
- Altmetric
Background
X‐linked dystonia‐parkinsonism is a rare neurological disease endemic to the Philippines. Dystonic symptoms appear in males at the mean age of 40 years and progress to parkinsonism with degenerative pathology in the striatum. A retrotransposon inserted in intron 32 of the TAF1 gene leads to alternative splicing in the region and a reduction of the full‐length mRNA transcript.
Objectives
The objective of this study was to discover cell‐based and biofluid‐based biomarkers for X‐linked dystonia‐parkinsonism.
Methods
RNA from patient‐derived neural progenitor cells and their secreted extracellular vesicles were used to screen for dysregulation of TAF1 expression. Droplet‐digital polymerase chain reaction was used to quantify the expression of TAF1 mRNA fragments 5′ and 3′ to the retrotransposon insertion and the disease‐specific splice variant TAF1‐32i in whole‐blood RNA. Plasma levels of neurofilament light chain were measured using single‐molecule array.
Results
In neural progenitor cells and their extracellular vesicles, we confirmed that the TAF1‐3′/5′ ratio was lower in patient samples, whereas TAF1‐32i expression is higher relative to controls. In whole‐blood RNA, both TAF1‐3′/5′ ratio and TAF1‐32i expression can differentiate patient (n = 44) from control samples (n = 18) with high accuracy. Neurofilament light chain plasma levels were significantly elevated in patients (n = 43) compared with both carriers (n = 16) and controls (n = 21), with area under the curve of 0.79.
Conclusions
TAF1 dysregulation in blood serves as a disease‐specific biomarker that could be used as a readout for monitoring therapies targeting TAF1 splicing. Neurofilament light chain could be used in monitoring neurodegeneration and disease progression in patients. © 2020 The Authors. Movement Disorders published by Wiley Periodicals LLC on behalf of International Parkinson and Movement Disorder Society.
X‐linked dystonia‐parkinsonism (XDP) is a rare neurological disease found predominantly in men with maternal ancestry linked to the island of Panay, Philippines. Dystonic symptoms typically appear in males at the mean age of 40 years and progress to parkinsonism with degenerative pathology in the striatum. 1 , 2 , 3 , 4 , 5 , 6 , 7 , 8 , 9 In the most recent study in 2010, 505 patients with XDP were registered in the Philippines, 312 of which were survivors. The reported prevalence rate in the country overall was 0.31 per 100,000, and for Panay Island, 5.74 per 1,000,000. 2 Recent efforts have narrowed down the causal mutation to a short interspersed elements (SINE) ‐ variable number of tandem repeats (VNTR) ‐ Alu (SVA) retrotransposon insertion in intron 32 of the TAF1 gene10, 11, 12, 13 with an increased number of repeat elements in this SVA associated with earlier disease onset. 14 , 15 TAF1 encodes the TATA‐box binding protein associated factor 1 (TAF1), the largest subunit of the multiprotein complex that makes up the general transcription factor complex, transcription factor II D (TFIID), which is critical to the formation of the RNA polymerase II preinitiation complex. 16 , 17 , 18 The 38 constitutive exons of TAF1 undergo various splicing events at the mRNA level, leading to differential expression of different isoforms combining exons 1‐38, or in some cases including alternative exons annotated as 32′, 34′, and 35′. 10 , 11 , 12
In XDP cells, recent transcriptome assembly analysis showed that exon usage progressively decreases 3′ to intron 32. 10 It is thought that the presence of the SVA in intron 32, possibly attributed to the formation of stacked guanine tetrads known as G4 motifs and/or reverse transcription of SVA‐derived sequences, is responsible for this transcriptional interference. 10 , 14 Recent studies have shown that the excision of the SVA rescues TAF1 expression levels 3′ to the SVA. 10 , 19 Earlier studies compared TAF1 expression in various tissues via reverse transcription polymerase chain reaction (RT‐qPCR) amplification using primers/probes that span the TAF1 transcript, including sets that amplify 5′ to the SVA insertion (TAF1‐5′) and others 3′ to it (TAF1‐3′). Additional primers/probes were designed to amplify alternatively spliced isoforms, 1 of which is specific to a neuronal TAF1 isoform (nTAF1), which includes a 34′ exon. 11 Consistent with the transcriptome assembly, TAF1‐3′ expression was lower than TAF1‐5′ expression in patient‐derived neuronal stem cells (NSCs) 10 and primary fibroblasts 20 , 21 as well as RNA extracted from whole blood of patients with XDP compared with controls. 20 The neuronal isoform, nTAF1, was shown to have lower expression in the brain of 1 patient with XDP 11 and in patient‐derived NSCs 21 based on RT‐qPCR amplification. In addition to decreased exon usage 3′ to intron 32, the presence of the SVA retrotransposon in TAF1 induces partial retention of the proximal segment of intron 32 as well as multiple aberrant splicing events that terminate immediately proximal to the SVA insertion site, the most abundant of which was annotated as TAF1‐32i. This TAF1 isoform is expressed at higher levels in XDP patient‐derived cell lines compared with controls, especially cells undergoing rapid division in the following rank order: induced pluripotent stem cells (iPSCs)>NSCs>fibroblasts>neurons. 10 Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR‐associated protein‐9 nuclease (CRISPR/Cas9) ‐based gene therapy on patient cell lines to selectively excise the SVA in intron 32 has normalized levels of TAF1‐32i. 10
