
- Altmetric
Interferon regulatory factor 4 (IRF4) is a key transcription factor (TF) in the regulation of immune cells, including B and T cells. It acts by binding DNA as both a homodimer and, in conjunction with other TFs, as a heterodimer. The choice of homo and heterodimeric/ DNA interactions is a critical aspect in the control of the transcriptional program and cell fate outcome. To characterize the nature of this interaction in the homodimeric complex, we have determined the crystal structure of the IRF4/ISRE homodimeric complex. We show that the complex formation is aided by a substantial DNA deformation with co-operative binding achieved exclusively through protein–DNA contact. This markedly contrasts with the heterodimeric form where DNA bound IRF4 is shown to physically interact with PU.1 TF to engage EICE1. We also show that the hotspot residues (Arg98, Cys99 and Asn102) contact both consensus and non-consensus sequences with the L1 loop exhibiting marked flexibility. Additionally, we identified that IRF4L116R, a mutant associated with chronic lymphocytic leukemia, binds more robustly to DNA thereby providing a rationale for the observed gain of function. Together, we demonstrate key structural differences between IRF4 homo and heterodimeric complexes, thereby providing molecular insights into IRF4-mediated transcriptional regulation.
INTRODUCTION
Interferon regulatory factors (IRFs) are a family of transcription regulators that mediate a multitude of functions including the differentiation and development of haematopoietic cells, regulation of apoptosis and host defence against pathogens (1–5). The family is composed of nine members (IRF1–IRF9) and typically recognize promoters consisting of the IRF consensus sequence 5′-GAAA-3′ (6). Amongst these members, IRF4 is considered unique due to its restricted expression in immune cells such as lymphocytes and dendritic cells. Moreover, IRF4 is the only IRF member that is not regulated by interferons (IFNs) (7). In B and T cells, IRF4 is expressed at multiple stages of their development, affecting differentiation, clonal expansion and cellular outcome (7–11). Due to its critical role in B-cell development, it is not surprising that IRF4 is linked directly to immune-related disease conditions including B cell-specific chronic lymphocytic leukemia (CLL) and multiple myeloma (MM). Indeed, genome-wide analysis of CLL patients has identified IRF4 as a strong candidate for disease susceptibility (12). In addition, several recurrent IRF4 somatic mutations that directly implicate IRF4 in CLL pathogenesis have been identified (13,14). Similarly, mutations in IRF4 have been found in rare patients with MM (15–17) with the malignant cells in MM found to be highly dependent on IRF4 (18). Together these observations make IRF4 an attractive target for the development of new therapies to treat these disease conditions. However, how these mutations impact IRF4 function and its role in CLL and MM disease development remains unresolved.
IRF4 consists of two structural domains: a highly conserved N-terminal DNA-binding domain (DBD) and variable C-terminal IRF association domain (IAD) joined by a flexible linker (7,19) (Figure 1A). The DBD is characterized by five conserved tryptophans enabling it to form a helix–loop–helix motif that facilitates DNA binding (20). IAD is a protein–protein interaction domain which mediates not only homo and heterodimeric interactions amongst IRFs but also association with multiple distinct transcription factors (TFs). Notably, IAD also contains a C-terminal auto-inhibitory region (AR) which directly binds the DBD and modulates its interaction with the target DNA (19,21,22).

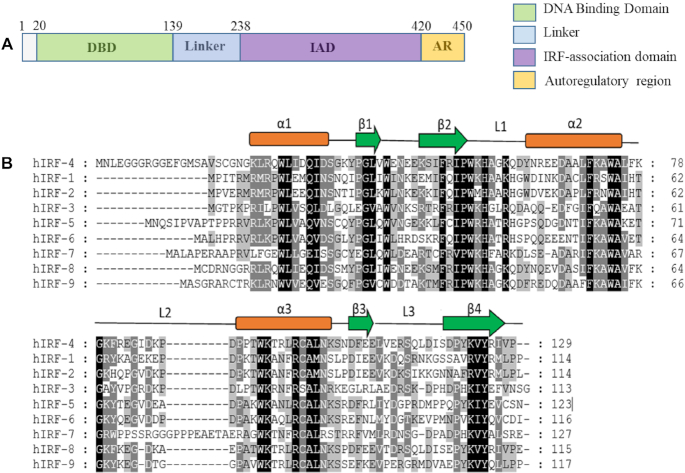
An overview of the IRF4 DNA-binding domain. (A) Schematic representation of the IRF4 DNA-binding domain. Panel (B) depicts the alignment of human IRF4 DNA-binding domain with its respective IRF family members. Shown above the sequences are the location of the secondary structure elements comprising α-helices (α1–α3), β-sheets (β1–β4) and connecting loops (L1–L3).
Due to its versatile function, it is not surprising that numerous DNA targets have been identified to interact with IRF4 (10). It binds the canonical interferon-stimulated response elements (ISRE) as a homodimer and regulates the activation of interferon-stimulated genes (ISGs). Conversely, it engages erythroblast transformation specific (Ets), interferon composite elements (EICE) and AP-1-IRF composite elements (AICE1 or 2) as a heterodimer and requires PU.1, SPIB or BATF TFs for its high-affinity interaction (7). Notably, the choice of heterodimeric complex formation depends largely on the target cell type and is essential for the cellular outcome. The binding of IRF4 with ETS TFs is largely restricted to B cells and dendritic cells, whereas the heterodimeric complex formed between IRF4 and AP-1 TFs is the main complex in T cells (23) but is also relevant during germinal centre B cell and plasma cell regulation.
