
- Altmetric
Similarly to enzymes, functionalized gold nanoparticles efficiently catalyze chemical reactions, hence the term nanozymes. Herein, we present our results showing how surface‐passivated gold nanoparticles behave as synthetic nanonucleases, able to cleave pBR322 plasmid DNA with the highest efficiency reported so far for catalysts based on a single metal ion mechanism. Experimental and computational data indicate that we have been successful in creating a catalytic site precisely mimicking that suggested for natural metallonucleases relying on a single metal ion for their activity. It comprises one Zn(II) ion to which a phosphate diester of DNA is coordinated. Importantly, as in nucleic acids‐processing enzymes, a positively charged arginine plays a key role by assisting with transition state stabilization and by reducing the pK a of the nucleophilic alcohol of a serine. Our results also show how designing a catalyst for a model substrate (bis‐p‐nitrophenylphosphate) may provide wrong indications as for its efficiency when it is tested against the real target (plasmid DNA).
Gold nanoparticles passivated with Zn(II) complexes and short peptides were transformed into nanozymes able to very efficiently cleave plasmid DNA with a mechanism not much different from that of mononuclear nucleases.
Introduction
Natural selection has reserved to the phosphate bond the role of holding together the backbone of the “molecules of life”, DNA and RNA. There is a reason for this: the phosphate bond is very stable towards hydrolytic cleavage (at 25 °C and pH 7 the half‐life is 31 000 000 years for DNA and “only” 110 years for RNA). [1] The mastering of the phosphate ester bond cleavage (as well as of the reverse reaction, ligation) is of paramount importance for controlling fundamental processes, like DNA cloning, for instance. In order to take part in this challenging endeavor, a scientist needs to understand the fundamental aspects of the reaction mechanism of these processes. [2] In spite of the many progresses made so far, [3] still many mechanistic aspects remain elusive and the challenge for obtaining efficient, synthetic catalysts for the hydrolytic cleavage of both RNA and DNA is far from having been accomplished.
In Nature, hydrolysis of DNA and RNA is efficiently catalyzed by nucleic acids‐cleaving enzymes (nucleases), which typically feature one or two metal ions in their catalytic site. [4] In such metal ion‐based nucleases, the metal(s) present in their reaction site aid catalysis by:[ 3 , 5 , 6 ] (a) activating the phosphoryl group towards nucleophilic attack, thus acting as Lewis acid(s); (b) increasing the leaving group ability; (c) providing a large fraction of an anionic nucleophile, by inducing deprotonation of hydrating water molecules or other coordinated alcoholic groups. Importantly, most of these nucleic acids‐processing metalloenzymes use positively charged side chains strategically located in the surrounding of the catalytic site. [7] These residues often serve to align the scissile phosphate for nucleophilic attack and stabilize the transition state towards the covalent intermediate or the final product.
The features of natural enzymes have been exploited by several groups to obtain artificial nucleases.[ 8 , 9 , 10 ] These synthetic catalysts mimic an enzyme catalytic site by placing together several functions supposed to cooperate in the catalytic process. [11] Quite interesting is the possibility to use multivalent systems [12] that exploit a platform that can be functionalized with several copies of catalytically active functional groups. They not only induce the cooperation of the reactive functions but also, since several catalytic sites are typically present on the same platform, they allow many catalytic events to occur simultaneously. [13] Examples are constituted by dendrimers, [14] micellar and vesicular aggregates, [15] carbon dots, [16] polyoxometalates, [17] or nanoparticles. [18] For instance, we have reported that gold nanoparticles (AuNPs) passivated with thiols functionalized with 1,4,7‐triazacyclonane (TACN) complexed with Zn(II) are among the most efficient catalysts for the cleavage of the RNA‐model substrate 2‐hydroxypropyl p‐nitrophenyl phosphate (HPNP) and dinucleotides as well. [19a] The impressive efficiency and the enzyme‐like kinetic profile they show in the catalytic process, led us to dub them “nanozymes”.[ 19 , 20 ]
All these nanoparticles are very simple as they feature several copies of the same metal complex. Their design paradigm is confinement, hence cooperation: far from the sophisticated arrangement of functional groups present in the catalytic site of an enzyme. Not surprisingly, while their efficiency with model substrates places them among the most powerful synthetic catalysts so far reported, the results with RNA and DNA are contradictory and less exciting. For instance, nanozymes based on the Zn(II) complex of ligand bis(2‐amino‐pyridinyl‐6‐methyl)amine (BAPA), (thiol BAPASH in Figure 1) cleave plasmid DNA with moderate efficiency, [21] those functionalized with the TACN‐Zn(II) complex are totally inactive. [22] The two metal complexes require a dinuclear mechanism for optimum performance. This is mandatory in the case of the TACN‐Zn(II)‐based catalyst while a mononuclear one, although less active, is possible for BAPA‐Zn(II)‐based nanoparticles. This is likely because the ancillary amino groups, present in BAPA, provide a substrate activation effect similar to that of a second metal ion. [23] This is a non‐existing option for TACN‐Zn(II).[ 24 , 25 ]

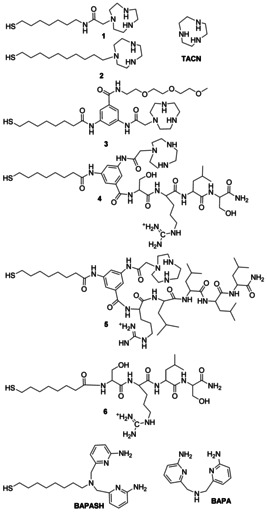
Chemical structure of the thiolated molecules 1–6 used for the passivation of AuNPs and ligands TACN, BAPA and BAPASH discussed in this work. All amino acids are the (L) enantiomers.
