
- Altmetric
The complexities of gene expression pose challenges for the clinical interpretation of splicing variants. To better understand splicing variants and their contribution to hereditary disease, we evaluated their prevalence, clinical classifications, and associations with diseases, inheritance, and functional characteristics in a 689,321-person clinical cohort and two large public datasets. In the clinical cohort, splicing variants represented 13% of all variants classified as pathogenic (P), likely pathogenic (LP), or variants of uncertain significance (VUSs). Most splicing variants were outside essential splice sites and were classified as VUSs. Among all individuals tested, 5.4% had a splicing VUS. If RNA analysis were to contribute supporting evidence to variant interpretation, we estimated that splicing VUSs would be reclassified in 1.7% of individuals in our cohort. This would result in a clinically significant result (i.e., P/LP) in 0.1% of individuals overall because most reclassifications would change VUSs to likely benign. In ClinVar, splicing VUSs were 4.8% of reported variants and could benefit from RNA analysis. In the Genome Aggregation Database (gnomAD), splicing variants comprised 9.4% of variants in protein-coding genes; most were rare, precluding unambiguous classification as benign. Splicing variants were depleted in genes associated with dominant inheritance and haploinsufficiency, although some genes had rare variants at essential splice sites or had common splicing variants that were most likely compatible with normal gene function. Overall, we describe the contribution of splicing variants to hereditary disease, the potential utility of RNA analysis for reclassifying splicing VUSs, and how natural variation may confound clinical interpretation of splicing variants.
Introduction
DNA variants that abolish, change, or create splice sites can disrupt messenger RNA splicing and adversely affect protein synthesis or structure, leading to impaired cellular function and consequent disease.1 Variants that may alter RNA splicing can be computationally predicted, and these predictions can be confirmed by RNA analysis. However, assessing the clinical consequences of abnormal splicing can be challenging because of an incomplete understanding of alternative splicing and normal RNA expression profiles across tissues.2 Some studies have revealed previously unrecognized variety in RNA transcript isoforms associated with well-studied genes, including BRCA1 (MIM: 113705) and BRCA2 (MIM: 600185), showing that our understanding of naturally occurring alternative splicing of disease gene transcripts is still evolving.3, 4, 5, 6 Recent studies have also illuminated how differential expression of transcript isoforms can influence whether certain sequence variants are tolerated.7,8 As a result of this underappreciated complexity, variants that allow biologically viable alternative splicing may be incorrectly classified as disease causing. Therefore, investigating both the spectrum of variants predicted or assumed to cause abnormal splicing across a broad variety of genes and their contribution to naturally existing genomic variation is essential to understanding their overall involvement in hereditary disease.
Several computational tools are used to predict the potential splicing effects of variants encountered during clinical genetic testing for hereditary disease.9,10 However, these tools often do not have a high positive predictive value when used individually, particularly for variants outside the essential splice site(s) (ESS).11,12 Therefore, their predictions are often not considered usable evidence for variant interpretation unless there is consensus among them.13 Direct analysis of RNA, through RNA sequencing or other methods, may provide evidence that corroborates computational predictions, but this is not yet routinely and broadly performed in clinical genetic testing. RNA analysis can be used to confirm the etiology of a hereditary disease through gene discovery, variant discovery in a known disease gene, or accurate interpretation of an observed variant.14, 15, 16, 17 In this article, we focus specifically on RNA analysis as a variant interpretation tool for confirming or refuting the splicing effects of computationally predicted splicing variants identified by DNA sequencing.18, 19, 20 We investigated splicing variants previously identified through clinical genetic testing and specifically the proportion of splicing variants of uncertain significance (VUSs) that could be reclassified to either pathogenic or benign categories via RNA analysis.
Although a few studies have shown the utility of targeted RNA analysis for confirming the effects of splicing variants identified through targeted gene sequencing, it remains unclear how often these variants explain the cause of suspected hereditary disease in individuals referred for genetic testing and whether their effects can be observed in disease-relevant tissues.14,18 In addition, results from RNA analysis can be misleading or equivocal,5 similar to other types of evidence considered during clinical variant interpretation. Further, the prediction of splicing effects can depend on the reference transcript used. Some clinical laboratories may choose a single transcript as a reference to report observed variants, and a splicing variant’s impact on alternative transcripts can vary significantly. Therefore, using a single reference transcript in some cases may lead to improper variant classification and result in missed molecular diagnoses.7,8
Professional laboratory practice standards are not yet available to provide detailed and specific guidance for consistently interpreting the clinical significance of splicing variants. Variant interpretation guidelines from the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) state that functional evidence such as RNA analysis may garner supporting, moderate, or strong evidence depending on performance of the assay and quality of the specimen.13 Per the guidelines, to use data from such complementary methods as strong evidence, the methods should be well established and used in assays that reflect the biological environment and designed to generate reproducible and robust data. In addition, the Clinical Genome Resource (ClinGen) Sequence Variant Interpretation Working Group recommends that the strength of a functional assay should be determined by validation metrics, including the use of known pathogenic and benign variants as benchmarks. The level of evidence awarded (supporting, moderate, or strong) should then be concomitant with the demonstrated validation metrics.21 To our knowledge, such thorough validations with clearly demonstrated high positive predictive values have not yet been described for RNA analysis in a clinical setting.
To better understand the spectrum of splicing variants in hereditary disease genes and the challenges of interpreting their clinical significance, we investigated data from a clinical cohort of nearly 700,000 individuals who underwent diagnostic genetic testing. We also surveyed splicing variants in two large public databases of human genomic variation. We considered three classes of splicing variants: (1) those at the highly conserved ESS (±1–2 intronic positions); (2) those in the splice region near the ESS (±3–8 bp intronic and ±1–3 bp exonic positions); and (3) within our clinical data only, exonic variants that were outside the consensus splice region but were algorithmically predicted to have a splicing effect. The first aim of this study was to examine the contribution of clinically reportable splicing variants to different types of hereditary disease detected with targeted gene panels. The second aim, taking a cautious approach to weighting RNA analysis evidence, was to estimate the proportion of splicing VUSs that could reach definitive clinical classification with RNA analysis. The last aim was to understand the prevalence and implications of splicing variants amid natural variation in healthy genomes.
