- Altmetric
Tropolone sesquiterpenoids (TS) are an intriguing family of biologically active fungal meroterpenoids that arise through a unique intermolecular hetero Diels–Alder (hDA) reaction between humulene and tropolones. Here, we report on the combinatorial biosynthesis of a series of unprecedented analogs of the TS pycnidione 1 and xenovulene A 2. In a systematic synthetic biology driven approach, we recombined genes from three TS biosynthetic gene clusters (pycnidione 1, xenovulene A 2 and eupenifeldin 3) in the fungal host Aspergillus oryzae NSAR1. Rational design of the reconstituted pathways granted control over the number of hDA reactions taking place, the chemical nature of the fused polyketide moiety (tropolono‐ vs. monobenzo‐pyranyl) and the degree of hydroxylation. Formation of unexpected monobenzopyranyl sesquiterpenoids was investigated using isotope‐feeding studies to reveal a new and highly unusual oxidative ring contraction rearrangement.
New bis‐ and mono‐ tropolono‐ and benzo‐pyranyl humulene meroterpenoids, priviledged structures with diverse potent bioactivities, were synthesised using a rational synthetic biology platform based on the heterologous expression of genes from three different natural product pathways in the fungal host Aspergillus oryzae. Control over number of additions to humulene, ring contractions and humulene hydroxylation was achieved.
Introduction
Tropolone sesquiterpenoids (TS) are fungal meroterpenoid natural products [1] that display a significant array of biological activities. For example: pycnidione 1 is an antiproliferative vs. human lung cancer cells (9 nM); [2] xenovulene A 2 inhibits the human γ‐aminobutyrate A (GABAA) benzodiazepine receptor (40 nM); [3] eupenifeldin 3 and neosetophome B 4 are potent antitumor agents (nanomolar activity towards human cancer cell lines);[ 1 , 4 ] while epolone A 5 selectively induces erythropoietin (EPO) expression in human cells in the μM range; (Scheme 1 A). [5]

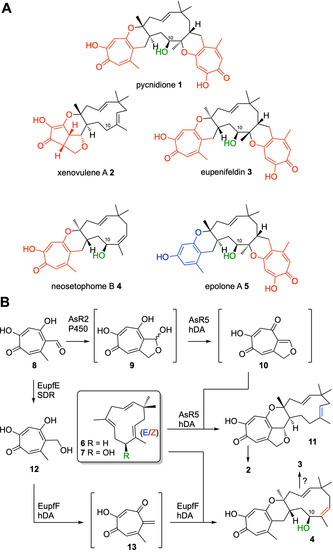
A, Selected tropolone sesquiterpenoids with key structural features highlighted. Red=polyketide derived tropolones; Blue=benzopyranyl moiety; Green=optional C‐10 hydroxylation; B, Hetero Diels–Alder reaction in the biosynthesis of xenovulene B 11 and neosetophome B 4; hDA=hetero Diels–Alderase; SDR=short‐chain dehydrogenase; P450=cytochrome P450.
All tropolone sesquiterpenoids share the structural motif of a core 11‐membered macrocycle (derived from humulene 6; Scheme 1 B) connected to one or two dihydropyran rings that link the macrocycle with polyketide‐derived tropolones. [6] The structural diversity of TS is further enhanced via: optional hydroxylation at the C‐10 position (e.g. eupenifeldin 3); [4] different olefin configurations of the central humulene macrocycle (e.g. xenovulene B 11 vs. neosetophome B 4); [6] replacement of one or two tropolone moieties by monobenzopyranyl moieties (e.g. epolone A 5); [5] or consecutive oxidative ring contractions of the polyketide (e.g. xenovulene A 2). [7]
Successful total syntheses of known TS natural products have not been reported in the literature,[ 8 , 9 , 10 , 11 ] suggesting that a biosynthetic approach might be more feasible to access TS scaffolds for biological testing. [6] Biosynthetically, TS are of significant interest due to the unusual enzymology involved in the formation of the core meroterpenoid skeleton.
Fungal biosynthetic gene clusters (BGC) have been linked to the production of the two tropolone sesquiterpenoids xenovulene A 2 (aspks1 BGC in Sarocladium schorii=Acremonium strictum) and eupenifeldin 3 (eup and eupf BGC in Phoma sp. and Penicillium janthinellum, respectively).[ 6 , 7 , 12 ] TS biosynthesis proceeds via initial formation of stipitaldeyhde 8 by cooperation of a non‐reducing PKS (nrPKS, TropA), an FAD‐dependent monooxygenase (FMO, TropB) and a non‐haem iron dioxygenase (NHI, TropC), analogous to early biosynthetic steps in the biosynthesis of stipitatic acid (Scheme 2 and Figure S14 in the Supporting Information).[ 6 , 7 , 13 ]

![Biosynthetic routes towards tropolone sesquiterpenoids isolated in this study: A, each route representing an individual expression experiment in A. oryzae NSAR1. Compounds in blue were newly isolated in this study. Compounds in brackets were not observed; B, Route to 6 and 24; C, incorporation of labelled sodium [1,2‐13C2] acetate and (methyl‐13C) methione into 21 and incorporation of labelled sodium [1,2‐13C2] acetate into 26.](/dataresources/secured/content-1765852128011-e2c87b87-2ea6-4f96-9917-c79732300c6b/assets/ANIE-59-23870-g006.jpg)
Biosynthetic routes towards tropolone sesquiterpenoids isolated in this study: A, each route representing an individual expression experiment in A. oryzae NSAR1. Compounds in blue were newly isolated in this study. Compounds in brackets were not observed; B, Route to 6 and 24; C, incorporation of labelled sodium [1,2‐13C2] acetate and (methyl‐13C) methione into 21 and incorporation of labelled sodium [1,2‐13C2] acetate into 26.
