 mutant. The colored bar indicates log2 (FPKM) values.
Interestingly, the sodium ion transporter (GO:000681) was specifically enriched among DEGs with NaCl treatment (Fig 2D), implicating sodium ion transporters might be involved in salt stress response in osprr73 mutant. Furthermore, eight DEGs in 4‐h NaCl treated osprr73 plants were overlapped with previously well‐characterized 157 salt stress responsive genes (Fig 2E), including two sodium transporter (LOC_Os06g48810 and LOC_Os01g20160). Next, heatmap analysis with the RNA‐seq data clearly demonstrated that transcript level of LOC_Os06g48810 encoding OsHKT2;1 was induced, while LOC_Os03g25500 encoding a cytochrome P450 was repressed (Fig 2F) in osprr73 mutant with 4‐h NaCl treatment. Together, these results implicate that abnormal Na+ and ROS homeostasis might contribute to the hypersensitive salinity response in osprr73 mutant.
OsPRR73 confers sodium ion stress response
To further investigate whether Na+ accumulation was the direct cause for hypersensitivity of osprr73 mutant to NaCl stress, we checked Na+ contents in both shoots and roots in osprr73 mutants. There was no obvious difference in Na+ content between osprr73 mutant and DJ under non‐NaCl stress condition (Fig 3A). However, with 180 mM NaCl treatment for 7 days, the Na+ content was significantly higher in both shoots and roots of osprr73 (Fig 3A). Similarly, the Na+ content in osprr73‐C lines of NIP background was also significantly higher (Appendix Fig S7A). Meanwhile, we measured K+ contents with the same samples and found there was no evident difference between osprr73 and DJ under either normal or NaCl stress condition (Fig 3B and Appendix Fig S7B). Thus, even though there was no difference or even lower in the Na+/K+ ratio of the plants under un‐stressed condition, the Na+/K+ ratio was increased in osprr73 mutant plants after 180 mM NaCl treatment (Fig 3C), which might contribute to their hypersensitivity to NaCl treatment (Zhu, 2016; Yang & Guo, 2018b). Together with the transcriptomic analysis results, we proposed that OsPRR73 might play a vital role in maintaining sodium ion homeostasis to confer salt stress tolerance.


The osprr73 mutant displays specific hypersensitivity to sodium ion stress
A, BThe Na+ and K+ contents in the shoots and roots of DJ and osprr73 mutant plants with or without 180 mM NaCl treatment for 7 days. Na+ (A) and K+ (B) contents were measured with ICP method. Data represent means ± SD, (n = 3, biological replicates). (**) P ≤ 0.001 and (*) P ≤ 0.001 indicate significant difference by Student’s t‐test. The abbreviation of n.s. stands for no significant.
CRatio of Na+ to K+ was calculated with their respective content in (A) and (B). Data represent mean ± SD (n = 3, biological replicates). For each of biological replicates, 6 individual plants were measured. The asterisk represents significant difference among means by Student’s t‐test with (*) P ≤ 0.05, and n.s. indicates no significant.
DFour‐week‐old seedlings of DJ and osprr73 were treated with 90 mM Na2SO4 for 14 days (middle panel) and recovered for additional 7 days (right panel). Scale bar, 5 cm.
EStatistical analysis of survival rates with Na2SO4 treatment in (D). Data represent means ± SD. n = 3 biological replicates, 24 plants for each biological replicate. (***) P ≤ 0.001 indicates significant difference by Student’s t‐test.
FThe seedlings of DJ and osprr73 mutant were treated with 90 mM MgCl2 for 14 days (middle panel) and recovered for 7 days (right panel). Scale bar, 5 cm.
GStatistical analysis of survival rates with MgCl2 treatment in (F). The survival rate of DJ and osprr73 plants in MgCl2 stress were calculated after recovery 7 days. Data represent means ± SD. n = 2 biological replicates, 24 plants of each replicate.
HThe seedlings of DJ and osprr73 mutant were treated with 180 mM mannitol for 36 days (middle panel) and recovered for 7 days (right panel). Scale bar, 5 cm.
IThe survival rate of DJ and osprr73 plants in mannitol stress. Data represent means ± SD. n = 3 biological replicates, 24 plants of each replicate. The abbreviation of n.s. stands for no significant generated by Student’s t‐test.
Source data are available online for this figure.
To further distinguish whether hypersensitivity in osprr73 mutant was caused by Na+ or Cl− accumulation, we utilized different salt solution including Na2SO4, and MgCl2 to treat osprr73 mutant and DJ plants. Interestingly, the osprr73 mutant plants displayed hypersensitivity to 90 mM Na2SO4 (Fig 3D and E), but not 90 mM MgCl2 treatment (Fig 3F and G), suggesting that OsPRR73 is required for Na+‐associated stress. As Na+ accumulation subsequently produces osmotic stress, we test the osmotic stress sensitivity by treating osprr73 mutant with 180 mM mannitol. As shown in Fig 3H and I, osprr73 plants displayed similar osmotic stress sensitivity to DJ. Hence, we concluded that the main cause for hypersensitivity of salinity stress in osprr73 may be due to the abnormal accumulation of Na+.