Subjects and methods
Clinical genetic testing and cohort analyses
Patient cohort data
We collected DNA sequencing results from 689,321 individuals who received genetic testing between January 21, 2014 and July 1, 2019 for a range of hereditary diseases related to nine clinical areas: cancer, cardiology, dermatology, hematology, immunology, metabolism, neurology, pediatrics, and ophthalmology. Data were de-identified and approved for use in this study by an independent institutional review board (Western IRB #20161796). Procedures followed were in accordance with the ethical standards of the IRB.
DNA sequencing
Next-generation sequencing (NGS)-based gene panels and customized gene sets were curated by clinical phenotype, clinical heterogeneity, age of disease onset, mode of inheritance, degree of penetrance, and other relevant information. As described previously, we targeted gene sequences with oligonucleotide baits (Agilent Technologies, Santa Clara, CA; Roche, Pleasanton, CA; Integrated DNA Technologies, Coralville, IA) to capture the exons, ±10–20 bases flanking intronic sequences, and certain non-coding regions of clinical interest.22,23 Targeted regions were sequenced to a minimum depth of 50× and an average depth of 350× read coverage at each nucleotide position in the reportable range. All sequencing was performed on Illumina HiSeq or NovaSeq instruments (Illumina, San Diego, CA).
Bioinformatics, predictive algorithms, and variant interpretation
The bioinformatics pipeline combined a suite of community-standard and custom-developed algorithms to simultaneously identify single nucleotide variants, small indels, large indels, structural variants that have breakpoints within targeted sequences, and exon-level copy number variants (i.e., deletions and duplications).22,23 Truncating variants comprised stop-gain and frameshift variants. Effects of missense variants were predicted by PolyPhen2, SIFT, and AlignGVGD. Loss of a canonical splice site (i.e., ≥15% decrease in splice site score compared with reference) was predicted by MaxEntScan (MES)24 and Splice Site Finder-like (SSF-like), per prior demonstrations of high sensitivity and specificity for these tools and thresholds.25,26 Gain of a splice site was predicted by the splicing module of Alamut (Interactive Biosoftware, Rouen, France) when two or more algorithms met the significance thresholds for the variant sequence but not for the reference sequence: MES score > 0, SSF-like score > 70, and Splice Site Prediction by Neural Network (NNSPLICE)27 score > 0.4. Cryptic site activation was predicted when two or more algorithms met the significance thresholds and scores for the variant sequence were >10% higher than for the reference sequence, per Alamut software documentation.28
For this study, we considered three classes of splicing variants: (1) variants at the ESS, including all variants found at the highly conserved dinucleotide splice sites flanking the beginning and end of each intron (±1–2 bp intronic); (2) variants located in the splice site region near the ESS, specifically all variants at ±3–8 bp intronic and ±1–3 bp exonic positions; and (3) clinically relevant splicing variants >8 bp into an intron or >3 bp into an exon that were either described in the literature or identified internally by MES, NNSPLICE, and SSF-like.
All pathogenic (P) variants, likely pathogenic (LP) variants, and VUSs identified in the clinical cohort were considered “observed” variants. Some variants were present in more than one individual and were therefore counted more than once in analyses of observed variants. “Unique” variant analyses removed any such duplicates.
ACMG/AMP sequence variant interpretation guidelines prescribe classifying variants into five tiers: P, LP, VUSs, likely benign (LB), and benign (B).13 We used Sherloc—a validated, semiquantitative score-based refinement of the ACMG/AMP guidelines—for clinical variant interpretation.29 Within Sherloc, we assigned each type of evidence (e.g., population frequency, functional data) predetermined points that were tallied to determine a variant’s clinical classification. Results from computational tools that predict the effects of variants on splice sites, or the effects of missense variants, were awarded a maximum of one point if there was consensus among the methods. P/LP classification required a minimum of four points accumulated from evidence indicating a pathogenic effect, and B/LB classification required a minimum of three points indicating a benign effect. Variants that did not reach the thresholds for LP or LB were categorized as VUSs.
For the purpose of this study, variants classified as P, LP, and VUSs were considered clinically reportable, but only those classified as P or LP were considered clinically significant.
Projection of reclassification rates following RNA analysis
As described above, Sherloc supports a semiquantitative approach to clinical interpretation of sequence variants by awarding points to each type of evidence applicable to a variant. After all available evidence is considered, some variants do not have sufficient points to reach classification as P/LP or B/LB but instead fall within a range of points that warrants a VUS classification. To estimate the potential for RNA analysis to provide useful evidence for clinical interpretation of variants within our large clinical cohort, we used a cautious approach for weighting evidence from RNA analysis. We awarded such evidence only supporting weight, which corresponded to one point in Sherloc. We identified all splicing variants classified as VUSs and separately determined the proportion that would reach a definitive classification (1) if one evidence point were added toward P/LP classification, to represent concordance between the DNA and RNA analyses, and (2) if one evidence point were added toward B/LB classification, to represent discordance. Projected evidence points were added separately in both directions because a given VUS may have evidence points on both the pathogenic and benign scales.
ClinVar analyses
ClinVar is a public database of sequence variants identified through literature review and clinical and research testing that typically have clinical classification(s) assigned by submitters.30 We evaluated ClinVar variant submissions from 95 clinical laboratories (excluding Invitae-only submissions) by using the data release from February 3, 2020. Because ClinVar-provided variant call format (VCF) annotations only include splicing variants at the ESS, we ran SnpEff to reannotate variants in ClinVar and identify splicing variants at both the ESS and non-ESS locations. We predicted splicing variants by location and gene annotation with SnpEff by using the Ensembl database GRCh38.86, filtering variant effect annotations to protein-coding transcripts specifically and identifying those annotated as “splice_donor_variant” (+1–2 intronic), “splice_acceptor_variant” (−1–2 intronic), or “splice_region_variant” (±3–8 bp intronic and ± 1–3 bp exonic positions). Resulting splicing variants were grouped by interpretation as (1) P/LP, when all entries were classified as P or LP; (2) B/LB, when all entries were classified as B or LB; (3) VUSs, when all entries were classified as VUSs; or (4) discordant, when ≥2 entries had conflicting interpretations.