In this paper a new generation of AuNPs is studied where several functional groups and not just metal ions form the catalytic site of the nanozyme. We show that cooperation between them in the confined space of a nanoparticle passivating monolayer is possible. This allowed us to obtain one of the most efficient DNA‐cleaving Zn(II)‐nanozymes reported so far. We also show how evaluating a catalyst against a model substrate (as bis‐p‐nitrophenylphosphate, BNP, in the case of DNA) may lead to wrong conclusions on its efficiency against the real biological target.
Results and Discussion
Our strategy towards AuNPs selection for efficient phosphate bond cleavage
In our investigation, we used six different AuNPs, 2‐nm in diameter that were very stable upon passivation with all ligands we used.[ 24 , 26 ] The TGA analyses revealed the presence of 65±7 ligands for each nanoparticle. The inspiring strategy for their design was to bring together in a combinatorial way metal complexes, cationic groups, and nucleophilic functions key players in the catalytic sites of natural nucleases. Nucleases use divalent metal ions such as Mg(II), Mn(II), or Zn(II) with Mg(II) the most frequent one. [27] We used Zn(II) since it binds very strongly to the TACN ligand thus allowing us to precisely control the metal ion location in the monolayer passivating the nanoparticles. Accordingly, all the thiols used for their passivation, but one, (Figure 1) share, as a common feature, a TACN ligand. Thiols 1 and 2 are linear molecules comprising TACN as the single functional group, while 3–5 are bifurcated molecules (Y‐shaped) presenting, in addition to the TACN macrocycle, additional chemical moieties. In the case of 3, this is a short polyether chain, while in the case of 4 and 5 it is the tetrapeptide H‐Ser‐Arg‐Leu‐Ser‐NH2 and the pentapeptide H‐Arg‐Leu‐Leu‐Leu‐Leu‐NH2, respectively. Notably, the pivotal 1,3,5 trifunctionalized benzene unit present in thiols 3–5 allows one to place different functional groups in close proximity to the metal complex formed, in situ, with TACN by addition of Zn(NO3)2 to the solution. This approach prevents the always possible sorting of different thiols on the monolayer whenever a mixture of them is used.[ 28 , 29 ] Should sorting occur, cooperativity between the functional groups could be partially or totally lost.
Thiols 1 and 2 differ on two counts: the presence of a longer hydrophobic chain in 2 and of an amide group in 1. The resulting passivating monolayers are more hydrophobic (with 2) or more tightly packed (with 1) because of the H‐bonds formed between the thiol chains within the monolayer, when an amide is present. With the RNA‐model substrate HPNP, both aspects were beneficial for these nanozymes.[ 30 , 31 ] In the case of thiol 3 the polyether chain is expected to be a neutral spectator while for 4 and 5 the two functionalizing peptides include one arginine that, with its guanidinium group, is a key component of several nucleases.[ 2 , 32 , 33 ] The two Ser present in the tetrapeptide of 4 may provide a nucleophile in the catalytic site, while the four Leu in the pentapeptide of 5 could contribute to render the catalytic site less hydrophilic. Finally, we also considered thiol 6, a linear thiol that features the same tetrapeptide present in thiol 4 but lacking the TACN ligand. Hence, being a TACN‐ and metal‐free analog of 4, it was studied to assess the role of the metal complex in the catalytic process. The syntheses of thiols 1–5 and of the 2‐nm AuNPs obtained upon passivation of the gold clusters (AuNP1–5) was performed according to previously reported procedures. [24] Thiol 6 and AuNP6 were prepared following similar protocols (see Supporting Information, SI).
AuNP1–5 were tested in the cleavage of BNP and, quite importantly, of the supercoiled plasmid pBR322. AuNP6 was tested only with the latter. BNP is a phosphate diester often used as a DNA model substrate and was used for a preliminary screening of our catalysts collection prior to venturing into the cleavage of the biological target. The plasmid pBR322 is a circular double‐strand DNA, 4361 base pairs (bp) in length. The cleavage of one single phosphodiester bond transforms the native supercoiled form (form I) into the nicked circular one (form II). This latter, if further cleaved within ca. 15 nucleobases in the complementary strand, is transformed into the linear form (form III). Throughout the paper, the concentration of nanoparticles is referred to the concentration of the TACN ligand present in their passivating monolayer. This allows one to assess quantitatively the role of the metal complexes once they are confined on the surface of the nanoparticles, independently of their number in each nanoparticle.
AuNP‐mediated cleavage of BNP requires two Zn(II) ions for catalysis
The reported rate constant for the spontaneous hydrolysis of BNP is very slow (k =2×10−10 s−1 at 50 °C). [34] In the presence of AuNP1–4 and a stoichiometric amount of Zn(II) (AuNP1–4‐Zn(II)) there is a substantial increase in the rate of cleavage as shown by the Michaelis–Menten‐like plots reported in Figure 2 A and the relevant kinetic data reported in Table 1. The hydrolysis stops after the release of a single p‐phenolate with the only formation of the phosphate monoester p‐nitrophenyl phosphate. AuNP4‐Zn(II) and AuNP5‐Zn(II) are not particularly soluble in an aqueous solution in the presence of the very lipophilic substrate BNP. The problem was solved in part for AuNP4‐Zn(II) by adding 5 % DMSO to the solution, but not at all for AuNP5‐Zn(II). Accordingly, the latter could not be studied due to the immediate precipitation of the highly lipophilic complex these nanoparticles form with BNP. The different nanoparticles we have tested showed similar catalytic activity in the presence of Zn(II) with AuNP1‐Zn(II)>AuNP3‐Zn(II)>AuNP2‐Zn(II). From the very few points we could collect for AuNP4‐Zn(II) we gathered it is a rather poor catalyst but no reliable Michaelis–Menten parameters could be determined for it.