OsHKT2;1 is a direct transcriptional target of OsPRR73
Since the osprr73 mutant is specifically sensitive to Na+ stress with abnormally high cellular Na+ accumulation, and transcript abundance of OsHKT2;1 was up‐regulated in osprr73 mutant, we were promoted to determine whether OsHKT2;1 is a direct transcriptional target of OsPRR73. Intriguingly, we found OsHKT2;1 displayed a robust oscillation pattern with an acrophase in the morning and trough level at evening (Appendix Fig S8), which was overall an antiphase with the temporal expression profile of OsPRR73 (Appendix Fig S8G and H). Next, we assessed the transcript levels of OsHKT2;1 in osprr73 mutant by RT–qPCR and found its levels were significantly higher in osprr73 mutant, especially with either short‐term (4 and 16 h) or long‐term (72 h) 180 mM NaCl treatment (Fig 4A). In addition, we detected the diurnal expression pattern of OsHKT2;1 in osprr73 and WT plants with or without NaCl stress presence. We found that the transcript levels of OsHKT2;1 are significantly higher in osprr73 mutant especially in the presence of salt stress (Appendix Fig S9), suggesting OsPRR73 can efficiently repress the expression of OsHKT2;1 upon salt treatment. We also determined the transcript levels of OsHKT1;5, encoding a Na+ exporter, and found its transcript level was only modestly altered in the short‐term NaCl treatment (Fig 4B). Together, these results implied that OsHKT2;1, but not OsHKT1;5, was a potential target of OsPRR73.


OsPRR73 binds OsHKT2;1 promoter to repress its transcription
A, BThe expression level of OsHKT2;1 and OsHKT1;5 in osprr73 mutant and the wild control Dongjin (DJ) plants with or without salinity stress in multiple time points. DJ Data represent means ± SD. n = 3, technical replicates. The representative data are from at least two individual biological replicates. The asterisks represent significant difference among means by Student’s t‐test with (*) P ≤ 0.05, (**) P ≤ 0.01 and (***) P ≤ 0.001.
CTransient expression assay in the infiltrated leaves of Nicotiana benthamiana showing OsPRR73 can repress OsHKT2;1 transcription. Left panel showing the bioluminescence signal of OsHKT2;1 and OsPRR73. Bioluminescence of co‐transformation of either GFP or 35S: GFP‐OsPRR73 (GFP‐OsPRR73) with OsHKT2;1pro: LUC in N. benthamiana.
DQuantification of bioluminescence intensity of OsHKT2;1pro: LUC shown in (C). Data represent means ± SD. n = 10. The asterisks indicate the significant difference by Student’s t‐test with (***) P ≤ 0.001. At least two biological replicates were conducted with similar results.
EChIP‐qPCR assay showing the enriched DNA fragments including the S3 and S4 regions of OsHKT2;1 promoter by OsPRR73, compared with wild‐type DJ controls. The amplicon of Ubiquitin was taken as a negative control. Two‐week‐old seedlings were harvested at ZT12. Data represent means ± SD (n = 3, technical repeats). The experiments were performed at least two biological replicates with similar result. Top scheme indicates the locations of amplicons for ChIP analysis. The asterisks represent significant difference among means by Student’s t‐test with (**) P ≤ 0.01.
FEMSA assay with the OsPRR73 incubated with the probes designed for the S4 region of OsHKT2;1 promoter. Unlabeled probes were used as competitor with indicated folds. MBP alone was used as negative control. Arrowhead marks the shifted positive bands.
Source data are available online for this figure.
To further determine whether OsHKT2;1 was a direct target of OsPRR73, we employed a transient co‐expression assay using N. benthamiana leaf infiltration. Evidently, OsPRR73 could significantly repress the transcriptional activity of OsHKT2;1 promoter (Fig 4C and D). As Arabidopsis ortholog of OsPRR73 mainly binds G‐box containing sequence, we performed cis‐element scanning within the ~ 1.5 Kb promoter region of OsHKT2;1 using PlantCARE database (Lescot et al, 2002) and found there were two potential G‐box elements at −456 bp (S3) and −1,298 bp (S4) upstream of start codon (Fig 4E). Chromatin immunoprecipitation (ChIP) assay using 35S:GFP‐OsPRR73 transgenic lines grown in 12‐h light/12‐h dark for 2 weeks and samples collected at ZT12. The two amplicons containing a potential G‐box element (S3 and S4) were significantly enriched (Fig 4E) in ChIP‐qPCR assay. In contrast, there were no significant enrichments for other regions and a negative control in selected ubiquitin promoter (Fig 4E). We then used electrophoresis mobility shift assay (EMSA) to determine whether OsPRR73 could directly bind the enriched region using OsPRR73‐CCT, the DNA‐binding domain, tagged by myelin basic protein (MBP). As shown in Fig 4F, a specific shifted band was observed when the labeled DNA probes, which contains an atypical G‐box element (CACGAC), were incubated with MBP‐OsPRR73‐CCT. No band shift was observed in the presence of non‐labeled or mutated G‐box probe, implicating that OsPRR73 could directly bind the promoter of OsHKT2;1. Together, our data support the notion that OsHKT2;1 is a direct transcriptional target of OsPRR73.
OsPRR73 directly interacts with rice histone deacetylase HDAC10
To further explore the mechanism by which OsPRR73 repressed OsHKT2;1 transcriptionally, we revealed the interactome of OsPRR73 through immunoprecipitation followed by mass spectrometry (IP‐MS). Two‐week‐old seedlings of T2 progeny of 35S: GFP‐OsPRR73 transgenic line in DJ background were used for IP‐MS as previously described (Wang et al, 2019). In total, we identified 19 nuclear proteins in OsPRR73 complex shared by two biological replicates (Appendix Table S1). Among them, SDG704 (LOC_Os11g38900), encoding a SET‐domain containing protein, and Histone Deacetlylase 10 (HDAC10, LOC_Os12g08220) were potential components related to transcriptional suppression. Since SDG704 was unable to methylate H3K9 (Ding et al, 2007), we determined whether HDAC10 were a bona fide interactor of OsPRR73.