Genome Aggregation Database analyses
Genome Aggregation Database (gnomAD) is an open-source database of 15,708 genomes and 125,748 exomes from sequencing studies of individuals without overt monogenic disease (v.2.0.2).31 The >8 million gnomAD variants included in this study comprised coding-region variants (including missense variants, truncating variants, silent variants, in-frame indels, and alterations to start and stop codons) and splicing variants in all canonical coding transcripts (i.e., CANONICAL = YES and BIOTYPE = protein_coding per the Ensembl Variant Effect Predictor, v.85).32 Variants of low quality (i.e., FILTER! = PASS) in either the exome data or the genome data were excluded from the analysis; only variants that were high quality in both datasets, or high quality in one dataset and absent from the other, were included. Variants with no population prevalence (i.e., allele count = 0) in both datasets were also removed from analysis.
Splicing variants analyzed in this study were based on existing annotations by the Ensembl Variant Effect Predictor and included “splice_donor” (+1–2 intronic), “splice_acceptor” (−1–2 intronic), and “splice_region” (±3–8 bp intronic and ±1–3 bp exonic positions) variant consequences. Predicted loss-of-function variants included stop-gain or frameshift variants and ESS variants. We computed the total number of variants at the ESS and non-ESS locations for all protein-coding transcripts as well for a restricted set of 5,951 genes currently associated with monogenic disease (i.e., the “Mendeliome”) as curated from the Online Mendelian Inheritance in Man (OMIM) database.33 In addition, we examined the distribution of splicing variants in genes in the Mendeliome with respect to modes of inheritance (i.e., autosomal dominant, autosomal recessive, autosomal dominant and recessive, X chromosome linked, and Y chromosome linked). Minor allele frequencies for splicing variants in gnomAD were determined by the maximum credible allele frequency via the “popmax” filter and grouped as common (>1%), rare (0.1%–1%), and very rare (<0.1%).
Statistical analyses
Comparisons were performed with Mann-Whitney tests (Wilcoxon rank sum tests) where applicable, and statistical significance was set at p < 0.05.
Results
Splicing variants in the clinical cohort
We first evaluated a cohort of 689,321 individuals who underwent clinical genetic testing and explored the distribution of observed splicing variants by location within genes, clinical interpretation, and association with a variety of hereditary diseases. The data in this study were derived from a combined equivalent of 26,893,248 single-gene tests. Among all observed clinically reportable variants (i.e., P, LP, or VUSs), 13.0% were splicing variants, 72.3% were missense variants, and 13.8% were truncating variants (Table 1). Among the observed splicing variants, 16% were at the ESS; the majority of splicing variants at non-ESS locations were within exons (Figures 1 and 2A). A vast majority of the 1,732 genes sequenced in the cohort had at least one splicing variant observed: 1,298 had at least one non-ESS variant, 721 had at least one ESS variant, 655 had at least one of each, and 368 had none.

| Variant class | Invitae—observed P/LP/VUS variants (N = 466,736), No. (%) | Invitae—unique P/LP/VUS variants (N = 149,139), No. (%) | Invitae—patients (N = 689,321), No. (%) | ClinVar P/LP/VUS variants (N = 229,329), No. (%) | gnomAD variants in protein-coding genesa (N = 8,795,492), No. (%) |
|---|---|---|---|---|---|
| Splicing variants | 60,807 (13.0) | 22,344 (15.0) | 52,047 (7.6) | 23,041 (10.1) | 825,992 (9.4) |
| Splicing VUSs | 42,534 (9.1) | 16,965 (11.4) | 37,064 (5.4) | 11,116 (4.8) | N/A |
| Splicing VUSs that RNA analysis may reclassify | 13,281 (2.8) | 5,200 (3.5) | 12,013 (1.7) | N/A | N/A |
| Missense variants | 337,649 (72.3) | 110,774 (74.3) | 219,515 (31.8) | 115,571 (50.4) | 5,152,451 (58.6) |
| Truncating variants | 64,472 (13.8) | 16,806 (11.3) | 58,815 (8.5) | 35,985 (15.7) | 396,944 (4.5) |
Columns do not add up to 100% because some variants that fit multiple categories are counted more than once, while other variants (e.g., copy number variants and in-frame indels) are only represented in the total N. ClinVar data include submissions with conflicting interpretations and exclude Invitae submissions. gnomAD, Genome Aggregation Database; P/LP, pathogenic/likely pathogenic; VUS, variant of uncertain significance.
a Includes missense variants, truncating variants, silent variants, in-frame indels, alterations to start and stop codons, and splicing variants (to ±8 bp intronic) in all canonical coding transcripts.

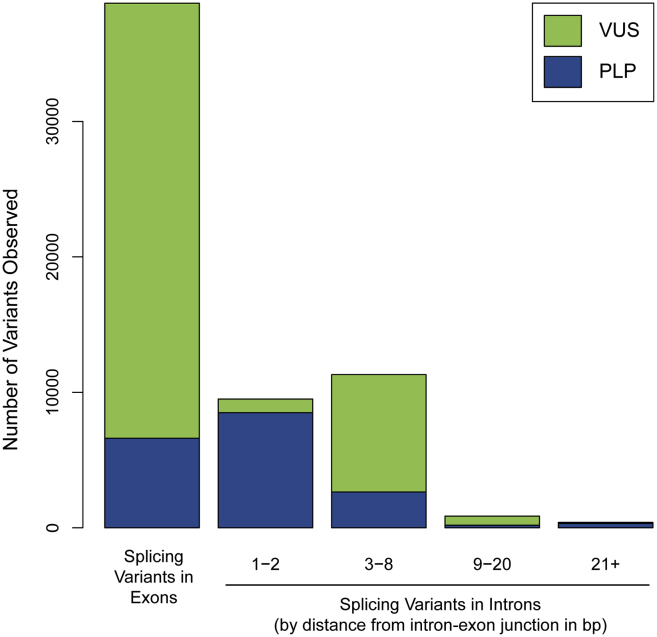
Clinically classified splicing variants in a large clinical cohort
Number of splicing variants at exonic or intronic positions indicated among 689,321 individuals tested for a variety of inherited diseases. All exonic splicing variants are grouped together; the intronic splicing variants are grouped by distance from the intron-exon junction in base pairs (bp). Note that intronic variants more than 10 bp from the intron-exon junction may not be detected because of reportable range of the sequencing assay; therefore, splice variants ±10 bp intronic are most likely underrepresented.