IRF4 is a key regulator for B cell fate dynamics upon antigen encounter. Notably, it plays an essential role in plasma cell differentiation which it does by interacting predominately with the Prdm1 locus encoding for Blimp1 transcription factor (24,25). It was shown that high expression of IRF4 in GC B cells leads to the upregulation of Blimp-1 and formation of plasma cells (25). However, another study has shown that Blimp-1 is upregulated even in the absence of IRF4 but is not sufficient for induction of the Blimp-1 dependent plasma cell program, suggesting a cooperation of Blimp-1 and IRF4 for plasma cell differentiation (26). Chromatin crosslinking and immunoprecipitation (ChiP) studies have confirmed that IRF4 binds the conserved noncoding sequence 9 (CNS-9) region of Prdm1 (encoding Blimp-1) locus and have identified 5′-CAACTGAAACCGAGAAAGC-3′ ISRE DNA as one of the over-represented target sequences (24,25). The study also shows that it engages the above-mentioned sequence as a homodimer with lower affinity than the heterodimer and that this interaction with ISRE is a key factor skewing the B-cell development program towards plasma cell differentiation (24).
Much of the work on the biochemical and structural basis for the co-operative binding between IRF4 and other TFs has been undertaken on the IRF4–DNA–PU.1 complex (21,27,28). These studies have identified two distinct protein–protein interaction networks for the heterodimeric complex formation—one between the DBDs of IRF4 and PU.1 and the other relies on the interaction between the PEST region of PU.1 and the IAD of IRF4. While PU.1 on its own can bind the composite element, the recruitment of IRF4 to PU.1-bound DNA is facilitated by the phosphorylated PEST region, which by interacting with the IRF4–IAD relieves auto-inhibition attributed to the direct binding of the AR to the IRF4–DBD. Likewise, the interaction between IRF4 with BATF/c-jun or BATF/JunB follows a similar partner dependent binding pattern wherein the BAFT leucine zipper region participates in the recruitment of IRF4 to AICE motif (29,30). Despite a detailed knowledge on IRF4 heterodimeric interactions, it is not known how IRF4 interacts with DNA as a homodimer to regulate the cellular outcome of ISGs. Key questions that remain include how both the IRF4 DNA-binding domains communicate with each other to facilitate DNA interaction and why the binding of the IRF4 homodimer to DNA is inherently weaker than the binding of its heterodimeric counterpart.
In order to explore the molecular basis of IRF4 homodimer/DNA interaction and to delineate the stereochemical differences between IRF4 homo and heterodimeric complexes, we have co-crystallized the DNA-binding domain of IRF4 with ISRE DNA (5′-CAACTGAAACCGAGAAAGC-3′) comprising two overlapping consensus IRF (GAAA) recognition sequences and determined the ternary complex structure. Our study shows that IRF4 binds DNA by substantially distorting its structure to accommodate the unique binding mode of the adjacent IRF4-binding sites. Furthermore, unlike the heterodimeric complex, no intermolecular interactions were observed between the interacting DNA-binding domains. The structural elucidation of the IRF4 homodimer/DNA complex provides a molecular basis for the functional effects of IRF4 mutations observed in CLL patients (13).
MATERIALS AND METHODS
Expression and purification
The codon-optimized IRF4 DBD gene constructs were cloned into pJ411KanR (ATUM) and overexpressed as an N-terminal His6 tag-fusion protein in Escherichia coli BL21(DE3) (Novagen) at 16°C following induction with 0.5 mM IPTG in 2X YT media. The cells were resuspended in 50 mM sodium phosphate buffer (pH 7.0), 500 mM NaCl and 30 mM imidazole (buffer A) with protease inhibitor cocktail (Sigma), 3 mM β-mercaptoethanol and lysed by French press (1500 psi). The lysate was centrifuged at 15 000 rpm for 30 min and both the WT and IRF4 mutant proteins were purified using a 5 ml HisTrap column (Cytiva) in buffer A with 30–500 mM imidazole gradient. To cleave the His tag, the eluted fractions were pooled and subjected to HRV3c digestion overnight at 4°C in buffer A. The His-tag cleaved IRF4 proteins were subsequently loaded and purified by passing through a HisTrap column (Cytiva). The Ni-NTA purified protein was dialysed into 20 mM Tris buffer (pH 7.4), 150 mM NaCl, 1 mM TCEP and subsequently purified by size exclusion chromatography (SEC) using a superdex 200 16/600 gel filtration column (Cytiva).
To form and purify intramolecular IRF4/DNA complexes, the SEC purified IRF4 WT was incubated with ISRE DNA (Integrated DNA technologies) in a 1:0.5 molar ratio overnight at 4°C. The IRF4 WT homodimer–ISRE complex was isolated and purified by injecting the sample onto a superdex 200 16/600 gel filtration column. In parallel, IRF4 WT in the absence of DNA was purified in an identical environment for the comparison of the SEC elution profile.
Crystallization and structural determination
The SEC purified IRF4 WT–ISRE complex was concentrated to 5–7 mg/ml and crystallized in 5–13% PEG 4000, 0.1 M Na acetate pH 4.6 at 18°C using the hanging drop vapour diffusion method. Diffraction quality crystals were obtained in 10% PEG 4000, 0.1 M Na acetate pH 4.6 at 18°C and cryoprotected using the mother liquor plus 15–20% PEG 4000, and flash-frozen in liquid nitrogen. X-ray diffraction intensity data for the crystals were collected at the MX2 beamline (Australian synchrotron). The dataset for IRF4WT–ISRE complex was processed with the XDS software package and scaled using Aimless (31,32) in the CCP4 suite (33). The crystal structure of the IRF4 WT DNA-binding domain–ISRE ternary complex was determined by molecular replacement using the Phaser-MR program with IRF2 DNA-binding domain from the structure of IRF2–DNA complex (34) and B-DNA of ISRE generated using COOT (35) as a separate search model [Protein Data Bank (PDB) ID code: 2IRF]. Iterative model building and subsequent refinement cycles were performed with the program COOT and Phenix refine, respectively (35,36). The quality of the structure was validated at the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB) Validation and Deposition Services. All presentations of molecular graphics were created with the programme PyMOL.