Stipitaldehyde 8 represents a branching point in TS biosynthesis (Scheme 1 B): in the case of xenovulene A 2, stipitaldehyde 8 is oxidised by the cytochrome P450 AsR2 to the corresponding hemiacetal 9 and subsequent elimination of water yields the reactive quino‐methide 10 that undergoes an enzyme‐catalyzed hetero Diels–Alder reaction with α‐humulene 6, synthesised by an unusual terpene cyclase, yielding xenovulene B 11 (Scheme 1 B).[ 6 , 7 ] However, during the biosynthesis of eupenifeldin 3, stipitaldehyde 8 is reduced by the short‐chain dehydrogenase (SDR) EupfE to the corresponding alcohol stipitol 12. The hetero Diels–Alderase EupfF then catalyses formation of the o‐quino‐methide 13 prior to hetero Diels–Alder reaction with 10‐hydroxy‐humulene 7, to give neosetophome B 4 (Scheme 1 B). [6] Notably, in vitro experiments with EupfF only gave rise to mono‐substituted tropolone sesquiterpenoids and formation of bistropolones such as 1 and 3 has not yet been achieved in vitro or in vivo. [6] Access to double hDA adducts thus represents an intriguing biosynthetic challenge; similarly, the origin and formation of TS natural products with benzene rings, such as epolone A 5,[ 1 , 14 , 15 , 16 , 17 ] has remained uninvestigated so far.
We decided to deploy a synthetic biology driven combinatorial heterologous biosynthesis approach to rationally expand the chemical space around TS natural products and to investigate key biosynthetic steps. Aspergillus oryzae has previously been established as an excellent host for the expression of biosynthetic gene clusters[ 18 , 19 , 20 , 21 ] and recently we reconstituted the total biosynthesis of the xenovulenes therein, granting rapid access to a variety of xenovulenes on a multi‐milligram scale. [7] We reasoned that rational extension and diversification of the xenovulene A 2 biosynthetic pathway by mixing and matching genes from different TS BGC should give rise to new TS natural products and further illuminate key biosynthetic steps in TS biosynthesis. Here, we identify BGC involved in pycnidione 1 and eupenifeldin 3 biosynthesis and generate a series of new, unnatural TS analogues. Labelling studies were deployed to investigate the origin of monobenzopyranyl moieties observed in several natural and unnatural compounds and the results shed light on a new ring‐contraction during their biosynthesis.
Results and Discussion
Phaeosphaeriaceae sp. CF‐150626 and Leptobacillium sp. CF‐236968 were obtained from Fundación MEDINA (Granada, Spain). Phaeosphaeriaceae sp. (formerly referred to as unidentified ascomycete F‐150626) was previously reported to produce the bistropolone‐humulene eupenifeldin 3 and the monotropolone‐monobenzopyranyl‐humulene noreupenifeldin 14 (Figure 1 C). [17] In our hands CF‐150626 produced eupenifeldin 3 (8.7 mg L−1; HRMS, [M]H+ calculated C33H41O7 549.2852, found 549.2856) as the major product, confirmed by full NMR characterization (Figure S24–S32 and Table S10). A second compound, satisfying the molecular weight of noreupenifeldin 14 (HRMS: [M]H+ calculated C32H41O6 521.2903, found 521.2907) was also produced. Purification to homogeneity (2.8 mg L−1) and NMR analysis revealed slight differences in 1H‐ and 13C‐NMR chemical shifts as compared to noreupenifeldin 14. [17] Full structure elucidation confirmed the compound to be a regioisomer of noreupenifeldin in which the tropolone and benzene rings are exchanged as compared to noreupenifeldin 14 (Figure S37–S45 and Table S13). Subsequent comparison with literature NMR data identified this compound as the previously described noreupenifeldin B 15 (Figure 1 A). [1] Dehydroxyeupenifeldin 16 (HRMS: [M]H+ calculated C33H40O6 533.2903, found 533.2912; Figure 1 A) was observed as a minor component and characterized by NMR analysis (Figure S50–S57 and Table S16).

