
AUTHOR CONTRIBUTIONS
A.M.C. and L.D.W. designed the study; G.H.B and T.O. synthesized, purified, and characterized the stapled peptides, and performed circular dichroism and peptide proteolysis assays; A.M.C. conducted all of the biochemical studies, except for the VLCAD enzyme assay and ubiquitin pathway reconstitution assays performed by M.S.P and H.D.H., respectively. H.D.H., respectively. H.D.H. performed the fluorescent bead binding assay, and U.A. contributed to peptide specificity-of-action assays. A.M.C., T.E.W., S.L., E.P.H., Z.J.H. and C.E.N. performed and analyzed the HXMS analyses under the supervision of J.R.E. L.D.W. and A.M.C. wrote the manuscript, which was reviewed by all co-authors.
- Altmetric
The ubiquitin-proteasome system (UPS) is a highly-regulated protein disposal process critical to cell survival. Inhibiting the pathway induces proteotoxic stress and can be effective cancer treatment. The therapeutic window observed upon proteosomal blockade has motivated multiple UPS-targeting strategies, including preventing ubiquitination altogether. E1 initiates the cascade by transferring ubiquitin to E2 enzymes. A small molecule that engages the E1 ATP-binding site and derivatizes ubiquitin disrupts enzymatic activity and kills cancer cells. However, binding-site mutations cause resistance, motivating alternative approaches to block this promising target. We identified an interaction between the E2 N-terminal alpha-1 helix and a pocket within the E1 ubiquitin-fold domain as a potentially druggable site. Stapled peptides modeled after the E2 alpha-1 helix bound to the E1 groove, induced a consequential conformational change, and inhibited E1 ubiquitin-thiotransfer, disrupting E2 ubiquitin-charging and ubiquitination of cellular proteins. Thus, we provide a blueprint for a distinct E1-targeting strategy to treat cancer.
INTRODUCTION
The ubiquitin-proteasome system (UPS) is essential to cellular homeostasis and is responsible for sensing, ubiquitin-tagging, and distributing proteins to discrete intracellular locations or eliminating them entirely through proteasomal degradation. The three steps of this biochemical process include (1) activation of ubiquitin by an E1 enzyme, (2) conjugation of ubiquitin to an E2 enzyme, and (3) transfer of ubiquitin to the target protein by an E3 ubiquitin ligase, which is often followed by proteosomal degradation of the ubiquitinated protein1–4. The remarkable array of proteins subject to UPS activity highlights its critical role in regulating fundamental signal transduction pathways, such as apoptosis, the cell cycle, transcription, stress responses, growth and differentiation. Deregulation of the UPS has been associated with a myriad of human pathologies, including cancer, neurogenerative disorders, immunological deregulation, and cardiovascular disease5,6.
Whereas inhibition of the proteasome would seem too blunt a tool to elicit a therapeutic response without undue toxicity, the success of bortezomib proved otherwise, and it was approved by the US Food and Drug Administration for the treatment of multiple myeloma and mantle-cell lymphoma7,8. Among the lessons learned from bortezomib is that cancer cells are more sensitive to UPS inhibition than normal cells due to a host of genetic defects and cellular demands that heighten UPS dependency9–14. Thus, targeting the UPS at a series of control points has become a major therapeutic goal. Although proteasome inhibition is the most clinically-validated strategy, diverse approaches to UPS modulation include targeting deubiquitinases and the E1-E2-E3 enzymes, and induction of disease-target degradation using PROTACs15. E1 emerged as an intriguing target because its inhibition is expected to have a pervasive effect. Relative to hundreds of E3 enzymes and dozens of E2s and deubiquitinases, there is one predominant mammalian E1 enzyme, called UBE1 (UBA1), which is responsible for the first step of the ubiquitination cascade. Blockade of UBE1 could shut down ubiquitin activation at its source and arrest the downstream cascade. Genetic knockdown of E1 in leukemia cell lines and patient samples decreases ubiquitination levels, upregulates stress markers, and triggers cancer cell death16.
UBE1 is a 118 kDa multi-domain enzyme that catalyzes ubiquitin adenylation and thioesterification. Initially, the X-linked UBE1 gene was believed to be the sole E1 enzyme in animals and fungi, but a second enzyme, UBA6, was later identified in vertebrates and the sea urchin17. UBE1 interacts with all identified E2s except for the UBA6-specific E2 USE1 and is estimated to activate >99% of cellular ubiquitin; in contrast, UBA6 interacts with a smaller subset of E2 enzymes including USE1, and overall activates <1% of ubiquitin14,17,18. Two covalent strategies for disrupting E1-E2 ubiquitin transfer focused on the ubiquitin-charging region. One approach used short heptapeptide ubiquitin mimics that become activated by E1 to form thioester conjugates, which compete with shuttling of native ubiquitin through the UPS19. The second strategy involved an adenosyl sulfamate molecule, TAK-243 (MLN7342), which occupies the UBE1 ATP binding site and forms a covalent adduct with the ubiquitin C-terminus. The TAK-243-ubiquitin conjugate remains tightly bound to UBE1 and prevents further enzymatic activity. TAK-243 is a first-in-class UBE1 inhibitor that advanced to clinical testing based on promising efficacy in mouse models of solid tumors and multiple myeloma20. In a small phase 1 trial of TAK-243 for advanced solid tumors (NCT02045095), one of twenty evaluable patients achieved a partial response (5%) and 11 patients experienced stable disease (55%)21.
A major cause of cancer cell resistance to ATP competitive inhibitors like TAK-243 derives from a mutation at or near the enzyme’s ATP-binding site22. Mutation of the highly-conserved UBE1 residue A580 conferred resistance to TAK-243 in cell-free assays, and A580S and Y583C mutations were identified in cultured leukemia cells chronically exposed to TAK-24320,23. Given the importance of E1-targeting as a potential cancer therapy, and that the singularly-vetted modality is vulnerable to mutation-based resistance, we sought to identify an alternative approach to UBE1 inhibition. We harnessed peptide stapling to identify and validate a novel and potentially druggable binding pocket within the E1 ubiquitin fold domain. Engagement of this site by a stapled alpha-1 helix of E2 induces a conformational change in E1, neutralizes its enzymatic activity, and disrupts the downstream ubiquitination of cellular proteins. Thus, we provide a blueprint for the development of therapeutic peptides and small molecules that target a distinct site on E1 for inhibiting the UPS in cancer.
RESULTS
Stapled E2 α1 peptides block E1 ubiquitin transfer
Hydrocarbon stapling is an effective strategy for structurally-stabilizing α-helices and has been used to target helix-in-groove interactions in vitro and in vivo24, with one agent currently in phase 1 and 2 clinical trials25. In evaluating crystal structures of E1 in complex with E2s, namely S. pombe Ubc4 and Ubc15, and S. cerevisiae Cdc3426–28, we noted a binding-interface between the N-terminal E2 alpha-1 helix 1 (E2h1) and an E1 surface groove (Fig. 1a). Of the ~3000 Å2 of surface area involved in E1-E2 interaction, ~1000 Å2 is buried by the interface between E2h1 and the E1 ubiquitin-fold domain (E1UFD)28. Biochemical studies showed that the E1UFD-E2h1 interaction is critical to forming the E1-E2 encounter complex and accounts for approximately half of the ΔΔG of E1-E2 binding26,29,30. Mutation or deletion of residues within E1UFD or E2h1 impair or abolish ubiquitin-charging of E2 by E126,28,30. These observations pointed to E2h1 as a promising candidate for stapled-peptide mimicry.
In assays that evaluated conjugation of ubiquitin by S. pombe E1 to wild-type and mutant E2 proteins, the lowest Km was observed for a Ubc15 E2-construct bearing an E7R mutation26. The E2-interacting surfaces and structure of the human and S. pombe ubiquitin fold domains (UFDs) are similar root mean square deviation (r.m.s.d.) 1.1–1.6 Å, corresponding to 110 of 119 equivalent Cα atoms superimposed, with the largest difference from a six-residue human UFD insertion located opposite to the E2-binding face)31. Thus, we selected Ubc15E7R (aa 1–18) as our initial template for staple-scanning. We installed i,i+4 or i,i+7 staples along the length of the Ubc15E7R α1 sequence (Fig. 1b-c), avoiding the initial four residues due to potential steric hindrance of surrounding interactions within a deep crevice on the E1 surface (Fig. 1a). We screened the panel of stabilized alpha-helices of Ubc15 (SAH-Ubc15) in an in vitro biochemical assay that measures UBE1-mediated thioester transfer of ubiquitin to the human UBE2D2 E2 enzyme. Whereas the unmodified Ubc15 template showed no inhibitory activity by thioester-transfer assay, consistent with its lack of α-helical structure (Extended Data Fig. 1a), constructs bearing staples opposite to the E1 UFD-binding interface (e.g. SAH-Ubc15 peptides 5, 11, 14) were most effective at inhibiting ubiquitin thioester transfer, with SAH-Ubc15–11 (staple spanning positions S6 and Q13) emerging as the most potent inhibitor (Fig. 1c-d). Those staples that replaced highly conserved hydrophobic E2h1 residues (Supplementary Fig. 1), such as L10 and L14, showed no activity (e.g. SAH-Ubc15 peptides 6, 9, 12, 15), whereas staples that reinforced the region of a conserved salt-bridge among E2s (positions 9 and 13) were especially effective (e.g. SAH-Ubc15 peptides 5, 11, 14), consistent with a structure-activity relationship.
Having identified a lead staple-position for SAH-Ubc15 inhibition of UBE1, we confirmed that α-helical structure was conferred by staple insertion (Extended Data Fig. 1b) and then conducted dose-responsive thioester-transfer assays to quantitate the inhibitory potency of SAH-Ubc15–11, a half-maximum inhibitory concentration (IC50) of 96.2±42 μM (Fig. 2a-c). Given the dozens of E2 substrates for E1 ubiquitin transfer activity, we evaluated alternate E2h1 templates to improve compound activity. We selected (1) UBE2A, which shares an identical E2h1 sequence with UBE2B, (2) UBE2G2, which of the two human Ubc15 homologs lacks negative charge at the N-terminus that could impair UFD acidic patch engagement (Fig. 1a), and (3) UBE2D2, which represents the large UBE2D family and has the lowest-reported Km for E1-mediated thioester transfer of any human E2 enzyme30. We installed the optimal i,i+7 staple from SAH-Ubc15–11 into these E2h1 templates to generate SAH-UBE2A, SAH-UBE2G2, and SAH-UBE2D2 peptides, and subjected them to thioester-transfer assays. SAH-UBE2A demonstrated the most potent inhibitory activity with an IC50 of 2.3±0.2 μM, reflecting a 40-fold improvement over SAH-Ubc15–11 (Fig. 2a-c).
Specificity of SAH-UBE2A activity
Before advancing SAH-UBE2A to structure-function analyses, we performed a series of orthogonal assays to confirm that SAH-UBE2A was operating through a specific mode-of-action32. First, we repeated the thioester-transfer assay with 0.01% Triton X-100, which typically attenuates non-specific activity of aggregator compounds32, and observed the identical dose-responsive inhibitory activity for SAH-UBE2A (Extended Data Fig. 2a). To evaluate the potential influence of osmolytes on SAH-UBE2A activity, we performed the thioester-transfer assay with 50 mM (gold-standard conditions) or 250 mM NaCl. Whereas the higher salt condition impaired UBE1 enzymatic activity, the ability of SAH-UBE2A to inhibit thioester transfer was unaffected (Extended Data Fig. 3b). We developed a fluorescence microscopy-based qualitative binding assay to directly observe SAH-UBE2A binding specificity. Fluorescein isothiocyanate (FITC)-labeled SAH-UBE2A was incubated with Ni-NTA beads pre-loaded with His-tagged UBE1, His-tagged p53 or no protein, and then the beads were isolated by centrifugation and visualized. Whereas the UBE1-coated beads showed robust FITC-SAH-UBE2A fluorescence, no binding activity was evident for the beads-alone sample or beads bound to the unrelated p53 protein; further, no fluorescent peptide aggregates were seen (Extended Data Fig. 3c). Another feature of non-specific, assay-interference compounds is their capacity to disrupt proteins unrelated to the target-of-interest. In a distinct assay where very-long-chain acyl-CoA dehydrogenase (VLCAD) time-responsively oxidizes a palmitoyl-CoA substrate, as evidenced by decreasing absorbance of the ferrocenium hexafluorophosphate electron acceptor33,34, we show that incubation with SAH-UBE2A at 30-fold excess has no inhibitory effect on VLCAD enzymatic activity (Extended Data Fig. 3d).