Surface plasmon resonance
The affinity measurements were performed at 20°C on a Biacore 8K (Cytiva) with HBS buffer (10 mM HEPES-HCl, pH 7.4, and 150 mM NaCl) supplemented with 3 mM ETDA and 0.05% P20 as a running buffer. The biotinylated DNA motifs (ISRE, EICE1, AICE1 and AICE2; Supplementary Table S1) (Integrated DNA technologies) were coupled (up to ∼2000 RU) onto a series S streptavidin (SA) chip (Cytiva) according to manufacturer’s instructions. The affinity measurement was performed by passing the serially increased concentrations of IRF4 WT, IRF4 L116R, IRF4 K59A and IRF4 Y62A (up to 5 μM) at the flow rate of 30 μl/min. The final response unit was calculated by subtracting the response unit obtained from the reference flow cell. The steady-state multicycle affinity data were fitted using the Biacore 8K BIAevaluation software. GraphPad Prism Version 8.0 was used for data presentation.
RESULTS
Co-complexation and structure determination
Size exclusion chromatography was used to unambiguously isolate and purify the IRF4/ISRE homodimer. The apo IRF4 DNA-binding domain (molecular mass of ∼13 kDa) began eluting at a volume of ∼90 ml, consistent with the expected elution profile of a monomer. Conversely, when the protein was incubated with DNA and separated using the same conditions, the IRF4/DNA started to elute at a volume of ∼80 ml, consistent with the molecular mass of ∼40 kDa protein (Supplementary Figure S1). The expected molecular mass of the IRF4 dimer bound to ISRE is ∼38 kDa, suggesting that as expected, the IRF4/ISRE complex elutes at this volume as a homodimer.
To determine the structural basis of IRF4/DNA homodimeric complex formation, we co-crystallized the DNA-binding domain of IRF4 comprising residues from 21 to 130 amino acids with ISRE DNA. The complex crystals diffracted to 2.95 Å and belonged to the space group of P31 2 1 with unit cell dimensions of a = 117.8, b = 117.8, c = 154.58 and α = 90°, β = 90°, γ = 120°. We determined the structure by molecular replacement and refined it to the final Rwork and Rfree of 18.5% and 20.5%, respectively (Table 1). In the asymmetric unit, the crystal structure contained four IRF4 DNA-binding domains, namely IRF4-A (21–129), IRF4-B (23–129), IRF4-G (21–129) and IRF4-H (21–128) and two DNA duplexes, respectively, representing two IRF4/DNA homodimeric ternary complexes. In addition, the DNA duplex in the crystal structure was stabilized by stacking interactions with the other symmetry related complexes to form a continuous DNA helix in the crystal. The two homodimeric complexes within the asymmetric unit were essentially identical (RMS deviation of 0.207 Å for all Cα-carbon atoms). Moreover, the interacting subunit of the homodimer displayed minimal structural changes with a RMSD of 0.305 Å for all Cα-carbon atoms and therefore, for subsequent analyses we used the homodimeric complex that comprised IRF4 (chain A and B) and DNA (Chain D and E).

| Wavelength | |
|---|---|
| Resolution range | 48.44–2.95 (3.055) 2.95) |
| Space group | P 31 2 1 |
| Unit cell | 117.802 117.802 154.579 90 90 120 |
| Total reflections | 295918 (26403) |
| Unique reflections | 26659 (2492) |
| Multiplicity | 11.1 (10.1) |
| Completeness (%) | 99 (100) |
| Mean I/sigma(I) | 19.23 (1.82) |
| Wilson B-factor | 76.67 |
| R-merge | 0.1054 (1.251) |
| R-means | 0.1106 (1.318) |
| CC1/2 | 0.999 (0.692) |
| CC* | 1 (0.904) |
| Reflections used in refinement | 26532 (2491) |
| Reflections used for R-free | 1320 (125) |
| R-work | 0.1848 (0.3318) |
| R-free | 0.2055 (0.3460) |
| CC (work) | 0.966 (0.719) |
| CC (free) | 0.960 (0.753) |
| Number of non-hydrogen atoms | 5204 |
| macromolecules | 5204 |
| Protein residues | 433 |
| RMS (bonds) | 0.004 |
| RMS (angles) | 0.90 |
| Ramachandran favoured (%) | 95 |
| Ramachandran allowed (%) | 4.7 |
| Ramachandran outliers (%) | 0 |
| Rotamer outliers (%) | 0 |
| Clashscore | 1.55 |
| Average B-factor | 76.66 |
| macromolecules | 76.66 |
| Number of TLS groups | 31 |
Statistics for the highest resolution shell are shown in parentheses.
Overall structure of IRF4/DNA ternary complex
The IRF4/DNA structure revealed a head to tail orientation with each IRF4 molecule binding the opposite face of the DNA. Similar to the previous IRF DNA-binding domain structures, the IRF4 DNA-binding domain retained a conserved α/β structural architecture comprising three α-helices (α1–α3) flanked by a four-stranded antiparallel β sheet (β1–β4) forming a helix–loop–helix motif found commonly in transcription factors such as catabolite gene activator protein (CAP)-related proteins and hepatocyte nuclear factor 3γ (HNF-3γ) (37,38). The IRF DNA-binding domain is also comprised of unusually long loops (Loop 1–Loop 3) and a cluster of five tryptophan residues—a feature characteristic of all IRFs members. These loops connect different parts of the secondary structure. Loop 1 and loop 2 connects β2 and α2 and α2 and α3 helix (recognition helix), respectively, whereas the loop 3 connects β3 and β4 (Figure 1B). In addition, IRF4 also contains a short 310-helix between β3 and loop L2. This region was previously described as a part of the connecting loop L2 in the structures of IRF1 and IRF2 DNA-binding domains (20,34). The electron density is well-defined throughout the complex, enabling us to unambiguously characterize the molecular interaction between IRF4 and DNA.