![Cleavage of BNP by the different AuNPs. A) Initial rate constants obtained for the cleavage of BNP by AuNP1‐Zn(II) (blue symbols), AuNP2‐Zn(II) (green symbols), AuNP3‐Zn(II) (red symbols). The points in gray are for AuNP4‐Zn(II). The solid lines represent the fitting with the Michaelis–Menten equation. Conditions: [AuNP]=3×10−5 M, pH 8, 40 °C; 5 % DMSO added to the AuNP4 solution. Error bars in Panel A refer to data collected in three independent experiments. B) Dependence of the catalytic activity of AuNP1–3 (color codes are the same as in Panel A) and AuNPBAPASH (black symbols) on the number of equivalents of Zn(II) added. The dashed lines have been drawn to guide the eye. Conditions are the same as in Panel A. Inset: double log plots of vi versus [Zn(II)] for AuNP1–3. Color code as in Panel A.](/dataresources/secured/content-1765822518437-102dc52a-a623-4d0a-8790-aba970890b02/assets/ANIE-60-1423-g002.jpg)
Cleavage of BNP by the different AuNPs. A) Initial rate constants obtained for the cleavage of BNP by AuNP1‐Zn(II) (blue symbols), AuNP2‐Zn(II) (green symbols), AuNP3‐Zn(II) (red symbols). The points in gray are for AuNP4‐Zn(II). The solid lines represent the fitting with the Michaelis–Menten equation. Conditions: [AuNP]=3×10−5 M, pH 8, 40 °C; 5 % DMSO added to the AuNP4 solution. Error bars in Panel A refer to data collected in three independent experiments. B) Dependence of the catalytic activity of AuNP1–3 (color codes are the same as in Panel A) and AuNPBAPASH (black symbols) on the number of equivalents of Zn(II) added. The dashed lines have been drawn to guide the eye. Conditions are the same as in Panel A. Inset: double log plots of vi versus [Zn(II)] for AuNP1–3. Color code as in Panel A.

|
Entry |
Catalyst |
k cat [s−1] |
K M [mM] |
k 2 [s−1 M−1][a] |
k rel |
Ref |
|---|---|---|---|---|---|---|
|
1 |
OH− |
– |
– |
2.4×10−5 [b] |
1 | |
|
2 |
TACN‐Zn(II) |
n.d. |
n.d. |
1.2×10−4 [c] |
5 | |
|
3 |
BAPA‐Zn(II) |
n.d. |
n.d. |
0.012 |
500 | |
|
4 |
AuNP1‐Zn(II) |
1.1×10−4 |
8.6×10−2 |
1.3 |
54 166 |
[f] |
|
5 |
AuNP2‐Zn(II) |
6.6×10−5 |
1.9×10−1 |
0.35 |
14 583 |
[f] |
|
6 |
AuNP3‐Zn(II) |
8.2×10−5 |
9.3×10−2 |
0.89 |
37 083 |
[f] |
|
7 |
AuNPBAPASH‐Zn(II)[d] |
3.8×10−5 |
2.1×10−2 |
1.5 |
62 500 | |
|
8 |
AuNPBAPASH‐Zn(II)[e] |
n.d. |
n.d. |
0.11 |
4 583 |
[a] For dinuclear catalysts the concentrations used in the calculations were half that of the Zn(II) complex present. [b] At 35 °C. [c] At 25 °C. [d] As a dinuclear catalyst, 10 % BAPASH in the monolayer, see ref. [20]. [e] As a mononuclear catalyst, 10 % BAPASH in the monolayer, see ref. [29]. [f] This work. Errors are within ±6 %.
In order to get clues on the mechanism of the catalytic reaction, we performed kinetic studies with increasing equivalents of Zn(II) with respect to the TACN (Figure 2 B). All three nanoparticles (AuNP1–3‐Zn(II)) gave a sigmoidal dependence when the initial rate was plotted against the equivalents of Zn(II) added (dashed red line of Figure 2 B). This behavior is typically associated with the cooperativity between Zn(II) ions which is only achieved when all TACN units present on the surface of a nanoparticle are saturated with the metal. [31] To assess the number of Zn(II) involved in the process we performed double log plots of v i vs. [Zn(II)] for AuNP1–3. They gave straight lines (inset of Figure 2 B) with slopes 2.0, 2.1, and 1.9, respectively. The slope indicates the number of ions involved in the catalytic process[ 14 , 35 ] which is ca. two for all three catalysts, confirming a dinuclear catalytic site. AuNP passivated with BAPASH showed a sizeable contribution by a mononuclear catalytic site [21] as it is shown in Figure 2 B for comparison. This mononuclear catalysis is less efficient than the dinuclear one (Table 1). By adjusting the concentration for the apparent dinuclear catalytic site for AuNP1–3‐Zn(II) we obtained the relevant kinetic and binding parameters reported in Table 1. Noteworthy, in terms of efficiency (k cat) all three nanocatalysts based on the TACN‐Zn(II) complex perform better than the corresponding dinuclear one based on the BAPA‐Zn(II) complex. However, the latter shows a higher apparent second order rate constant (k 2), as a consequence of the stronger affinity for BNP.