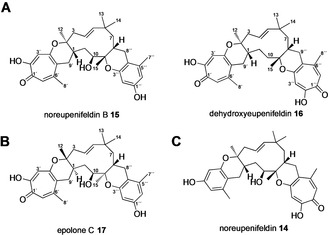
A, tropolone sesquiterpenoids isolated from Phaeosphaeriaceae sp.; B, tropolone sesquiterpenoids isolated from Leptobacillium sp.; C, Structure of noreupenifeldin 14.
The hitherto undescribed fungus CF‐236968 produced pycnidione 1 (2 mg; HRMS: [M]H+ calculated C33H41O7 549.2852, found 549.2853) as the dominant product, confirmed by full NMR characterization (Figure S58–S65 and Table S17). Additionally, a related compound 17 with the molecular weight of 520 (HRMS: [M]H+ calculated C32H41O6 521.2903, found 521.2905) was observed and purified to homogeneity. Overall, the obtained NMR data was similar, but not identical, to the previously reported epolone A 5. [5]
Analysis of COSY and HMBC data for 17 established the carbon skeleton of the humulene, tropolone and monobenzopyranyl moieties (Figure S66–S73 and Table S18). Among others, key HMBC/COSY correlations between H2‐9′ and C‐6′ and between H‐9′ to H‐1 established the tropolone ring at the western side of humulene; 1H‐1H COSY coupling of H‐8 to H2‐8′′ together with 3 J coupling of H2‐8′′ to C‐5′′ placed the benzopyranyl moiety at the eastern side of humulene and confirmed the regioselectivity of the fused ring‐systems as opposite to those encountered in epolone A 5, thus establishing 17 as a novel TS that we name epolone C (Figure 1 B).
Biosynthetic gene clusters involved in formation of pycnidione 1 and eupenifeldin 3, were identified by standard Illumina paired‐end sequencing which afforded high quality draft genome sequences for both fungi (e.g. CF‐150626, 44.7 MBp and N50 164515; CF‐236968 28.6 MBp and N50 466766; Table S3). TS BGC were identified by search for nrPKS 3‐methylorcinaldehyde synthase (AsPKS1) homologs, since 3‐methylorcinaldehyde is the precursor of 8. [7] A single aspks1‐like gene cluster was revealed in each fungal genome (here named eup2 BGC [CF‐150626] and pyc BGC [CF‐236968]; Figure 2 A). The Artemis comparison tool was used to visualize homologies between the two clusters and the previously reported aspks1 BGC from Acremonium strictum (Figure 2 A). [23] As expected all clusters share core genes necessary for stipitaldeyhde 8 formation (aspks1, asL1, asL3; eupPKS, eupL1, eupL5; pycPKS, pycL1, pycL3 respectively) and additionally homologous copies of a hetero Diels–Alderase (asR5; eupR1; pycR1) and a humulene synthase (asR6; eupR3; pycR6, Table S9).

![A, Artemis comparison between aspks1 BGC (Acremonium strictum), eup2 BGC (Phaeosphaeriaceae sp.) and pyc BGC (Leptobacillium sp.); B, isotopic labelling of key tropolone sesquiterpenoids with sodium [1‐13C, 2‐13C, 1,2‐13C2]‐labelled acetate and/or [methyl‐13C]‐labelled methionine. Labelling pattern of xenovulene B 11 as described by Simpson and co‐workers.
[22]](/dataresources/secured/content-1765852128011-e2c87b87-2ea6-4f96-9917-c79732300c6b/assets/ANIE-59-23870-g002.jpg)
A, Artemis comparison between aspks1 BGC (Acremonium strictum), eup2 BGC (Phaeosphaeriaceae sp.) and pyc BGC (Leptobacillium sp.); B, isotopic labelling of key tropolone sesquiterpenoids with sodium [1‐13C, 2‐13C, 1,2‐13C2]‐labelled acetate and/or [methyl‐13C]‐labelled methionine. Labelling pattern of xenovulene B 11 as described by Simpson and co‐workers. [22]
Unique to the eup2 BGC and pyc BGC are short‐chain dehydrogenase encoding genes (eupL4 and pycL2; homologous to eupfE) and a cytochrome P450 (eupR6, pycR5) homologous to eupfD, previously shown to be responsible for C‐10 hydroxylation of the terpene moiety.[ 6 , 12 ] Noticeably, the ring‐contraction enzymes asL4 and asL6 only have a single homologue in these clusters (eupR5 and pycR4 respectively).
Comparison of the eup2 BGC to the previously reported eup cluster in Phoma sp. [12] revealed a high degree of similarity and this very strongly supported the eup2 cluster to be responsible for the production of 3 in CF‐150626. (Table S9 and Figure S15). RT‐PCR further confirmed upregulation of the eup2 BGC only under 3‐producing conditions (Figure S16).
Pycnidione 1 has been isolated from several fungal sources,[ 16 , 24 , 25 ] but, to‐date no biosynthetic gene cluster has been reported. In order to link the putative pyc BGC to the production of 1 we tried to genetically manipulate CF‐236968. However, the fungus proved resistant to common gene disruption technologies. Furthermore, attempts to isolate RNA from the fungus failed, preventing a similar analysis by gene expression profiling as done for CF‐150626. However, the high degree of homology to the eup2 cluster and absence of any other aspks1‐like gene cluster in the genome strongly supports the pyc BGC to be responsible for 1 formation. Additionally, recombinant production of the putative humulene synthase PycR6 and incubation with farnesylpyrophosphate afforded α‐humulene 6 as confirmed by GC/MS analysis (Figure S17–S19).