We next examined SAH-UBE2A specificity in blocking UBE1 thioester transfer in a reconstituted system that incorporates all enzymatic components of the ubiquitination pathway (Extended Data Fig. 3e). When p53 protein is added to a mixture of E1 (UBE1), E2 (UBE2A), E3 (HDM2), ubiquitin, and ATP, in vitro ubiquitination of p53 is blocked by SAH-UBE2A. If E1-mediated ubiquitin transfer to E2 is bypassed by including pre-ubiquitinated E2 rather than uncharged E2 and free ubiquitin, SAH-UBE2A is unable to block p53 ubiquitination. p53 deubiquitination upon addition of a deubiquitinating enzyme (DUB) such as USP7 is also unaffected by SAH-UBE2A. These data demonstrate that SAH-UBE2A only blocks the enzymatic activity of UBE1 (Extended Data Fig. 3e). Even within the context of UBE1 activity, we found that SAH-UBE2A inhibits thioester transfer but not ubiquitin self-charging, the enzymatic step preceding thioester transfer to E2 (Extended Data Fig. 3f). We verified that SAH-UBE2A activity was not dependent on ubiquitin discharge from E2 onto the stapled peptide itself, which bears a lysine at position 14, by showing that SAH-UBE2A K14R had identical inhibitory activity in the thioester-transfer assay despite lysine mutagenesis (Extended Data Fig. 3g). Taken together, these data demonstrate that SAH-UBE2A exerts a highly-selective mode of action in inhibiting UBE1 thioester-transfer activity.
Binding Determinants of SAH-UBE2A Interaction with E1
We sought to evaluate the structure-function relationship of SAH-UBE2A, quantitate its direct interaction with UBE1, and define the binding determinants. As for the structure-function correlation of Ubc15–11 and SAH-Ubc15–11, we found that insertion of the corresponding i,i+7 staple into the UBE2A sequence conferred α-helical structure (Fig. 2d) and UBE1-inhibitory activity (Fig. 2e). The impact of stapling on peptide shape and activity was further evident upon protease-resistance testing, whereby α-helical reinforcement conferred a 23-fold prolongation in half-life for SAH-UBE2A compared to the unstructured UBE2A template upon exposure to proteinase K (t1/2: SAH-UBE2A, 390 min; UBE2A, 17 min) (Fig. 2f). Thus, we find that i,i+7 stapling enhances α-helicity of the E1-interacting UBE2A peptide and, in doing so, confers protease resistance and UBE1-inhibitory activity.
We next examined the influence of UBE1 on the deuterium-exchange profiles of UBE2A and SAH-UBE2A in solution by hydrogen-deuterium exchange mass spectrometry (HXMS) as a measure of UBE1 interaction. HXMS probes changes in peptide or protein conformation based on differences in hydrogen-bonding and solvent exposure, as assessed by deuterium exchange of backbone-amide protons35. Incubation with UBE1 had no meaningful effect on the deuterium exchange of the UBE2A peptide, which is indicative of no peptide/protein interaction; in contrast, SAH-UBE2A demonstrated striking protection from deuterium exchange upon exposure to UBE1 compared to the peptide uptake observed in aqueous solution alone (Fig. 2g). The HXMS data are consistent with the comparative circular dichroism, protease resistance, and thioester-transfer assay analyses that collectively demonstrate the correlation between peptide stapling and structural stabilization, UBE1 interaction, and UBE1 inhibition (Fig. 2d-g). Using orthogonal binding assays, we further demonstrate that (1) a C-terminally biotinylated SAH-UBE2A peptide directly and robustly binds to recombinant UBE1, as assessed by streptavidin pull-down and silver stain (Fig. 2h) and that (2) a C-terminally FITCylated peptide binds to UBE1 by fluorescence polarization (FP) assay with a calculated Kd of 384±43 nM, an affinity similar to those reported for interactions between full-length E2 enzymes and UBE130,36 (Fig. 2i). Notably, the presence or absence of ubiquitin and ATP had little to no effect on the binding interaction between UBE1 and SAH-UBE2A, as assessed by HXMS and FP assay (Extended Data Fig. 3).
Having demonstrated the importance of SAH-UBE2A α-helical structure for its functional interaction with UBE1, we sought to define the sequence determinants by synthesizing and testing an alanine-scanning library of SAH-UBE2A (Fig. 3a). Each amino acid of SAH-UBE2A was sequentially replaced with alanine, and the resultant stapled peptides tested for UBE1 inhibitory activity by thioester-transfer assay. Whereas alanine mutagenesis of N-terminal residues B1 (where B represents norleucine that replaces methionine to maximize ruthenium catalysis of the stapling reaction), S2, T3, P4, and R6, along with R8, R11, K14, R15, and Q17, had little to no effect on E1 inhibition, mutation of R7, L9, B10, F13, and L16 substantially reduced inhibitory activity, revealing the key residues for peptide function (Fig. 3b-c). Docking the UBE2A alpha-1 helix onto human UBE1 (PDB: 6DC6) revealed that disruptive alanine mutants localized to the predicted binding groove on the UBE1 surface, consistent with a defined structure-activity relationship (Fig. 3d). Because previous E1-E2 interaction studies emphasized the importance of E2h1 N-terminal basic residues and a complementary acidic patch on the E1 UFD26,28,31, we further evaluated the functional relevance of electrostatic interactions by synthesizing and testing a series of R-to-E charge-reversal mutants at the N-terminus of SAH-UBE2A (Fig. 3e). Whereas R6E and R8E mutagenesis was tolerated, R7E mutation significantly reduced peptide activity, as assessed by thioester-transfer assay (Fig. 3f-g). We confirmed that the selective impairment of SAH-UBE2A R7E was dependent on amino acid sequence rather than peptide structure, as all R-to-E mutants demonstrated a similar degree of high α-helicity, as assessed by circular dichroism (R6E, 83%; R7E, 97%; R8E, 98%) (Extended Data Fig. 4). The docked structure of UBE2Ah1 places R7 within 3.1 Å of UBE1 E1049, suggesting that loss of electrostatic complementarity and charge-charge repulsion between E7 of SAH-UBE2A R7E and the native E1049 of UBE1 could account for the impaired activity (Fig. 3h).
Conformational Effect of SAH-UBE2A Inhibition of UBE1
To assess the impact of SAH-UBE2A interaction on the conformational dynamics of UBE1, we performed HXMS analyses that compared the difference in deuterium uptake by UBE1 in the presence and absence of SAH-UBE2A. We analyzed the deuterium uptake of full-length recombinant UBE1 (sequence coverage of ~82.5%) in aqueous solution containing excess ATP and ubiquitin at 10-second, 1-minute, and 10-minute deuterium-labeling time points, and then compared the HXMS profile of protein alone and when combined with SAH-UBE2A (Supplementary Table 1 and Data File 1). The deuterium-uptake profile of UBE1 over time reflects structural dynamics consistent with previous reports of the structural remodeling that occurs in E1 during its catalytic cycle31,37,38 (Extended Data Fig. 5). High deuterium uptake at the earliest labeling time point (10 seconds) generally reflects solvent-exposed regions not involved in intramolecular hydrogen bonds39,40. In UBE1, these regions include the N-terminal extension, cysteine cap, and crossover loop. These domains are either solvent exposed or missing from E1 crystal structures23,26,31,37,41,42. In contrast, increasing exchange over time represents structurally-dynamic protein regions39,40. For example, peptides of the N-terminal alpha-helices, adenylation active site, second catalytic cysteine half-domain (SCCH) (excluding the cysteine cap), re-entry loop, and UFD linker hinge demonstrate time-dependent deuterium uptake, consistent with the dynamism of these regions during the E1 catalytic cycle38. In contrast, α-helices and beta sheets located in the interior of the protein were not deuterated at any time point, indicating that the core of the inactive adenylation domain (IAD) and active adenylation domain (AAD) remains structurally-stable over time.
Upon addition of SAH-UBE2A, the most persistent effect on UBE1 across all three time points is deprotection of residues 936–951 (peptide 198), which corresponds to the UFD linker hinge (Fig. 4a-d, Supplementary Data File 1). The UFD is the anticipated binding site for SAH-UBE2A on UBE1 and, notably, the linker hinge lies immediately subjacent to the binding groove, serving as a flexible β-hairpin that connects the UFD to the E1 protein core (Fig. 4d). Crystal structures of E1 alone and in complex with E2 proteins demonstrate that flexion or bending of the UFD linker hinge around a tryptophan anchor (W955) causes the UFD to undergo a 25° rigid-body rotation with respect to the the E1 protein core26,28,31,41 (Fig. 4e). This twisting motion causes the UFD linker hinge to lose contacts with the E1 core and become more solvent accessible28 (Fig. 4f). The UFD conformational change from a distal to proximal position is believed to have functional relevance in that rotation closes a gap of 10–25 Å between the E1 and E2 active cysteines, allowing for thioester transfer of ubiquitin from E1 to E228. Since UFD rotation into the proximal position is accompanied by an increase in solvent accessibility of the UFD linker hinge, its deprotection and higher deuterium level upon UBE1 interaction with SAH-UBE2A suggests that peptide binding stabilizes the proximal UFD conformation. A second region of deprotection, which becomes more prominent with time, corresponds to residues 409–422 (peptides 64–70) that are located at the C-terminal end of UBE1 helix 11 within the IAD of the E1 core. Thus, it is plausible that SAH-UBE2A engagement of the E1 UFD-binding groove induces the proximal UFD conformation and, in doing so, causes further allosteric changes that propagate across the adjacent AAD and IAD (Fig. 4d).
Importantly, we documented the specificity of our HXMS observations by repeating the study using the SAH-UBE2A R7E single point mutant control, which decreases (1) peptide-inhibitory activity, (2) interaction with UBE1, and (3) the conformational changes observed by HXMS (Extended Data Fig. 6). To evaluate whether combinatorial mutagenesis based on the alanine scan results could yield an even more impaired construct, we generated SAH-UBE2A bearing R7A, L9A, and B10A mutants, all of which as single mutants partially inhibited peptide activity (Fig. 3b). The triple alanine mutant essentially abrogated UBE1-inhibitory activity, markedly reduced UBE1 interaction, and had little to no effect on UBE1 conformation, as assessed by HXMS (Extended Data Fig. 6). These data further reinforce the structure-activity relationship for SAH-UBE2A interaction at the UFD surface groove of UBE1, with consistent correlation between peptide sequence, deuterium exchange of both peptide and protein, and functional inhibition of thioester-transfer activity.