The comparison between AuNP1 and AuNP2 indicates that the amide functional group present in thiol 1 plays some role in the optimization of the catalytic site, while a hydrophobic hydrocarbon chain does not provide a significant kinetic advantage. H‐bonding within the monolayer [37] may result in its better packing allowing a more precise control on the relative position of the Zn(II)‐TACN with enhanced cooperativity. The low solubility of AuNP4 did not allow to know whether it behaves as a mononuclear or dinuclear catalyst. Judging from its sluggish reactivity, it is more likely that the cooperativity between two metal ions is not attained and that the presence of the flanking guandinium of arginine is not mechanistically relevant, in line with the modest rate acceleration reported for a catalyst exploiting both a TACN‐Zn(II) complex and a guanidinium on a calix[4]arene rim. [38]
Cleavage of pBR322 plasmid DNA is more efficiently catalyzed by AuNP4.
Incubation of pBR322 (19.3 μM bp−1) at pH 7.5 and 37 °C with AuNP1–6 (45 μM) with and without added Zn(II), gave the results reported in Figure 3 and Table 2 (see also Figures S8–S10 of SI). Unexpectedly, the obtained picture totally reversed what we observed in the cleavage of BNP: the worst catalyst (AuNP4‐Zn(II)) became the best, while the best (AuNP1‐Zn(II)) became the worst. Notably AuNP4 and AuNP5 are significantly active even in the absence of Zn(II). Addition of EDTA (12.5 μM) to AuNP4 resulted in a small (6 %) decrease in activity indicating the effect is not due to adventitious metal ions presence. AuNP6 featuring the same peptide of AuNP4 but lacking the Zn(II) chelating moiety is not active. One should infer that the protonated TACN present in the monolayer passivating AuNP4 has a beneficial effect for catalysis even in the absence of the metal. The absence of the Ser in the monolayer of AuNP5 is likely responsible for the lower nuclease activity observed with respect to AuNP4 upon Zn(II) coordination. Figure S11 (SI) reports the time course of the reaction of cleavage of supercoiled pBR322 by AuNP4‐Zn(II). It shows the disappearance of form I of the plasmid that converts into form II and, eventually, to form III. Using a 35 μM concentration of catalyst the half‐life (t 1/2) of supercoiled pBR322 DNA is 50 min. For comparison, the enzyme BamHI, a Type II dinuclear restriction endonuclease, cleaves the same plasmid ([enzyme]=6 nM, pH 8.0, 37 °C) with t 1/2=4 min, [39] while that estimated for AuNPBAPASH‐Zn(II) is 90 h ([catalyst]=15 μM, pH 7.0, 37 °C). [21] Thus, taking into account the different concentrations, AuNP4‐Zn(II) is ca. 7×104‐fold less active than BamHI but ca. 40‐fold better than AuNPBAPASH‐Zn(II). However, at variance with the BAPASH‐based nanoparticle, there is not a preference for double strand cleavage and the product composition follows what is statistically expected for accumulation of single strand cuts, as in the case of the enzyme (Figure S11). [40] No further cleavage in smaller fragments was indeed spotted in the gel plates in the time frame of our studies, at variance with what was observed with micellar or vesicular catalysts. [15] The comparison with the free ligands not bound to the gold cluster (entries 2–5 of Table 2 and Figure S15) shows the reactivity is a property of the entire ensemble (the nanozyme) and not of the isolated ligands. Ligand 4‐Zn(II) is at least >200‐fold less active than nanozyme AuNP4‐Zn(II). This number is likely even larger considering a ca. 3 % background cleavage and the deviation from linearity of the kinetic profile with AuNP4‐Zn(II) already observed after 1 h (see Figure S11).

![Percentage of plasmid pBR322 cleaved by AuNP1–6. Data in the presence of 1 equivalent of Zn(II) (red bars) and without it (blue bars). Lighter symbols represent data collected after 1 h; darker ones after 24 h. Conditions: [AuNP]=45 μM; [pBR322]=19.3 μM (bp); pH 7.5; 37 °C; Original gel electrophoresis data are in Figures S8‐S10 and S16 of the SI.](/dataresources/secured/content-1765822518437-102dc52a-a623-4d0a-8790-aba970890b02/assets/ANIE-60-1423-g003.jpg)
Percentage of plasmid pBR322 cleaved by AuNP1–6. Data in the presence of 1 equivalent of Zn(II) (red bars) and without it (blue bars). Lighter symbols represent data collected after 1 h; darker ones after 24 h. Conditions: [AuNP]=45 μM; [pBR322]=19.3 μM (bp); pH 7.5; 37 °C; Original gel electrophoresis data are in Figures S8‐S10 and S16 of the SI.