P/LP, pathogenic/likely pathogenic; VUS, variant(s) of uncertain significance. Colors within each bar indicate the number classified as P/LP (blue) or VUSs (green).

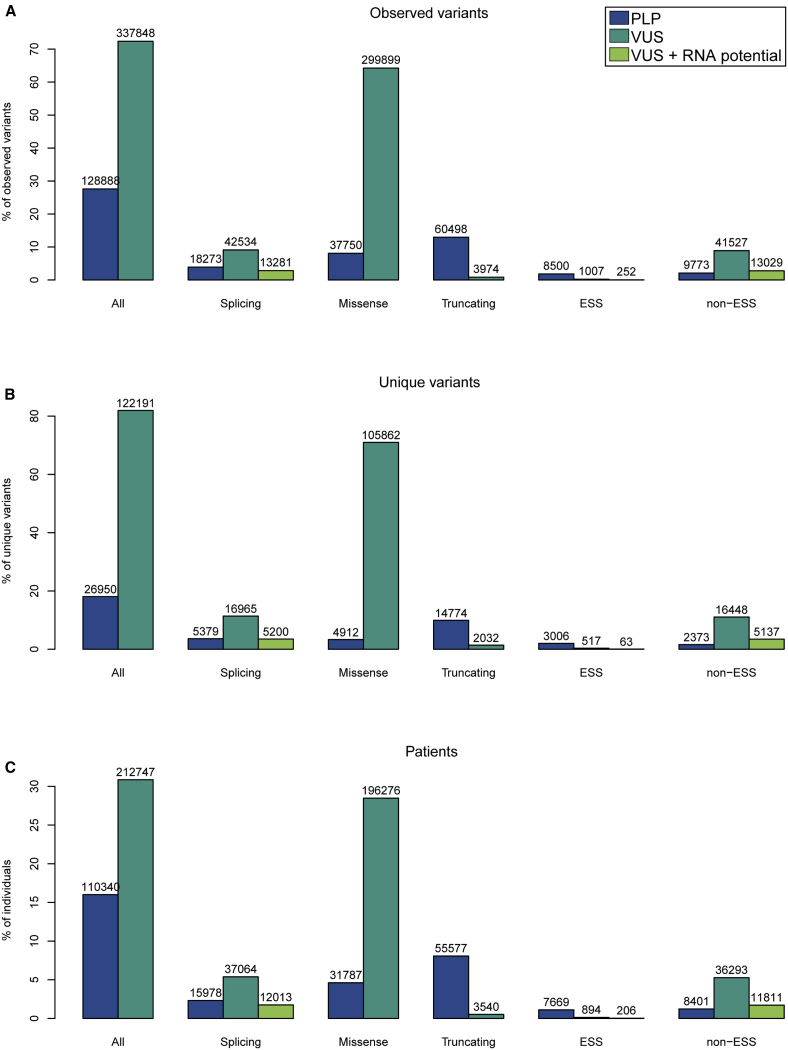
Distribution of variant types and their clinical classifications in a clinical cohort of 689,321 individuals tested for genetic disease
(A–C) Number and proportion of variants by type and clinical classification among (A) all observed variants, (B) unique variants, and (C) patients. Splicing variants are shown both as a group and split into ESS and non-ESS variants. VUS + RNA potential indicates splicing VUSs that have the potential to be reclassified with the addition of evidence from RNA analysis; these are included in the splicing VUSs total.
ESS, essential splice site.
We next examined 22,344 unique reportable splicing variants in the clinical cohort. One-fourth (24.1%) were classified as P/LP and the remaining 75.9% were VUSs (Figure 2B). In contrast, the vast majority (88%) of truncating variants and only a small minority (4.4%) of missense variants were classified as P/LP. Most of the unique splicing variants (84%) were outside the ESS. The majority (85%) of unique ESS variants were classified as P/LP and the majority (87%) of non-ESS variants were classified as VUSs, consistent with the ACMG/AMP guidelines specifying that variants at the ESS should be awarded weight as very strong evidence toward pathogenicity, while splicing variants outside the ESS warrant only supporting weight as evidence in the absence of relevant functional data. However, we also observed a small subset of 517 unique ESS variants that were classified as VUSs (Figure 2B) and, in a separate analysis of the clinical cohort, another ten that were classified as B/LB (data not shown). In general, these classifications were due to relatively high allele frequency in the general population or known or inferred functional effects such as in-frame changes, alternate skipping of exons, or escape from nonsense-mediated decay.
Potential reclassifications with RNA analysis
To estimate the proportion of unique splicing VUSs that could potentially be reclassified to LP or LB with additional evidence from RNA analysis, we established that such data would be awarded one point in Sherloc. Most splicing VUSs in our clinical cohort were not at the ESS and therefore, even if they were to correlate with abnormal results from RNA analysis, various considerations related to quantitative and tissue-specific expression precluded awarding more weight toward pathogenicity by default. On the basis of this approach, we projected that up to 5,200 (31%) of the 16,965 unique splicing VUSs in our cohort could reach LP or LB classifications with evidence from RNA analysis (Table 1). Given that RNA analysis may either corroborate or refute a computational prediction of altered splicing, we evaluated what proportion of splicing VUSs would be reclassified to LB or LP if an evidence point toward pathogenicity and, separately, an evidence point toward benign effect were applied. Data from our clinical cohort suggested that 4,851, or 93%, of the reclassifications would result in LB classifications and the remaining 389, or 7%, would be LP classifications. Notably, evidence from RNA analysis would not be sufficient to reclassify most splicing VUSs (69%) because of the absence of other types of applicable evidence for 11,765 of the 16,965 unique splicing VUSs.