SAH-UBE2A Impairs E2-Charging and Protein Ubiquitination
In order to effectively block the ubiquitin pathway by targeting UBE1, an inhibitor must broadly disrupt UBE1 interactions with its spectrum of E2 partners. To assess the generalized inhibitory capacity of SAH-UBE2A, we performed thioester-transfer assays with SAH-UBE2A or the SAH-UBE2A R7E mutant control for twenty E2 ubiquitin-conjugating enzymes (Fig. 5a). SAH-UBE2A impaired thioester transfer to each E2 tested, with R7E point mutagenesis blunting the inhibitory effect to various extents. As a further measure of specificity-of-action, we found that excess E2 protein could outcompete SAH-UBE2A for UBE1 interaction, as measured by fluorescent-bead assay (Extended Data Fig. 7). We next tested the ability of SAH-UBE2A to inhibit UBE1 paralogs by replacing UBE1 with the alternate ubiquitin-activating enzyme UBA6 or the heterodimeric NEDD8-activating enzyme NAE1/UBA3, and assessing thioester transfer to the E2 enzyme USE1 or UBE2M, respectively (Fig. 5b-c). SAH-UBE2A was an even more potent inhibitor of UBA6 than UBE1 by 5-fold, but displayed 6-fold less inhibitory activity toward UBA3, indicating that SAH-UBE2A is a relatively selective inhibitor of E1 enzymes of the ubiquitin-conjugation pathway.
Finally, we examined whether SAH-UBE2A could compete with the full complement of native E2 enzymes to inhibit endogenous UBE1 and block the ubiquitin cascade. We treated HeLa cell lysate with SAH-UBE2A in the presence of excess ubiquitin and an ATP regeneration system to stimulate ubiquitin-pathway flux, followed by anti-ubiquitin western blot under non-reducing conditions to evaluate peptide-inhibitory activity (Fig. 5d). Indeed, SAH-UBE2A inhibited ubiquitin-pathway flux, as demonstrated by a dose-responsive decrease in bulk polyubiquitin chain derivatization of proteins within the HeLa cell lysate. These data suggest that targeting the E2h1 interaction surface of E1 ubiquitin-activating enzymes is a viable strategy for blockade of the ubiquitin pathway.
DISCUSSION
The relative dependence of cancer cells on reducing proteotoxic stress to maximize pathologic survival has provided a therapeutic window for targeting the UPS in cancer patients. Modulating the levels of oncogenic and tumor suppressor proteins by pharmacologic engagement of the UPS underlies the promise of PROTACs and E3-ubiquitin ligase inhibitors, such as dBET and idasanutlin respectively43,44. Given the unexpected safety and efficacy of proteasome inhibitors like bortezomib45, an alternative small-molecule approach to generalized disruption of protein degradation through E1 inhibition has been developed and validated in preclinical models20. Based on encouraging phase 1 testing of this molecule21, TAK-243, a second trial in hematologic malignancies is planned (NCT03816319). The mechanism involves engaging the UBE1 ATP-binding site and covalent reaction with the ubiquitin C-terminus, rendering the molecule vulnerable to interaction-site mutagenesis, as observed in vitro and in cellulo20,23. Given the benefits of E1 inhibition in preclinical models and select patients in phase 1 trials, we developed an alternative approach to E1 targeting.
The diversity of E2 enzymes provides a reservoir of N-terminal α1-sequences for exploring the feasibility of targeting the helix-in-groove interaction between E2h1 and the E1 UFD, and its impact on the structure and function of E1. Applying a staple-scanning and template-optimization workflow, we identified a lead i,i+7 stapled peptide that directly bound to E1 with nanomolar affinity and impaired E1 thioester transfer with single-digit micromolar activity. Alanine scanning, charge-reversal mutagenesis, and triple-alanine mutagenesis defined a structure-activity relationship consistent with the corresponding E1/E2 binding interface demonstrated by x-ray crystallography and docking analyses. Whereas crystallography successfully captured snapshots of E1 at various enzymatic stages, including those stabilized by covalent small molecules14,23,26,28,31,37,38,41,42,46, the large and dynamic nature of the E1 protein makes it a challenging target for conformational analysis. We applied HXMS to characterize the unbound state of the protein and interrogate the structural impact of E1 interaction with an E2h1-based stapled peptide, SAH-UBE2A. Peptide-induced exposure of the UFD linker hinge is consistent with an inhibitory mechanism whereby SAH-UBE2A locks E1 into its proximal UFD conformation. A prior FTmap hot-spot analysis of E1 implicated a deep crevice between the UFD and E1 core, which is naturally engaged by the extended N-termini of E2 proteins (PDB: 5KNL), as a potential ligand-binding site31. This hot spot was only detected when E1 was in the proximal UFD conformation (PDB: 4II2), suggesting that the pocket is not fully formed in distal conformations that position UFD further away from the AAD. The E2h1 sequences of SAH-UBE2A include both the α1 portion, which interacts with the UFD groove, and the N-terminal extension that inserts into the crevice.
A strategic benefit of E1-targeting is the capacity to broadly inhibit E2-charging and suppress ubiquitin-mediated protein degradation at the most apical step of the UPS pathway. SAH-UBE2A impaired ubiquitin thioester transfer between E1 and a variety of E2 proteins, and suppressed native-protein ubiquitination in cellular lysates. This suggests that the E1UFD-E2h1 interface represents a potentially druggable binding-site for E1 inhibition. Compared to TAK-273, SAH-UBE2A targets a distinct surface and has a unique mechanism, which appears to involve stabilization of a particular E1 conformer. Thus, viable drug development approaches for UBE1 and UBA6 could include (1) transforming the SAH-UBE2A prototype into a clinical candidate, as accomplished for a staple peptide dual inhibitor of HDM2 and HDMX25,47; (2) harnessing SAH-UBE2A in a competitive screen48 to identify small-molecule inhibitors of the SAH-UBE2A/UBE1 interaction; or (3) pursuing in silico or de novo small-molecule drug design. Advancing a diversity of pharmacologic approaches to E1 inhibition could expand the arsenal of UPS-modulating agents for potential clinical benefit in human cancer.
ONLINE METHODS
Stapled Peptide Synthesis and Purification
Stapled peptides were generated as described49 using solid phase Fmoc chemistry on a Symphony X peptide synthesizer (Gyros Protein Technologies), with amino acids sequentially added to rink amide AM resin. Two S-pentenyl alanine, or one S-pentenyl alanine and one R-octenyl alanine, replaced two native amino acids at the i, i+4 or i, i+7 positions, respectively. The all-hydrocarbon staple was formed by olefin metathesis using the Grubbs first-generation ruthenium catalyst, followed by peptide deprotection and cleavage from the resin. Peptides were purified by reverse phase high performance liquid chromatography and mass spectrometry (LC/MS) and quantified by amino acid analysis (Supplementary Table 2).
E1-E2 Thioester Transfer Assay
Thoiester transfer assays were performed using 10 nM UBE1, 135 nM UBE2D2, 500 nM ubiquitin, and 20 μM ATP, in assay buffer containing 50 mM NaCl, 5 mM MgCl2, 20 mM HEPES, pH 7.4. The Km of UBE2D2 was experimentally determined to be 135 nM and assay conditions were conducted near the UBE2D2 Km in the presence of excess ubiquitin and ATP. For each assay, recombinant human UBE1 protein (Boston Biochem E-304) was mixed with ubiquitin (R&D Systems U100H10M), ATP (Sigma A9187), and peptide or vehicle (<2% DMSO). For the initial staple-scanning screen of SAH-UBC15E7R, peptide concentration was 150 μM, whereas alanine-scanning, charge-reversal, triple point mutant SAH-UBE2A and unstapled UBE2A testing was conducted at 10 μM dosing. E1-E2 thioester transfer was initiated by addition of recombinant human UBE2D2 (Boston Biochem E2–622) and reactions allowed to proceed for 45 min at room temperature, followed by quenching with non-reducing LDS loading buffer. Proteins were separated by SDS-PAGE gel electrophoresis and visualized by silver stain (Pierce 24612). Three independent experiments were quantified by densitometry analysis using ImageJ software. Percent inhibition was calculated as percent conversion relative to vehicle:
IC50 values were determined by nonlinear regression analysis using Prism software (Graphpad). For evaluation of SAH-UBE2A inhibitory activity in the presence of detergent, the assay was performed as described with addition of the indicated amount of non-ionic detergent (0.01% Triton X-100) in the reaction buffer. For evaluation of a potential SAH-UBE2A effect on UBE1~Ub thioester formation, the assay was performed as described with longer electrophoresis running time to resolve silver stain bands corresponding to UBE1 and UBE1~Ub. For the evaluation of SAH-UBE2A inhibitory activity across a panel of E2 enzymes, the indicated E2 enzyme (final concentration, 135 nM; E2 Select Ubiquitin Conjugation Kit, Boston Biochem K-982) was substituted for UBE2D2. UBA6 thioester transfer assays were performed as above, substituting 10 nM GST-UBA6 (Boston Biochem E-307) for UBE1 and 135 nM His6-USE1 (Boston Biochem E2–677) for UBE2D2. For UBA3 thioester transfer assays, 15 nM NAE1/UBA3 was substituted for UBE1 (Boston Biochem E-313), 135 nM UBE2M for UBE2D2 (Boston Biochem E2–656), and 1 μM NEDD8 (Boston Biochem UL-812) for ubiquitin. SAH-UBE2A inhibitory activity was compared across thioester transfer assays bearing the distinct protein components after 15 min incubation at 37°C (NEDD8 pathway) or 45 min at room temperature (ubiquitin pathway).
Circular Dichroism
Acetylated peptides were dissolved in 10% (vol/vol) acetonitrile in water to achieve a concentration of 50 μM. Circular dichroism (CD) spectra were obtained on a spectropolarimeter (Aviv) using standard measurement parameters of temperature, 37°C; wavelength, 190–260 nm; step resolution, 0.5 nm; speed, 20 nm min−1; accumulations, 1049. Helical content was calculated from the mean residue molar ellipticity value at 222 nm, ([θ]222)50.
Peptide Proteolysis Assay
In vitro proteolysis was measured by LC/MS (Agilent 1200) using the following parameters: 20 μl injection, 0.6 mL flow rate, 20-min run time consisting of a gradient of water (0.1% formic acid) to 20%–80% acetonitrile (0.75% formic acid) over 15 min, 4-min wash to revert to starting gradient conditions, and 0.5-min post-time. The MSD was set to scan mode with one channel at (M+2H)/2, ± 1 mass units, one channel at (M+3H)/3, ± 1 mass units, and the other at (M+4H)/4, ± 1 mass units. Integration of each MSD signal yielded areas under the curve of >108 counts. Reaction samples were composed of 5 μl peptide in DMSO (1 mM stock) and 195 μl buffer consisting of 50 mM Tris HCl pH 7.4. Upon injection of the zero-time sample, 6 μl of 20 ng/μL proteinase K (New England Biolabs) was added, and the amount of intact peptide quantitated by serial injection over time. A plot of MSD area versus time yielded an exponential decay curve, and half-lives were determined by nonlinear regression analysis using Prism software (GraphPad).
Streptavidin Capture
Recombinant human UBE1 (10 pmol) was incubated with C-terminally biotinylated SAH-UBE2A (5 nmol) or vehicle (0.5% DMSO) in a total volume of 1 mL binding buffer (50 mM NaCl, 20 mM HEPES pH 7.4, 5 mM DTT) for 1.5 hours at 4°C. Pre-equilibrated streptavidin agarose beads (30 μL) were then added and incubated for an additional 1.5 hours at 4°C. The beads were pelleted by centrifugation and washed three times with NP-40 buffer (20 mM HEPES, pH 7.4, 50 mM NaCl, 5 mM DTT, 0.5% [v/v] NP-40) before eluting the protein sample from the beads by heating at 70°C for 10 minutes in 3x SDS loading buffer. Samples were subjected to electrophoresis and silver staining (Pierce 24612).
Fluorescence Polarization Assay
Fluorescence polarization (FP) assays were performed with 50 nM C-terminally FITCylated SAH-UBE2A incubated with a serial dilution of recombinant UBE1 in binding buffer (50 mM NaCl, 20 mM HEPES pH 7.4, 5 mM DTT) in the presence or absence of 10 μM ubiquitin and 20 μM ATP. Fluorescence polarization was measured on a Spectramax M5 Microplate Reader (Molecular Devices) and the data plotted and Kd values calculated using Prism software (Graphpad).