|
Entry |
Catalyst |
% Cleaved plasmid[a] | |
|---|---|---|---|
|
|
|
After 1 h |
After 24 h |
|
1 |
none |
<3 |
<3 |
|
2 |
1‐Zn(II) [b] |
n.d. |
12±3 |
|
3 |
1 [b] |
n.d. |
5±2 |
|
4 |
4‐Zn(II) [b] |
n.d. |
9±4 |
|
5 |
4 [b] |
n.d. |
4±2 |
|
6 |
AuNP1 |
9±3 |
8±3 |
|
7 |
AuNP1‐Zn(II) [c] |
5±2 |
6±3 |
|
8 |
AuNP2 |
13±4 |
18±2 |
|
9 |
AuNP2‐Zn(II) [c] |
15±3 |
19±3 |
|
10 |
AuNP3 |
8±1 |
11±3 |
|
11 |
AuNP3‐Zn(II) [c] |
12±3 |
15±2 |
|
12 |
AuNP4 |
25±3 |
47±4 |
|
13 |
AuNP4‐Zn(II) [c] |
68±4 |
99±1 |
|
14 |
AuNP5 |
20±4 |
38±3 |
|
15 |
AuNP5‐Zn(II) [c] |
22±3 |
41±4 |
|
16 |
AuNP6 |
<3 |
<4 |
[a] Conditions: pH 7.5, 37 °C, [AuNP]=45 μM, [pBR322]=19.3 μM bp−1. [b] Thioacetylated. [c] One equivalent of Zn(NO3)2 added with respect to TACN.
The rate of cleavage of pBR322 by AuNP4‐Zn(II) is maximum in the pH range 6.5–7.5 and decreases sharply at lower pH. At higher pH (up to pH 9) the decrease is modest, Figure 4 A and S12 (SI). The progressive addition of Zn(II) ions to AuNP4 results in the sigmoidal type of profile shown in Figure 4 B and S13 (SI). This type of profile hints to some sort of cooperativity in the process.

![Cleavage of plasmid pBR322 by AuNP4‐Zn(II). A) Dependence of the cleavage of BNP (red symbols, AuNP1‐Zn(II)) and pBR322 (blue symbols, AuNP4‐Zn(II)) on pH. All points have been normalized with respect to maximum efficiency (% cleavage for DNA, v
i for BNP). The dashed line connecting the points for DNA was drawn to guide the eye; the solid one for BNP represents the best fitting for the two kinetically relevant pK
a (7.6 and 9.1). Conditions: for DNA, 37 °C, [AuNP4‐Zn(II)]=35 μM, [pBR322]=19.3 μM bp−1, 3.5 h incubation time; conditions for BNP: [AuNP1‐Zn(II)]=3×10−5 M, [BNP]=1×10−4 M, 40 °C. B) Percentage of pBR322 cleaved by AuNP4 upon addition of increasing equivalents of Zn(II). Conditions: pH 7.5, 37 °C, [AuNP4]=35 μM, [pBR322]=19.3 μM bp−1, 3 h incubation time. Error bars refer to data collected in three independent experiments.](/dataresources/secured/content-1765822518437-102dc52a-a623-4d0a-8790-aba970890b02/assets/ANIE-60-1423-g004.jpg)
Cleavage of plasmid pBR322 by AuNP4‐Zn(II). A) Dependence of the cleavage of BNP (red symbols, AuNP1‐Zn(II)) and pBR322 (blue symbols, AuNP4‐Zn(II)) on pH. All points have been normalized with respect to maximum efficiency (% cleavage for DNA, v i for BNP). The dashed line connecting the points for DNA was drawn to guide the eye; the solid one for BNP represents the best fitting for the two kinetically relevant pK a (7.6 and 9.1). Conditions: for DNA, 37 °C, [AuNP4‐Zn(II)]=35 μM, [pBR322]=19.3 μM bp−1, 3.5 h incubation time; conditions for BNP: [AuNP1‐Zn(II)]=3×10−5 M, [BNP]=1×10−4 M, 40 °C. B) Percentage of pBR322 cleaved by AuNP4 upon addition of increasing equivalents of Zn(II). Conditions: pH 7.5, 37 °C, [AuNP4]=35 μM, [pBR322]=19.3 μM bp−1, 3 h incubation time. Error bars refer to data collected in three independent experiments.
Cleavage of pBR322 and BNP by AuNP4 occurs with different mechanisms.
The striking reversal of the activity between AuNP1‐Zn(II) and AuNP4‐Zn(II) in the cleavage of BNP and pBR322 plasmid DNA calls for a difference in the catalytic mechanism. The kinetic evidence points to a cooperative mechanism between two Zn(II) in the hydrolysis of BNP only for AuNP1‐Zn(II). The limited data obtained for AuNP4‐Zn(II) suggest the lack of cooperation between metal ions. The accepted mechanism for the cleavage of a phosphate diester by TACN‐Zn(II)‐based catalysts unambiguously requires a dinuclear catalytic site. [31] This is also the case for catalysts AuNP1–3 for the cleavage of BNP (see inset of Figure 2 B). The data hence suggest a dinuclear hydrolytic mechanism for BNP and a mononuclear one for the plasmid. Nevertheless, the kinetic profile of Figure 4 B suggests cooperativity.
It is possible to reconcile a mononuclear catalytic site with a cooperative process if we consider that cooperativity may originate also from the binding between the catalyst and the substrate. This is particularly true for multivalent species like the DNA and the Zn(II)‐nanoparticle. [41] It has been reported that multiple metal ions drive the association of DNA to an endonuclease in a cooperative manner. [42] The sigmoidal profile of Figure 4 B could be hence the result of cooperative binding and not cooperative catalysis between metal ions. In this scenario each metal complex would be involved in the binding of a different, single phosphate with a zipper‐like interaction. [43] The coordination of two Zn(II) ions to a single phosphate, as required by a cooperative, dinuclear catalytic mechanism, would be hence impossible. This could explain why AuNP1–3‐Zn(II) show very low or no activity at all. All of them require a dinuclear catalytic site for activity. Conversely, catalyst AuNPBAPASH‐Zn(II) is still active under these conditions because, although its best performance is observed with a dinuclear mechanism, the mononuclear contribution is significant as can be seen from the data of cleavage of BNP (entry 8 of Table 1 and Figure 2 B). [21] The kinetic evidence is compelling but not conclusive in this regard. Accordingly, this point will be specifically addressed in the computational analysis described below.