While fungal tropolone formation is well‐understood, the origin of the monobenzopyranyl moiety in noreupenifeldin B 15 and epolone C 17 poses an intriguing biosynthetic question. Chen and co‐workers as well as Zhang and co‐workers recently proposed a hetero Diels–Alder reaction between humulene and an o‐quinomethide (derived from orsellinaldehyde) to explain the presence of the benzopyranyl moiety.[ 26 , 27 ] Orsellinaldehyde is a common fungal metabolite and has been identified as the tetraketide product of an nrPKS. [28] However, no biosynthetic evidence to support such an hDA reaction has been reported so far. To establish the biosynthetic origin of the benzopyranyl moiety we performed incorporation studies with 13C‐labelled acetates. In separate experiments we fed CF‐150626 with [1‐13C]‐ and [2‐13C]‐labelled acetates (Figure 2 B). 13C‐NMR of purified 3 showed that 31/33 carbon signals were enhanced, and the obtained labelling pattern of terpene and tropolone parts was identical as previously reported for xenovulene B 11 (Figure S33–S36 and Table S11+S12). [22] Only the NMR signals for carbons C‐3′ and C‐3′′ (within the two tropolone rings) were not enhanced, in agreement with their proposed origin from methionine. [29]
In the case of noreupenifeldin B 15 30/32 carbon signals were enhanced (Figure 2 B). The observed pattern of label incorporation in the tropolone and humulene was the same as in eupenifeldin 3: all carbon signals except the signal for C‐3′ of the tropolone were enhanced (Figure S46–S49 and Table S14+S15). Surprisingly, carbons C‐6′′ and C‐1′′ were both derived from [2‐13C]‐acetate, while the signal for C‐2′′ was not derived from acetate. This suggests that C‐2′′ is derived from methionine. C‐6′′ and C‐1′′ being both derived from [2‐13C]‐acetate indicate that one acetate was disrupted during the biosynthesis of the benzopyran moiety of noreupenifeldin B 15. A putative orsellinaldehyde precursor would show intact labelling for four sequential intact acetate units and is thus eliminated as a possible precursor to 15.
Contrary to previous suggestions, the labelling data suggests that 15 is derived from a ring‐contraction of a tropolone precursor (probably eupenifeldin 3), instead of originating from an hDA reaction between humulene and a benzylic o‐quinomethide. Notably, both the eup2 and pyc BGC each include a gene homologous to asL4 and asL6 (eupR5, pycR4). AsL4 and AsL6 are FAD‐dependent oxygenases known to catalyze regioselective oxidative ring contractions during the biosynthesis of xenovulene A 2, and thus represent possible candidates to catalyze such a reaction. [7] Attempts to probe the role of PycR4, AsL4 and AsL6 in vitro were prevented in this study (and previously) by the inability to obtain soluble protein preparations. [7] Furthermore, knockout and silencing experiments to probe the role of these genes is not currently possible since the host organisms cannot yet be transformed.
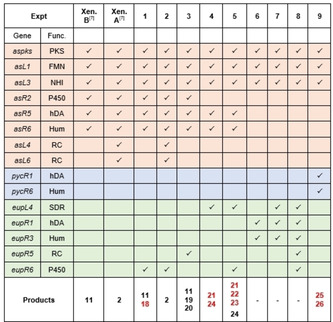
We next devised expression experiments in an attempt to generate novel unnatural derivatives (Table 1). Previously we reported on the heterologous production of xenovulene B 11 and xenovulene A 2 in A. oryzae NSAR by co‐expressing six or eight biosynthetic genes, respectively, using the modular expression system established by Lazarus and co‐workers (Table 1, Exp. Xen B and Xen A respectively).[ 7 , 30 ] A series of additional A. oryzae NSAR1 expression plasmids comprising key biosynthetic genes from the eup2 and pyc BGC were generated using standard yeast homologous recombination/Gateway technology. Ectopic integration of biosynthetic genes into A. oryzae NSAR1 gDNA was confirmed by PCR analysis (Figure S5–S13).