Fluorescent Bead Binding Assay
The indicated His-tagged protein (25 μg) or vehicle was incubated with Ni-NTA agarose beads (100 μL) (Invitrogen R90110) in PBS for 30 min. The beads were then incubated with FITC-labeled peptide (10 μM) for 30 min, isolated by benchtop centrifugation (2000xg), resuspended in PBS for plating in 386-well plate format (10 μL/well) (Corning 384-well High Content Imaging Glass Bottom Microplate), and imaged using an Olympus wide-field epifluorescence microscope, a 63× LCPlanFL NA 0.7 objective and a CoolSNAP DYNO camera. For E2 competition experiments, the assay was performed as above except that the His-UBE1 coated beads were incubated with the indicated E2 proteins (25 μM; Boston Biochem) for 30 min, followed by addition of FITC-SAH-UBE2A peptide (0.5 μM) for an additional 30 min incubation.
VLCAD Enzyme Activity Assay
Recombinant VLCAD protein was generated and enzymatic activity assays performed as previously described33. VLCAD protein (1.44 μM) was incubated with palmitoyl-CoA (600 μM) and either vehicle (0.43% [v/v] DMSO) or SAH-UBE2A (43.2 μM) in a total volume of 337.5 μL of assay buffer (100 mM potassium phosphate pH 7.2, 0.1 mM EDTA) for 45 min at room temperature. Ferrocenium hexafluorophosphate (Sigma-Aldrich, Cat# 388297) was dissolved in 10 mM HCl to a concentration of 1 mM and then diluted to 300 μM in assay buffer. Each reaction was initiated by adding an equal volume of protein mix to 300 μM ferrocenium, resulting in the dilution of each reaction component by half. The decrease in ferrocenium absorbance at 300 nm was measured as a function of time on a Spectramax M5 Microplate Reader (Molecular Devices).
In Vitro Ubiquitin Pathway Reconstitution Assay
Reactions containing recombinant His-p53 (0.1 μM), in the presence or absence of SAH-UBE2A (30 μM), UBE1 (0.1 μM), UBE2A (1 μM), HDM2 (1 μM), ubiquitin (1 μM) and ATP (10 mM) (all Boston Biochem K-200B), preactivated Ub-UBEA2 (3 μM) (Boston Biochem E2–802), and USP7 (1 μM) (Boston Biochem E-519), as indicated, were mixed and incubated for 2 h at 37ºC in a final volume of 30 μL. The samples were then subjected to boiling in SDS-containing loading dye for 10 min at 90ºC, followed by agarose gel electrophoresis and transfer to nitrocellulose membrane. The membranes were blocked with 3% milk for 1 h, incubated overnight in PBS containing 3% BSA and mouse p53 antibody (Boston Biochem K-200B) at a 1:1000 dilution, washed in PBS containing 0.1% Tween-20, and then incubated in PBS containing 3% BSA and anti-mouse secondary antibody (BioRad AAC10P) at a 1:5000 dilution for 1 hour at room temperature. The membranes were again washed in PBS containing 0.1% Tween-20 and imaged using Amersham ECL Prime (GE Life Sciences) on an Amersham Imager 680 (GE Life Sciences).
Docking Analysis
The UBE2A helix 1 sequence MSTPARRRLMRDFKRLQ was docked to the crystal structure of human UBE1 (PDB: 6DC6) using the PEPFOLD3.5 webserver, allowing peptide residues to be completely flexible and maintaining receptor residues as completely rigid51–53. The active receptor patch was defined as M989, S991, V1035, G1037, and C1039, based on previous reports26. The top-scoring cluster was visualized in Pymol (Schrödinger, LLC).
Hydrogen Deuterium Exchange Mass Spectrometry
For analysis of deuterium exchange into UBE2A, SAH-UBE2A, SAH-UBE2AR7E, and SAH-UBE2AR7A/L9A/B10A, the peptide was incubated alone or with the indicated molar ratio of recombinant human UBE1 for 10 min on ice in buffer containing 20 mM HEPES pH 7.4, 50 mM NaCl, 5 mM MgCl2, 0.5 μM ubiquitin, 20 μM ATP, and 5 mM DTT. Deuterium labeling was initiated with an 18-fold dilution into D2O buffer (20 mM HEPES pD 8, 50 mM NaCl). After 10 sec of labeling, the reaction was quenched with the addition of an equal volume of quench buffer (0.8 M guanidinium chloride, 0.8% [v/v] formic acid, pH 2). Samples were then injected onto a VanGuard Pre-Column trap (2.1 mm × 5 mm, ACQUITY UPLC BEH C8, 1.7 μm) for peptide trapping and desalting for 3 min. A Waters nanoACQUITY LC was used to elute the peptide and UBE1 from the trap with an 8%–70% gradient of acetonitrile over 5 min at a flow rate of 100 μL/min. Eluant was directed into a Waters Synapt G2Si mass spectrometer operated in TOF-only mode for mass analysis. Data were analyzed as described54. All mass spectra were processed manually using MagTran. The relative amount of deuterium in the indicated peptide was determined by subtracting the centroid mass of the undeuterated form from the deuterated form, at each condition. Deuterium levels were not corrected for back exchange and thus reported as relative.
For conformational analysis of UBE1, recombinant full-length UBE1 (5 μM purified protein in 20 mM HEPES pH 8.0, 50 mM NaCl) was combined with 50 μM ATP and 50 μM ubiquitin, and then mixed with either vehicle (0.5% DMSO) or separately with SAH-UBE2A, SAH-UBE2AR7E, or SAH-UBE2AR7A/L9A/B10A (125 μM peptide in 0.5% DMSO) for a final molar ratio of 25:5:5:1 peptide:ATP:ubiquitin:UBE1 and equilibrated on ice for 5 min. Deuterium labeling was initiated with an 18-fold dilution with D2O buffer (20 mM HEPES pD 8.0, 50 mM NaCl, 99.9% D2O). After the indicated labeling time at 21°C, the reaction was quenched with the addition of an equal volume of ice-cold quench buffer (0.8 M guanidinium chloride, 0.8% (v/v) formic acid, H2O, pH 2.0). Deuterated and control samples were digested online at 15°C using an in-house packed pepsin column (2.1 mm x 50 mm, pepsin immobilized on POROS-20AL beads). The cooling chamber of the UPLC system, which housed all of the chromatographic elements, was held at 0.0 ± 0.1°C for the entire measurement time. Peptides were trapped and desalted on a VanGuard Pre-Column trap (2.1 mm × 5 mm, ACQUITY UPLC BEH C18, 1.7 μm for 3 min at 100 μL/min). Peptides were then eluted from the trap using a 5%–35% gradient of acetonitrile over 10 min at a flow rate of 100 μL/min, and separated using an ACQUITY UPLC HSS T3, 1.8 μm, 1.0 mm × 50 mm column. The error of determining the deuterium levels was ± 0.15 Da in this experimental setup. To eliminate peptide carryover, a wash solution of 1.5 M guanidinium chloride, 0.8% formic acid and 4% acetonitrile was injected over the pepsin column during each analytical run. Mass spectra were acquired using a Waters Synapt G2-Si HDMSE mass spectrometer in ion mobility mode. The mass spectrometer was calibrated with direct infusion of a solution of glu-fibrinopeptide (Sigma, F3261) at 200 femtomole/μL at a flow rate of 5 μl/min prior to data collection. A conventional electrospray source was used, and the instrument was scanned over the range 50 to 2000 m/z. The instrument configuration was the following: capillary was 3.2 kV, trap collision energy at 4 V, sampling cone at 40 V, source temperature of 80°C and desolvation temperature of 175°C. All comparison experiments were done under identical experimental conditions such that deuterium levels were not corrected for back-exchange and are therefore reported as relative. Peptides were identified using PLGS 3.0.1 using 4 replicates of undeuterated control samples. Raw MS data were imported into DynamX 3.0 and filtered as follows: minimum consecutive products of 1; minimum number of products per amino acid of 0.25; maximum mass error of 10 ppm. Those peptides meeting the filtering criteria were further processed automatically by DynamX followed by manual inspection of all processing. The relative amount of deuterium in each peptide was determined by subtracting the centroid mass of the undeuterated form of each peptide from the deuterated form, at each time point, for each condition. These deuterium uptake values were used to generate uptake graphs and difference maps. The HXMS data have been deposited to the ProteomeXchange Consortium via the PRIDE55 partner repository with the dataset identifier PXD016014.
Native E1 Inhibition Assay
HeLa lysate assays were performed with a 1 mg/mL final concentration of S100 lysate fraction (R&D Systems F-372–05M) and the indicated concentration of peptide or vehicle control (0.25% DMSO) in assay buffer containing 1x ATP regeneration solution (Abcam ab139471), 20 mM HEPES, 50 mM NaCl, 5 mM MgCl2, 2 mM DTT, 100 nM ubiquitin aldehyde as a DUB inhibitor (Boston Biochem U-201), and a protease-phosphatase inhibitor cocktail (complete Mini, Roche 11836170001) at pH 7.4. Reactions were initiated by addition of ubiquitin (R&D Systems U100H10M) to a final concentration of 100 μM and allowed to proceed for 30 min at room temperature, followed by quenching with non-reducing LDS loading buffer. Proteins were separated by SDS-PAGE gel electrophoresis and detected by western blot using a poly-ubiquitin antibody (Cell Signaling Technology CST3936).
Statistical Methods
Prism software (Graphpad) was used for data analysis, error calculations, and determination of dissociation constants. Data are expressed as mean ± s.e.m. or mean ± s.d., as calculated in Excel (Microsoft Office) or Prism (Graphpad) software.
Data Availability Statement
All data generated or analyzed during this study are included in this manuscript and its supplementary information. HXMS data have been deposited to the PRIDE database with identifier PXD016014. Structures corresponding to PDB IDs 6DC6, 5KNL, 4II2, 4II3, and 3CMM were used in this study.
Extended Data

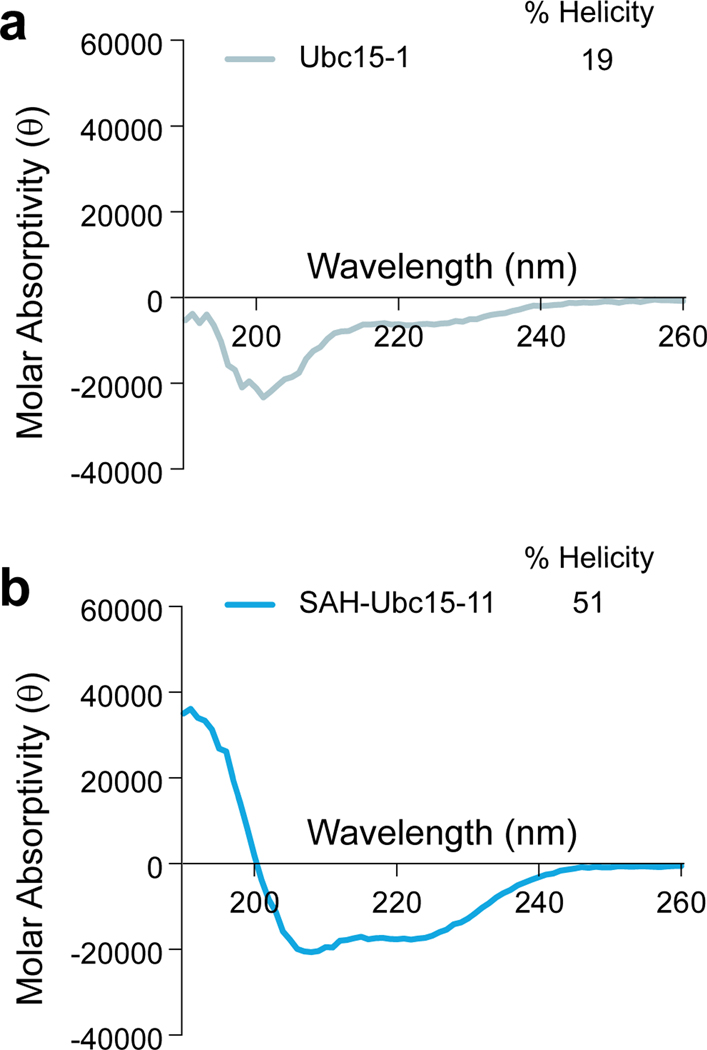
Circular dichroism analysis of Ubc15–1 and SAH-Ubc15–11.