Interestingly, the analysis of the data reported in Figure 3 and Table 2 indicates that the best results in terms of DNA hydrolysis are obtained when the monolayer passivating the AuNP comprises: a TACN‐Zn(II) complex (compare entry 13 with entry 14); a guanidinium ion (from Arg; compare entry 13 with entries 7, 9, and 11) and a primary alcohol (from Ser; compare entry 13 with entry 15). All these components must be present at the same time (compare entry 13 with all the others). This means that they cooperate to assist the DNA‐cleavage mechanism by AuNP4‐Zn(II). The guanidinium could help in stabilizing the developing charge on the phosphate and also decrease the pKa of one of the two Ser present in the peptide flanking the metal complex. The comparison between the activity of AuNP4‐Zn(II) and AuNP5‐Zn(II) (entries 13 and 15 of Table 2) suggests that Ser plays a critical role in enhancing the activity of the nanozyme in the presence of the metal ion, likely acting as a nucleophile. The involvement of a Ser as a nucleophile in nucleases is not uncommon. [44] The ability of guanidinium ions in the presence of a primary alcohol to cleave plasmid DNA has been recently demonstrated. [45]
In the cleavage of phosphate diesters by dinuclear catalysts, the dependence of the reaction rate from pH presents a bell‐shaped profile. This is the case also for AuNP1‐Zn(II) with substrate BNP as shown in Figure 4 A. The ascending part of the curve represents the deprotonation of a Zn(II)‐bound water molecule (that would act as a general base or nucleophile) while the descending one represents the depletion by OH‐ of the substrate from the coordination sphere of a second metal complex. The apparent pKa of these groups involved in these two processes is 7.6 and 9.1, respectively (see SI). The pH dependence of the cleavage of pBR322 plasmid by AuNP4‐Zn(II) goes also through a maximum (Figure 4 A) but its profile is substantially different from that of AuNP1‐Zn(II) with BNP. In this case, too, a nucleophilic species is generated but at a lower pH (ca. 6.5), while the decrease of efficiency at the highest pH studied (9.0) is too low to determine at what pH occurs the deprotonation of the species that negatively affect the cleavage process. Two aspects should be taken into consideration. First, the multivalent interaction DNA‐AuNP4‐Zn(II) is so strong that hardly OH‐ can replace a phosphate coordinated to Zn(II). Second, the presence of the guanidinium ions, like in cationic micelles [46] or in cationic catalytic sites of enzymes, [47] may rise the local pH more than two units. This would affect the deprotonation of both the nucleophile (a Zn(II)‐bound Ser ‐OH) as well as the guanidinium ion. This is in line with the lower pKa observed for the nucleophilic species (compare red and blue traces of Figure 4 A) as well as the partial loss of activity at high pH. All these results are consistent with a mononuclear mechanism were the metal ion is flanked by a guanidinium similarly to what is observed with many nucleic acids‐processing enzymes. [7]
Modeling the binding of AuNP4 to DNA and identification of pre‐catalytic binding sites in the nanozyme monolayer
The above experimental results provide strong indication that the best nanozyme (AuNP4) operates as a mononuclear catalyst and that in its catalytic site the guanidinium of Arg and the hydroxyl of a Ser play relevant roles. To lend further support to the conclusions drawn from the above kinetic results we used molecular dynamic (MD) simulations to model the interaction of AuNP4‐Zn(II) with dsDNA and find possible pre‐catalytic binding arrangements of the functional groups present in our nanozyme. The computational model of AuNP4 was based on the three‐dimensional Au144(SR)60 structure [48] that matches the dimension of the nanoparticles of our experiments. For the nucleic acid counterpart, we used a system based on the dsDNA structure retrieved from the enzymatic Topoisomerase II/DNA binary complex (PDB code: 3L4K). [49] In this complex, the DNA comprises 11 double stranded base pairs and 4 single stranded bases (822 atoms, see SI).
Visualizing AuNP4‐Zn(II) and its monolayer for metal‐aided catalysis. Next, MD simulations were used to evaluate, at the atomic level, the formation of pre‐catalytic complexes with the substrate. For AuNP4‐Zn(II), the root‐mean‐square deviation (RMSD) values of all nanoparticle's atoms reached a plateau at 0.61±0.05 nm after the first 25 ns of simulations (Figure 5 A). The overall structure relaxed and all Zn(II) atoms remained chelated by the TACN moiety of the ligand (Figure 5 B). We first analyzed the possible formation of such pre‐catalytic conformational states with AuNP4‐Zn(II) without the bound DNA, in water. In our analysis, a pre‐catalytic binding site is defined by a distance metric that ensures to find one Zn(II) atom, the C atom of arginine's guanidinium group, and the O atom of the hydroxyl group of the serine, in close proximity, within a given cut‐off (details in Figure 5 C). Accordingly, the systems were allowed to equilibrate without any bias deriving from the experimental results. While no pre‐catalytic states were observed using a cut‐off value <0.5 nm, a significant number of them (up to ca 25) was observed when the cut‐off was set at 0.8 nm (see Figure 5 A and Figure S15 of SI for details). However, it should be noted that this metric does not strictly guarantee to pick only geometries of proper pre‐catalytic binding sites, i.e., geometries properly formed for efficient substrate recognition for catalysis. It is well known that even in natural enzymes, the binding of a substrate may induce changes in the spatial arrangement of the functional groups present in the catalytic site.[ 50 , 51 , 52 , 53 ] This is even more likely to occur in the passivating monolayer of a nanoparticle where the conformational freedom of the molecules is higher than in a protein.