|
|
We began by introducing modifications into the existing biosynthetic route to xenovulene B 11. Hydroxylation at C‐10 of α‐humulene is a recurring feature in bistropolones[ 4 , 25 ] but has never been observed in the xenovulenes. Co‐expression of the xenovulene B 11 producing genes with the humulene hydroxylase encoding gene eupR6 (Exp.1, Table 1) and analysis by LCMS showed production of xenovulene B 11 and a new compound 18 (Figure 3, Exp. 1; Scheme 2 A), with a 16 amu difference relative to xenovulene B 11 (nominal mass 382) as observed by HRMS ([M]H+ calculated C24H31O4 399.2171 found 399.2174). As expected, NMR structure elucidation revealed the C‐10 methylene observed in xenovulene B 11 (δ H 2.12/2.26; δ C 38.0) to be replaced by a downfield shifted oxygenated carbon (δ H 4.36; δ C 77.1) in 18 and confirmed 18 as 10‐hydroxyxenovulene B (Figure S74–S82 and Table S19). The relative stereochemistry of 18 was determined by NOESY‐NMR; absence of correlation between H3‐12 and H‐1 established these to be trans. H3‐12 NOE correlation to H‐9′, H2‐3 and H2‐11, but not H‐10, places H3‐12 and OH‐10 on the same face.

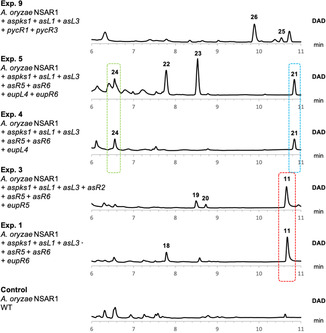
Heterologous expression of key biosynthetic gene combinations in Aspergillus oryzae NSAR1. Shown are LC/MS diode array (DAD) traces of extracts of representative transformants.
Surprisingly, inclusion of the ring‐contraction encoding genes asL4 and asL6 in the expression system (Exp. 2, Table 1) led only to the production of xenovulene A 2 (Figure S20) but not to production of any hydroxylated analogue of 2, suggesting that ring‐contraction might out‐compete hydroxylation of the humulene moiety and that the fully ring‐contracted scaffold of xenovulene A 2 is not a possible substrate for hydroxylation.
The known ring‐contraction enzymes AsL4 and AsL6 from the aspks1 BGC show 34.8 % and 31.8 % sequence identity to EupR5 encoded in the eup2 cluster. Recently, Che and co‐workers proposed the EupR5 homologue EupH to be a putative redox partner of the humulene hydroxylase present in all eupenifeldin BGC. [12] However, the distinct sequence homology of EupR5 to the known ring‐contraction enzymes AsL4 and AsL6 prompted us to hypothesise that EupR5 might catalyse a similar reaction in CF‐150626—given the observation of a ring‐contraction during 15 biosynthesis.
To probe the biosynthetic role of EupR5 we co‐expressed the xenovulene B 11 producing genes with eupR5 in A. oryzae (Exp. 3, Table 1). Analysis of transformants revealed the formation of two compounds, 19 and 20, both having the nominal mass of 370 (HRMS: [M]H+ calculated C23H31O4 371.2222 found 371.2218 and 371.2227; Figure 3; Exp. 3). A 12 amu difference compared to xenovulene B 11 (382) was consistent with a ring contraction. Purification to homogeneity by preparative LCMS of both compounds individually failed as the difference in retention time was too small. However, NMR characterization of a mixture of 19 and 20 was sufficient to quickly identify 19 and 20 as the previously reported products of AsL4 (19) and AsL6 (20, previously only available in trace amounts) observed during the biosynthesis of xenovulene A 2 (Scheme 2 A, Figure S83‐S92 and Table S20). [7] Exp. 3 clearly demonstrates the ability of EupR5 to catalyse oxidative ring‐contractions in vivo.
Based on existing biosynthetic knowledge, the biosynthesis of xenovulene A 2 and eupenifeldin 3 diverges after formation of stipitaldehyde 8 (Scheme 1 B). We hypothesised that replacement of asR2 by the SDR gene eupL4 might redirect xenovulene A 2 biosynthesis in the direction of mono‐ or bistropolones lacking the characteristic tetrahydrofuran ring present in all xenovulenes (Exp.4, Table 1). Analysis of transformants by LCMS analysis confirmed their inability to produce any xenovulenes as was expected by omission of asR2, halting the biosynthesis prior to the hDA reaction. Instead, a single new compound 21 was produced in excellent titres. HRMS analysis of 21 (HRMS: [M]H+ calculated C23H33O2 341.2481 found 341.2481) suggested a molecular formula of C23H33O2. Surprisingly, the nominal mass of 340 was too small to correspond to either a mono‐ or bistropolone TS. Purification to homogeneity (3.4 mg) and subsequent analysis by full NMR spectroscopy elucidated the structure of 21.
Key tropolone NMR signals (e.g. characteristic aromatic protons at 6.9–7.2 ppm; aromatic methyl singlet at 2.4 ppm; carbonyl signal at 170 ppm) were replaced by aromatic protons at 6.18 ppm (H‐2′) and 6.25 ppm (H‐6′) and together with an aromatic methyl singlet at 2.19 ppm (H3‐7′) indicated substitution of the usual tropolone by a benzene. Key HMBC correlations from H2‐8′ to C‐11, C‐3′ and C‐5′ respectively further corroborated the structure of 21. Selective 1D‐NOE experiments confirmed absence of coupling between H3‐12 and H‐1 and, together with 2D‐NOESY data, established the relative stereochemistry at the humulene/dihydropyran ring junction as trans, in agreement with biosynthetic considerations (Figure S93–S103 and Table S21).