Overall, of the 122,191 unique VUSs in our cohort that could be studied further to resolve their clinical significance, 16,965 (13.9%) were splicing VUSs, 105,862 (86.6%) were missense VUSs, and 2,032 (1.7%) were truncating VUSs (Figure 2B). Given these proportions, the 5,200 unique splicing VUSs in this cohort that could potentially be reclassified with RNA analysis accounted for 4.3% of unique VUSs of all types.
Given that reclassification of each unique splicing VUS could impact genetic testing results for multiple people, we also estimated the impact of RNA analysis across individuals. Among the 689,321 individuals in the cohort, 37,064 (5.4%) had a splicing VUS, whereas 196,276 (28.5%) had a missense VUS and 3,540 (0.5%) had a truncating VUS (Figure 2C). If RNA analysis were to provide informative data to reclassify the splicing VUSs, we estimated that 917, or 0.1%, of all 689,321 individuals tested would receive an updated result from VUS to LP and 11,273, or 1.6%, would receive a downgraded result from VUS to LB (Table S1). When only the 37,064 individuals with splicing VUSs were considered, we found RNA analysis could reclassify variants to LB or LP for up to 12,013 (32%) (Table S1). Thus, RNA analysis could potentially result in a clinically significant reclassification (VUS to LP) for 917 (2.5%) of individuals with a splicing VUS (Table S1).
Distribution of splicing variants across clinical areas
Individuals in the clinical cohort were tested for different sets of genes depending on their clinical presentation, which allowed us to examine the distribution of splicing VUSs across nine clinical areas to gain insight into the extent to which RNA analysis may be useful for different disease genes. The percentage of individuals with clinically reportable splicing variants ranged from 2.2%–18.2% (mean, 7.6%) across the clinical areas (cancer, cardiology, dermatology, hematology, immunology, metabolism, neurology, ophthalmology, and pediatrics); splicing VUSs specifically were found in 0.9%–14.3% (mean, 5.4%) of individuals (Table S1). Within genes related to cancer (the clinical area with the most individuals tested), splicing variants were found in 5.9% of individuals. In comparison, a higher proportion of individuals (12.8%–18%) who were tested in the clinical areas of pediatrics, neurology, and immunology had splicing variants, most likely because considerably more genes (and intron-exon junctions) were tested. The proportion of individuals harboring splicing VUSs that could be potentially reclassified with RNA analysis also varied by clinical area, ranging from 0.2% in hematology to 5.6% immunology (Table S1).
ClinVar splicing variants
We next queried ClinVar to explore the characteristics of splicing variants reported by other clinical genetic testing labs (Table S2). Splicing variants accounted for 10.1% of all clinically reportable entries in ClinVar, distributed across >11,000 genes and reported by 95 clinical laboratories, excluding our own submissions (Table 1). Of the 23,041 splicing variants, 38.1% were at the ESS, while the remaining 61.9% were at non-ESS locations. ClinVar P/LP splicing variants were mostly within the ESS (84.7%), while splicing VUS were mostly at non-ESS locations (69.6%) (Tables S2 and S3). About half (48.2%) of all splicing variants in ClinVar were VUSs and another 7.3% had discordant interpretations (Table S2).
When all 229,329 clinically reportable variants in the ClinVar dataset were considered, only 11,116 (4.8%) were splicing VUSs and thus potentially eligible to test for reclassification with RNA analysis (up to 12,799, or 5.6%, could be eligible if discordant interpretations were also considered eligible). However, the majority of these are unlikely to be resolved with RNA analysis unless yet another category of evidence is applied. Among the limited B/LB submissions to ClinVar (laboratories do not routinely submit B/LB variants), roughly a quarter were splicing variants and the vast majority were outside the ESS (Table S4).
Splicing-related natural variation in the human genome
In some instances, splicing variants may be rare polymorphisms that create viable alternative transcripts that have not yet been recognized, or they may be clinically significant variants present in healthy individuals as carrier or low-penetrance alleles. To investigate splicing variants within naturally existing variation in the human genome, we examined their occurrence in exome or genome sequences from healthy individuals in gnomAD and compared their prevalence to that of other variant types commonly identified in clinical testing. Splicing variants accounted for 9.4% of all variants in protein-coding genes in gnomAD (Table 1), and there was a distribution of 0–1,594 (mean = 42.0, median = 29) splicing variants per gene. In comparison, missense variants accounted for 58.6% of all coding variants, and truncating variants accounted for 4.5% (Table 1). Thus, the majority of variants were missense variants (Figure 3A). With respect to location, 11.6% of splicing variants in gnomAD were at the ESS and 88.4% were at non-ESS locations. The overwhelming majority (92.3%) of splicing variants were very rare with allele frequencies < 0.1% among different subpopulations represented in gnomAD (Figure 3B) (the majority of missense variants [93%] and truncating variants [96%] were also very rare). Splicing variants at the ESS, which typically act as loss-of-function changes, comprised 19.4% of all predicted loss-of-function variants (including nonsense and frameshift variants) in gnomAD, although some annotations of predicted loss-of-function effects (i.e., splice_donor, splice_acceptor, stop_gain, and frameshift) in gnomAD may be due to sequencing artifacts or may indicate variants that are already known not to have loss-of-function effects.7 Those classified as B/LB in ClinVar were also present in gnomAD and uniformly had high allele frequencies.