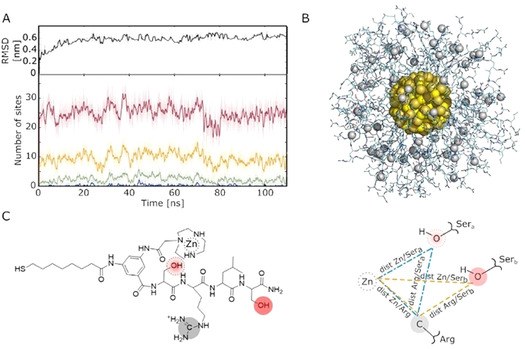
Molecular dynamic simulations for AuNP4. A) Top panel: Convergence of the RMSD value for the AuNP4‐Zn(II) MD simulations, calculated using as a reference the structure at time t=0 (i.e., the minimized model). Bottom panel: Number of pre‐catalytic binding sites on AuNP4‐Zn(II) over simulation time, calculated for different cut‐offs (0.8 nm in red, 0.7 nm in yellow, 0.6 nm in green, and 0.5 nm in blue). B) Fully equilibrated model of AuNP4‐Zn(II) (gold core in yellow and Zn Ions as gray spheres). C) The formation of pre‐catalytic binding sites is defined by a metric where at least two out of the following three distances are below a defined cut‐off distance: i) dist Zn/Arg, between the Zn ion and the C atom of arginine's guanidinium group; ii) dist Zn/Sera or dist Zn/Serb, between the Zn ion and the O atom of the hydroxyl group of the serine; iii) dist Arg/Sera or dist Arg/Serb, between C atom of arginine's guanidinium group and the alcoholic O atom of the serine side chain. In this way, the binding sites are defined so to contain one Zn atom, the guanidinium group of an arginine, and the side chain of at least one serine fragment (Sera or Serb, in the blue and orange binding site, respectively).
The nanozyme/DNA interaction to form pre‐catalytic states. To address specifically this point we assembled the AuNP4‐Zn(II)/dsDNA complex by placing the substrate dsDNA in four different conformations at a distance of 0.5 nm from the Zn(II) atom (distance Zn⋅⋅⋅P of the fissile phosphate in the dsDNA) of a pre‐catalytic binding site (identified as defined above, see Figure 5 C). These four different AuNP4‐Zn(II)/dsDNA binary complexes were minimized, equilibrated, and further relaxed with additional >35 ns of MD runs (see SI). During these steps, the dsDNA molecule bound to the nanoparticle's monolayer, forming a large number of interactions (Figure S16 of SI). To quantify these interactions in the binary AuNP4‐Zn(II)/dsDNA complex, we used three Zn.P cut‐offs (0.6, 0.5, and 0.4 nm). Using the cut‐off of 0.6 nm, we found 11 phosphates (out of the 24 available for binding along the dsDNA) bound to the Zn(II). At times, with this cut‐off, we also noticed the presence of a Zn(II)‐bound water molecule bridging the Zn(II) and the P atom. Using the cut‐off of 0.5 nm, we found 7 phosphates bound to the Zn(II), while a cut‐off of 0.4 nm captured only 4 conformations characterized by a direct Zn‐P interaction (Table S1 of SI). We found Complex1 (Figure 6 A) as the most frequently formed pre‐catalytic site (i.e., this pre‐catalytic state is found in ≈17 % of the simulations time). In this complex, one phosphate group is coordinated on top of the Zn(II) ion, electrostatically stabilized by the arginine, with the nucleophilic serine properly oriented in order to perform nucleophilic attack (see Figure 6 A). Notably, a few water molecules solvate the catalytic site in proximity of the Zn ion, which suggests that these waters may be acting as proton acceptors for nucleophile formation or proton donors to the leaving group.[ 54 , 55 ] Their involvement as nucleophiles instead of the hydroxyl of Ser cannot be ruled out. This could be the mechanism of action of AuNP5‐Zn(II) which is devoid of any Ser in the peptide sequence.

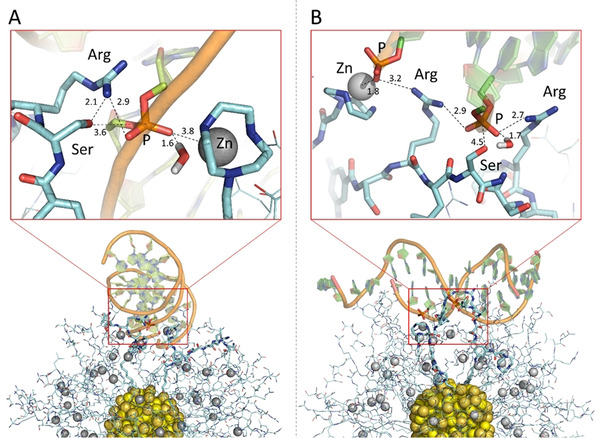
Pre‐catalytic binding complexes formed between AuNP4 and dsDNA during the MD simulations. A) Pre‐catalytic binding Complex1 formed by the coordination of the hydrolytic phosphate group (P) on top of the Zn ion (P‐O.Zn distance of 3.8 Å and via Zn‐bound water at the distance of 1.6 Å), while stabilized by one arginine Arg (2.9 Å) with nucleophilic serine (Ser) in close proximity (3.6 Å). The dsDNA is anchored to the monolayer by another phosphate‐Zn and phosphate‐arginine interactions along the backbone. B) Pre‐catalytic binding Complex2, where Zn atom coordinates one phosphate (P‐O.Zn distance of 1.8 Å), while the hydrolytic phosphate (P) is chelated by two arginine residues (at distances of 2.9 Å and 2.7 Å) with serine (Ser) in close proximity (4.5 Å) and water at the distance of 1.7 Å.