The DNA-complexed IRF4 DNA-binding domain has a buried surface area of approximately of 1.796 Å2. This value is significantly greater than in the classical homeodomain–DNA complex structure (39) (1128 Å2) and reflects the extensive contact IRF4 DNA-binding domains make with DNA. Indeed, the presence of lengthy loops, especially the connecting loop L1 enables the DNA backbone interactions to contribute to this extended interaction footprint. We also identified a substantial bend in the DNA duplex induced predominantly by the binding of the IRF4 DNA-binding domain. This resulted in the DNA adopting an unusually S-shaped structure. A quantitative analysis of the DNA conformational parameters was performed using the nucleic acid-based package, Curves+ (40) showed that both the IRF4 DNA-binding domains distorted the DNA backbone by approximately 15° relative to an ideal B DNA structure which is larger than that observed for the PU.1/IRF4/DNA heterodimer (8°). The homodimer IRF4 structure comprised a mean axial rise per turn of 3.41 Å that was comparable to that of B-DNA ISRE (3.36 Å). However, an obvious difference was observed in their respective base pair tilt with IRF4 bound DNA having a tilt of 1.5 Å. The helical twist per base pair varied from 21.2° to 46.3° with an average helical twist of 31.8 °. Likewise, the difference was also observed in their respective average propeller twist with -11.7° and -14.6°, respectively for the IRF4 bound and ideal B DNA. In addition, engagement with IRF4 reduces the overall length of the DNA by 5.7 Å with respect to the vertical axis of an ideal B DNA. The deformation has not only enables the optimal positioning of the DNA-binding domains but also facilitates the accommodation of α3 recognition helix in the major groove for the direct recognition of IRF consensus sequences. The overall structural features were nevertheless comparable to PU.1/IRF4/DNA ternary complex (28) suggesting that IRF4 adopts a conserved DNA-binding mode when recognizing its DNA targets regardless of its dimeric composition (Figure 2A and B).

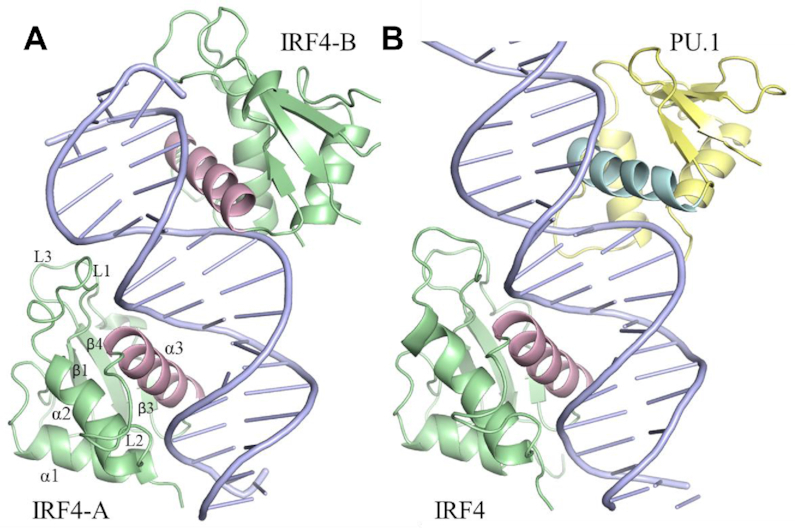
An overview of the IRF4 homo and heterodimer complexes. (A) Overall structure of the IRF4/DNA homodimer complex. IRF4 is coloured green, α3-recognition helix; pink, DNA; light blue. (B) PU.1/IRF4/DNA heterodimer complex. IRF4 and PU.1 are coloured as green and yellow, respectively. The α3-recognition helix of IRF4 and PU.1 are coloured pink and pale cyan, respectively.
IRF4 exhibits minimal structural plasticity upon binding
The NMR structure of the apo IRF4 DNA-binding domain (PDB: 2DLL) has enabled us to compare the conformational rearrangements of the DNA-binding domain upon DNA interaction. Superposition of the lowest energy model of the IRF4 NMR structure with our homodimeric structure gave an RMS deviation of ∼1.2 Å for all the atoms. Since IRF4 intimately interacts with the DNA consensus sequence through the α3 recognition helix, conformational changes in this helix were specifically compared between the DNA bound and apo IRF4 DNA-binding domain, resulting in an RMS deviation of ∼0.623 Å. Notably, apart from the connecting loop L1 (∼2.8 Å) (Supplementary Figure S2), the other loops only showed subtle differences in their orientation with no major structural rearrangements (RMS deviation <1.5 Å). The conformational rearrangements in the connecting loop L1 were also compared in both the IRF4 homo and heterodimer complexes and were found to have an RMS deviation of ∼3.49 Å. This suggests that the connecting loop L1 is inherently flexible and is discussed in detail below. Taken together, these data indicate that except for the connecting loop L1, the IRF4 DNA-binding domain undergoes minimal structural rearrangement upon engagement with the target DNA with minimal entropic cost.