Further inclusion of the humulene hydroxylase gene eupR6 in the expression system (Table 1, Exp. 5) led to formation of two additional compounds (22, 23), both having a nominal mass of 356 respectively (HRMS [M]H+ calculated C23H33O3 357.2430 found 357.2428 (22) and 357.2431 (23)) consistent with 22 and 23 being hydroxylated derivatives of 21. Indeed, purification to homogeneity (2.8 mg, 4.4 mg) and subsequent full NMR characterization revealed that the C‐10 methylene group in 21 (δ C 37.9 ppm; δ H 1.83 and 2.10 ppm) was replaced by downfield shifted oxygenated carbons in both 22 (δ C 78.4 ppm; δ H 3.99 ppm) and 23 (δ C 73.9 ppm; δ H 4.35 ppm, Figure S105–S131 and Table S23+S24). 22 and 23 comprise the same structural skeleton as the previously reported Pughiinin A, isolated from the fungus Kionochaeta pughii BCC 3878. [16] However, small differences in 1H and 13C NMR shifts suggest 22 and 23 to be stereoisomers of Pughiinin A. A series of 1D‐NOE experiments was performed to establish the relative stereochemistry of 22 and 23. For 22, correlation of H‐10 to H‐1 and H‐8′b and vice versa suggested H‐1, H‐8′b and H‐10 to be on the same face. Absence of correlation from H3‐12 to either H‐1, H‐10 or H‐8′b confirmed trans‐fusion of the humulene/dihydropyran ring (Figure 4). Compared to 22, compound 23 displayed significant differences in 1H‐NMR chemical shift: most notably both H‐10 (δ H‐10 4.35 ppm in 23; δ H‐10 3.99 ppm in 22) and H‐1 (δ H‐1 2.05 ppm in 22; δ H‐1 1.69 ppm in 23) were shifted downfield by 0.36 ppm. Absence of nOe correlations between H3‐12 and H‐1, H‐10 and H‐8′b confirmed trans‐fusion of the humulene/dihydropyran ring. Contrary to 22, H‐10 did not correlate to H‐1 whereas H‐1 still correlated to H‐8′b and vice versa. Together this nOe data suggests that in 23 the 10‐hydroxy moiety faces in the opposite direction as observed in 22. This finding is further supported by careful comparison of 1H‐NMR shifts of H‐1 in 1, 3, 15–21. Not surprisingly in all compounds with H‐1 and OH‐10 on the same face δ H‐1 was found to be >2.00 ppm whereas in compounds with H‐1 and OH‐10 on different faces δ H‐1 was typically found to be <1.90 ppm.

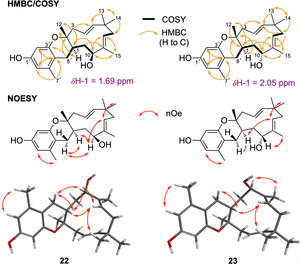
Structure elucidation of 22 and 23. See Supporting Information for detailed structure elucidation of all other compounds. 3D model structures of 22 and 23 were calculated using Spartan 18 and minimised using molecular mechanics.
As a co‐metabolite, humulene derivative 24 (Scheme 2 B) was concomitantly produced with 22 and 23 and purified to homogeneity (4 mg; HRMS: [M‐H2O]H+ calculated C15H23O 219.1749 found 219.1749). NMR analysis confirmed 24 to be the 1,2‐epoxy‐10‐hydroxy derivative of humulene 6 (Figure S132–S140 and Table S25) and was similar to data previously reported for phomanoxide (Figure S133). [31] Contrary to compound 24, phomanoxide harbours an additional epoxide at C‐4/C‐5. We thus name compound 24 phomanoxide B. Interestingly, trace amounts of 22, 23 and 24 were also observed in Exp. 4, lacking the co‐expressed humulene hydroxylase eupR6. Given the wealth of native cytochromes P450 present in Aspergillus oryzae NSAR1 (ca. >150) it seems likely that a native oxygenase can hydroxylate the TS scaffold albeit to a significantly lower degree. [32]
The results of experiments 1–5 (Table 1) demonstrated the feasibility to engineer the biosynthesis of xenovulenes, resulting in the successful generation of 18–24. Notably, despite introduction of humulene hydroxylase EupR6, and short‐chain dehydrogenase EupL4, all transformants solely produced mono‐substituted Diels–Alder adducts, suggesting that the xenouvlene A 2 hDA enzyme AsR5 is limited in regard to the number of hDA reactions it catalyses. We reasoned that exchange of AsR5 for an hDA enzyme from the eupenifeldin (EupR1) or pycnidione (PycR1) pathway might redirect the pathway to the production of bistropolones. However, omission of asR2 and replacement of asR5 and asR6 for eupR1 and eupR3 did not lead to production of any tropolone sesquiterpenoids (Table 1, Exp. 6; Figure S21). Further inclusion of short‐chain dehydrogenase encoding gene eupL4 (Table 1, Exp. 7; Figure S22) and humulene hydroxlyase eupR6 and FAD‐dependent monooxygenase eupR5 (Table 1, Exp. 8; Figure S23) in the expression also did not lead to production of the desired meroterpenoids.
