
The authors have declared that no competing interests exist.
Current address: Department of Microbiology and Immunology, Vagelos College of Physicians and Surgeons, Columbia University, New York City, New York, United States of America
Current address: Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, United States of America
- Altmetric
Malaria remains a major global health problem, creating a constant need for research to identify druggable weaknesses in P. falciparum biology. As important components of cellular redox biology, members of the Thioredoxin (Trx) superfamily of proteins have received interest as potential drug targets in Apicomplexans. However, the function and essentiality of endoplasmic reticulum (ER)-localized Trx-domain proteins within P. falciparum has not been investigated. We generated conditional mutants of the protein PfJ2—an ER chaperone and member of the Trx superfamily—and show that it is essential for asexual parasite survival. Using a crosslinker specific for redox-active cysteines, we identified PfJ2 substrates as PfPDI8 and PfPDI11, both members of the Trx superfamily as well, which suggests a redox-regulatory role for PfJ2. Knockdown of these PDIs in PfJ2 conditional mutants show that PfPDI11 may not be essential. However, PfPDI8 is required for asexual growth and our data suggest it may work in a complex with PfJ2 and other ER chaperones. Finally, we show that the redox interactions between these Trx-domain proteins in the parasite ER and their substrates are sensitive to small molecule inhibition. Together these data build a model for how Trx-domain proteins in the P. falciparum ER work together to assist protein folding and demonstrate the suitability of ER-localized Trx-domain proteins for antimalarial drug development.
One of the leading and persistent causes of childhood mortality in the world is malaria, which is caused by parasites from the genus Plasmodium. Unfortunately, the parasite has developed resistance to all available drugs, making the discovery of new drug targets and potential small molecule inhibitors of essential parasite biology a top priority. A critical pathway required for many different biological processes in the parasite is oxidative folding which requires members of the Thioredoxin (Trx) superfamily of proteins. But we know almost nothing about the function and essentiality of Trx-domain proteins that localize to the endoplasmic reticulum, the origin of the secretory pathway, within P. falciparum. Here we show that a network of Trx-domain containing proteins function together and are essential for parasite survival within human red blood cells. Further, we identify a small molecule inhibitor of the redox activities of these Trx-domain containing proteins. This study demonstrates the suitability of this pathway for future antimalarial drug development.
Introduction
Today, the majority of the world’s population lives at risk for contracting malaria, a disease caused by eukaryotic parasites of the genus Plasmodium, with P. falciparum causing the most severe forms of the disease [1]. In 2018, the world saw approximately 228 million cases of malaria resulting in more than 400,000 deaths. These numbers reflect a concerted effort to combat malaria in the past few decades, but progress has stagnated, with the numbers of malaria cases/deaths largely unchanged in recent years. A major impediment in the fight against malaria is the rise of drug-resistant—including multidrug-resistant—P. falciparum parasites, highlighting the continuous need for research into the biology of this major human pathogen and identification of promising drug targets.
The thioredoxin system and members of the Thioredoxin (Trx) superfamily of proteins have received interest as potential drug targets in Apicomplexan parasites, including in Plasmodium [2–5]. Members of the Trx superfamily typically contain at least one Trx domain with a “CXXC” active site. An oxidized Trx domain in which the active site cysteines have formed a disulfide bond can accept electrons to oxidize other proteins, and the sulfhydryl groups of a reduced Trx domain can donate electrons to reduce other proteins. As modulators of protein redox states, members of the Trx superfamily regulate diverse aspects of cellular biology. In Plasmodium, members of this superfamily are found in several cellular compartments [6]. Of these, the cytoplasmic Trx system has been most characterized in Plasmodium [7]. Within the parasite cytoplasm, Trx Reductase reduces Trx1, which in turn serves as a reductase for other proteins potentially involved in protein synthesis and folding, anti-oxidant stress response, carbohydrate and lipid metabolism, and several other processes [8,9].