Structural comparison between IRF4 homo and heterodimeric complexes
Despite IRF4’s conserved overall DNA-binding mode, we observed some key differences that distinguish between the homo and heterodimeric IRF4/DNA complexes. We have used the IRF4-A structure of the homodimeric complex for comparison with the IRF4 DNA-binding domain of the heterodimer and we have used the IRF4-B structure for any comparison with the DNA-binding domain of PU.1 in the heterodimeric complex. The IRF4 and PU.1 DNA-binding domains share a low sequence identity of ∼ 30% that is also reflected in their distinct structural recognition (Supplementary Figure S3A and B). One striking difference between the homo and heterodimeric complexes lies in the distinct orientation of their respective α3 recognition helix. In the homodimeric complex, the α3 recognition helix of both IRF4 DNA-binding domains sit in the major groove almost in a parallel orientation to the sugar-phosphate backbone of the DNA axis. Remarkably, this has resulted in the connecting loops between the monomers extending away from both the DNA and the other interacting IRF4 monomer, thus completely abolishing the likelihood of intermolecular interactions between the monomers with the closest distance between them of 14 Å (Figure 3A). This contrasts with the heterodimeric complex, where the centre of mass of the superimposed DNA-binding domain of PU.1 and IRF4 showed an overall shift of 5.5 Å. This has resulted in the α3 recognition helix of PU.1 adopting a perpendicular orientation while the equivalent helix of the interacting partner IRF4 lies almost in a parallel orientation to the sugar-phosphate backbone of the DNA axis. This parallel-perpendicular orientation has enabled PU.1 to drape over the minor groove of the DNA to form a protein–protein mediated intermolecular interaction with the DNA-binding domain of IRF4 (Figure 3B).

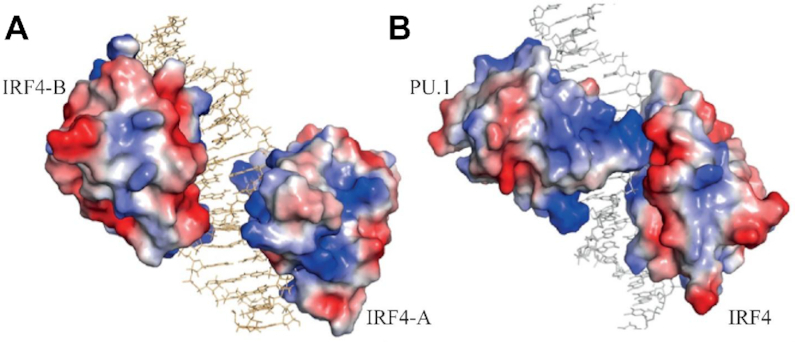
Overall structural difference between the IRF4 homo and heterodimer complexes. Panel (A) shows an electrostatic map of IRF4-DNA homodimer complex with DNA coloured in light brown. Panel (B) depicts the electrostatic map highlighting the IRF4 and PU1 interaction. In both the figures, DNA is depicted as stick figures.
While the overall structure of the IRF4 DNA-binding domain between the homo and heterodimeric complex showed no substantial structural changes with an RMSD of 0.430 Å, a significant rearrangement was observed in their respective connecting loop L1. Notably, L1 loops in the homodimeric complex are extended by three amino acids in the C-terminus when compared to the heterodimeric complex and traverse the minor groove of the DNA. Also, conformation of some of the residues within this loop deviated significantly with that observed in the heterodimeric complex. For instance, the Cα of Lys 59 is shifted by 7.3 Å resulting in the side chain of Lys 59 pointing inwardly to contact the GAAA (the recognized bases are underlined) recognition sequence. Specifically, Lys 59 forms a hydrogen-bonded contact with the second adenine which is further enhanced by a van der Waals interactions with the first guanine. In contrast, the equivalent Lys 59 residue in the IRF4 heterodimeric complex contacts the DNA exclusively through the phosphate backbone via electrostatic networks. Likewise, the position of the Cα of the adjacent residues also varies considerably. For example, Tyr 62 is displaced by 5.3 Å, enabling the aromatic sidechain to contact the phosphate backbone of the terminal adenine of the consensus GAAA via a hydrogen bond which is further enhanced by van der Waals mediated interactions. (Figure 4A). To test the effects of Lys 59 and Tyr 62 of connecting loop L1 on ISRE DNA binding, we mutated these residues to alanine and measured its binding strength using surface plasmon resonance (SPR). In comparison to WT (KD = 0.25 ± 0.15 μM, see below), K59A was found to bind with a KD of 0.66 ± 0.10 μM which is ∼3-fold lower than the observed affinity of the WT. Mutation of Tyr 62, on the other hand, had the opposite effect on DNA binding with a KD of >5 μM observed (Figure 4B). Together, our affinity data suggest that these residues play a crucial role for the ISRE interaction and corroborate the structural data.