Using transient co‐expression of GFP‐OsPRR73 and HDAC10‐FLAG proteins in N. benthamiana leaves, we observed strong co‐immunoprecipitation of HDAC10 with GFP‐OsPRR73 (Fig 5A). A split‐luciferase imaging analysis confirmed the interaction between OsPRR73 and HDAC10 in vivo (Fig 5B). Moreover, bimolecular‐fluorescence complementation (BiFC) analysis revealed positive signal in the nucleus of leave epidermal cells when YFPN‐OsPRR73 and YFPC‐HDAC10 were co‐expressed (Fig 5C). We also found that the truncated OsPRR73 containing PR (PSEUDO‐RESPONSE) domain and without CCT (CONSTANS, CONSTANS‐like, and TOC1) domain (1–711 amino acids, OsPRR73‐F1) was capable of interacting with HDAC10. Deletion of both PR and CCT domain (200–711 amino acids, OsPRR73‐F2) almost abolished the interaction with HDAC10 (Fig 5D), suggesting that the N‐terminal PR domain of OsPRR73 was required for interacting with HDAC10. To determine whether the interaction between OsPRR73 and HDAC10 could affect the level of H3ac on the promoter of OsHKT2;1, we further performed ChIP‐qPCR assay with H3K9ac antibody. We found that the levels of H3K9ac on the promoter regions of S3 and S4 were significantly increased in osprr73 mutant compared with DJ (Fig 5E), while H3 levels were about the same between osprr73 mutant and DJ (Fig 5F), indicating that the interaction between OsPRR73 and HDAC10 could affect H3ac. To further examine the regulatory role of HDAC10 on OsHKT2;1, we conducted a transient co‐expression assay using N. benthamiana leaf infiltration. As expected, HDAC10 could modestly repress the transcriptional activity of OsHKT2;1. Noticeably, the repressive effect of OsPRR73 on OsHKT2;1 transcription was augmented by co‐transforming with HDAC10, indicating that the interaction between OsPRR73 and HDAC10 can inhibit the transcription of OsHKT2;1 more efficiently (Appendix Fig S10A and B). Together, our results suggested that OsPRR73 might recruit the histone decatylase HDAC10 to repress the transcription of OsHKT2;1.


OsPRR73 physically interacts with HDAC10 in nucleus
ACo‐immunoprecipitation assay showing the physical interaction between OsPRR73 and HDAC10 in vivo. CoIP was conducted with GFP‐Trap beads, and GFP and FLAG antibodies were used to detect OsPRR73 and HDAC10 in immunoprecipitants as indicated.
BSplit‐luciferase complementation (SLC) assay showing OsPRR73 interacts with HDAC10 in the co‐infiltrated N. benthamiana leaves.
CBimolecular‐fluorescence complementation (BiFC) assay shows the interaction between OsPRR73 and HDAC10 occurs in nucleus. BF represents bright field. Scale bar, 40 µm.
DN terminus of OsPRR73 containing PR domain is required and sufficient for interacting with HDAC10. Upper panel shows the schematic illustration of full‐length and the truncated OsPRR73 proteins. CoIP was performed with GFP‐Trap beads, and immunoprecipitants were detected by FLAG for HADC10 and GFP for truncated proteins of OsPRR73, respectively.
E, FChromatin Immunoprecipitation‐qPCR (ChIP‐qPCR) assay to detect the enriched promoter regions of OsHKT2;1 by using the H3K9ac (E) and H3 (F) antibodies. Two‐week‐old seedlings were harvested at ZT12. The locations of amplicons were described in Fig 4E. Data represent means ± SD. n = 3 technical replicates. The asterisk indicates the significant difference by Student’s t‐test with (*) P ≤ 0.05. The experiments were repeated at least twice with similar result.
Source data are available online for this figure.
OsHKT2;1 negatively regulates rice salinity tolerance
Previously, OsHKT2;1 was shown to be responsible for Na+ uptake in K+ starved rice plants (Horie et al, 2007) and had an impact on potassium use efficiency (Hartley et al, 2020). However, the role of OsHKT2;1 in salinity stress tolerance is unclear. As OsHKT2;1 is a direct transcriptional target of OsPRR73 (Fig 4), and higher level of Na+ was accumulated in osprr73 mutant plants (Fig 3A), we hypothesized that increased transcript level of OsHKT2;1 in osprr73 caused the higher Na+ accumulation, which might result in salinity hypersensitivity phenotype of osprr73 mutant. Hence, we were promoted to investigate whether OsHKT2;1 was required for rice salinity stress tolerance. By employing CRISPR/Cas9 technology, we generated the oshkt2;1 mutant in NIP background. Sanger sequencing confirmed that the first exon of OsHKT2;1 had 1 base pair insertion in oshkt2;1 mutant (Fig 6A and B), resulted in a premature stop codon (Fig 6C). The Cas9‐free homozygous seedlings of F3 progeny were used for examining salinity stress tolerance phenotype. Before NaCl treatment, the morphology of oshkt2;1 was indistinguishable compared with NIP plants; however, after salinity stress, the survival rate of oshkt2;1 was higher than that of NIP (Fig 6D and E), suggesting OsHKT2;1 was indeed involved into salinity stress tolerance.


OsHKT2;1 negatively regulates salinity tolerance in rice
Nucleotide sequence showing the mutated site of OsHKT2;1 generated via CRISPR/Cas9‐based genome editing. Green box indicates the CRISPR target sequence, which locates on the first exon of OsHKT2;1. PAM sequences were labeled with underline within the green boxes.