An important subset of Trx superfamily members that has received little study in Plasmodium are those that localize to the endoplasmic reticulum (ER). Classically, Trx domains in the ER are used to regulate the redox state of cysteines in other proteins, and to facilitate oxidative folding (the formation, reduction, and isomerization of disulfide bonds in newly synthesized proteins) [10]. The P. falciparum ER functions in many essential processes during the asexual replication cycle—particularly by serving as the root of the parasite’s complex secretory pathway—and ER-localized members of the Trx superfamily likely play critical roles in supporting these functions. P. falciparum encodes an ER-localized Hsp40 chaperone with a C-terminal Trx domain known as PfJ2, as well as four members of the Protein Disulfide Isomerase (PDI) family of proteins, which typically use their Trx domains as oxidoreductases to assist folding proteins form their correct disulfide bonds [11,12]. PfJ2 bares homology to the mammalian chaperone ERdj5, whose Trx domains serve primarily to reduce other disulfide bonds in the ER, and exogenous overexpression shows ER localization of PfJ2 [13–16]. One member of the PDI family, PfPDI8, has been localized to the ER and found able to form and reduce disulfide bonds in folding proteins in vitro, including the parasite protein Erythrocyte-Binding Antigen 175 (EBA175) [12,17]. However, these proteins remain largely unstudied in the parasites, and their essentiality for P. falciparum survival within the host RBC and functions within the ER have not been investigated.
The ability to reduce disulfide bonds is critical within the oxidative environment of the ER, both for regulating protein function and for allowing correct disulfide pairs to form as proteins are folding [18]. We therefore chose the putative reductase PfJ2 for study, and we report here that PfJ2 is essential for the P. falciparum asexual lifecycle. We identify PfJ2 interacting partners and use a chemical biology approach to specifically identify those proteins which may be reduced by PfJ2. Surprisingly, these were found to be other Trx-domain proteins: PfPDI8 and PfPDI11. We demonstrate that PfPDI8 is also essential for the asexual lifecycle, and our data suggests that PfJ2 and PfPDI8 may work together with the Hsp70 PfBiP to promote protein folding in the ER. Additionally, we demonstrate that the redox interactions between these essential proteins and their substrates are disrupted by a small molecule inhibitor. Together, these data suggest a model in Plasmodium for oxidative folding, in which Trx-domain proteins and PfBiP cooperate to ensure proteins reach their native states, and we propose that the oxidative folding process of the P. falciparum ER is an exploitable drug target.
Results
PfJ2 is an essential, ER-resident Hsp40
PfJ2 is a putative ER-resident Hsp40 with a C-terminal thioredoxin (Trx) domain (Fig 1A and 1B). It has similarity to a reductase in the mammalian ER which has Trx domains following an Hsp40 J-domain, suggesting PfJ2 may catalyze disulfide bond reduction of client proteins [13,15,16]. To investigate PfJ2 function and its potential role in P. falciparum oxidative folding, we generated a PfJ2 conditional knockdown parasite line using the TetR-PfDOZI aptamer system (referred to as PfJ2apt hereafter). In this knockdown system, protein expression is regulated by the presence of anhydrotetracycline (aTc), with knockdown induced by removal of aTc [19] (Fig 1C). Using CRISPR/Cas9 genome editing, we modified the pfj2 locus to encode a 3xHA-tag immediately upstream of the ER-retention signal, as well as the regulatory aptamer sequences and a cassette to express the TetR-PfDOZI fusion protein (Fig 1D). Correct integration of the construct into the pfj2 locus was determined by PCR, and expression of HA-tagged PfJ2 was demonstrated via western blot (Fig 1D).

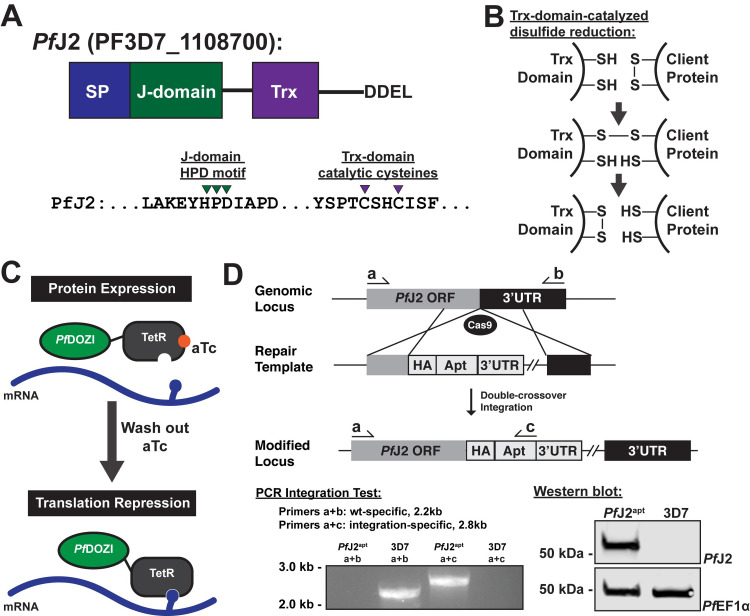
Generation of PfJ2 (PF3D7_1108700) conditional knockdown mutants using CRISPR/Cas9.