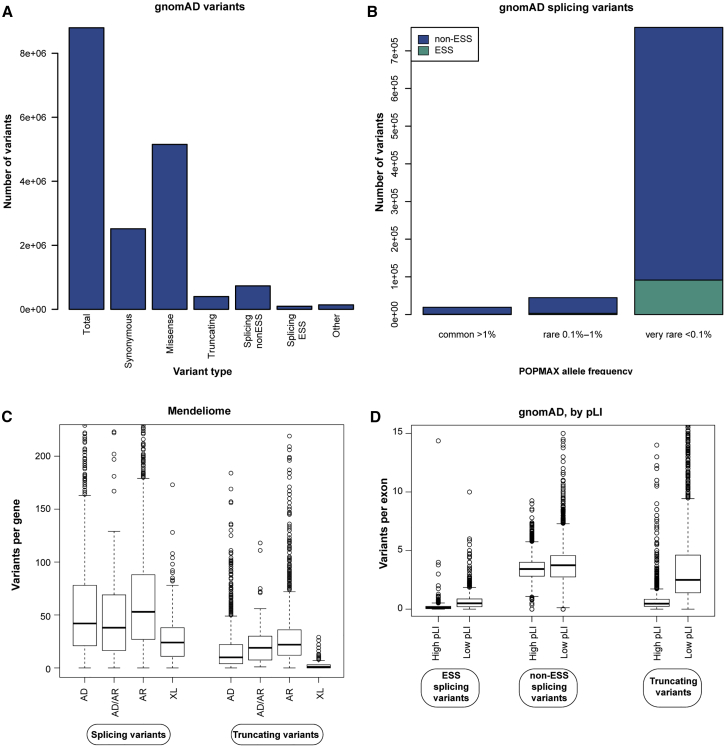
Frequencies of splice variants in the healthy human genome
Splicing variants were identified in gnomAD (v.2.0.2) via the Ensembl Variant Effect Predictor (v.85).
(A) Bar graph indicating the absolute number of variants identified in gnomAD within coding regions and ±8 bp of intronic sequence. Splicing variants include variants at the ESS (at intronic positions ±1–2) and at non-ESS locations (at intronic positions ±3–8 bp and exonic positions ±1–3 bp). Other includes in-frame indels and alterations to stop and start codons.
(B) Allele frequencies for splicing variants as determined by “popmax” and grouped as common (>1%), rare (0.1%–1%), and very rare (<0.1%).
(C) Outlier boxplot showing the distribution of splicing and truncating variants among hereditary disease genes by inheritance patterns.
(D) Outlier boxplot showing the distribution of splicing and truncating variants in exons of all gnomAD genes with high pLI scores (pLI > 0.9) and low pLI scores (pLI ≤ 0.9).
AD, autosomal dominant; AR, autosomal recessive; ESS, essential splice site; gnomAD, Genome Aggregation Database; pLI, probability of loss-of-function intolerant; XL, X-linked.
We also examined splicing variants in gnomAD across 5,951 genes associated with monogenic disease (referred to as the Mendeliome). Roughly a quarter (25.8%) of all splicing variants in gnomAD were in the Mendeliome and the vast majority (89.5%) were at non-ESS locations. Further, splicing variants accounted for 9.1% of Mendeliome variants, whereas missense and truncating variants accounted for 50.9% and 3.4%, respectively (Table S5). In addition, splicing variants at the ESS represented 21.9% of all predicted loss-of-function variants in the Mendeliome. The vast majority (92.6%) of splicing variants in the Mendeliome were very rare with frequencies < 0.1% among all subpopulations. Splicing variants within the Mendeliome were enriched in genes associated with autosomal recessive inheritance when compared with genes associated with autosomal dominant inheritance (mean splicing variants per gene = 67.8 versus 60.1, Wilcoxon p value < 0.01). Relatively few splicing variants were found in X-linked disease genes (mean splicing variants per gene = 30.5, Wilcoxon p value < 0.01). Lastly, splicing variants in the Mendeliome were uniformly more prevalent than truncating variants, and both were more common in genes associated with recessive disorders than those associated with dominant or X-linked disorders (Figure 3C).
High probability of loss-of-function intolerant (pLI) scores in gnomAD indicate that a gene has a high likelihood of being intolerant to loss-of-function mutations.34 As expected, splicing variants appeared to be depleted in genes with high pLI scores (mean = 3.6 per exon for pLI > 0.9, scale 0–1) when compared with those with moderate or low pLI scores (mean = 4.2 per exon for pLI < 0.9, Wilcoxon p value < 0.01) (Figure 3D), consistent with findings in a recent study.7 Regardless of whether a variant was at an ESS or had a truncating effect, genes with high pLI scores had fewer of these variant types than genes with low pLI scores (Figure 3D). Among genes with high pLI scores and associated with autosomal dominant disorders, 12 had ≥1 ESS variants but no truncating variants. All ESS variants were very rare (i.e., <0.1% population frequency); upon further inspection, the majority appeared to be flagged in gnomAD as “LC_LoF” by LOFTEE, suggesting that they were most likely artifactual annotations. Finally, ten genes that were annotated as high pLI and associated with autosomal dominant inheritance had no ESS variants or truncating variants at all.
Discussion
The large clinical cohort in this study revealed the prevalence of splicing variants relative to other variant types encountered in genetic testing, highlighted the extent to which clinical interpretation can be ambiguous among all types of variants, and specifically helped estimate the proportion of splicing VUSs that may reach a definitive clinical classification with the addition of RNA analysis as interpretation evidence. RNA analysis will be increasingly used to uncover clinically important variants that remain elusive through traditional clinical DNA sequencing. As an early step toward better understanding its use in germline genetic testing, this study quantified how and when RNA analysis can be useful. Our results suggest that RNA analysis will be especially useful for resolving splicing VUSs outside the ESS because any consensus predictions of abnormal splicing for these variants would only be considered supporting, but not strong, evidence in the variant interpretation process, per ACMG/AMP standards. RNA analysis could provide additional corroborating evidence to definitively classify these variants as disease causing or not. By contrast, splicing VUSs found at the ESS already have a high probability of pathogenicity because they affect a critical target of the splicing machinery and are therefore awarded greater weight toward pathogenicity. RNA analysis will largely serve to confirm or refute splicing defects associated with variants at these highly conserved sites.
Although computational tools may be able to predict splicing changes caused by novel variants, how those variants alter protein function and explain disease is often unclear. In our analysis, most splicing variants in gnomAD had very low allele frequencies, which would preclude their unambiguous classification as benign based on population frequencies alone. In addition, in both the clinical cohort and gnomAD, most splicing variants were outside the ESS; thus, their functional effects were not as predictable as those of variants at the highly conserved ESS. As a result, most splicing variants in our clinical cohort had been classified as VUSs because the available evidence did not clearly support or refute involvement in hereditary disease. Even some variants at the ESS can be classified as VUSs (such as the 517 unique variants at the ESS in our clinical cohort) if sufficient protein function appears to be retained through in-frame exon skipping or other mechanisms. In other cases, variants at the ESS can even be classified as benign; within our clinical cohort, at least ten unique ESS variants had been classified as B/LB because of evidence such as premature truncation near the end of the gene that would allow escape from nonsense-mediated decay (e.g., in CARD9 [MIM: 607212]), exon skipping consistent with known alternative splicing (e.g., in PKP2 [MIM: 602861]), and high population allele frequency that exceeds disease prevalence (e.g., in DMD [MIM: 300377]) (internal observations).