Considerable evidence supports neurofilament light chain (NfL) as a biomarker for neurodegeneration. 22 , 23 , 24 , 25 , 26 Through the development of a highly sensitive fourth‐generation single molecule array, SiMoA (Quanterix Corporation, Billerica, MA), for detection in blood, 27 , 28 NfL has been reported in different neurodegenerative diseases as a blood‐based biomarker correlating with disease status, progression, and outcomes in different neurological diseases. 23 , 28 Specifically, it has been shown to be useful in the differential diagnoses of parkinsonian disorders. 24 , 29 , 30
Clinically, XDP is diagnosed in patients with signs of dystonia and parkinsonism who have a positive history of affected relatives and maternal ancestry from Panay island. 2 Cell‐based and biofluid‐based disease‐specific biomarkers will be needed to understand disease mechanisms, predict progression, and serve as noninvasive readouts for novel therapeutics. To identify a potential biomarker in XDP, we focused on RNA in extracellular vesicles (EVs). EVs are found within various biofluids, including blood, cerebrospinal fluid, and urine, and provide a protective environment for different RNA species, including mutant mRNAs, miRNAs, and splice variants. 31 , 32 , 33 , 34 In this study, we were able to quantitate levels of TAF1‐5′ and TAF1‐3′ as well as TAF1‐32i RNA expression in neural progenitor cells (NPCs), NPC‐derived EVs, and whole‐blood RNA from patients with XDP, female carriers, and healthy controls. We found that TAF1 RNA species are differentially expressed among these study groups. In addition, we found that NfL is increased in the plasma of patients with XDP, adding XDP to the list of parkinsonian disorders and other neurodegenerative diseases with this biomarker feature. Overall, these studies implicate whole blood as a feasible biofluid to detect disease‐specific peripheral (and potentially brain derived) biomarkers in XDP, including differential TAF1 transcript expression and increased NfL levels.
Methods
Participant Recruitment
Standardized tissue samples (blood and urine) and phenotype data (Table 1, Table S1, Table S2) were obtained from tissue and data banks, including the Dystonia Partners Research Bank approved by the Partners Human Research Committee (Boston, MA) and the XDP‐Partners Research Bank approved by Jose Reyes Memorial Medical Center (Manila, Philippines). All participants provided written informed consent for their samples and data to be used for genetic and cellular analyses. All participants were of Filipino decent.

| P Value | |||||
|---|---|---|---|---|---|
| Patients with XDP | Female Carriers | Controls | Presymptomatic | ||
| Plasma samples | |||||
| n | 43 | 16 | 21 | 2 | |
| Age, y (SD) | 46.53 (9.37) | 45.19 (24.04) | 43.38 (14.46) | 23.50 (7.78) | 0.17 |
| Sex, M/F | 43/0 | 0/16 | 12/9 | 2/0 | |
| Dystonia at presentation, n (%) | 36 (83.72) | – | – | – | |
| Parkinsonism at presentation, n (%) | 7 (16.27) | 4 (25.00) | 3 (13.28) | ‐ | |
| Whole‐blood RNA samples | |||||
| n | 44 | 17 | 18 | 3 | |
| Age, y (SD) | 47.70 (10.55) | 48.59 (24.19) | 41.44 (14.63) | 32.67 (16.80) | 0.19 |
| Sex, M/F | 44/0 | 0/17 | 7/11 | 3/0 | |
| Dystonia at presentation, n (%) | 35 (79.54%) | – | – | – | |
| Parkinsonism at presentation, n (%) | 9 (20.45%) | 4 (23.53) | 2 (11.11) | ‐ | |
Plasma samples were used for plasma neurofilament light chain assays. Whole‐blood RNA samples were used for droplet‐digital polymerase chain reaction expression analysis of TAF1‐3′/5′ and TAF1‐32i.XDP, X‐linked dystonia‐parkinsonism; SD, standard deviation; M, male; F, female.
Sample Collection
To obtain plasma, whole blood from patients with XDP, female carriers, and controls was collected in 10 mL ethylenediaminetetraacetic acid tubes (BD Vacutainer Plastic Blood Collection Tubes with K2EDTA; ThermoFisher Scientific, Waltham, MA). Within 2 hours of collection, tubes were centrifuged at 1100g at room temperature for 10 minutes. Plasma was removed from the upper layer and then filtered through a Millex‐AA Syringe Filter Unit, 0.8 μm (MilliporeSigma, Burlington, MA), and aliquoted into cryovials and stored at −80°C. For blood RNA, whole blood from participants was collected using PAXgene tubes (Qiagen, Germantown, MD). Urine samples were collected using a sterile technique in 200 mL plastic bottles. Urine was centrifuged at low speed (2000g) with the supernatant passed through a Millex‐AA Syringe Filter Unit, 0.8 μm, and 50 mL aliquots were frozen at −80°C. Samples were shipped on ice or immediately processed whenever possible.