A) Predicted domain structure of PfJ2 showing signal peptide (SP), Hsp40 J-domain, thioredoxin domain (Trx), and C-terminal ER retention signal. Essential, conserved residues are shown: the J-domain HPD motif is required for Hsp40 activity (i.e., stimulation of Hsp70 ATPase activity), and the Trx-domain CXXC motif is required for redox activity. B) Mechanism of disulfide bond reduction catalyzed by Trx-domain active site cysteines. C) Regulation of protein expression using the TetR-PfDOZI knockdown system. TetR binds to aptamer sequences present in the mRNA, and PfDOZI localizes the complex to sites of mRNA sequestration, repressing translation. Anhydrotetracycline (aTc) blocks TetR-aptamer interaction. D) Schematic of CRISPR/Cas9-mediated introduction of the TetR-PfDOZI knockdown system into the pfj2 locus. A linearized repair template was transfected, along with a plasmid to express Cas9 and a gRNA, to introduce sequences for a 3xHA tag alongwith PfJ2’s DDEL ER retention signal, stop codon, and a 3’UTR. Included in the repair template was a cassette to express the TetR-PfDOZI fusion protein and blasticidin deaminase for drug selection. Bottom left: two PCR integration tests were used to amplify either a sequence from only wild-type locus (primers a+b) or the modified locus (a+c). Bottom right: anti-HA western blot. Representative western blot of three biological replicates shown.
Using an indirect immunofluorescence assay (IFA), we assessed the localization of PfJ2 throughout the asexual lifecycle (Fig 2A). Co-localization between PfJ2 and the ER-marker Plasmepsin V (PfPMV) revealed that PfJ2 is in fact an ER-resident protein, consistent with the localization of episomally overexpressed PfJ2 previously reported (Fig 2A) [14]. Using highly synchronized parasites, we showed that PfJ2 is primarily expressed in the trophozoite and schizont stages, and that during knockdown conditions (removal of aTc), PfJ2 expression is reduced (Fig 2B). Importantly, knockdown of PfJ2 was found to inhibit expansion of parasites in culture (Fig 2C). Consistent with peak PfJ2 expression during the trophozoite/schizont stages, we observed normal development of knockdown parasites in the beginning of the asexual life cycle, but the development of these parasites began to slow in the trophozoite stage and they failed to complete schizogony and produce new daughter parasites (Figs 2D and S1). These data demonstrate that PfJ2 is a bona fide ER-resident protein essential for progression through the P. falciparum asexual lifecycle.

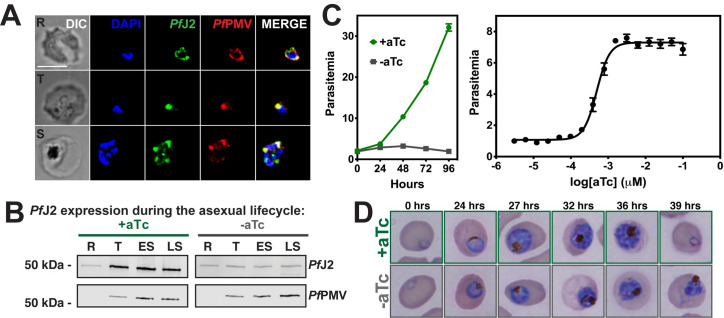
PfJ2 is an essential, ER-resident protein.
A) PfJ2apt parasites were fixed and stained with DAPI (blue) and with antibodies against HA (green) and the ER-marker PfPMV (red). Ring (R), Trophozoite (T), and Schizont (S) stage parasites are shown. Z-stack Images were deconvoluted and shown as a single, maximum intensity projection. Scale bar represents 5 μm. B) Parasites were tightly synchronized to the ring stage (0–3 hours) and split into two conditions: +aTc and–aTc. Samples were taken for western blot analysis at various time points in the life cycle (R = Ring, T = Trophozoite, ES = Early Schizont, LS = Late Schizont). Equal parasite equivalents were loaded into each lane, and membranes were stained with antibodies for HA and PfPMV. Shown is a representative experiment of two biological replicates. C) Left: asynchronous parasites were grown in normal (+aTc) or PfJ2 knockdown (-aTc) conditions, and parasite growth was monitored daily for 96 hours via flow cytometry. Right: asynchronous parasites were grown in a range of aTc concentrations and growth measured at 72 hours via flow cytometry. The aTc EC50 was determined to be 0.5 nM. Representative of three biological replicates shown for each growth curve. Each data point represents the mean of three technical replicates; error bars represent standard deviation. D) Parasites were tightly synchronized to the ring stage (0–3 hours) and split into two conditions: normal (+aTc) and PfJ2 knockdown (-aTc). Samples from each condition were smeared and field-stained at time points throughout the lifecycle. A representative experiment from three biological replicates is shown.