Sanger sequencing confirmed the oshkt2;1 mutant harbor a 1‐bp insertion (pointed by red arrowheads).
Alignment the amino acid sequences of OsHKT2;1, and its mutated version showing a premature stop codon occurred in the ORF of OsHKT2;1.
The oshkt2;1 seedling is more tolerant to the treatment of NaCl. The oshkt2;1 and NIP, its wild‐type, 4‐week‐old seedlings grown under NaCl conditions for 0 days (left panel), then were transferred to 200 mM NaCl for 28 days (middle panel), and recovered for 7 days (right panel). Scale bar, 5 cm.
Statistical analysis of survival rate in (D). Data represent means ± SD. n = 3 biological replicates, 24 plants of each replicate. (*) P ≤ 0.05 was generated by Student’s t‐test.
Source data are available online for this figure.
OsPRR73 may indirectly inhibit the accumulation of reactive oxygen species
ROS accumulation is considered as a secondary response of salt stress. High ROS contents in plant cells can damage proteins, lipids, and other cellular components (Yang & Guo, 2018a). Our RNA‐seq data showed that the genes related to oxidative stress were enriched in GO analysis; we thus explored if ROS was abnormally acumulated in osprr73 mutant upon salt stress. We measured the malondialdehyde (MDA) contents, an indicator of membrane lipid peroxidation (Ouyang et al, 2010), and found its level in osprr73 mutant was very similar to DJ under normal condition. However, upon NaCl treatment, MDA content was dramatically increased in osprr73 mutant, much higher than that in DJ plants (Fig 7A). In addition, diaminobenzidine (DAB) staining and 2′, 7′‐dichlorodihydrofluorescein diacetate (H2DCFDA) fluorescent signal also confirmed the high ROS contents in shoots and roots of osprr73 mutant only under salinity stress (Fig 7B–D). Similar results were obtained in osprr73 null mutant generated by CRISPR/Cas9 (Appendix Fig S11A and B) in NIP background. Consistently, the ROS‐scavenging enzyme activities of superoxide dismutase (SOD; EC 1.15.1.1) and catalase (CAT, EC1.11.1.6) were significantly reduced in osprr73 mutants under salinity stress condition (Fig 7E and F, and Appendix Fig S11C and D). Taken together, OsPRR73 appeared to negatively regulate accumulation of ROS caused by salinity stress. Finally, application of H2O2 did not affect the expression of OsPRR73 under either short or long time treatment (Appendix Fig S12A and B), which implicated that OsPRR73 might affect the oxidative stress response indirectly. In addition, we also detected the accumulation of ROS in oshkt2;1 mutant with both DAB staining and H2DCFDA fluorescent signal as an indicator. We found that the ROS content was significantly reduced in both leaves and roots of oshkt2;1 mutant in the presence of NaCl stress (Appendix Fig S13), indicating OsHKT2;1 may negatively regulate ROS accumulation in response to salt stress.


ROS was highly accumulated in osprr73 mutant under salinity stress
AMalondialdehyde (MDA) content in leaves of 14‐day‐old DJ and osprr73 plants treated by 180 mM NaCl for 3 d and normal condition. Data represent means ± SD. n = 6, biological replicates. The asterisks indicate the significant difference (***) P ≤ 0.001 by student’s t‐test.
BDAB staining showing the greater ROS accumulation in the leaves of osprr73 mutant treated by 180 mM NaCl treatment for 3 days. Scale bar, 1 cm.
CH2O2 concentration was detected by using H2DCFDA in the root tip of DJ and osprr73 mutant plants. Scale bar, 5 mm. −NaCl and +NaCl represented the untreated or treated plants by NaCl, respectively.
DQuantitative analysis of H2O2 concentration in the root of DJ and osprr73 mutant. Data represent means ± SD. n ≥ 6. The asterisks indicate the significant difference (***) P ≤ 0.001 by Student’s t‐test.
E, FMeasurement of SOD (Superoxide dismutase) (E) and CAT (Catalase) (F) enzymes activity in shoots of DJ and osprr73 seedlings with or without NaCl stress. Data represent means ± SD. n = 3 technical replicates. Data are representative from two independent biological replicates with similar result. The asterisks indicate the significant difference (*) P ≤ 0.05 by Student’s t‐test.
Source data are available online for this figure.
Discussion
It has been implicated that clock components could be involved in the regulation of salinity stress tolerance in Arabidopsis (Nakamichi et al, 2009; Kim et al, 2013). Meanwhile, circadian clock was shown to act as a crucial regulator to enhance fitness under unfavorable conditions, by providing temporal regulatory mechanisms for the sessile organisms (Sanchez & Kay, 2016). However, whether rice clock component contributes to salt tolerance remains unclear yet. Here, we found that the null mutants of OsPRR73 generated either by T‐DNA insertion or CRISPR/Cas9 approach, displayed hypersensitive to salt stress, which could be detrimental to rice grain yield. We further revealed the underlying molecular mechanism, which was in part due to the de‐repression of OsHKT2;1 that subsequently caused enhanced Na+ uptake followed by secondary ROS accumulation. Moreover, we found that HDAC10 could physically interact with OsPRR73 to form a transcriptional repression complex to inhibit OsHKT2;1 expression. Consistently, the null mutant of OsHKT2;1 was more tolerant to salt stress. Moreover, the oshkt2;1 osprr73 double mutant also displayed the salt‐tolerant phenotype with much less ROS accumulation, similar to that of oshkt2;1 plant (Fig EV5), suggesting that OsHKT2;1 is a major downstream component to mediate the salt hypersensitivity of osprr73 plants. Our findings thus revealed an OsPRR73‐OsHKT2;1 molecular module that confers the salt tolerance in rice via regulating Na+ homeostasis (Fig 8), which represents a novel molecular link between circadian clock and salt tolerance in rice.