(a-b) Circular dichroism spectra of Ubc15–1 (BPSSASRQLLRKQLKEIQ) (a) and SAH-Ubc15–11 (b), demonstrating induction of α-helicity by insertion of the i, i+7 staple at positions S6 and Q13. B, norleucine (replacement for methionine, whose sulfur residue decreases the efficiency of ruthenium-catalyzed olefin metathesis). Source data for CD spectra are available online.

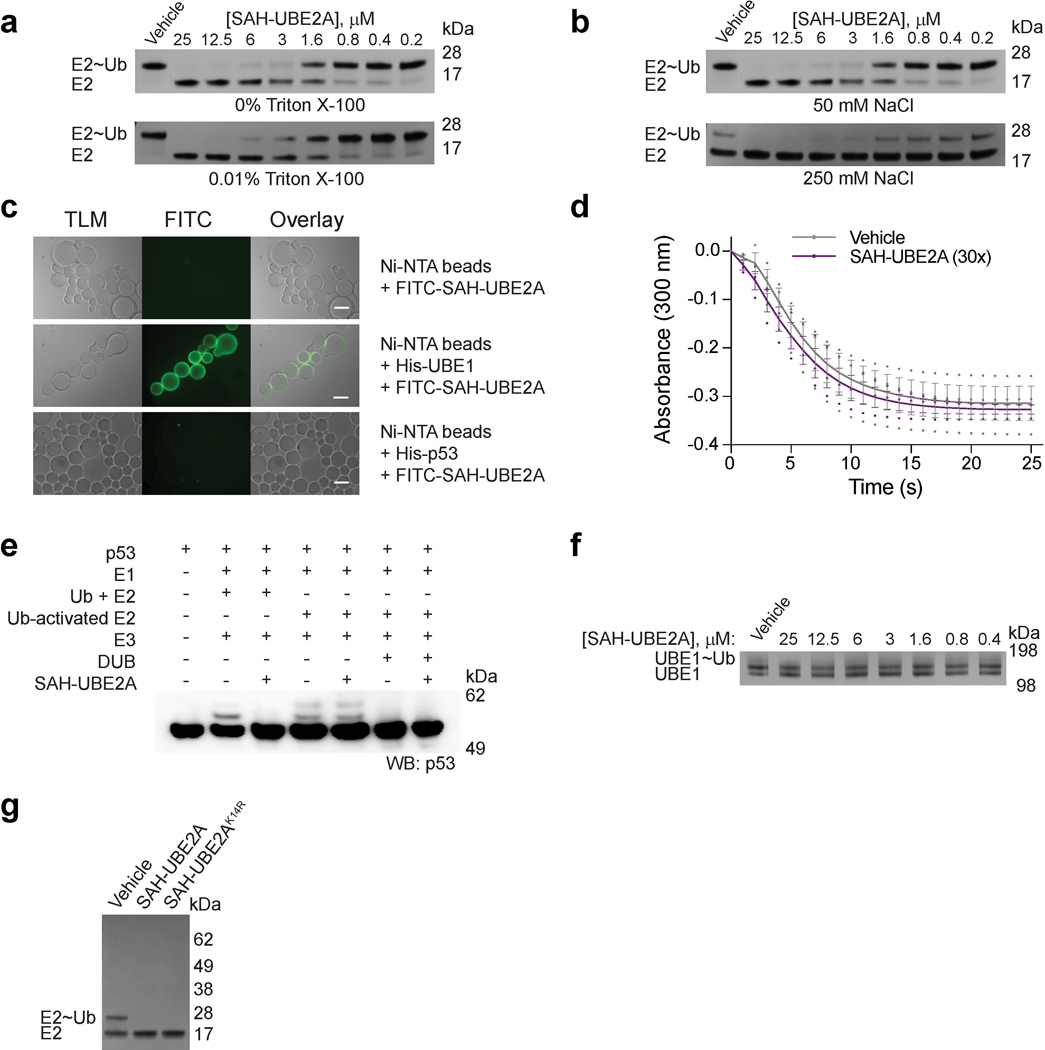
Specificity of action of SAH-UBE2A.
(a) Dose-responsive inhibition of UBE1 by SAH-UBE2A in the presence and absence of 0.01% Triton X-100, as monitored by thioester transfer assay, gel electrophoresis, and silver stain. (b) Dose-responsive inhibition of UBE1 by SAH-UBE2A using reaction buffer containing 50 mM (standard; Fig. 2b) or 250 mM (elevated) NaCl, as monitored by thioester transfer assay, gel electrophoresis, and silver stain. (c) Fluorescent bead binding assay showing that FITC-SAH-UBE2A exhibits no non-specific binding to Ni-NTA beads (top row), selective binding to His-UBE1 coated beads (middle row), and no non-specific binding to beads coated with a negative control protein, such as His-p53 (bottom row). Scale bars, 200 μm. (d) VLCAD enzymatic assay performed in the presence of vehicle or SAH-UBE2A (30:1 peptide:enzyme ratio, final peptide concentration 43.2 μM), demonstrating no interference by SAH-UBE2A on the capacity of VLCAD to oxidize its palmitoyl-CoA substrate. Data are mean ± s.e.m. for experiments performed in technical triplicate. (e) In vitro ubiquitin pathway reconstitution assay showing that SAH-UBE2A (30 μM, excess) inhibits ubiquitination of p53 when the cascade relies on E1 thioester transfer activity but not when the E1 step is bypassed by the addition of E2 pre-charged with ubiquitin. In addition, SAH-UBE2A has no effect on the capacity of the DUB USP7 to deubiquitinate p53. The entire experiment was performed twice using independent preparations of reagents with similar results. (f) SAH-UBE2A has no effect on the capacity of UBE1 to form a covalent UBE1~ubiquitin thioester adduct, the enzymatic step of the UBE1 catalytic cycle that precedes thioester transfer to E2, as demonstrated by the presence of a UBE1 and UBE1~Ub doublet (silver stain) across all SAH-UBE2A concentrations of the thioester transfer assay. (g) To verify that the E1-inhibitory mechanism was not dependent on ubiquitin discharge from E2 onto SAH-UBE2A, a lysine-free K14R mutant was synthesized and tested in the thioester transfer assay. Both SAH-UBE2A and SAH-UBE2AK14R (10 μM dosing) showed equivalent E1-inhibitory activity, as reflected by blockade of E2~Ub formation. Uncropped gels for panels a, b, e-f are available online. Source data for the VLCAD enzymatic assay plot are available online.

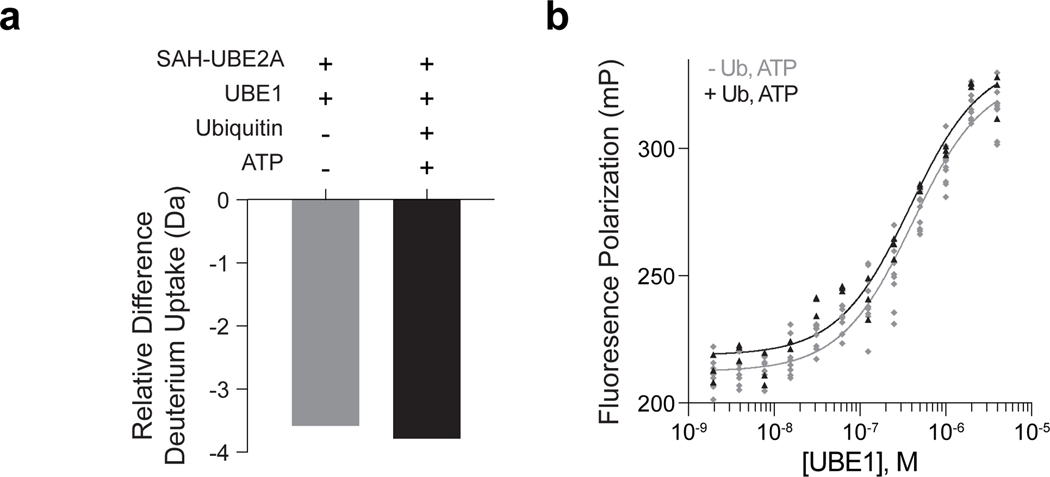
SAH-UBE2A binds to UBE1 in the presence and absence of ubiquitin and ATP.
(a) Similar protection of SAH-UBE2A from deuterium exchange upon incubation with UBE1 (2:1 protein:peptide) in the presence (black) or absence (light gray) of ubiquitin and ATP. The relative difference in deuterium uptake after 10 sec of labeling is shown. The difference in uptake was calculated from the mean deuterium level for SAH-UBE2A in the presence of UBE1 minus that of the peptide alone. Mean deuterium levels were obtained from at least two independent biological replicates of each condition. (b) C-terminally FITCylated SAH-UBE2A bound to UBE1 with respective Kds of 384 ± 43 nM and 359 ± 60 nM in the absence (light gray) and presence (black) of ubiquitin and ATP, as assessed by fluorescence polarization assay. Kds were determined by nonlinear regression analysis of experiments performed in technical triplicate (with ubiquitin/ATP) and plotted alongside the data shown in Figure 2i (without ubiquitin/ATP). The two curve fits have R2 values of 0.9491 (without ubiquitin/ATP) and 0.9636 (with ubiquitin/ATP). Source data for the HXMS and FP plots are available online.

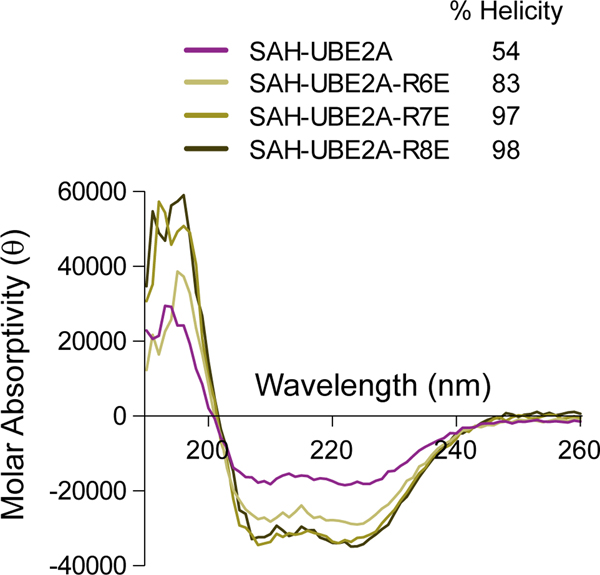
Circular dichroism analysis of SAH-UBE2A and its R-to-E point mutants.
Circular dichroism spectra of SAH-UBE2A and its R6E, R7E, and R8E single point mutants, showing a similar degree of high α-helicity for the R-to-E mutants. Source data for the CD spectra are available online.