PfJ2 interacts with essential ER chaperones and proteins in the secretory pathway
As a putative chaperone possibly involved in ER oxidative folding, we reasoned that the essentiality of PfJ2 is likely related to its ability to interact with other proteins in the ER. We therefore took a co-immunoprecipitation (coIP) approach to identify PfJ2 interacting partners. PfJ2 was immunoprecipitated from PfJ2apt parasite lysates using anti-HA antibodies, and co-immunoprecipitating proteins were identified by tandem mass spectrometry (MS/MS) analysis. Control parental parasites (lacking HA-tagged PfJ2) were also used for immunoprecipitation and analyzed in the same manner. Each co-IP experiment was performed in triplicate, and the abundance of each identified protein was calculated by summing the total MS1 intensities of all matched peptides for each selected protein, and normalizing by the total summed intensity of all matched peptides in the sample (Fig 3A) [20,21]. Because PfJ2 is an ER-localized protein, we further filtered our list of interacting partners to those containing a signal peptide and/or at least one transmembrane domain (i.e. proteins putatively localized to the ER or trafficked through the secretory pathway). The filter ensures that our list is comprised of proteins that potentially localize or traverse via the cellular compartment where PfJ2 is located. We identified a stringent list of biologically relevant interacting partners as those proteins which were present in all three PfJ2apt coIP experiments, and were at least 5-fold more abundant compared to the controls, as previously described [21] (Fig 3B and 3C). A complete list of all identified proteins is provided in S1 Table.

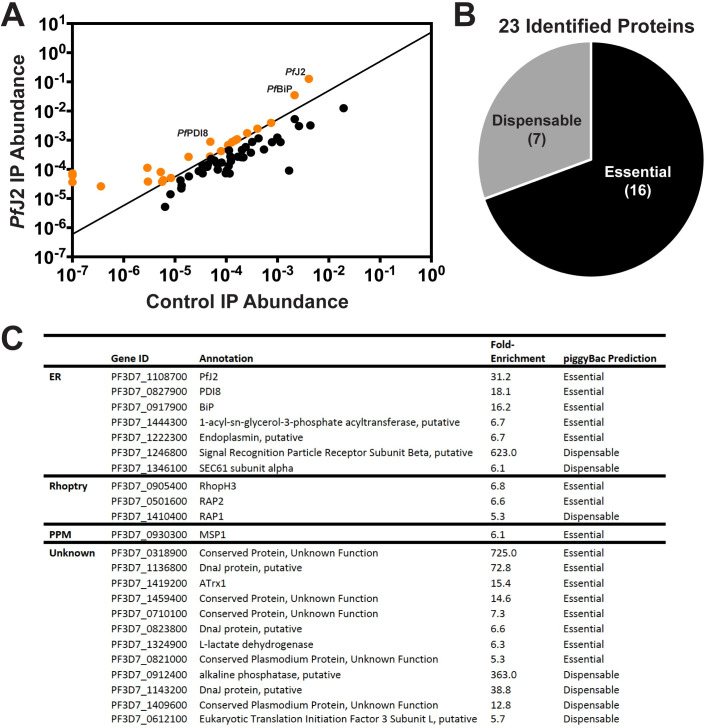
PfJ2 interacts with other essential chaperones, proteins in the secretory pathway.