OsPRR73 regulates OsHKT2;1‐mediated sodium uptake to conferred salinity tolerance in rice
Salt tolerance phenotype of WT and oshkt2;1, osprr73 and oshkt2;1 osprr73 plants. Three‐week‐old plants were treated with 200 mM NaCl for 14 days, followed by 8‐day recovery. Scale bar, 5 cm.
Survival rate of WT and oshkt2;1, osprr73 and oshkt2;1 osprr73 plants in (A) after recovery 8 days (n = 3 biological replicates, 16 plants for each biological replicate). Data represent means ± SE. P‐values were generated by Student’s t‐test.
DAB staining showing ROS accumulation in the leaves of WT and oshkt2;1, osprr73 and oshkt2;1 osprr73 plants treated with 200 mM NaCl for 7 days. Scale bar, 1 cm.
Source data are available online for this figure.


Proposed working model for OsPRR73 conferred salinity tolerance in rice
OsPRR73 binds the promoter of OsHKT2;1, by recruiting HDAC10, to negatively regulate the expression of OsHKT2;1 to reduce the import of cellular sodium ion, which subsequently reduced ROS accumulation, thus conferring salinity stress (left panel). In the absence of OsPRR73, the transcription inhibition of OsHKT2;1 was released, hence resulted in sodium ion accumulation. The excessive sodium ion subsequently accumulation caused ROS accumulation, to forming a Na+‐ROS stress cascade, leading to decreased survival.
The expression pattern of OsPRR73 was previously shown to be quite similar to that of OsPRR37 in both diurnal and constant light condition (Murakami et al, 2003). Here, we found that OsPRR73, but not OsPRR37, was specifically required for salt stress tolerance. Previous findings demonstrated that OsPRR37 was a major factor to determine rice heading time in the long day condition (Murakami et al, 2005; Koo et al, 2013; Yan et al, 2013; Gao et al, 2014), suggesting that despite OsPRR73 and OsPRR37 share similar temporal expression pattern, their physiological functions are distinct. Our time course RT–qPCR assay with plants grown in both free‐running and natural diurnal conditions suggested that OsPRR73 is involved into the regulation of circadian clock. In Arabidopsis, PRR proteins had been shown to form heterodimers either with other PRRs or with key regulators of plant growth and development such as CONSTANS protein (Wang et al, 2010; Hayama et al, 2017). Interestingly, we also identified OsPRR59 as an interacting protein of OsPRR73 in our IP‐MS assay (Appendix Table S1), suggesting that rice OsPRR proteins likely form heterodimer as well. Hence, it will be fascinating to completely decipher the underlying mechanisms of OsPRR73 in regulating rice circadian clock in the future, at both transcript and post‐translational levels. Nonetheless, our findings collectively provide a molecular link between circadian clock and salt tolerance in rice, through regulating sodium homeostasis, which paved a way for further deciphering regulatory network of rice circadian clock‐regulated abiotic stress response.
It has been long known that OsHKT2;1 can uptake Na+, but is rapidly repressed by salt stress (Horie et al, 2007), thus to maintain Na+ homeostasis. However, the molecular mechanism for salt stress‐induced repression of OsHKT2;1 remains unknown. In our study, we found that OsPRR73 could be rapidly induced by 4‐h NaCl treatment. Meanwhile, the transcript level of OsHKT2;1 was also induced by 4‐h NaCl treatment, but was severely reduced in 8‐h NaCl treated wild‐type plants, which was 4 h lagged behind the peak expression of OsPRR73, suggesting that the transcriptional repression role of OsPRR73 might be subjected to post‐transcriptional regulation. In Arabidopsis, the peaks of PRRs protein levels are usually delayed 2–3 h relative to their respective transcript peaks, and PRR proteins could be modified at post‐translational level (Fujiwara et al, 2008; Wang et al, 2010). It would be imperative to determine the mechanisms underlying OsPRR protein peaks lagging behind their transcript peaks in the future. The mechanism for rapid induction of OsPRR73 by NaCl treatment also remains to be elucidated. Importantly, the OsHKT2;1 transcript level was constantly higher in osprr73 mutant, especially in the presence of salt stress (Fig 4A), consistent with OsHKT2;1 is a transcriptional target of OsPRR73. In addition, OsHKT2;1 transcript level was slightly higher in the absence of NaCl treatment in osprr73 mutant, while it was much higher upon salt treatment, suggesting that there might be other unidentified factors responsible for the acute induction of OsHKT2;1 at the early stage of NaCl treatment.