Omission of asR2 and introduction of hetero Diels–Alderase and terpene cyclase encoding genes pycR1 and pycR3 from the pycnidione pathway instead of eupR1 and eupR3 proved more successful (Exp. 9). Analysis of transformants by LCMS analysis identified the production of two new compounds (25, 26) compared to a WT control (Figure 3). The nominal mass (532, Figure S142) and UV spectrum of minor component 25 was consistent with a bistropolone lacking the C‐10 hydroxyl group. Indeed, purification of compound 25 and NMR characterization established the structure of 25 (Figure S141–S147; Table S26): key aromatic 1H‐NMR signals at 7.33/7.27/7.17/7.15 ppm together with two aromatic methyl group signals at 2.50 ppm and 2.43 ppm were characteristic for the presence of two tropolone rings. Key COSY and HMBC correlations further confirmed the structure of 25.
Purification of major component 26 to homogeneity afforded 4 mg ([M]H+ calculated C32H41O5 505.2954 found 505.2941) and subsequent NMR analysis confirmed its structure. Key tropolone NMR signals including aromatic protons at 7.08 ppm and 7.22 ppm and a methyl singlet at 2.41 ppm established the presence of one tropolone ring; key HMBC correlations from H2‐9′′ to C‐7, C‐9, C‐4′′ and C‐6′′ placed the tropolone ring at the eastern side of humulene. Additional aromatic proton signals at 6.14 and 6.25 ppm together with an aromatic methyl group at 2.21 ppm confirmed replacement of the second tropolone ring by a benzene. HMBC correlations of H2‐8′ to C‐2, C‐11, C‐3′ and C‐5′ further corroborated the structure and placed the benzene ring at the western face of humulene (Figure S148‐S156 and Table S27). Absence of NOESY correlations between H‐1 and H3‐12 as well as between H‐8 and H3‐15 together with other NOESY correlations established the relative stereochemistry at the ring‐junctions to be trans, in agreement with biosynthetic considerations.
Surprisingly, both Exp. 4 and Exp. 9 afforded TS natural products (21, 26) with a benzene ring instead of the expected tropolone moiety, despite the reconstituted pathways containing no ring‐contraction enzyme. In order to establish the biosynthetic origin of these 6‐membered rings in A. oryzae we performed labelling experiments using [1,2‐13C2]‐labelled acetate. Labelled 21 and 26 were analysed by 13C‐NMR (Figure S104+S157–S158 and Table S22+S28). Analysis of coupling constants quickly identified integration of intact acetate units. For both compounds the labelling pattern of the benzopyranyl moieties were identical, comprising three intact acetate units (C7′‐C5′, C6′‐C1′ and C4′‐C8′). Instead of a fourth intact acetate unit, both C‐2′ and C‐3′ did not show coupling to any other carbon atom. We reasoned that the obtained labelling pattern was in agreement with the six‐membered ring in 21 and 26 being derived from a ring‐contraction of a tropolone precursor, resulting in rearrangement of one acetate unit and C‐2′ to be derived from methionine. To probe this hypothesis, we fed [methyl‐13C]‐labelled methionine to the 26 producing strain.
Purification of labelled 26 and subsequent analysis by 13C‐NMR showed signal enhancement for two carbon signals, corresponding to C‐3′′ (within the tropolone ring) and C‐2′ (within the benzene ring, Figure S159–S161). The obtained labelling pattern thus demonstrates that 26 (and 21 accordingly) are derived from a ring‐contraction of a tropolone precursor—it is therefore highly likely that 26 is derived from 25. Benzopyran 21 is therefore most likely derived from an unobserved tropolonopyran precursor 27 (Scheme 2 A).
Significantly, the labelling pattern of the six‐membered ring in 21 and 26 differs from labelling pattern of the six‐membered ring in noreupenifeldin B 15 (Scheme 3). For 21 and 26, ring contraction in A. oryzae must proceed via excision of the C‐2′ carbon in 27 and 25 respectively, whereas ring contraction of 3 in CF‐150626 must proceed via excision of the C‐1′′ (Scheme 3 A). In the absence of a transformed ring‐contraction enzyme in Exp. 5 and Exp. 9 we reasoned that Aspergillus oryzae NSAR1 itself must contain a putative ring‐contraction enzyme, but the identity of this enzyme remains unknown. Notably, the ring‐contraction by A. oryzae is only observed for compound 21 and 26 but not for xenovulene B 11, which is stable in A. oryzae albeit it also comprises an intact tropolone ring. Structurally, xenovulene B 11 differs from 21 and 26 by the tetrahydrofuran ring characteristic of all xenovulenes and it appears that the putative ring‐contraction enzyme in A. oryzae does not accept these as substrates. Furthermore, the observed ring‐contractions in A. oryzae and also in Phaeosphaeriaceae sp. appear to be highly regioselective as in A. oryzae only tropolone rings on the western face of humulene are contracted whereas in Phaeosphaeriaceae sp. the ring‐contraction occurs on the eastern side of humulene. These ring contractions also differ from those observed to be performed by AsL4 and AsL6 in the xenovulene pathway, and EupR5 observed here (Scheme 2). AsL4, AsL6 and EupR5 catalyse an oxidative ring contraction which leaves a hydroxyl group on the benzene ring, for example, 19 and 20, whereas 15, 21 and 26 do not contain additional oxygen.