A) PfJ2 was immunoprecipitated from PfJ2apt parasites using anti-HA antibodies, and co-immunoprecipitated proteins were identified by tandem mass spectrometry analysis. Control, parental parasites were also used for immunoprecipitation and analyzed in the same manner. Each coIP experiment was performed in triplicate, and the abundance of each identified protein was calculated as previously described in Boucher et al. 2018 and Florentin et al. 2020. Biologically relevant candidate proteins of interest were further identified as those potentially in the secretory pathway (predicted to contain a signal peptide and/or transmembrane domains) and those which were present in all three PfJ2apt replicates and demonstrated a 5-fold enrichment compared to control experiments (shown in orange). B) The 23 proteins meeting our strict criteria were assessed against the piggyBac mutagenesis screen performed in Zhang et al. 2018, and 16 were predicted to have essential functions in the P. falciparum asexual stages. C) Identified proteins were categorized by subcellular localization (ER, Rhoptry, Parasite Plasma Membrane [PPM], or Unknown). Also shown are GeneIDs and annotations from PlasmoDB.org, calculated fold-enrichment compared to control experiments, and essentiality as predicted by the piggyBac mutagenesis screen performed by Zhang et al., 2018.
We identified other conserved proteins classically involved in essential ER processes—such as the Hsp70 Binding immunoglobulin Protein (BiP), the Hsp90 Endoplasmin, and the oxidoreductase Protein Disulfide Isomerase (PDI). We further identified proteins that are trafficked through the ER late in the parasite lifecycle and are required for egress and invasion, including PfMSP1 and proteins destined for rhoptries [22–25]. Nearly half of the identified proteins lack empirical evidence for their subcellular localization, and many have no known function. But, given the presence of a signal peptide and/or transmembrane domains, these proteins likely have localizations in the ER, parasite plasma membrane, apicoplast, and other destinations that are part of the secretory pathway. Also of note, approximately two-thirds of the identified proteins are predicted to have essential functions [26]. These data together suggest that PfJ2 may work with other ER-resident chaperones to ensure proper folding/functioning of proteins that have essential roles throughout the parasite.
PfJ2 is a redox-active protein in the P. falciparum ER
We next sought to determine whether PfJ2 participates in disulfide exchanges with other proteins by chemically trapping these redox partnerships. To this end we employed the bifunctional, electrophilic crosslinker divinyl sulfone (DVSF), which shows remarkable specificity for nucleophilic cysteines, like those present in Trx domain active sites [27]. In Saccharomyces cerevisiae, DVSF was used to trap cytosolic Thioredoxin to two proteins known to exchange electrons with the CXXC active site, validating the compound’s ability to covalently and irreversibly trap Trx domains to their redox substrates [28]. DVSF was subsequently used to identify substrates of other redox-active proteins containing hyper-reactive cysteines in S. cerevisiae and human cells [29,30]. To determine whether DVSF was capable of trapping redox partnerships in P. falciparum, we treated PfJ2apt parasite cultures with DVSF and isolated proteins for western blot analysis. In the absence of DVSF, PfJ2 was detected at approximately 50 kDa, while the addition of DVSF resulted in additional bands containing PfJ2 to appear between 100–150 kDa (Fig 4A). To demonstrate specificity of DVSF for nucleophilic cysteines participating in disulfide exchanges, the ER-resident protein PfPMV—which contains 16 cysteines after its signal peptide—was probed for in the same samples and its migration pattern was found to be unaffected by DVSF treatment (Fig 4A). As an additional control, PfJ2apt parasite cultures were treated with the sulfhydryl-blocking compound N-ethylmaleimide (NEM) prior to the addition of DVSF. Pre-treatment with NEM resulted in the blockage of cross-linking between PfJ2 and its redox partners (Fig 4B). These results indicate that PfJ2 likely does participate in disulfide exchange, and that DVSF is a useful chemical tool for trapping redox interactions in the ER of P. falciparum.

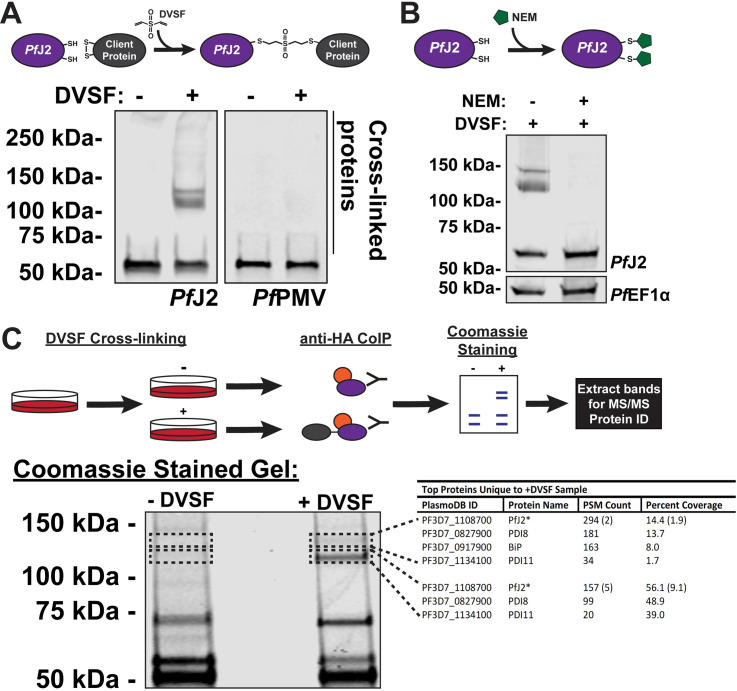
PfJ2 redox partners identified as PfPDI8 and PfPDI11.