Cell Culture
Stable and highly expandable NPCs were derived from previously established iPSCs of patients with XDP and controls. 10 NPC differentiation was performed based on a previously reported protocol, 35 with several in‐house modifications. Sorting steps and subsequent culturing were followed as described in ref. 35 on Geltrex‐coated plates. NPCs at passage >9 were used for experiments. Subsequently, NPCs were grown for no more than 20 passages on Geltrex‐coated (ThermoFisher Scientific) tissue cultureware using Dulbecco's Modified Eagle Medium/Nutrient Mixture F‐12 (DMEM/F12 media) (ThermoFisher Scientific) supplemented with 2% B27 (ThermoFisher Scientific), 20 ng/mL of epidermal growth factor (EGF) (PeproTech, Rocky Hill, NJ), 20 ng/mL fibroblast growth factor (FGF) (MilliporeSigma), 5 ng/mL heparin (Sigma‐Aldrich), and 1% penicilin‐streptomycin (Corning, Corning, NY [catalog number 30‐002‐CI]).
EV Isolation from Culture Media, Plasma, and Urine Samples
Conditioned culture media from NPCs was collected after 24 to 48 hours of incubation. Using Amicon Ultra‐15 Centrifugal Filter Units, 100 kD (MilliporeSigma), 60 to 90 mL of the NPC‐conditioned media or 150 to 200 mL of the urine samples were concentrated to a final volume of 500 μL. The concentrated sample was then added to “qEV” columns (IZON Science, Medford, MA) for size exclusion chromatography to separate EVs from free protein according to size. Plasma samples were filtered through a 0.8 μm pore‐size membrane (Millex‐AA Syringe Filter Unit, MilliporeSigma) and then 500 μL were added directly to “qEV” columns for EV isolation. For all samples, the IZON “qEV” automatic fraction collector was used to collect fractions 7 to 11 containing EVs. 36 Using Amicon Ultra‐0.5 Centrifugal Filter Units, 30 kD (MilliporeSigma), a total of 2.5 mL EV‐enriched filtrate was then concentrated to 100 to 200 μL for downstream RNA isolation.
RNA Extraction
RNA was extracted from NPC cell pellets and EV concentrate from NPC, plasma, and urine by adding QIAzol (Qiagen); samples were then mixed with one‐fifth volume of chloroform with brief centrifugation at 12,000g to allow for phase separation. The aqueous phase was processed using miRNeasy spin columns (Qiagen) with on‐column DNase digestion as recommended. RNA was extracted from whole blood using the PAXgene Blood RNA Kit according to the manufacturer's protocol (Qiagen). The resulting RNA samples were quantified by Nanodrop (ThermoFischer Scientific) and Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). RNA from NPCs (40 ng), NPC EVs (2–10 ng), plasma EVs (0.5–3.5 ng), urine EVs (0.5–3.5 ng), and whole blood (40 ng) were reverse transcribed using the SuperScript VILO cDNA Synthesis Kit (ThermoFisher Scientific).
RT‐qPCR