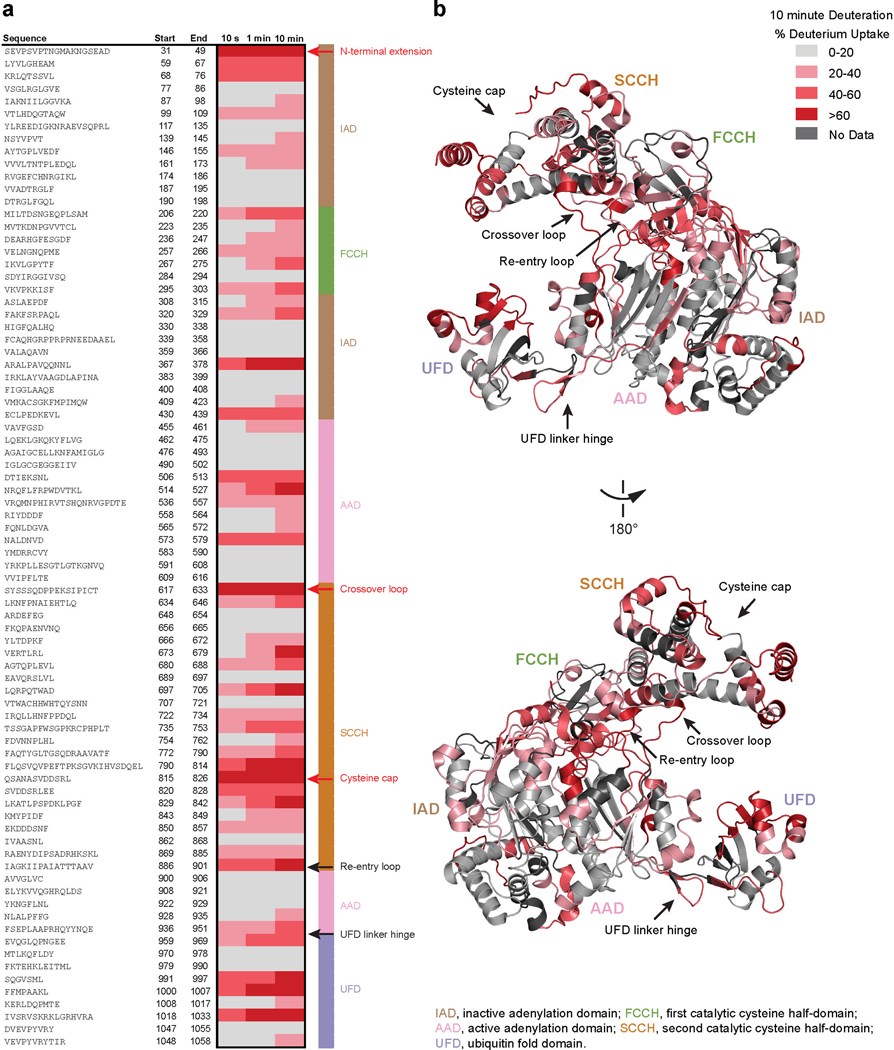
Deuterium exchange profile of UBE1.
(a) The hydrogen-deuterium exchange profile of UBE1 was measured at 10 sec, 1 min, and 10 min of deuterium labeling and the average deuterium uptake (numerical values are found in Supplementary Data File 1) over time for three independent biological replicates displayed with a chiclet plot (see also Supplementary Table 1). Exemplary regions of early uptake are labeled with red arrows and text, whereas exemplary areas that show gradual uptake over time are labeled with black arrows and text. The structurally-defined regions of E1, including the SCCH, FCCH, IAD, AAD, and UFD, are shown in the key to the right of the chiclet plot. (b) The regions and extent of deuterium uptake are mapped onto the structure of E1 (PDB: 6DC6) in accordance with the color scale shown. Regions of high deuterium uptake at the earliest 10 sec time point, reflective of solvent exposure, include, for example, the N-terminal extension, cysteine cap, and crossover loop. Regions that demonstrate increasing exchange over time, consistent with dynamic structures, include N-terminal alpha-helices, adenylation active site, SCCH (excluding the cysteine cap), re-entry loop, and UFD linker hinge. Conformationally stable regions that show no deuterium uptake (light gray) at any of the time points include α-helices and beta sheets of the IAD and AAD protein core.

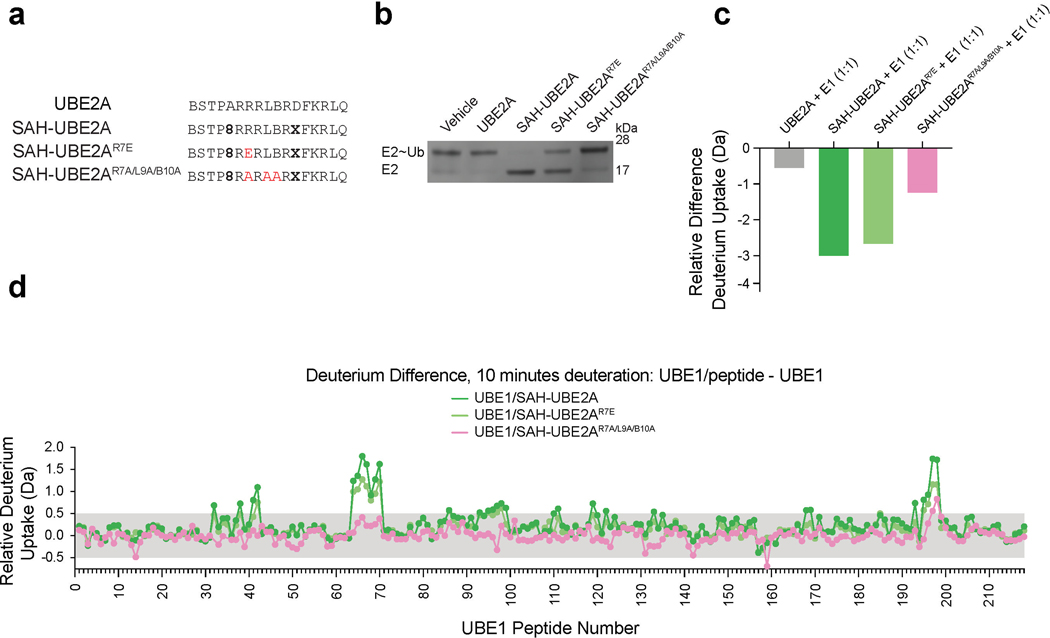
Single R7E and triple R7A/L9A/B10A mutagenesis progressively inhibit the functional and conformational effects observed upon SAH-UBE2A/UBE1 interaction.
(a) Sequence compositions of the unmodified UBE2A template peptide and stapled constructs SAH-UBE2A, SAH-UBE2AR7E, and SAH-UBE2AR7A/L9A/B10A. (b) Comparative effects of UBE2A, SAH-UBE2A, SAH-UBE2AR7E, and SAH-UBE2AR7A/L9A/B10A on UBE1-mediated thioester transfer of ubiquitin to the E2 enzyme UBE2D2. Whereas SAH-UBE2A (10 μM) completely inhibits UBE1 activity, triple mutagenesis abrogates the inhibitory effect and single R7E mutagenesis has an intermediate effect. (c) Progressive loss of UBE1 protection from deuterium exchange into SAH-UBE2A upon single R7E and triple R7A/L9A/B10A mutagenesis. The relative difference in deuterium uptake after 10 sec of labeling is shown. The difference in uptake was calculated from the mean deuterium level for each SAH-UBE2A peptide in the presence of UBE1 minus that of the peptide alone. Mean deuterium levels were obtained from at least two independent biological replicates of each condition. (d) Difference in deuterium uptake plots demonstrate the relative deuterium incorporation of SAH-UBE2A/UBE1, SAH-UBE2AR7E/UBE1, or SAH-UBE2AR7A/L9A/B10A/UBE1 minus the relative deuterium incorporation of UBE1 at 10 min of deuterium labeling. The shaded gray region indicates differences in deuteration that are below the meaningful differences threshold. Consistent with results of both the thioester transfer assay (b) and peptide deuterium exchange (c), and the predicted SAH-UBE2A/UBE1 binding mode, triple mutagenesis essentially abrogates the effect of SAH-UBE2A on UBE1 conformation, with single R7E mutagenesis having an intermediate influence. Data are representative of three independent replicates for SAH-UBE2A, and two independent replicates for SAH-UBE2AR7E and SAH-UBE2AR7A/L9A/B10A. Each peptide, from N- to C-terminus, was given a peptide number to simplify the presentation. The peptide list and residue identity of each peptide can be found in Supplementary Data File 1. Uncropped gel for panel b is available online. Source data for the HXMS analyses are available online.

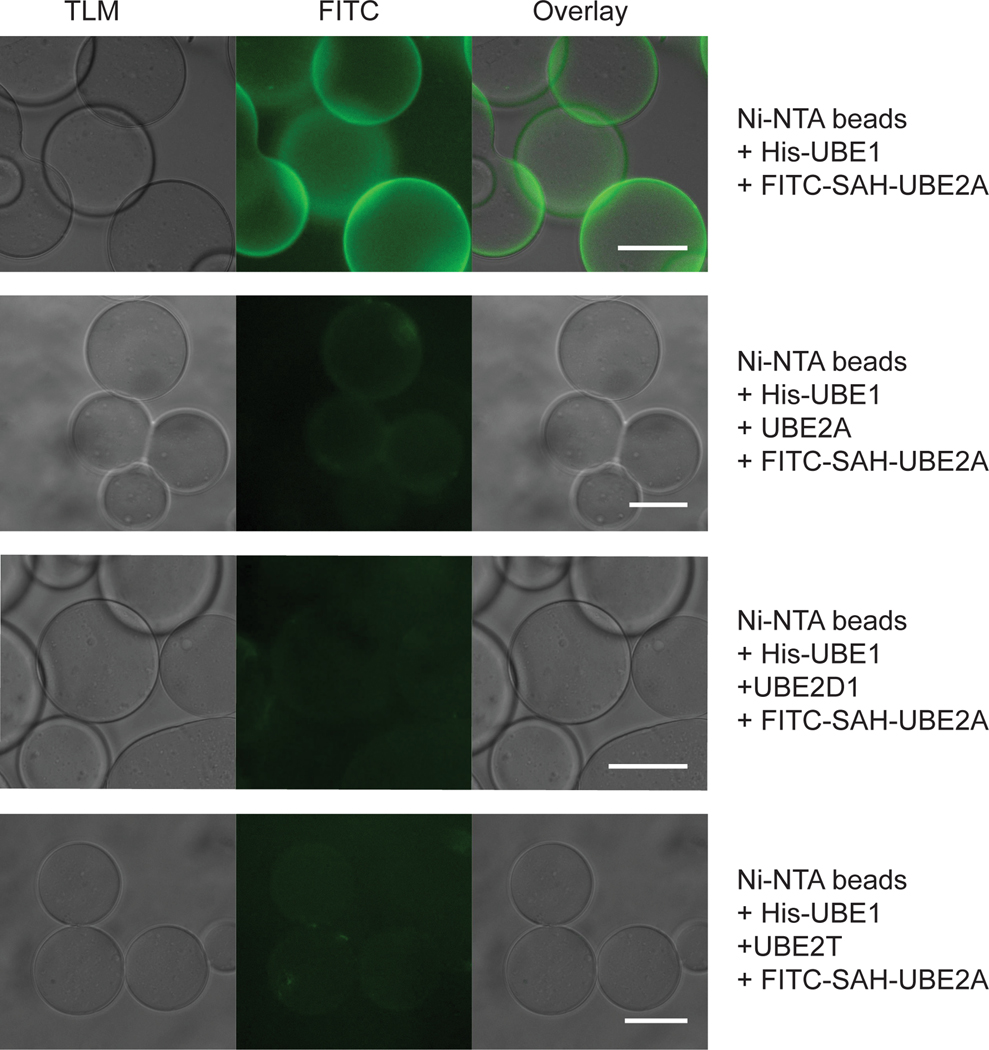
E2 proteins compete with SAH-UBE2A for UBE1 interaction.
Fluorescent bead binding assay showing that the binding of FITC-SAH-UBE2A (0.5 μM) to His-UBE1 coated Ni-NTA beads (top row) is blocked by preincubation of the UBE-1 beads with UBE2A, (second row), UBE2D1 (third row), and UBE2T (fourth row) proteins (25 μM). Scale bars, 200 μm.