The molecular mechanisms by which circadian clock regulates abiotic stresses, including cold and drought stress, have been extensively documented. However, the detailed mechanisms for the roles of core clock components in salt stress response are unclear, especially in rice and other monocot crops. Among the Na+ transporters, HKT has been shown to play critical roles in both monocots and dicots in the salinity stress tolerance, via maintaining Na+ and K+ homeostasis (Horie et al, 2009; Almeida et al, 2013). Unlike in Arabidopsis, there is only one HKT gene (Uozumi et al, 2000); rice HKT gene family contains nine members. Among them, OsHKT1;1 and OsHKT2;4 are high‐affinity potassium transporters. In this study, K+ content was not significantly changed; instead, Na+ was excessively accumulated in osprr73 mutants, indicating that OsPRR73 is majorly involved in regulating Na+ homeostasis. Besides OsHKT2;1, we found that transcript levels of OsHKT1;5, OsHKT2;3, and OsNHX2 also displayed robust rhythmic patterns (Appendix Fig S8). Nevertheless, OsHKT1;5 was only marginally decreased in osprr73 mutant after NaCl treated 4 h and maintained at relatively similar levels to the wild‐type plants, suggesting that it was unlikely a direct target of OsPRR73. NHX2 encodes a putative Na+/H+ exchanger which presumably transports Na+ from cytoplasm to vacuole to reduce cellular Na+ toxicity (Drozdowicz & Rea, 2001). The transcript level OsNHX2 was not significantly altered in our RNA‐seq data, suggesting other core clock components might be responsible for shaping their temporal transcription pattern. Hence, as plant circadian clocks frequently integrate multiple mechanisms to regulate a crucial physiological process, it is imaginable that OsPRR73 positively regulated salt tolerance may through both directly repressing OsHKT2;1 transcription and indirectly regulating the transcriptional abundance of other transporters, which jointly confers rice salt tolerance. Further efforts to comprehensively uncovering additional underlying pathways and identifying the superior haplotypes will surely be beneficial for breeding the salt‐tolerant rice cultivars.
Materials and Methods
Plant materials and growth condition
The T‐DNA insertion rice mutant of osprr73 (3A_12296) and its wild‐type Dongjin (DJ) were obtained from RiceGE (Rice Functional, http://signal.salk.edu/cgi‐bin/RiceGE) and identified by the OsPRR73‐specific primers and T‐DNA right board primers which were listed in the Appendix Table S2. Then, the mutant plants were backcrossed with DJ, and the F3 homozygous progeny was used for consequent research. For obtaining OsPRR73pro: GFP‐OsPRR73 transgenic plant in DJ background, the fragment of OsPRR73 promoter (−1,665 to −1 bp, upstream of the start codon) was firstly subcloned into EcoR I and Kpn I sites of p1300 promoterless vector, then the GFP‐OsPRR73 fragment was inserted into Xba I and Pst I sites. To make 35S: OsPRR73 constructs, the fragment of GFP‐OsPRR73 was inserted into the EcoRI and XhoI sites of the pENTR2B vector and then was subcloned into 35S: GFP vector via LR recombination. All the primers used are listed in Appendix Table S2. OsPRR73 mutants in Nipponbare (NIP) were generated with CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas9 technology according to previously described method (Ma et al, 2015). Briefly, the single‐guide RNAs (sgRNAs) were designed to target the first exon or crucial domain of OsPRR73 and other family members to generate allelic mutants (see Appendix Table S2 for a list of sgRNAs). Then, the sgRNAs were cloned into the pYLCRISPR/Cas9Pubi‐MH vector (Ma et al, 2015). These constructs were transformed into Agrobacteria tumefaciens EHA105 for inoculation with rice callus as previously reported (Hiei et al, 1994). To generate OsPRR73pro: GFP‐OsPRR73 osprr73 complemented plants, osprr73 plants were crossed with the transgenic plant OsPRR73pro: GFP‐OsPRR73. To make oshkt2;1 osprr73 double mutant, osprr73 plant was crossed with oshkt2;1. The seeds from self‐pollinated and F3 homozygous plants after genotyping were used in this study.
For physiological analysis, seeds were immersed in water at 37°C for 2 days; then, the germinated seeds were placed into the bottomless 96‐wells plate that containing liquid Yoshida’s solution. For seedling stage of NaCl or other chemicals treatment, 4‐week‐old seedlings were transferred to the Yoshida’s nutrient solution containing 180 mM NaCl for growing 21 days. Subsequently, they were moved to normal Yoshida’s nutrient solution for recovery. After 7 days or alternative duration as noted, the survival rate was calculated according to the percentage of alive seedlings. For mature stage of NaCl treatment, the plants were grown in trays filled with soil and irrigated with 75 mM NaCl in water. Unless the specifically described, all the rice seedlings were growing under 12‐h light/12‐h dark, 30 ± 2°C conditions.
Measurements of the chlorophyll contention and membrane ion leakage
Measurement of total chlorophyll content was performed as described previously with minor modification (Sakuraba et al, 2014; Qi et al, 2015; Zhang et al, 2018b). Briefly, the leaves of 14‐day‐old seedlings treated with or without 180 mM NaCl for 7 days were used for the measurement of chlorophyll content. The weight of detached leaves was recorded as fresh weight. For chlorophyll extraction, the leaves were firstly incubated in 80% acetone (v/v) in the darkness for 48 h at room temperature, and the volume was correspondingly recorded as V for calculation as the below formula. Absorbance was measured at OD645 and OD663 nm with a spectrophotometer. The chlorophyll contents were calculated according to the formula (8.02 A663 + 20.21 A645) × V/W.
The relative membrane ion leakage was measured according to the method described previously (Cao et al, 2007; Zhang et al, 2018b). In brief, seven leaves of each treatment were incubated in deionized water with gentle shaking overnight. The conductivity was measured before (C1) and after boiling for 15 min (C2) with an electro‐conductivity meter. The ratio of C1:C2 represents the membrane ion leakage rate.