RT‐qPCR was performed using custom TAF1 TaqMan primers (ThermoFisher Scientific) as previously described 11 to detect TAF1‐5′ (forward 5′ ➔ 3′: GACTGACGGTGCCTTGGT, reverse 5′ ➔ 3′: GTCTGAATAGTCCACAGCATCTTCT, reporter probe 5′ ➔ 3′: ACCCACCCTTCATCATTT, amplicon size 70 bp) and TAF1‐3′ (forward 5′ ➔ 3′: ACCTTATTCTGGCCAACAGTGTT, reverse 5′ ➔ 3′: ACAATCTCCTGGGCAGTCTTAGTAT, reporter probe 5′ ➔ 3′: ACTCTCAGGTCCATTATAC, amplicon size 73 bp) (Fig. 1D). The TAF1‐32i Taqman primer set (TaqMan Custom Assay identification number: AJWR28J; ThermoFischer Scientific) was used to detect the presence of the exon 32/intron 32 splice site such that the forward primer bound to exon 32 and the reverse primer bound to the spliced region of intron 32, named here as exon 32i (Fig. 2D), and the probe spanned the splice junction. 10

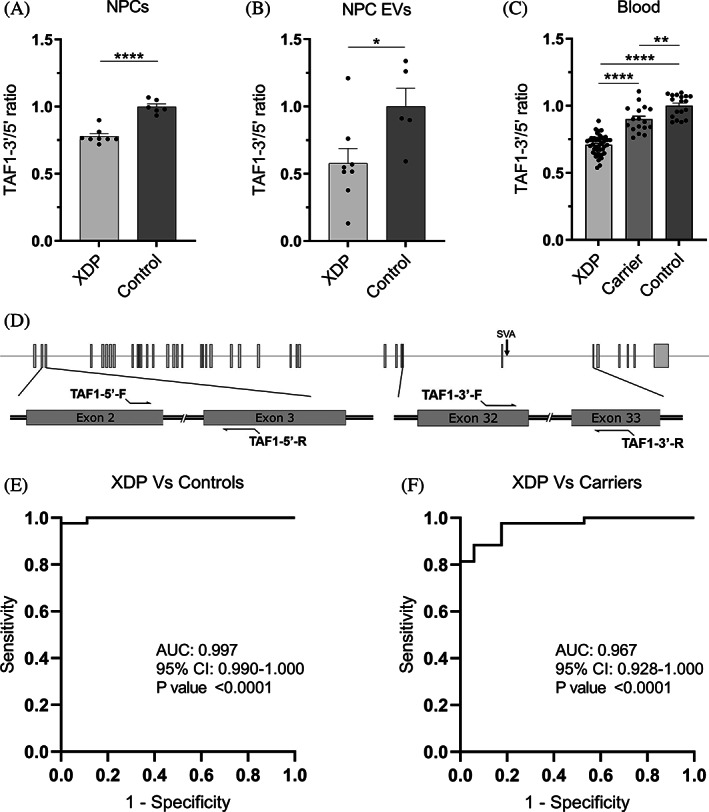
TAF1‐3′/5′ ratio expression in NPCs, NPC EVs, and peripheral blood. (A) RT‐qPCR expression of TAF1‐3′/5′ ratio in XDP (n = 8) and control (n = 6) NPCs. (B) Droplet‐digital polymerase chain reaction expression of TAF1‐3′/5′ ratio in XDP (n = 8) and control (n = 5) EVs isolated from NPCs in culture. (C) Droplet‐digital polymerase chain reaction expression of TAF1‐3′/5′ ratio in patients with XDP (n = 43), female carriers (n = 17), and controls (n = 18) in whole‐blood RNA. Each dot represents a unique sample. Mean and standard error are represented. (D) Schematic for TAF1‐5′ primers relative to TAF1 exons 2 and 3 and TAF1‐3′ primers relative to the SVA and TAF1 exons 32 and 33. (E) Receiver operating characteristic analysis of TAF1‐3′/5′ ratio in whole‐blood RNA in XDP versus control samples. (F) Receiver operating characteristic analysis of TAF1‐3′/5′ ratio in whole‐blood RNA in XDP versus female carrier samples. AUC, area under the curve; CI, confidence interval; EV, extracellular vesicle; NPCs, neural progenitor cells; XDP, X‐linked dystonia‐parkinsonism. *P < 0.05, **P < 0.01, ****P < 0.0001.

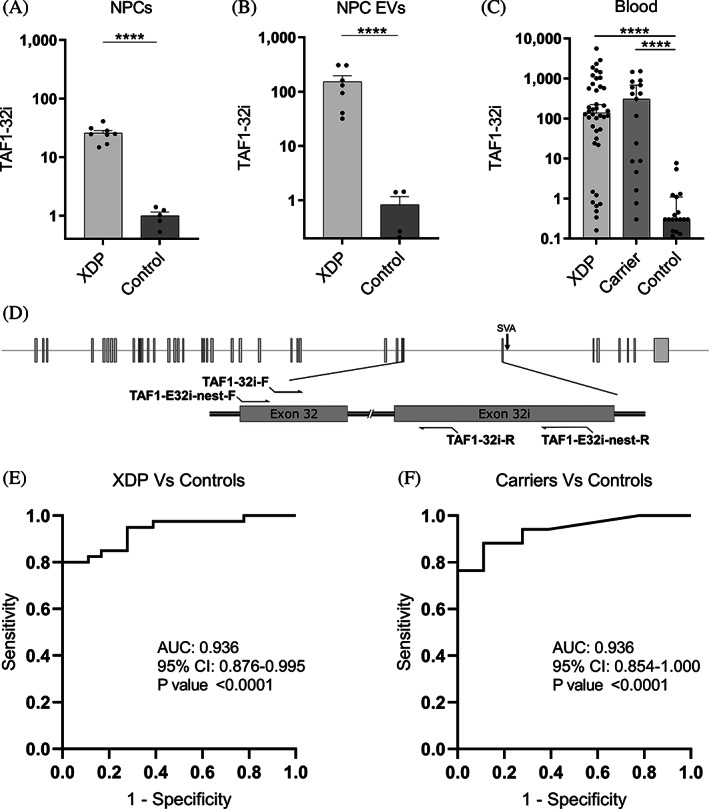
TAF1‐32i expression in NPCs, NPC EVs, and peripheral blood. (A) RT‐PCR expression of TAF1‐32i in XDP (n = 8) and control (n = 5) NPCs. (B) Droplet‐digital polymerase chain reaction expression of TAF1‐32i in XDP (n = 8) and control (n = 5) EVs isolated from NPCs in culture. (C) Droplet‐digital polymerase chain reaction expression of TAF1‐32i ratio in XDP (n = 40), female carriers (n = 17), and controls (n = 18) in whole‐blood RNA. Dotted line shown at y = 5 to separate background expression seen in all 17 controls, in 8 XDP samples, and 3 carrier samples (y < 5). Each dot represents a unique sample. Means with standard errors are represented. Data are logarithmically transformed. (D) Schematic for TAF1‐32i and TAF1‐32i preamplification primers (TAF1‐E32i‐nest) relative to the SVA and TAF1 exons 32 and 32i. (E) receiver operating characteristic analysis of TAF1‐32i expression in whole‐blood RNA in XDP versus control samples. (F) Receiver operating characteristic analysis of TAF1‐32i expression in whole‐blood RNA in female carrier versus control samples. AUC, area under the curve; CI, confidence interval; EV, extracellular vesicle; NPCs, neural progenitor cells; XDP, X‐linked dystonia‐parkinsonism. ****P < 0.0001.