ACKNOWLEDGMENTS
We thank E. Smith for graphics support and assistance with figure preparation, and D.T.C. for helpful discussions. The study was funded in part by NIH grants T32GM007753, T32GM008313, and 5F30CA221087 to A.M.C.; NIH grant R50CA211399 to G.H.B.; Lauri Strauss Leukemia Foundation Discover grant to H.D.H.; NIH grant 5F31CA210592 to E.P.H.; NIH grant 5F31CA210590 to Z.J.H.; NIH grant T32GM007753 to C.E.N.; Landry Cancer Biology Research Fellowship and Chleck Family Scholarship to U.A.; NSF predoctoral fellowship to M.S.P.; NIH grant R01GM101135 to J.R.E., and NIH grant R35CA197583 to L.D.W. Additional support was provided by a research collaboration between J.R.E. and the Waters Corporation. We are also thankful to the Wolpoff Family Foundation, J. and L. LaTorre, the family of I. Coll, and the Todd J. Schwartz Memorial Fund for their financial contributions to our cancer chemical biology research.
L.D.W. is a scientific co-founder and shareholder in Aileron Therapeutics.
REFERENCES
1.
2.
3.
4.
5.
6.
7.
8.
9.
10.
11.
12.
13.
14.
15.
16.
17.
18.
19.
20.
21.
22.
23.
24.
25.
26.
27.
28.
29.
30.
31.
33.
34.
35.
36.
37.
38.
39.
40.
41.
42.
43.
44.
45.
46.
47.
METHODS REFERENCES
49.
50.
51.
52.
53.
54.
Floating objects

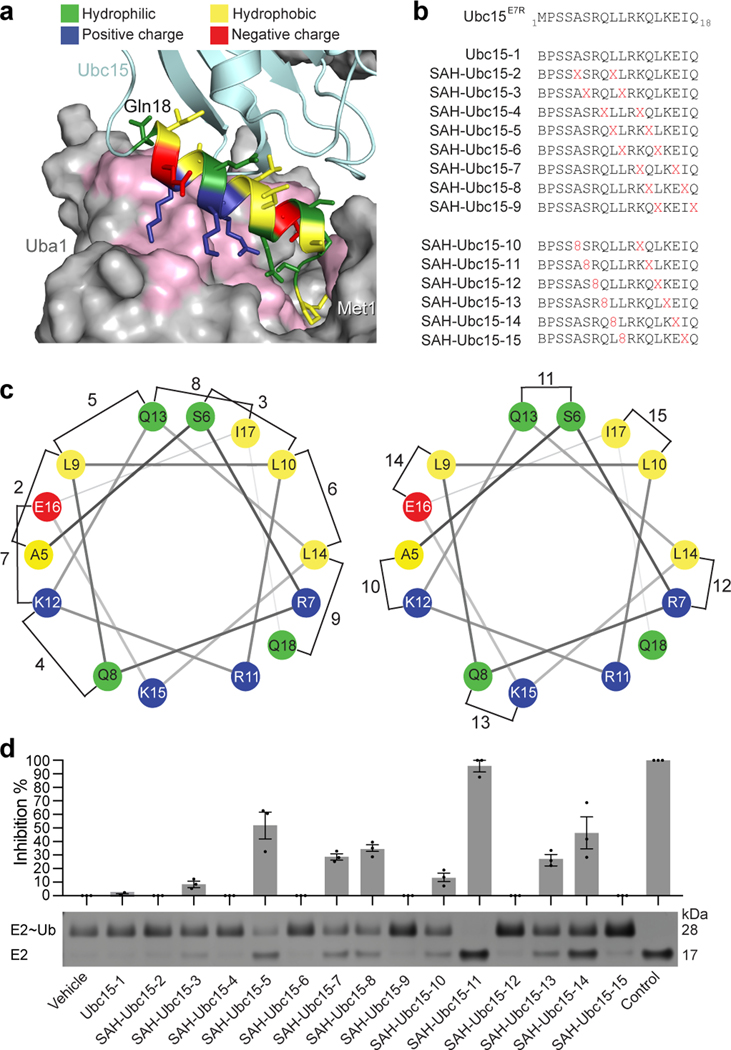
Stapled peptides modeled after Ubc15 helix 1 inhibit E1-mediated thioester transfer of ubiquitin to E2.
(a) Structure of the binding interface between the UFD of S. pombe Uba1 (interacting residues colored pink) and the alpha-1 helix (aa 1–18, colored by amino acid property) of the E2 Ubc15 (cyan), as revealed by the x-ray structure of the S. pombe Uba1/Ubc15 complex (PDB: 5KNL)26. (b) Amino acid sequences of the SAH-Ubc15E7R staple-scanning library, which was generated by inserting all-hydrocarbon i, i+4 or i, i+7 staples sequentially along the length of the Ubc15E7R peptide. X, S5-pentenyl alanine; 8, R5-octenyl alanine; B, norleucine (replacement for methionine, whose sulfur residue decreases the efficiency of ruthenium-catalyzed olefin metathesis). (c) Helical wheel depiction of SAH-Ubc15E7R peptides and their staple positions (helical residues span from aa 5–18). (d) Inhibition of UBE1-mediated thioester transfer of ubiquitin to the E2 enzyme UBE2D2 by the indicated SAH-UBC15E7R stapled peptides. Thioester transfer activity was monitored by the shift in E2 molecular weight from 17 kDa (free UBE2D2) to 25 kDa (UBE2D2~ubiquitin conjugate), as detected by gel electrophoresis and silver stain. Vehicle lane contained 1.5% DMSO and control lane lacked UBE1 protein. The bar graph represents mean inhibition relative to vehicle control ± s.e.m., as quantified and averaged across three independent biological replicates. Uncropped gel for panel d is available online. Source data for the thioester transfer assay plot is available online.

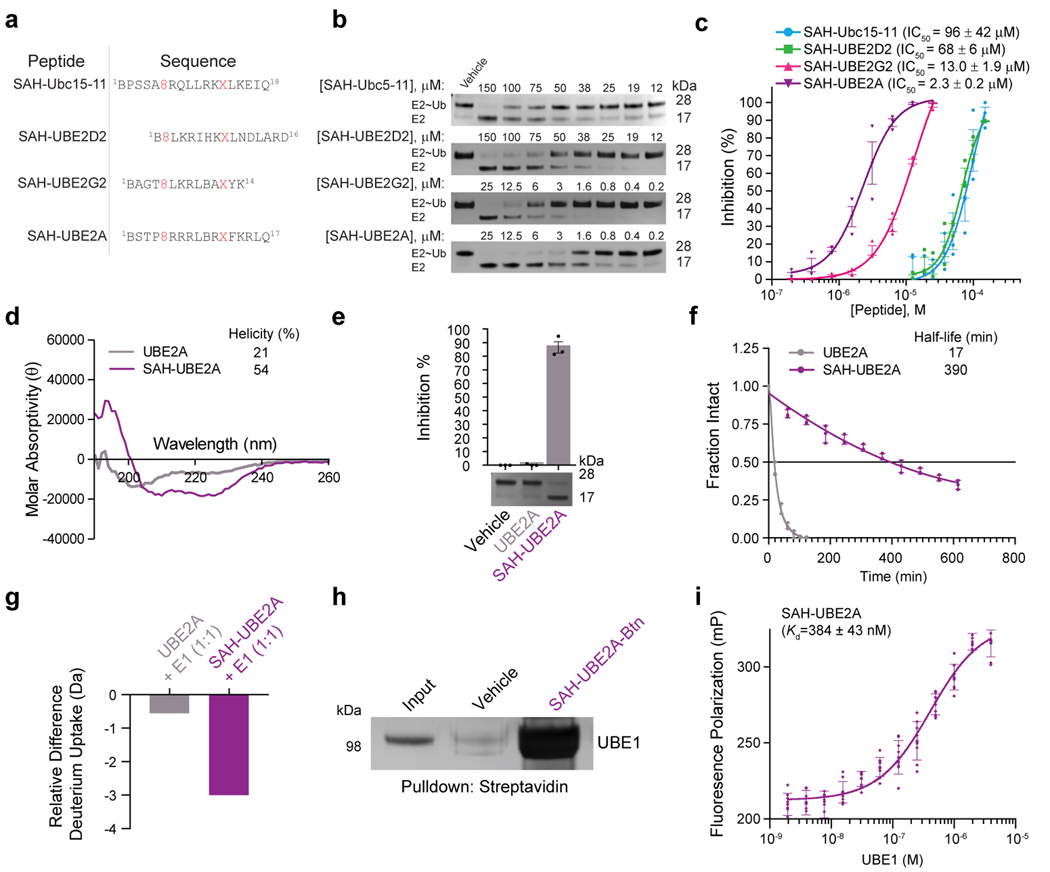
Biophysical and biochemical properties of an optimized stapled peptide mimetic of E2h1 that binds and inhibits E1.
(a) Amino acid sequences of stapled peptides corresponding to the N terminus and alpha-1 helix of UBE2D2 (aa 1–16), UBE2G2 (aa 1–14), and UBE2A (aa 1–17), which incorporate the optimal staple position identified for SAH-Ubc15–11. X, S5-pentenyl alanine; 8, R5-octenyl alanine; B, norleucine. (b) Comparative dose-responsive inhibition of UBE1 by the indicated SAH-E2h1 peptides, as monitored by thioester transfer assay, gel electrophoresis, and silver stain. (c) Quantitation of the data in b by ImageJ analysis. Data are mean percent inhibition ± s.e.m. for three independent biological replicates. (d) Circular dichroism spectra of UBE2A (BSTPARRRLBRDFKRLQ, where B is norleucine) and SAH-UBE2A, demonstrating induction of α-helicity by insertion of the i, i+7 staple. (e) Inhibition of UBE1-mediated thioester transfer of ubiquitin to the E2 enzyme UBE2D2 by SAH-UBE2A but not the corresponding unmodified template peptide, UBE2A. Data are mean inhibition ± s.e.m. for three independent biological replicates. (f) Comparative protease resistance of SAH-UBE2A and UBE2A upon treatment with proteinase K, as measured by HPLC analysis. SAH-UBE2A displays a 23-fold longer half-life than UBE2A. Data are mean ± s.d. for experiments performed in triplicate. (g) SAH-UBE2A but not UBE2A was significantly protected from deuterium exchange upon incubation with UBE1. The relative difference in deuterium uptake after 10 sec of labeling is shown. The difference in uptake was calculated from the mean deuterium level for UBE2A or SAH-UBE2A in the presence of UBE1 minus that of the peptide alone. Mean deuterium levels were obtained from at least two independent biological replicates of each condition. (h) C-terminally biotinylated SAH-UBE2A directly bound to recombinant human UBE1 (MW 118 kDa), as demonstrated by streptavidin capture, gel electrophoresis, and silver stain. (i) C-terminally FITCylated SAH-UBE2A bound to UBE1 with a Kd of 384 ± 43 nM, as assessed by fluorescence polarization assay. Error bars are s.d. for assays conducted in technical triplicate and performed three times with independent preparations of peptide and protein (all 9 points for each dosing level are plotted). Uncropped gels for panels b, e, and h are available online. Source data for thioester transfer assays, CD spectra, protease resistance testing, HXMS analyses, and FP plot are available online.