Determination of Na+ and K+ concentration
The relative ion accumulation of Na+ and K+ was measured according to the method described previously (Liu et al, 2017) with slight modification. Roots and shoots of seedlings grown in the presence or absence of NaCl treatment were separately harvested and dried before weighed. All the samples were digested with nitric acid overnight and then heated to 200°C about 8 h. All samples were diluted with MilliQ water and filtrated. The Na+ and K+ contents in solutions were detected by inductively coupled plasma optical emission spectrometer (ICAP6300).
Determination of the malondialdehyde
Malondialdehyde (MDA) content was determined as previously described (Zhou et al, 2018). Briefly, about 0.1 g of rice leaves were homogenized in 3 ml of 10% trichloroacetic acid (TCA) and centrifuged at 1,503 g for 10 min. Two milliliters of the supernatant were reacted with 2 ml of 0.6% (w/v) thiobarbituric acid (TBA) (made in 10% TCA). The mixture was then boiled for 15 min and centrifuged at 13,523 g for 10 min. The absorbance of the supernatant was read at OD450, OD532, and OD600 nm. The MDA content was estimated using the extinction coefficient of (nmol/l/cm) and expressed as μmol/g FW.
Detection of reactive oxygen species and root H2O2 production
The formation of hydrogen peroxide was detected by 3,3‐diaminobenzidine (DAB) staining, as described before (Qiao et al, 2010). In brief, rice leaves from triplicate biological replicates of the samples were first cut into sections and then immersed in 10 ml staining solution with 1 mg/ml DAB containing 10 mM MES (pH 6.5). The incubation was conducted in room temperature in the dark overnight and shaking at 100 rpm. The reactions were stopped by transfer to 90% ethanol at boiling water bath until the chlorophyll was completed removed. The H2O2 production in roots of wild types and osprr73 mutations under normal and NaCl stress were detected using H2DCFDA (Molecular Probes, Thermo Fisher Scientific) based on the protocol described previously (Huang et al, 2009).
Measurement of peroxidase, superoxide dismutase, and catalase enzyme activity
Approx. 0.1 g rice leaves were ground with a cold mortar and pestle in 10 mM potassium phosphate buffer (pH 7.2). The homogenate was centrifuged at 1,503 g for 10 min at 4°C. The supernatant was crude enzyme extraction. The superoxide dismutase (SOD) and catalase (CAT) were measured using the manufacturer’s protocol (Nanjing Jiancheng Bioengineering Institute) followed the instruction.
Subcellular localization observation
To detect the subcellular localization of OsPRR73, the coding sequence of OsPRR73 was amplified and subcloned into pMDC45‐GFP vector, where GFP sequence was fused to OsPRR73 at its N terminus. The Agrobacteria harboring the corresponding construct and H2B‐mCherry were co‐infiltrated into the leaves of N. benthamiana. After 3 days, the fluorescence signals were observed by using confocal laser scanning microscope (Olympus FV 1000).
RNA‐sequencing analysis
For RNA‐sequencing experiments, plants were grown under 12‐h light/12‐h dark conditions at 30 ± 2°C for 14 days. The shoots of seedlings were harvested at the expression peak of OsPRR73 under NaCl condition 0, 4, and 28 h; the materials were frozen immediately in liquid nitrogen. The shoots from at least three seedlings were pooled to form one biological replicate, and three biological replicates were generated for each sample. RNA was extracted using a TRIzol reagent (Invitrogen). Sequencing libraries were generated using NEB Next UltraTM RNA Library Prep Kit for Illumina (NEB, USA) following manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. The library preparations were sequenced on an Illumina Hiseq X Ten platform and paired‐end reads were generated. The RNA‐seq clean reads mapped to the rice genome (MSU_v7.0) using TopHat (v2.1.1). DEseq (v1.10.1) was applied for differential gene expression analysis. Genes with q ≤ 0.05 and |log2_ratio| ≥ 1.5 were identified as DEGs. The GO enrichment of DEGs was performed by agriGO V2.0 (http://systemsbiology.cau.edu.cn/agriGOv2/).
RT–qPCR assay
The penultimate leaf blades of 14‐day‐old seedlings were harvested every 3 h during a 24‐h period from WT and osprr73 plants, respectively, under continuous light to examine the expression pattern of OsHKT2;1 and OsPRR73. To measure the transcript levels of OsPRR73 under NaCl stress, the 14‐day‐old seedlings were treated with 180 mM NaCl, and then, the shoots and roots were sampled at every 4 h starting from treatment. Total RNA was isolated using the TRIzol reagent (Invitrogen) and reverse‐transcribed into the first‐strand cDNA with a PrimeScript® RT Reagent Kit (Takara). Real‐time PCR was performed in an optical 96‐well plate with an Applied Biosystems StepOne™ Real‐Time PCR system. Each reaction contained 7.5 μl of 2 × SYBR Green Master Mix reagent, 3 μl of cDNA samples that was diluted 10‐folds, and 0.3 μl of 10 μM gene‐specific primers in a final volume of 15 μl. The following PCR program was used: 95°C for 2 min, followed by 40 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 15 s, followed by a melting‐curve program. Gene expression was normalized with the transcript level of Ubiquitin. Two‐week‐old seedlings were treated with 180 mM NaCl to examine the gene expression levels under salt stress.
Transcriptional activity assay
For transcriptional activity assay, the method was described previously (Zhang et al, 2018b), Agrobacterium tumefaciens AGL carrying the fusion expression vectors effector and reporter (effector: GFP‐OsPRR73 or GFP; reporter: OsHKT2;1pro:LUC‐1300) were co‐transformed into the leaves of N. benthamiana. The luciferase signal was detected using a CCD camera at 3rd day after infiltration. The bioluminescence intensity of LUC signals was quantified by Metamorph software.