However, there are less frequent and less structured complexes that may still lead to catalysis. For example, in Complex2 (present for ≈11.7 % of the simulations time), the Zn ion anchors one phosphate of the substrate, while one serine is properly oriented for nucleophilic attack at the proximal phosphate, which is chelated by two arginine residues (Figure 6 B). This complex highlights the role of Zn(II) ions in anchoring the DNA as we have pointed out in discussing the kinetic results. It also indicates that a catalytic site might be formed with the sole Arg and Ser residues. This is in full accord with the experimental evidence indicating a non‐negligible contribution to catalysis by AuNP4 in the absence of metal ions. A small fraction of the simulation time (2.7 %) converges to Complex3 (see Figure S17 of SI). In this complex a second Zn(II) ion might provide some sort of “remote” assistance as we have recently demonstrated in model systems. [56] In any case, this observation lends further support to the experimental data ruling out a dinuclear catalytic site.
Conclusion
By properly designing, for the first time, a multifunctional catalytic site in the monolayer of AuNP4‐Zn(II) we were able to transform this nanosystem in the best monometallic Zn(II)‐based catalyst reported for the cleavage of DNA. Its reactivity rivals that of the most effective bimetallic artificial systems based on unnatural metal ions like lanthanides [57] or CuII.[ 9 , 10 ] In the latter case the oxidative pathway may become very important. Table 3 shows comparative reactivities of reported nanozymes that operate hydrolytically under physiological conditions. AuNP4‐Zn(II) stands out as the best catalyst.

|
Nanozyme |
Substrate |
t 1/2 [min] |
Ref |
|---|---|---|---|
|
AuNP4‐Zn(II) |
pBR322 plasmid |
50[a] |
This work |
|
AuNPBAPASH‐Zn(II) |
pBR322 plasmid |
5700[b] | |
|
Cyclen‐Zn(II) vesicles |
pBR322 plasmid |
240[c] | |
|
CeO2 nanoparticles |
single strand 24‐mer DNA |
300[d] | |
|
Carbon Dots |
pRSET‐eGFP plasmid |
1200[e] |
[a] At [catalyst]=45 μM. [b] At [catalyst]=15 μM. [c] At [Zn(II) complex]=1×10−4 M. [d] With 100 μg mL−1 of CeO2 nanoparticles. [e] With 30 mg mL−1 of carbon dots.
The experimental evidence supported by the MD calculations, points to a mechanism in which the phosphate diester is coordinated to the metal ion while the positively charged arginine flanks it to assist with transition state stabilization and to depress the pK a of the hydroxyl group of serine (Figure 7 A). The role played by the metal ion and the guanidinium is consistent with the observation that in phosphate‐cleaving enzymes basic groups of amino acids are not typically found to coordinate directly the reactive phosphate. [7] This also true for model catalysts. [34] However, in the absence of the metal ion a second guanidinium appears to be able to replace it (Figure 7 B), although with a lower efficiency.

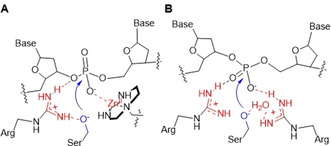
Suggested mechanism for the cleavage of a phosphate bond of plasmid DNA by AuNP4‐Zn(II) (A); and in the absence of the metal ion (B). Functions involved in acid catalysis (Lewis or proton) are in red; nucleophilic catalysis in blue.
The widely accepted mechanism for the DNA cleavage by type IA and II topoisomerases requires two Mg(II) ions in the catalytic site.[ 5 , 58 , 59 ] However, this conventional wisdom has been questioned by Berger et al. [49] who argued that these enzymes might operate by directly involving one metal ion in transition state stabilization by promoting the leaving and attacking of the ribose 39‐OH during DNA cleavage and religation. The other one would play the role of anchoring the DNA. This mechanistic suggestion is not much different from what we propose here. In spite of their outstanding efficiency, AuNP4‐Zn(II) are still four orders of magnitudes less efficient than a nuclease. Admittedly, nucleases are among the most efficient enzymes known. They had the advantage of several million years of evolution to optimize their catalytic sites, a luxury we could not afford.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
T.D., F.M., and P.S. thank the EU, Marie Curie program MSCA‐ITN‐2016, project MMBio (grant 721613) for financial support. Fellowships for J.C. and S.Z. were paid by the MMBio project. A.P. and M.D.V. thank the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska‐Curie grant agreement No. 843117 (project CompNanozymes). A.P. gratefully acknowledges Sebastian Franco‐Ulloa for technical support in using NanoModeler. [60] C.S. acknowledges the University of Padova for grant SISS‐SID19‐01.
References
3
5
6
8
9
11
12
13
14
15
16
16
17
18
19
19a
19a
19c
19d
20
21
22
23
24
26
28
33
34
35
35
37
38
39
41
41
43
43
43a
45
46
47
47
48
50
55
56
57
 A Gold Nanoparticle Nanonuclease Relying on a Zn(II) Mononuclear Complex
A Gold Nanoparticle Nanonuclease Relying on a Zn(II) Mononuclear Complex