PCR Preamplification
TAF1‐E32i‐preamplification primers (forward 5′ ➔ 3′: ACATCTCCAAGCACAAGTATCA) and (reverse 5′ ➔ 3′: GTAATGTACCAATATAAATTTCCTGGTTT, amplicon size 208 bp) were used to amplify a region of 208 bp spanning the TAF1‐32i splice variant site (Fig. 2D). Reverse‐transcribed cDNA was used as a template to run 10 to 15 cycles of PCR with Phusion Hot Start Flex DNA Polymerase according to the manufacturer's protocol (New England Biolabs, Ipswich, MA). The amplified cDNA mixture was then cleaned and concentrated using DNA Clean and Concentrator‐5 Kit (Zymo Research, Irvine, CA) according to the manufacturer's protocol.
Droplet‐Digital PCR
Gene expression was analyzed via droplet‐digital PCR using the same Taqman probes used for RT‐qPCR. Using the protocol listed by the manufacturer, the droplets were generated with the DG32 Cartridge using the Automated Droplet Generator QX200 AutoDG Droplet Digital PCR System from Bio‐Rad (Hercules, CA), and PCR was performed with thermal cycling conditions as described by the manufacturer. QX200 Droplet Reader and QuantaSoft Software (Bio‐Rad) were used to analyze gene expression.
NfL SiMoA
NfL concentrations were measured using the HD‐X NfL kit (catalog number 103186) on the SiMoA HD‐X Analyzer (Quanterix Corporation). Plasma samples were centrifuged at 10,000g for 10 minutes and diluted 1:4 in sample buffer (100 μL plasma). All samples were run in duplicates. The assay was run according to manufacturer's protocol. This assay has a lower limit of quantification of 0.174 pg/mL, a limit of detection of 0.038 pg/mL (range 0.003–0.079 pg/mL), and a dynamic range in serum/plasma of 0 to 1800 pg/mL.
Statistical Analysis
GraphPad Prism version 8.0.0 (GraphPad Software, San Diego, CA) was used to analyze RNA expression, NfL plasma quantification, receiver operating characteristic curves, and linear regression analysis. The Student t test was used to compare 2 means from data sets that exhibited normal distribution (Figs. 1A,B and 2A,B). One‐way analysis of variance was used to compare 3 or more means for normally distributed data (Fig. 1C). Data from TAF1‐32i expression (Fig. 2) were logarithmically transformed. The Kruskal‐Wallis test was used to compare 3 or more medians from data sets that were not normally distributed (Figs. 2C, 3A,C,D,E). Statistical significance was considered for P value <0.05.

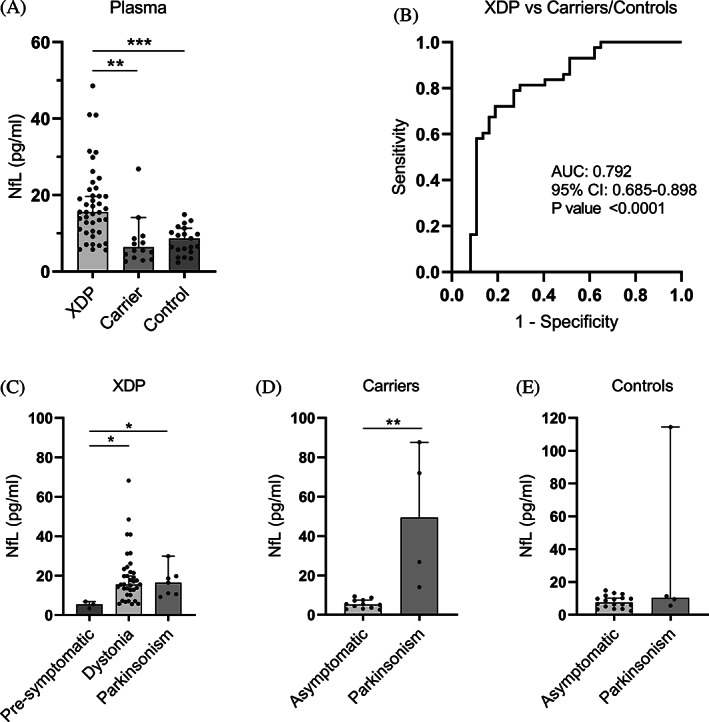
NfL in plasma. (A) NfL protein concentration (pg/ml) in plasma from patients with XDP (n = 43), female carriers (n = 16), and healthy controls (n = 21) using SiMoA. Medians with 95% CIs are represented. (B) Receiver operating characteristic analysis of plasma NfL in patients with XDP versus female carriers and controls. (C) Subgroup analysis of plasma NfL in patients with XDP according to the type of symptom, if any, at disease onset. (D) Subgroup analysis of plasma NfL in female carriers according to the type of symptom at presentation, if any. (E) Subgroup analysis of plasma NfL in controls according to the type of symptom at presentation, if any. Medians with 95% CIs represented. AUC, area under the curve; CI, confidence interval; NfL, neurofilament light chain; XDP, X‐linked dystonia‐parkinsonism. *P < 0.05, **P < 0.01, ***P < 0.001.