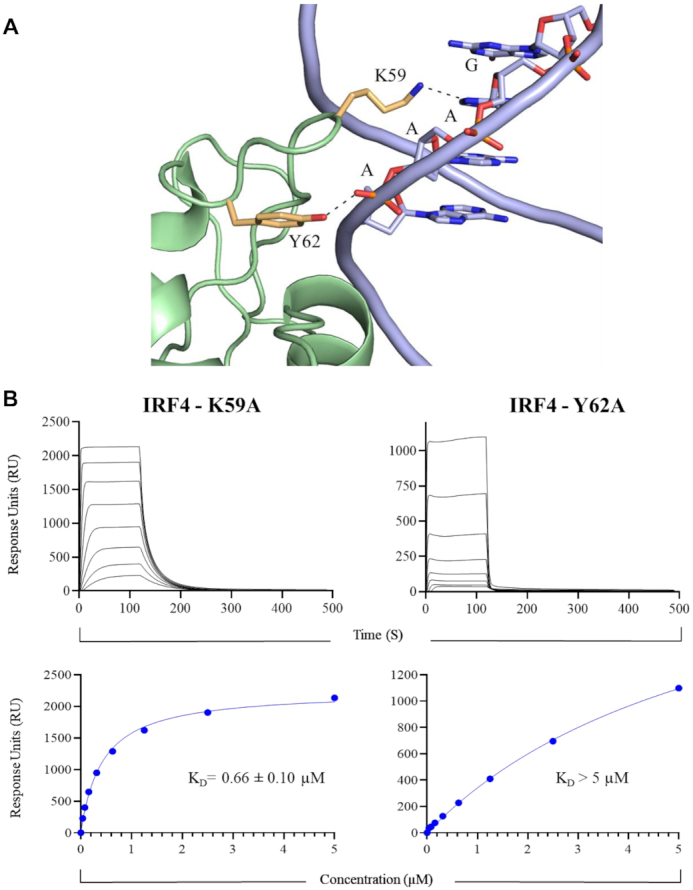
Connecting Loop L1–DNA interaction. (A) Cartoon representation of the interaction between connecting Loop L1 and DNA. IRF4 (green), interacting residues of IRF4 (pale yellow) and DNA (blue). (B) Affinity measurements of the IRF4 Loop 1 mutant/ISRE interaction. Top panel corresponds to typical sensograms for the interaction of IRF4 K59A and IRF4 Y62A with ISRE DNA. Bottom panel represents the affinity curves for IRF4 K59A and IRF4 Y62A interacting with ISRE DNA. Data are representative of three independent experiments.
IRF4–DNA interactions
The IRF4 DNA-binding domain predominantly engages the DNA through a series of phosphate backbone contacts enabling the positioning of the α3 helix in the major groove and connecting loop L1 in the minor groove (Figure 5A). Engagement with both the IRF recognition sequences (GAAA) arise from the C-terminal region of the α3 helix. Specifically, for the B chain of IRF4, these interactions are mainly mediated by Arg 98, Cys 99, Asn 102 and Lys 103. Briefly, Arg 98 interacts extensively with the first guanine base by contacting the base through a hydrogen bond. The recognition of the second base is facilitated by Cys 99 through which a sulphydryl group forms a hydrogen bond with the N6 of the adenine base. In addition, it also interacts with both the second and third base via van der Waals contacts. The contact for Asn 102 is primarily mediated by a hydrogen bond with the OP2 of the first base. The presence of Lys 103, which is restricted to a few IRF members, interacts with the fourth base of the recognition sequence predominately by forming a van der Waals mediated contact. Interestingly, Lys 123 which forms the part of β4 also contacts the guanine base through a salt bridge to the phosphate backbone as well as a van der Waals interaction. Notably, a mutation in Lys 123, (K123R) has been associated with significant pathology (14) (Figure 5B). Considering the A chain of IRF4, the recognition of the GAAA sequence is only through Lys 103 which makes a hydrogen bond with the N7 and O6 of the guanine base. The contact also extends to the second base (adenine) and is mediated by network of van der Waals interactions. Notably, the two bases upstream of the recognition sequence (GAGAAA) mimic a part of the IRF recognition sequence and have contacts with Arg 98, Cys 99, Asn 102 and Lys 103. More specifically, a salt bridged link is formed between the guanidinium side chain of Arg 98 and phosphate backbone of the guanine. The adenine base is recognized by Cys 99 which forms a hydrogen-bonded contact with Cys99. Asn 102 also forms a hydrogen bond with the guanine base. Several of these interactions are comparable to the conserved interactions observed in IRF4 chain B suggesting some degree of ‘molecular mirroring’ in DNA recognition (Figure 5C).

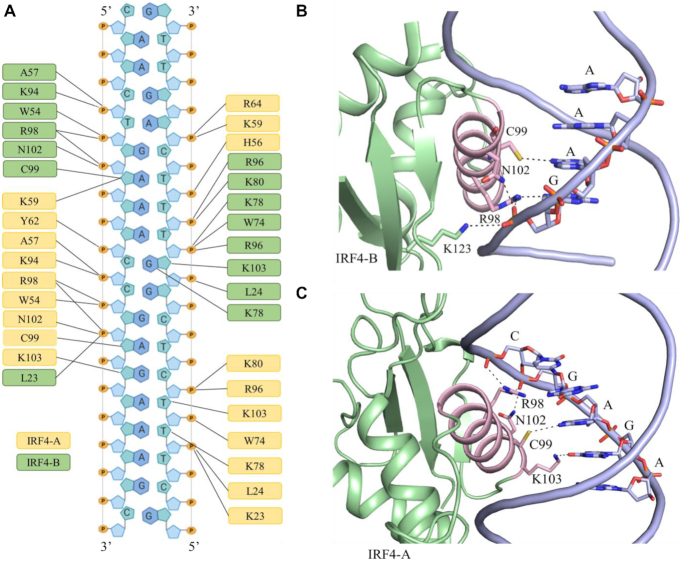
An overview of the IRF4–DNA interaction. (A) Schematic diagram of IRF4–DNA interaction. IRF4 chain A and chain B are coloured green and orange, respectively. Lines represents hydrogen bonds. (B) Cartoon representation showing IRF4 chain A recognition helix/DNA interaction. (C) Cartoon representation depicting IRF4 chain B recognition helix/DNA interaction. IRF4; green, recognition helix; pink, and DNA; blue.