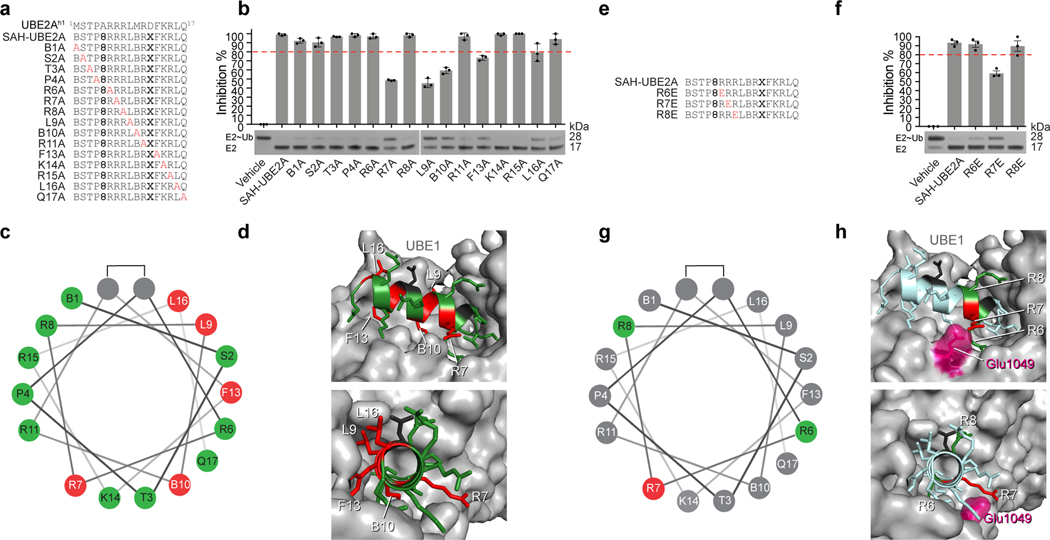
Sequence determinants for SAH-UBE2A inhibition of E1.
(a) Sequence compositions of the alanine scanning library of SAH-UBE2A peptides. (b) The influence of alanine mutagenesis at each position of the SAH-UBE2A sequence on E1-inhibitory activity, as assessed by thioester transfer assay. Data are mean inhibition ± s.e.m. for three independent biological replicates. (c) Helical wheel depiction of the location of alanine mutations in SAH-UBE2A. Alanine mutations that reduced inhibitory activity to <80% (dotted red line in b) compared to wild-type SAH-UBE2A peptide are colored red on the helical wheel, whereas constructs demonstrating ≥80% of wild-type inhibitory activity are colored green. (d) Structure of the UBE2A alpha-1 helix (aa 1–17) docked onto the human UBE1 protein (PDB: 6DC6). Residues are colored as in c, according to the effect of alanine mutagenesis on thioester transfer activity. Those mutations that reduced inhibitory activity map to the UBE1-binding interface of the UBE2A alpha-1 helix in the docked structure. Residues replaced by the i, i+7 hydrocarbon staple are indicated in black. (e) Sequence compositions of the R-to-E charge reversal mutants of SAH-UBE2A. (f) The influence of R6E, R7E, or R8E mutagenesis on the E1-inhibitory activity of SAH-UBE2A was assessed by thioester transfer assay. Data are mean inhibition ± s.e.m. for three independent biological replicates. (g) Helical wheel depiction of the location of glutamic acid mutations in SAH-UBE2A. The glutamic acid mutation that reduced inhibitory activity to <80% (dotted red line in f) compared to wild-type SAH-UBE2A peptide is colored red on the helical wheel, whereas the constructs demonstrating ≥80% of wild-type inhibitory activity are colored green. (h) Structure of the UBE2A alpha-1 helix (aa 1–17, light blue) docked onto the human UBE1 protein (PDB: 6DC6). Residues are colored as in g, according to the effect of R-to-E mutagenesis on thioester transfer activity. The R7 residue (red) of UBE2A is predicted to engage in a complementary electrostatic interaction with E1049 (pink) of UBE1. Uncropped gels for panels b and f are available online. Source data for thioester transfer assays are available online.

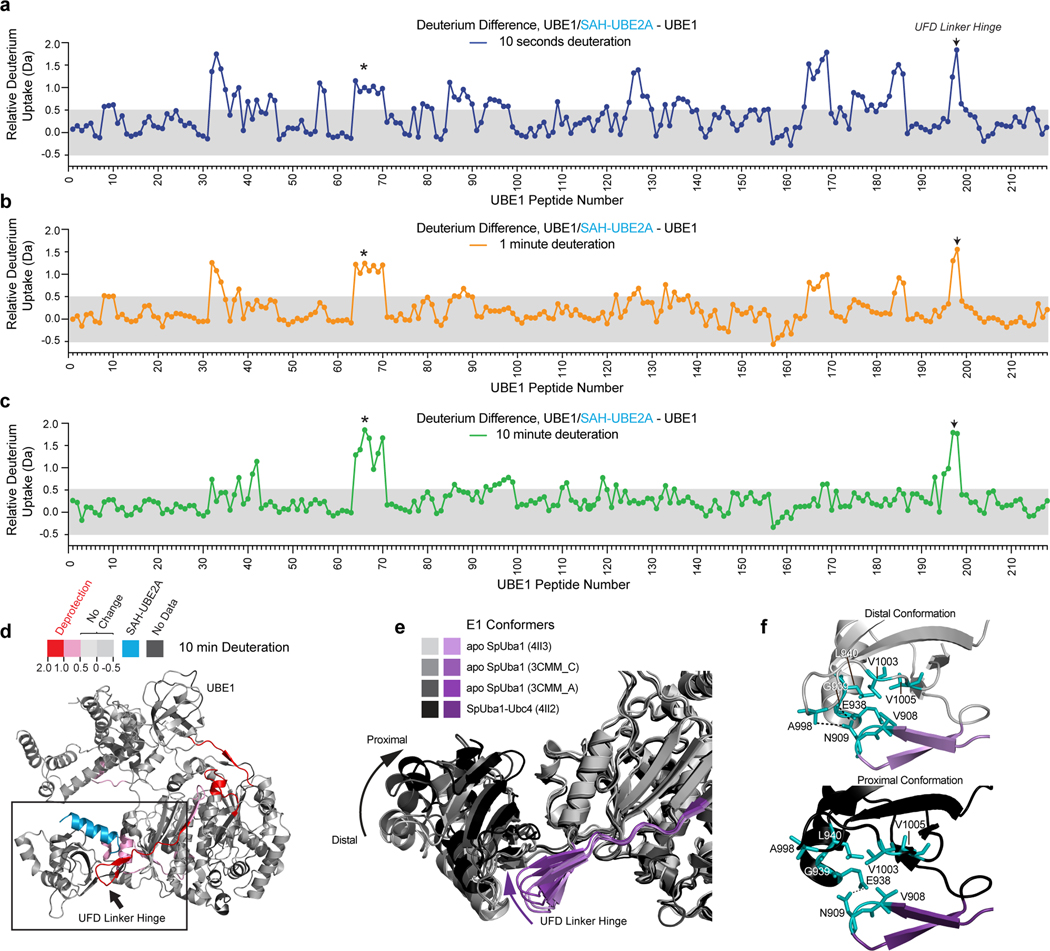
Influence of SAH-UBE2A on the conformational dynamics of E1.
(a-c) Difference in deuterium uptake plots reflect the relative deuterium incorporation of SAH-UBE2A/UBE1 minus the relative deuterium incorporation of UBE1 at 10 sec (a), 1 min (b), and 10 min (c) of deuterium labeling. Structural regions corresponding to the h11 helix of the IAD and the UFD linker hinge are indicated by an asterisk and arrow, respectively. The shaded gray region indicates differences in deuteration that are below the meaningful differences threshold. Data are representative of two independent biological replicates. Each peptide, from N- to C-terminus, was assigned a peptide number to simplify the presentation. The peptide list and residue identity of each peptide can be found in Supplementary Data File 1. (d) The regions of SAH-UBE2A-induced deprotection are mapped onto the ribbon diagram of UBE1 (PDB: 6DC6) according to the indicated color scale, with the UBE2A alpha-1 helix (cyan) docked at the UFD groove. The boxed region highlights the observed deprotection in the region adjacent to the putative SAH-UBE2Ah1/UBE1UFD binding interface, with the UFD linker hinge indicated by an arrow. (e) Flexion of the UFD linker hinge (shades of purple) causes a 25° rotation of the UFD with respect to the core of the E1 protein, as reflected by the indicated structures that depict the transition from the distal conformation in the apo structure (PDB: 4II3) to the proximal positioning of the UFD in the E2-bound complex (PDB: 4II2). Of note, only in the UFD proximal conformation is the active cysteine residue of E2 brought sufficiently close to the E1 catalytic cysteine to accept thioester transfer of ubiquitin. (f) UFD rotation from the distal (top) to the proximal (bottom) position causes loss of contacts between the linker hinge and the E1 protein core, which increases the solvent accessibility of the linker hinge.

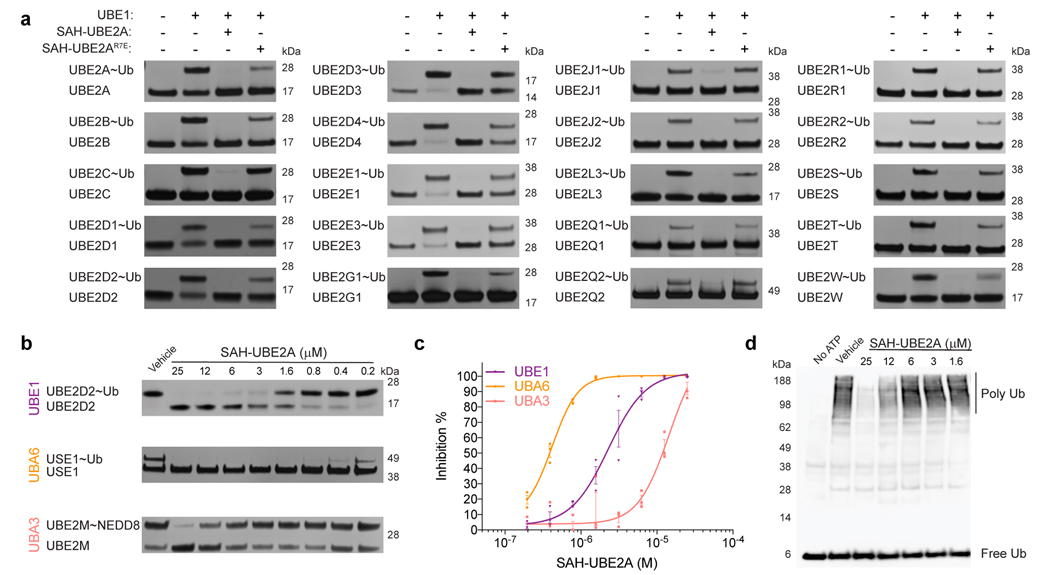
SAH-UBE2A is a pan-inhibitor of ubiquitin E1 thioester transfer that targets native E1 to achieve blockade of protein ubiquitination.
(a) SAH-UBE2A inhibited E1 thioester transfer to each of 20 E2 enzymes tested. R7E mutagenesis impaired inhibitory activity, highlighting the sequence specificity of SAH-UBE2A function. (b) SAH-UBE2A demonstrates even more potent inhibition of UBA6 (minor ubiquitin pathway) than UBE1 (major ubiquitin pathway; Fig. 2b), but markedly less of an effect on UBA3 (NEDD8 pathway), highlighting the relative specificity of SAH-UBE2A for inhibiting the ubiquitin pathway, as assessed by thioester transfer assay. IC50s: UBE1, 2.3 ± 0.2 μM; UBA6, 0.42 ± 0.02 μM; UBA3, 14.5 ± 5.4 μM. (c) Quantitation of the data in b by ImageJ analysis. Data are mean percent inhibition ± s.e.m. for three independent biological replicates. (d) The cytoplasmic fraction of HeLa cell lysates was incubated with SAH-UBE2A or vehicle (1% DMSO) in the presence of excess ubiquitin (Ub) and an ATP regeneration system, followed by non-reducing gel electrophoresis and ubiquitin western blot. SAH-UBE2A dose-responsively inhibited bulk polyubiquitin (Poly Ub) chain derivatization of native proteins in the HeLa cell lysate. Uncropped gels for panels a, b, and d are available online. Source data for thioester transfer assays are available online.
 Targeting a Helix-in-Groove Interaction Between E1 and E2 Blocks Ubiquitin Transfer
Targeting a Helix-in-Groove Interaction Between E1 and E2 Blocks Ubiquitin Transfer