Results
TAF1 Expression in NPCs, NPC EVs, and Peripheral Blood
To accurately compare TAF1 RNA dysregulation in XDP using a variety of sources (cell lysates, EVs, and whole blood), we evaluated TAF1 transcript expression as a ratio of TAF1‐3′/5′ using TAF1‐5′ expression for normalization, as the expression of classic housekeeping genes was found to vary greatly among tissues. In our NPC cell model, TAF1‐5′ expression showed no significant difference between XDP and control lines when corrected to the housekeeping gene GAPDH, whereas TAF1‐3′ expression was significantly lower (data not shown) in XDP samples, consistent with decreased TAF1 transcription expression 3′ to intron 32 in other cell types including iPSCs, NSCs, and fibroblasts. 10 , 20 , 21 We confirmed that TAF1‐3′/5′ ratio reflects the expected dysregulation in NPCs, with 22% lower expression (P < 0.0001) in XDP NPCs (n = 8) relative to controls (n = 6) (Fig. 1A). In NPC EVs, TAF1‐3′/5′ expression was 42% lower (P < 0.05) in XDP samples (n = 8) compared with controls (n = 5) (Fig. 1B). In peripheral blood, XDP samples (n = 3) showed a 29% decrease (P < 0.0001) in TAF1‐3′/5′ ratio relative to controls (n = 18), and its expression in female carriers (n = 17) was on average 19% higher (P < 0.0001) than patient samples and only 10% lower (P < 0.01) than controls (Fig. 1C). TAF1‐3′/5′ expression can accurately differentiate patients with XDP from control samples (area under the curve (AUC) = 0.997, P < 0.0001) (Fig. 1E) and patients with XDP from carrier samples (AUC = 0.967, P < 0.0001) (Fig. 1F). As for the 3 males with presymptomatic XDP, their TAF1‐3′/5′ ratios were 25%, 26%, and 19% lower than the average of the control samples (Table S1). Linear regression analysis between TAF1‐3′/5′ expression and subject parameters, including, age, age at disease onset, duration of disease, repeat size, and symptoms, were not revealing (data not shown).
TAF1‐32i Expression in NPCs, NPC EVs, and Peripheral Blood
qPCR analysis of the TAF1‐32i splice variant in NPCs derived from iPSCs showed on average a 25‐fold increase (P < 0.0001) in levels in XDP cells (n = 8) relative to controls (n = 6) (Fig. 2A). These data support a similar expression signature shown previously in other XDP‐derived cell models, including fibroblasts, iPSCs, and NSCs. 10 To increase our detection limit of this lowly expressed splice variant, we performed preamplification of the region in cDNA via PCR then used droplet‐digital PCR for quantitative detection using TaqMan primer/probes. Using this method, we found that the TAF1‐32i splice variant is abundant in EVs derived from XDP cell lines (n = 8), with a 184‐fold increase (P < 0.0001) in expression relative to EVs derived from control NPCs (n = 5) (Fig. 2B).
Next, we quantified TAF1‐32i expression in whole‐blood RNA from PAX tubes from patients with XDP (n = 40), female carriers (n = 17), and controls (n = 18) (Tables S1 and S2) using the same preamplification methods as previously. We found high expression levels of the TAF1‐32i splice variant in peripheral blood from the patients with XDP (P < 0.0001) and female carriers (P < 0.0001) compared with controls (Fig. 2C). Similar to the TAF1‐3′/5′ ratio, TAF1‐32i expression can accurately differentiate patients with XDP from control samples (AUC = 0.939, P < 0.0001) (Fig. 2E) and female carriers from control samples (AUC = 0.936, P < 0.0001) (Fig. 2E). Two males with presymptomatic XDP had TAF1‐32i expression 24 and 2300 times higher than the average of the control samples (Table S1).Thus, for the first time, we demonstrated evidence of high levels of the aberrant splice variant TAF1‐32i in peripheral blood samples in patients with XDP and female carriers. Linear regression analysis between TAF1‐32i expression and subject parameters were not revealing (data not shown).