IRF4 L116R results in enhanced DNA binding
IRF4 L116R is a well-recognized recurring heterozygous gain of function mutation observed in the DNA-binding domain of CLL patients (13). How this mutation accelerates IRF4 function and its implication in CLL development remains unknown. The availability of the homodimer complex structure has enabled us to map the location and rationalize the cause for the gain of function incurred due to this mutation. Leu 116 forms part of the connecting loop L3 and lies close to the DNA with its sidechain pointing towards the phosphate backbone (Figure 6A). This indicates that substitution with an arginine residue could potentially result in a tighter interaction with the negatively charged phosphate backbone via an electrostatic interaction. To test this hypothesis, we interrogated the binding strength of IRF4 WT and IRF4 L116R for the known DNA target motifs (ISRE, EICE1, AICE1, AICE2) using SPR. Both IRF4 WT and IRF4 L116R proteins bound the DNA variants with different KD value ranging from low nanomolar to high micromolar affinities. IRF4 WT bound ISRE DNA with a binding affinity (KD) of 0.25 ± 0.15 μM while it bound EICE1, AICE1 and AICE2 with affinities of 0.26 ± 0.07 μM, 2.8 ± 0.48 and 1.38 ± 0.51 μM, respectively. However, when similar affinity measurements were performed with IRF4 L116R, the mutant bound more robustly to all the DNA targets with binding affinities of 0.06 ± 0.04 μM for ISRE and 0.06 ± 0.06 μM, 2.21 ± 0.03, 0.63 ± 0.06 μM for EICE1, AICE1 and AICE2, respectively (Figure 6B–E). This indicated that the leucine to an arginine substitution in this position results in 2–4-fold tighter binding than the wild-type, as predicted from the structure.

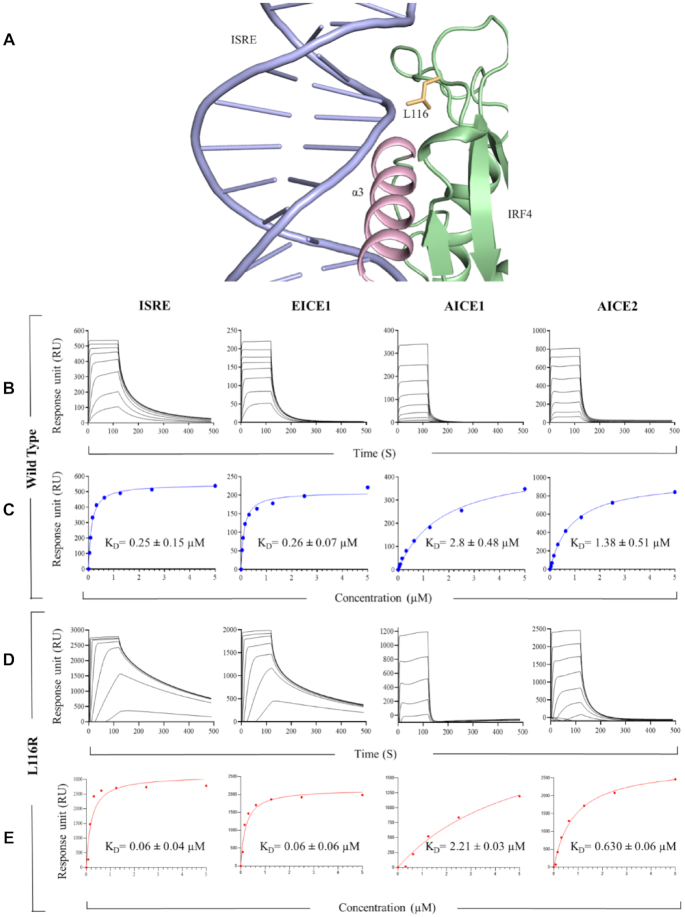
Impact of the IRF4 L116R mutation on DNA interactions. (A) Cartoon representation showing IRF4 L116 residue. IRF4; green, L116 residue; orange, DNA; light blue, α3-recognition helix; pink. (B–E) Affinity measurements of IRF4–DNA interaction. Panels (B) and (D) correspond to typical sensograms for IRF4 WT and IRF4 L116R, respectively, for specific DNA targets. Panels (C) and (E) represent affinity curves for IRF4 WT and IRF4 L116R, respectively. Data are representative of three independent experiments.
DISCUSSION
The co-operative binding of transcription factors is critical for the functional outcome of the target gene. IRF4, is a lymphoid transcription factor that engenders its function both as a homodimer, or as a heterodimer in combination with other DNA-binding proteins. The molecular switch between homo/heterodimeric–DNA interaction is critical for regulating the cell-fate outcome of the target cell. In B cells, for example, IRF4 binding of ISRE has been shown to shift the transcriptional program towards plasma cell differentiation, while its co-binding with PU.1 and EICE motifs facilitates B-cell activation and germinal center (GC) B-cell response (24). Given that the choice of homo and heterodimeric/DNA interaction is a key determinant of IRF4 mediated cell fate outcome, it is pivotal that we understand the underlying structural differences between homo and heterodimeric IRF4–DNA complexes. The structure we have reported here provides a glimpse of the molecular basis for the assembly of the IRF4/DNA homodimeric complex. Through this structure, key molecular differences that distinguish the homodimeric complex from its heterodimeric counterpart were mapped. Our study shows that unlike the heterodimeric complex, the IRF4/DNA homodimeric complex formation is restricted exclusively to protein–DNA contacts. A similar pattern in co-operative binding has also been observed in several other well-characterized bipartite DNA binding proteins. For example, the POU DNA-binding domain of transcription factor Oct-1 and its interacting partner POU-specific domain, displayed co-operative binding independent of protein-protein contacts (41). Likewise, the crystal structure of ATF-2/c-Jun and IRF3 bound to interferon-β enhancer showed no direct contact between the interacting IRF3 domains (42). Taken together, this demonstrates that co-operativity in binding is driven largely through the allosteric effects transmitted through the DNA with no contribution arising from the direct protein-protein interaction by the interacting DNA binding domains. Notably however, the absence of a direct protein–protein interaction has been shown to compromise the gross overall binding affinity of the complex. For instance, in the case of PU.1/IRF4/DNA interaction, the protein–protein contact between PU.1 and IRF4 DNA-binding domain was shown to contribute to the overall binding of DNA by 20- to 40-fold (27,28). Since the IRF4/DNA homodimeric interaction is devoid of any protein–protein interaction, our study provides a plausible explanation for the lower binding affinity that is usually observed for ISRE DNA (43).