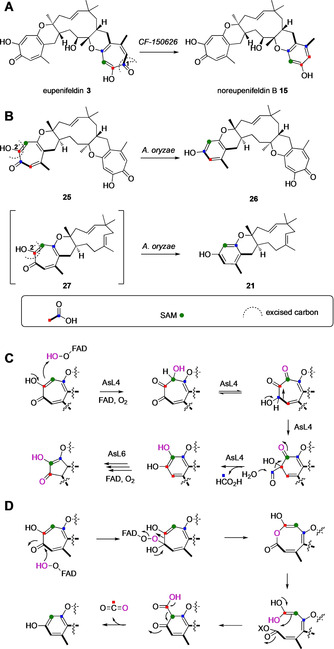
Proposed ring‐contraction in the biosynthesis of noreupenifeldin B 15, 26 and 21 based on observed labelling patterns: A, ring‐contraction in Phaeosphaeriaceae sp. CF‐150626 proceeding via excision of carbon C‐1′′ in 3; B, ring‐contraction in A. oryzae proceeding via excision of carbon C‐2′ in 25 and 27 respectively; C, proposed mechanism for AsL4/AsL6/EupR5; D, possible process for the formation of 15, 21 and 26 which is consistent with the observed labelling pattern, X=unknown activation.
We previously suggested a mechanism which would explain the retention of oxygen (Scheme 3 C) in the case of xenovulene A 2 biosynthesis. [7] It is possible that the production of 15, 21 and 26 could proceed via a similar mechanism to 19 and 20, followed by a reductive step. However, we did not observe any intermediates which might support this possibility. Alternatively, a different FAD‐dependent mechanism could be in play (Scheme 3 D), involving a ring‐opening‐ring‐closing sequence which would explain the observed labelling and oxygenation patterns in A. oryzae. A similar mechanism involving initial attack at C‐2′′ of 3 would explain the observed labelling pattern in 15. Tropolone ring‐opening mechanisms have been suggested during the biosynthesis of phomanolides C‐F. [27] Chemical investigations by Ito support the initial oxidative ring expansion in this possibility. [33] However, in the absence of additional information the precise origin and mechanism of these new transformations remains to be determined.
Conclusion
Here, we report on the identification of the eup2 and pyc BGC, responsible for formation of eupenifeldin 3 and pycnidione 1 in CF‐150626 and CF‐236968 for the first time. We successfully engineered A. oryzae NSAR1 for the heterologous production of seven new unnatural tropolone sesquiterpenoids in good yields, by exploiting a systematic combinatorial biosynthesis approach, demonstrating the power of heterologous expression in fungi for the rational creation of new compounds. In this case synthetic biology out‐performs synthetic chemistry which has not yet been used for the synthesis of these or related natural products. Furthermore, the heterologous expression system deployed was successfully used to determine the biosynthetic function of proteins with previously unknown activity (e.g. EupR5). This work also shows that the hDa enzymes differ in their ability to produce mono (e.g. AsR5) and di‐tropolone (e.g. PycR1) meroterpenoids. We also showed that benzopyranyl moieties are derived via a ring‐contraction of a tropolone precursor instead of the previously suggested hDA reaction with an orsellinaldehyde. Surprisingly, Aspergillus oryzae NSAR1 is capable of catalyzing a similar ring‐contraction, although the conducted labelling studies demonstrate, that the mechanism must differ from that previously observed during the biosynthesis of xenovulene A catalysed by AsL4/AsL6, or by EupR5. Given the wealth of potent biological activity present in naturally occurring TS natural products, this work paves the way to systematically assess TS natural products and to construct a compound library to be tested for additional/improved biological activity.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
CS thanks Leibniz University for funding. LL thanks the Chinese Scholarship Council for funding (CSC 201704910820). Fundación MEDINA (Olga Genilloud/Víctor González Menéndez) are thanked for strains and culturing conditions. Jörg Fohrer and Luca Codutti are thanked for assisting with 2D‐nOe data. DFG is thanked for the provision of NMR and LCMS equipment (INST 187/621‐1, INST 187/686‐1). Open access funding enabled and organized by Projekt DEAL.
References
1
2
3
4
5
6
8
10
12
13
14
15
16
17
18
19
19
20
21
22
23
24
25
26
27
28
30
31
33
 Synthetic Biology Driven Biosynthesis of Unnatural Tropolone Sesquiterpenoids
Synthetic Biology Driven Biosynthesis of Unnatural Tropolone Sesquiterpenoids