ChIP (Chromatin immunoprecipitation)‐PCR
ChIP assay was performed based on the previous report (Lu et al, 2013) with 2‐week‐old GFP‐OsPRR73 transgenic seedlings, in which a mGFP coding sequence was fused in frame to the 5′‐terminal of the OsPRR73 gene in transgenic line, and the expression is driven by CaMV35S promoter. WT was used it as negative control to erase the background noise, as it did not contain GFP. The leaves of 2‐week‐old seedlings grown in normal nutrient solution were harvested and were cross‐linked with 1% (v/v) formaldehyde under vacuum for 30 min and then ground with a cold mortar to extract the nuclear proteins for immunoprecipitation by using GFP‐Trap® magnetic agarose (ChromoTek, Germany). The ChIPed DNA fragments were then used for quantitative PCR.
EMSA
The EMSA assay was conducted with Thermo Fisher kit according to the protocol described previously (Lu et al, 2013). The labeled probes (0.5 μl of each) were incubated with purified proteins (5 μg fusion protein per reaction) in 20‐μl mixtures at 4°C for 1 h. After adding 2 μl of loading buffer, samples were loaded to electrophoresis with 0.5 × TBE buffer at 4°C for 1 h. Labeled probe and the shifted DNA–protein complexes were visualized by chemiluminescence apparatus (Tanon‐5200).
Immunoprecipitation‐mass spectrometry (IP‐MS) assay
Two‐week‐old rice transgenetic seedlings of 35S:GFP‐OsPRR73 grown under 12‐h light/12‐h dark, 30 ± 2°C conditions were collected at ZT12 (Zeitgeber time 12). Then, 3 ml of liquid nitrogen ground tissue powders of this sample was extracted protein and performed according to the method described previously (Wang et al, 2019), with two biological replicates.
Biomolecular fluorescence complementation assay
Agrobacteria containing 35S:OsPRR73‐nYFP (1–158) and 35S:HDAC10‐cYFP (159–238) were co‐infiltrated into N. benthamiana. Agrobacteria containing H2B‐mcherry was used as a nuclear marker. After 3 days, the confocal laser scanning microscope (Olympus FV1000) was used to observe the fluorescent signal of epidermal cells.
Co‐immunoprecipitation assays
The coding sequence of OsPRR73 and HDAC10 separately were, respectively, fused to the downstream of the N‐GFP or upstream of C‐Flag tags. Then, the agrobactria containing 35S:GFP‐OsPRR73 or 35S:HDAC10‐FLAG were co‐infiltrated into leaves of N. benthamiana. After 3 days, the materials were harvested and ground into power in liquid nitrogen. The protein extraction and co‐immunoprecipitation assays were conducted according to the method described previously with GFP‐Trap beads (ChromoTek) (Wang et al, 2013). GFP‐tagged OsPRR73, its deletions and FLAG tagged HDAC10 were detected by Western blotting using GFP antibody (ab6556, Abcam) or FLAG antibody (M2008, Abmart) as noted.
Split‐luciferase assay (Split‐LUC)
The coding sequence of OsPRR73 was subcloned into the pCAMBIA1300‐nLUC vector with Kpn I and Sal I sites, while the ORF of HDAC10 was subcloned into pCAMBIA1300‐cLUC vector. Then, pairwise constructs were transiently co‐infiltrated into N. benthamiana leaves. After 3 days, the leaves were briefly immersed into the luciferin buffer for 1 min and then the LUC signals were detected by the chemiluminescence apparatus (Tanon‐5200).
Accession number
Sequence data from this article can be found in the Rice Genome Annotation Project Database (http://rice.plantbiology.msu.edu/) under the following the accession numbers: OsPRR1 LOC_Os02g40510; OsPRR37 LOC_Os07g49460; OsPRR59 LOC_Os11g05930; OsPRR73 LOC_Os03g17570; OsPRR95 LOC_Os09g36220; LOC_Os03g17560; LOC_Os03g17580; OsHKT2;1 LOC_Os06g48810; OsHK1;5 LOC_Os01g20160; HDAC10 LOC_Os12g08220; SDG704 LOC_Os11g38900; OsCCA1 LOC_Os08g0157600; OsLHY LOC_Os06g0728700; OsGI LOC_Os01g0182600; OsFKF1 LOC_Os11g0547000; OsLUX LOC_Os01g0971800; OsELF3.1 LOC_Os06g0142600; OsLKP2 LOC_Os02g0150800.
Author contributions
Research design: HW and LW; Research: HW, XW, YH, and HX; Data analysis and discussion: HW, XW, YH, HX, and LW; Manuscript writing: HW and LW.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
We thank Prof. JC Jang (Ohio State University) for his constructive comments on the manuscript, and Dr. Zhuang Lu and Jingquan Li from Plant Science Facility of the Institute of Botany, Chinese Academy of Sciences for their excellent technical assistance on mass spectrometry and confocal microscopy, respectively. This work was supported by the National Key Research and Development Program of China (2016YFD0100604), National Natural Science Foundation of China (No. 31770287), and Strategic Priority Research Program of the Chinese Academy of Sciences, Grant No. XDB27030206.
Data availability
The RNA‐seq raw data have been deposited in NCBI SRA database with Bioproject number PRJNA602982 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA602982).
 Clock component OsPRR73 positively regulates rice salt tolerance by modulating OsHKT2;1‐mediated sodium homeostasis
Clock component OsPRR73 positively regulates rice salt tolerance by modulating OsHKT2;1‐mediated sodium homeostasis