The IRF4 structure reported here also revealed structural similarities with its apo form with exception of the connecting loop L1 which showed the greatest RMS deviation of all the structural components. This enabled the Lys 59 and Tyr 62 to directly contact the consensus sequence, a feature to our knowledge not observed in other IRF/DNA complexes. Mutation of these residues indicates that their interactions play an essential part in DNA recognition. Surprisingly, substitution of Tyr 62 with alanine had a marked impact on the binding affinity compared to the equivalent mutation of Lys 59 suggesting that the hydrophobic side chain may contribute to binding possibly through an allosteric effect. The structure of IRF3 and IRF7 also shows similar flexibility in the connecting loop L1, which together with our structure reveals that this loop is inherently flexible. Notably, this inherent flexibility has been shown to have a direct effect on DNA binding for IRF3 and IRF7 (44). Collectively, this indicates that the flexible nature of loop L1 may provide an insight into how different IRFs can control DNA specificity.
The other key finding from our structure is the ability of the highly conserved residues (Arg 98, Cys 99 and Asn 102) in the α3 recognition helix to interact with both consensus and non-consensus DNA sequence elements. Traditionally, these residues are typically known to specifically recognize the consensus GAAA sequence of the target DNA. While one of the IRF4-binding domain follows a conventional pattern of DNA recognition, the binding to the other DNA binding domain is mediated by a non-consensus sequence located upstream to the second recognition sequence (GAGAAA). These upstream nucleotides (underlined GA) were shown to play an essential role in IRF4 interaction. Interestingly, these upstream guanine and adenine bases have been shown to play an essential role in IRF4 mediated ISRE recognition and homodimerization. Notably, mutation of GA to CG has been shown to block IRF4 dimerization (24), thus validating our structure.
This atypical mode of interaction can be attributed to the unusual spacing of bases between the consensus sequence where four bases separate these sequences (GAAACCGAGAAA). Typically, the IRF recognition sequences are separated by two bases; however, it is now well-recognized that this spacing varies amongst the DNA targets. Notably, several naturally occurring IRF binding sites with atypical spacing have been identified (34) and shown to greatly influence the manner in which hot-spot residues contribute to the consensus sequence interaction. For example, the 3-base spacing in the positive regulatory domains (PRDs) of the IFN-β enhancer has enabled one of the IRF3 DNA-binding domains to bind to a similar non-consensus sequence with specific interactions arising from these hot spot residues (45). Likewise, the IRF7 DNA-binding domain in the crystal structure of the IRF-3/IRF-7/NFkB complex bound to PRDs of IFN-β enhancer exhibits a similar interaction where the conserved Arg 98 (Arg 96 in IRF7) interacts with bases upstream to the consensus sequence (46). Together, this study further reinforces the idea that IRF transcription factors are remarkably versatile in binding to their target DNA.
The homodimer structure also provides a structural insight into the gain of function of the IRF4 L116R mutation identified in CLL patients. While the biochemical and cellular basis of this mutation are well established, it is unclear if this enhanced function can be attributed to increased DNA binding and/or enhanced structural stability. The structure shows that DNA deformation enables Leu 116 to sit close to the DNA. Mutation of Leu 116 to arginine resulted in a 3–4-fold higher DNA interaction suggesting that a gain of function is most likely linked to enhanced binding to the target DNA. However, it is not known if this increased binding also applies to the heterodimeric form. Also, the allosteric effects of arginine substitution on nearby residues and its impact on the overall DNA-binding affinity remains unknown.
Collectively, this study has provided valuable structural insights on the molecular basis of IRF4/DNA homodimeric complex formation. It has shown that co-operative binding is driven exclusively by DNA conformational adaptability and shows that loop L1 is highly flexible, a feature inherent in several IRF transcription factors. We have also shown that hot spot residues in the IRF4 DNA-binding domain interact with both consensus and non-consensus DNA sequences. Based on the affinity studies, we have also found that the gain of function in IRF4 L116R can be attributed to its tighter DNA binding. Given that the molecular switch between homo and heterodimeric interaction defines the developmental program and cell fate outcome of the B cells, our study paves the way for a better understanding of IRF4 in B-cell regulation and related disease conditions.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structure have been deposited with the Protein Data bank under accession number 7JM4.
ACKNOWLEDGEMENTS
This research utilized the MX2 beamline at the Australian Synchrotron (ANSTO) as well as the Australian Cancer Research Foundation (ACRF) detector. The authors also thank Carlos R. Escalante (Virginia Commonwealth University) for providing the PU.1/IRF4/DNA complex PDB coordinates. We thank Professor Philip Board (Australian National University) for critically proofreading the manuscript and Dr Anand Ramakrishnan, Cytiva for technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NHMRC[GNT1079648to A.E.]. Funding for open access charge: Australian National University.
Conflict of interest statement. None declared.
REFERENCES
1.
2.
3.
5.
6.
7.
9.
10.
11.
12.
13.
14.
15.
16.
17.
18.
19.
20.
21.
22.
23.
24.
25.
26.
27.
28.
29.
30.
32.
33.
34.
35.
36.
37.
38.
39.
40.
41.
42.
43.
44.
45.
 Structural determinants of the IRF4/DNA homodimeric complex
Structural determinants of the IRF4/DNA homodimeric complex