This study adds to a growing awareness of the significant challenges of clinical interpretation of splicing variants observed in individuals with suspected hereditary disease. For instance, recent reports describe downgrading splicing variants at the ESS in BRCA1 and BRCA2 from P/LP to VUSs or B/LB because of evidence from the Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA) consortium and functional studies showing that some ESS variants result in viable, functional transcript isoforms.6,35, 36, 37 In addition, two clinical laboratories recently reported discordant interpretations of an intronic variant in BRCA2. One lab classified the variant as LP because of its low prevalence, in silico predictions of splicing defects, and RNA analysis demonstrating altered splicing (but without quantification).18 The other lab recently downgraded this same variant from VUS to B on the basis of functional evidence of a partial splicing defect, clinical and family histories of individuals with this variant, and co-occurrence with other known pathogenic variants in trans in BRCA2. Both labs classified the variant in accordance with ACMG/AMP guidelines.5,18 Although such examples are expected to be infrequent, they nevertheless have significant implications for individuals who carry these types of variants and may undertake irreversible clinical actions. Furthermore, the challenge of interpreting splicing variants is not limited to hereditary cancer and has been noted in other disease areas, such as inherited retinal diseases.38 To avoid negative outcomes, it is essential to consider a variety of types of evidence for variant interpretation and to use appropriate control samples during RNA analysis.
Compounding the challenge of accurately interpreting the effects of a splicing variant is the conundrum of defining which one or a subset of transcript isoforms may be affected. This has implications for identifying molecular etiologies of disease through genetic testing because clinical laboratories may sometimes choose a single reference transcript when reporting observed variants. In some cases, the chosen reference transcript may not be fully relevant to the disease in question (and the prediction of splicing effects can be dependent on the transcript chosen), leading to missed diagnoses.8 It is expected that clinical interpretation of variants identified during genetic testing for inherited disease will eventually consider the individual expression patterns of specific transcripts for each disease gene and how those patterns may affect the manifestation of disease. Various studies have shed light on the complexity of gene expression through discovery of novel exons,39 complex interactions between splicing variants and tissue-specific effects,40 the effect of cellular context on splicing in the absence of variants that affect splicing (e.g., in the progeria-related LMNA gene [MIM: 150330]),41 and quantitative effects of transcript isoforms on clinical phenotypes.7,42
Although individual discoveries and databases such as the Genotype-Tissue Expression Project (GTEx) are greatly improving our understanding of the qualitative and quantitative nature of gene expression, that understanding still remains far from complete.43 Given this and the aforementioned challenges, our Sherloc variant interpretation framework based on the ACMG/AMP guidelines awards a single point to such data and only considers them complementary to other lines of evidence in supporting a final clinical classification for sequence variants. From exploring the utility of adding RNA analysis as a routine component of clinical testing, we estimated that if data from RNA analysis were to complement clinical DNA sequencing of gene panels to reclassify a splicing VUS on the basis of ACMG/AMP guidelines, an additional 0.1% of individuals in our clinical cohort would receive a clinically significant result and an additional 1.6% would receive a negative result.
Our estimate of the potential utility of RNA analysis closely matched that of a recent study reporting that RNA sequencing (RNA-seq) would change VUSs to P/LP or B/LB in 0.7% of 1,000 individuals undergoing diagnostic testing of 18 hereditary cancer genes.44 Our methods produced a strikingly similar result: among 244,871 individuals in our clinical cohort who were tested for the same 18 genes, we projected that RNA analysis would change VUSs to P/LP or B/LB in 0.68% of them (data not shown).
Another challenge in interpreting splicing variants is the uncertainty of their effects in ostensibly healthy populations. Public sequence repositories such as gnomAD provide a critical source of evidence for interpreting variants in clinical laboratories. The observation that a variant is rare or absent from gnomAD is typically used as evidence supporting its pathogenicity when observed in an individual with suspected hereditary disease. However, our gnomAD analyses suggest that some ESS variants, like other loss-of-function variants, may be tolerated because they are observed in healthy individuals (although others may be challenging to differentiate from annotation errors). We observed a dozen rare ESS variants in presumably dosage-sensitive genes (i.e., those with high pLI and autosomal dominant inheritance) in gnomAD v.2.0.2. When inspecting these variants further, we found that many were secondarily labeled as low-confidence by the gnomAD tool LOFTEE, which can indicate an annotation error. Moreover, within the gnomAD web interface (v.2.1.1), additional variants had been flagged by LOFTEE or other quality metrics, and some had been removed when compared with the v.2.0.2 dataset, supporting our suspicion that most of these variants are tolerated. These observations fit with the results of a recent study that used quantitative tissue expression profiling to identify falsely annotated loss-of-function variants in haploinsufficient disease genes in gnomAD.7 These findings emphasize the need for clinical genetic laboratories to be aware of potential shortcomings in large, public control datasets and to proceed cautiously with clinical interpretation of splicing variants, even if they are in dosage-sensitive genes. Tools such as LOFTEE and expression-aware annotation of gnomAD variants may significantly aid in this effort.