NfL in XDP Plasma
Using the SiMoA platform, we were able to assay NfL protein levels in plasma from patients with XDP (n = 43), female carriers (n = 16), and healthy controls (n = 21) (Table 1). We detected abnormally high levels of plasma NfL in patients with XDP with a median of 15.57 pg/mL compared with 6.38 pg/mL in female carriers (P < 0.001) and 8.66 pg/mL in healthy controls (P < 0.01) (Fig. 3A). Plasma NfL is capable of differentiating samples from patients with XDP from female carriers and control samples together with a high AUC of 0.792 (P < 0.0001) (Fig. 3B). Three males with presymptomatic XDP aged between 40 and 51 years showed low levels of plasma NfL at a median of 5.49 pg/mL versus a median of 15.54 (P < 0.05) and 16.42 pg/mL (P < 0.05) in patients with XDP with dystonia and parkinsonism symptoms at disease onset, respectively (Fig. 3C). Four female carriers, aged between 77 and 84 years, had parkinsonism symptoms, but no specific clinical diagnosis. Their NfL levels were high at a median of 49.42 pg/μL versus 5.26 pg/μL in asymptomatic female carriers (P < 0.01) (Fig. 3D). Four male controls, aged between 32 and 53, also had parkinsonism symptoms with no specific clinical diagnosis, but did not carry the XDP‐specific SVA. Unlike the female carriers, only 1 symptomatic control had high NfL levels, and the median level (9.47 pg/mL) was not significantly higher than asymptomatic controls (8.66 pg/mL) (Fig. 3E). Linear regression analysis between plasma NfL levels and subject parameters were not revealing (Fig. S1).
Discussion
Our results confirm transcriptional dysregulation of TAF1 in our NPC cell model, in NPC‐secreted EVs, and in peripheral blood in patients with XDP and female carriers compared with neurologically healthy controls. Dysregulation manifested as a decreased ratio of TAF1‐3′/5’ mRNA and increased expression of the aberrant splice variant TAF1‐32i. This expression signature for TAF1 transcripts can be used as an XDP‐specific biomarker. In addition, we showed evidence of increased NfL protein levels in XDP plasma, which can be used as a nonspecific disease biomarker of neurodegeneration.
Previous work on TAF1 transcripts has consistently shown decreased exon expression 3′ to the SVA insertion in intron 32. 10 , 11 , 20 , 21 Most work was done on XDP patient‐derived cell lines, including fibroblasts, 10 , 20 iPSCs,10 and NSCs. 10 , 21 In addition, TAF1‐32i splice variant expression has been quantified in the same patient cell lines and was directly linked to the presence of the SVA in intron 32. 10 As expected, our NPC cells derived from patients with XDP showed similar TAF1 dysregulation, reflected by the decreased ratio of TAF1‐3′/5′ transcripts and the increased expression of the aberrant splice variant TAF1‐32i. Furthermore, we showed that TAF1 fragments, including 5′, 3′, and 32i, are secreted in EVs isolated from the NPC culture media and that the expression of these fragments in EVs reflected the dysregulation occurring in XDP NPC lines. These findings shed light on potential cellular processing mechanisms for TAF1 RNA in XDP. Cells could be using EVs to actively discard the aberrant splice variant TAF1‐32i. 37 , 38 , 39 , 40 Alternatively, cells may indiscriminately package RNA into EVs, 37 , 38 , 40 , 41 which would explain why NPC EVs reflect the TAF1 dysregulation seen in the source cell. It is important to note that the expression of the TAF1‐3′/5′ ratio in NPC EVs is highly variable relative to its expression in NPCs. This could be attributed to the extensive sample handling necessary for our analysis or to an underlying biological process pertaining to the cellular handling of different TAF1 fragments as mentioned previously. Irrespective of the secretion mechanism, EVs have been implicated in the pathogenesis of neurological disorders 42 , 43 , 44 and can be used as disease biomarkers when isolated from biofluids. 33 , 45 , 46 , 47 , 48 , 49 , 50 We assayed extracellular RNA from plasma and urine EVs from patient and control samples using size exclusion chromatography. 36 Our low RNA yields (0.5–3.5 ng total per sample) did not allow us to amplify TAF1‐32i in any of these samples (data not shown). With the improving techniques in isolating and assaying EV contents, our findings indicate that EVs from biofluids may be useful in assaying TAF1 transcripts and splice variants from patient samples.
We also interrogated TAF1 dysregulation in whole‐blood RNA. A previous study showed a reduction in TAF1‐3′ expression in XDP whole‐blood RNA compared with TAF1‐5′, which remained unchanged relative to control samples. 20 Our results support those findings by showing that the TAF1‐3′/5′ ratio is effective in distinguishing patients with XDP, asymptomatic female carriers, and healthy controls. Interestingly, 3 presymptomatic males with the XDP haplotype, aged 18, 29, and 51, had TAF1‐3′/5′ levels similar to patients with XDP. We hypothesize that this marginal yet consistent decrease in TAF1‐3′/5′ may correlate with disease status and could be related to an active underlying pathological process in both presymptomatic and symptomatic patients with XDP. TAF1‐32i XDP‐specific splice variant was discovered by transcriptome assembly and shown to be expressed in dividing cells, including iPSC, NSCs, and fibroblasts. 10 Ours is the first study to show evidence of TAF1‐32i expression in whole‐blood RNA, shedding light on a systemic dysregulation in TAF1 expression and the potential use of TAF1 RNA fragments in biofluids as an XDP disease‐specific biomarker.