In the context of interpreting novel sequence variants predicted to affect splicing in known disease genes, we awarded only modest weight to RNA analysis evidence in Sherloc. This is because transcript data may not always reflect actual protein function and because other types of evidence should also be considered in final variant classification. More weight can be granted to functional evidence generated from rigorously designed methods, such as well-controlled saturation mutagenesis studies, or from methods that reflect the biological environment, such as enzymatic activity assays or in vivo animal models of disease.45,46 The uniform use of data from RNA analysis as reliably strong evidence in variant interpretation will require a comprehensive and quantitative atlas of tissue and temporal gene expression patterns and a thorough understanding of the redundancy between transcript isoforms in healthy individuals and those affected by hereditary disease. This would improve interpretation of variant types beyond those affecting splicing because even disease-causing missense or other types of variants can be misidentified or missed altogether when the full complement of alternative transcripts and tissue expression patterns are not considered, and this can lead to false-negative or false-positive reports.8 This challenge can also be further compounded by difficulties in accessing relevant samples for RNA analysis, such as from brain tissue. Finally, until RNA analysis becomes standardized in clinical genetic testing and until professional practice guidelines specify how to weigh the evidence, laboratories will most likely vary in how they incorporate RNA data into variant interpretation.5
Notwithstanding the challenges mentioned above, RNA analysis is a tool that will be increasingly and necessarily used in germline genetic testing. Diagnostic and screening sequencing panels continually expand in gene content, increasing the number of VUSs overall and the number of splicing VUSs detected specifically, thus offering more opportunities for using RNA analysis to resolve their clinical significance. RNA analysis may also help resolve the clinical significance of some missense variants that have a more adverse effect on gene function through a splicing effect rather than through an amino acid change. Moreover, RNA analysis can identify deep-intronic variants that may be missed by standard clinical DNA sequencing, which typically captures only 10–20 bp of noncoding sequence beyond the intron-exon junction. As a result, some clinically relevant, deep-intronic variants have been reported. By our analysis, across 84 hereditary cancer genes in ClinVar, 15 P/LP intronic variants were between 10–20 bp into an intron and 12 P/LP intronic variants more than 20 bp into an intron, together accounting for 0.04% of all P/LP variants. Deep-intronic variants that affect splicing are not commonly reported in other types of genes because those regions are not routinely sequenced and methods to interpret their effects are not yet robust.12 Still, these variants are important for molecular diagnosis and attendant clinical management for some patients.14,47, 48, 49 RNA analysis can also be useful for determining the clinical significance of variant types beyond splicing, such as some truncating sequence variants and even intragenic copy number variants and other structural rearrangements.
Leveraging data from our nearly 700,000-person clinical cohort, this study collectively provides a clearer view of the frequencies of splicing variants across a wide range of genes in clinically affected individuals, the extent to which RNA analysis may provide useful evidence toward understanding the clinical significance of splicing variants, and an estimate of their contribution to normal variation in the human genome. Our projection of the utility of RNA analysis to reclassify splicing VUSs was aided by Sherloc’s semiquantitative, points-based interpretation framework, which objectively incorporates several types of evidence (i.e., population, clinical, computational, and functional). There are, however, limitations to our study that should be noted. First, we chose to restrict our analyses to only those sequence variants with computational predictions of abnormal splicing effects. This underestimates the utility of RNA analysis because experimental discovery of deleterious intronic variants in inherited genetic diseases is still in an early stage and because evidence is mounting that databases of clinically relevant genomic variation are depleted at present for intronic variants that flank the ESS or are farther away in deeper intronic sequences.12 Indeed, the clinical data presented in this manuscript do not broadly address splicing variants found more than 10 bp from intron-exon junctions because, as is typical in clinical genetic testing laboratories, our standard sequencing assay used for the majority of individuals in our cohort only addressed variants within 20 bp of exons for hereditary cancer genes and 10 bp of exons for other genes. Further, computational tools for predicting splicing variants can occasionally yield false negatives, so some true splicing variants may have been missed.11 In addition, although RNA analysis can help determine the consequences of other types of variants, our study did not address this. A second limitation of our analyses was that we assumed that each computational prediction of a splicing variant will be supported by reliable data from RNA analysis. As a result, our calculations most likely overestimate the utility of RNA analysis for resolving known splicing VUSs. In contrast, at least for some splicing variants and within specific genes, RNA analysis will be validated to yield reproducibly reliable information; evidence from that analysis will be given more weight in clinical variant interpretation, thus raising the number of instances in which RNA analysis can facilitate reclassification from VUS to LP or LB. A third limitation of this study was the use of an unselected cohort of clinically affected individuals referred for testing, which may have inherent demographic or other biases. For instance, some clinical areas such as cancer were overrepresented in the sample set and some genes were tested many more times than others; data from these genes should therefore be interpreted in that context. Finally, we only considered RNA analysis as a complementary method and not a primary testing method. As a result, splicing variants outside the typical sequencing range, as noted above, were not addressed in the projection. Future studies using whole-genome sequencing will help us better understand the true prevalence of these variants and address this limitation.
The expanding use of RNA analysis in hereditary disease testing will power both discovery and diagnosis, therefore improving the clinical sensitivity of genetic testing. For individuals that could benefit from RNA analysis, reaching a definitive diagnosis can be a profound outcome, particularly when it immediately improves clinical management. On a relative scale, because resolving the clinical significance of missense VUSs, rather than splicing VUSs, will naturally have a greater impact on reducing uncertainty in clinical genetic testing (our clinical cohort had more than five times more individuals with missense VUSs than those with splicing VUSs), several novel methods are being developed to understand the consequences of protein sequence changes. Likewise, a better understanding of transcript isoforms and their expression patterns will improve our ability to capture a broader spectrum of clinically relevant splicing variants. These advances have to occur within a guidance framework developed together by clinical laboratories and professional societies to support consistency in the methodology used to detect and clinically interpret splicing variants.
Declaration of interests
All authors are employees and stockholders of Invitae Corporation.
References
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
44
45
46
47
48
Data and code availability
The variants observed in the clinical cohort are all available in ClinVar: https://www.ncbi.nlm.nih.gov/clinvar/submitters/500031/.
Web resources
Genome Aggregation Database, https://gnomad.broadinstitute.org/
Online Mendelian Inheritance in Man, https://omim.org
Acknowledgments
We are grateful to Heidi Rehm and Beryl Cummings for their thoughtful and constructive feedback on this manuscript. We also thank Kerry Aradhya for scientific editing of this manuscript.
 Spectrum of splicing variants in disease genes and the ability of RNA analysis to reduce uncertainty in clinical interpretation
Spectrum of splicing variants in disease genes and the ability of RNA analysis to reduce uncertainty in clinical interpretation