TAF1 expression in asymptomatic female carriers had not been studied before, and these are the first experiments to show TAF1 dysregulation in whole‐blood RNA from carriers. Most female carriers of the disease‐causing haplotype are asymptomatic, with only 14 symptomatic cases reported to date of more than 500 male patients. 51 , 52 , 53 , 54 Although skewed X‐chromosome inactivation has been reported as the underlying mechanism in at least 1 case of an XDP‐carrier symptomatic female, 52 it has also been shown to be the underlying protective mechanism against X‐linked neurological diseases caused by TAF1 coding mutations. 55 , 56 , 57 We hypothesize that skewed X‐chromosome inactivation may attenuate TAF1 dysregulation, which in turn protects female carriers from the downstream effects of a significant decrease in normal TAF1 expression.
To the best of our knowledge, there are no published studies measuring plasma proteins in XDP. We used SiMoA technology to assay NfL, a nonspecific marker of neurodegeneration, 23 , 25 , 28 in the plasma of patients with XDP, males with presymptomatic XDP, female carriers, and healthy controls. We show for the first time evidence of abnormally high levels of NfL in XDP plasma, reflecting a neurodegenerative process occurring in the brains of patients with XDP, but not in males with presymptomatic XDP or asymptomatic female carriers. Our results are consistent with the knowledge that XDP is a disease of basal ganglia neurodegeneration. 1 , 2 , 3 , 4 , 9 This adds XDP to the list of atypical parkinsonian syndromes with increased plasma NfL. 24 , 29 , 30
Although NfL may be a nonspecific biomarker of neurodegeneration in XDP, it may still be useful in monitoring disease onset and progression in presymptomatic and symptomatic patients under therapeutic treatment. TAF1 dysregulation acts as a disease‐specific biomarker that makes it an attractive readout for target engagement by new therapies, particularly those targeting TAF1 expression and splicing regulation. Analyzed together, these nonspecific and specific biomarkers may reflect disease progression and serve as robust readouts for targeted therapeutics. Future longitudinal studies are needed to follow cohorts of XDP presymptomatic and symptomatic patients and female carriers to better detect clinical correlations between our developed biomarkers and different disease parameters. Such studies are also key in determining the usefulness of NfL in detecting evidence of neurodegeneration and disease activity in males and females with presymptomatic XDP.
Author Roles
(1) Research Project: A. Conception, B. Organization, C. Execution; (2) Statistical Analysis: A. Design, B. Execution, C. Review and Critique; (3) Manuscript: A. Writing of the First Draft, B. Review and Critique.
J.A.A.: 1A, 1B, 1C, 2A, 2B, 2C, 3A, 3B
C.A.V.: 1A, 1B, 1C,2C, 3A, 3B
S.S.: 1A, 1B, 1C, 2C, 3B
L.C.: 1A, 1B, 1C, 2C, 3B
A.H.: 1A, 1B, 1C, 2C, 3B
M.L.S.: 1A, 1B, 1C, 2C, 3B
P.A.: 1A, 1B, 1C, 2C, 3B
G.A.: 1A, 1B, 1C, 2C, 3B
T.M.‐B.: 1A, 1B, 1C, 2C, 3B
N.G.M.G.: 1B, 1C, 2C, 3B
J.B.B.L.: 1B, 1C, 2C, 3B
J.K.D.G.: 1B, 1C, 2C, 3B
C.G.: 1B, 1C, 2C, 3B
B.C.: 1A, 1C, 2C, 3B
B.T.: 1A, 1C, 2C, 3B
P.K.W.: 1A, 1C, 2C, 3B
M.T.: 1A, 1B, 2C, 3B
S.E.A.: 1A, 1B, 2C, 3B
P.S.C.: 1A, 1B, 2C, 3B
N.I.: 1A, 1B, 2B, 3A, 3B
N.S.: 1A, 1B, 2B, 3A, 3B
D.C.B.: 1A, 1B, 2B, 3A, 3B
L.O.: 1A, 1B, 2B, 3A, 3B
X.O.B.: 1A, 1B, 2A, 2B, 3A, 3B
Full financial disclosures for the previous 12 months:
N.S. reports grants from the National Institutes of Health and funding from the Massachusetts General Hospital Collaborative Center for X‐Linked Dystonia‐Parkinsonism. D.C.B. reports grants from the National Institutes of Health and funding from the Massachusetts General Hospital Collaborative Center for X‐Linked Dystonia‐Parkinsonism. L.O. reports grants from the National Institutes of Health and funding from the Massachusetts General Hospital Collaborative Center for X‐Linked Dystonia‐Parkinsonism. X.O.B. reports grants from the National Institutes of Health and funding from the Massachusetts General Hospital Collaborative Center for X‐Linked Dystonia‐Parkinsonism. All other authors have nothing to disclose.
Acknowledgments
We thank Mrs. Suzanne McDavitt for skilled editorial assistance. We also thank all the patients and their family members who participated in these studies. This work was supported by Massachusetts General Hospital Collaborative Center for X‐Linked Dystonia‐Parkinsonism.
References
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
 TAF1 Transcripts and Neurofilament Light Chain as Biomarkers for X‐linked Dystonia‐Parkinsonism
TAF1 Transcripts and Neurofilament Light Chain as Biomarkers for X‐linked Dystonia‐Parkinsonism