A) PfJ2apt parasites were incubated with 3 mM divinyl sulfone (DVSF) in 1x PBS for 30 minutes at 37°C, then samples were taken for western blot analysis. Membranes were incubated with antibodies against HA and PfPMV. Representative western blot of three biological replicates is shown. B) PfJ2apt parasite cultures were incubated with 1 mM N-ethylmaleimide (NEM) for 3 hours prior to removal of NEM and addition of 3 mM DVSF as described above. Samples were taken for western blot analysis. Membranes were incubated with antibodies against HA and PfEF1α. Representative western blot of two biological replicates is shown. C) PfJ2apt parasite cultures were evenly split into two conditions: 3 mM DVSF or PBS only for 30 minutes at 37°C, after which parasite lysates were used for anti-HA immunoprecipitation. Immunoprecipitated proteins were separated via SDS-PAGE and visualized using Coomassie staining. Bands unique to the DVSF-treated sample were extracted, along with the corresponding section of gel in the untreated sample. Proteins were identified by tandem mass spectrometry, and proteins identified in both plus and minus DVSF samples eliminated for further study. The GeneID, protein name, PSM count, and percent coverage for all proteins with more than 1% coverage are shown in the table. A small amount of PfJ2 was identified in the -DVSF samples, and the PSM count and percent coverage is shown in parentheses. One of two biological replicates shown.
PfPDI8 and PfPDI11 are PfJ2 redox partners
Having shown the redox activity of PfJ2, we next sought to specifically identify those redox partnerships. To identify the proteins trapped to PfJ2 by DVSF, cultures were treated with the compound and immunoprecipitation of PfJ2 was performed (Fig 4C). As a control, the immunoprecipitation was also performed in parallel using cultures that had not received DVSF treatment. The immunoprecipitated proteins were subjected to separation by SDS-PAGE and visualized using Coomassie (Fig 4C). Two bands between 100–150 kDa, corresponding to those previously detected by western blot, were extracted from the DVSF-treated sample, along with the corresponding areas of the gel in the untreated samples (Fig 4C, perforated boxes). Proteins present in these gel slices were identified via MS/MS analysis. By analyzing the untreated control samples, we were able to remove background proteins and found that the top gel slice primarily contained PfJ2, PfPDI8, PfPDI11 and PfBiP, and the bottom slice contained PfJ2, PfPDI8, and PfPDI11 (Fig 4C, table). The complete list of proteins identified in all samples can be found in S2 Table. Because alterations to PfBiP migration during SDS-PAGE after DVSF treatment were not detected (S2 Fig), we chose to focus our attentions on PfPDI8 and -11.
PfPDI8 and PfPDI11 are redox-active ER proteins
Like PfJ2, PfPDI8 and -11 are both predicted members of the Trx superfamily (Fig 5A). PfPDI8 appears to be a canonical PDI, with two Trx domains containing CXXC active sites that likely allow it to carry out disulfide oxidoreductase/isomerase activity. PfPDI11 also has two Trx domains, but each contains an unusual CXXS active site. PfPDI8 has been characterized recombinantly in vitro, but the functions of both PfPDI8 and -11 remain unstudied in parasites [12,17]. In order to validate interaction between these PDIs and PfJ2, and to understand their roles in the P. falciparum ER, we used the glmS ribozyme to create conditional knockdown parasite lines for each protein in the background of PfJ2apt parasites [31] (Fig 5B). Using CRISPR/Cas9 genome editing, we introduced sequences for a 3xV5 tag and the glmS ribozyme into the pfpdi8 or the pfpdi11 locus (PfJ2apt-PDI8glmS and PfJ2apt-PDI11glmS, respectively) (Fig 5C). Correct modifications of the loci were validated by PCR integration test, and V5-tagged proteins were visualized by western blot (Fig 5D and 5E).

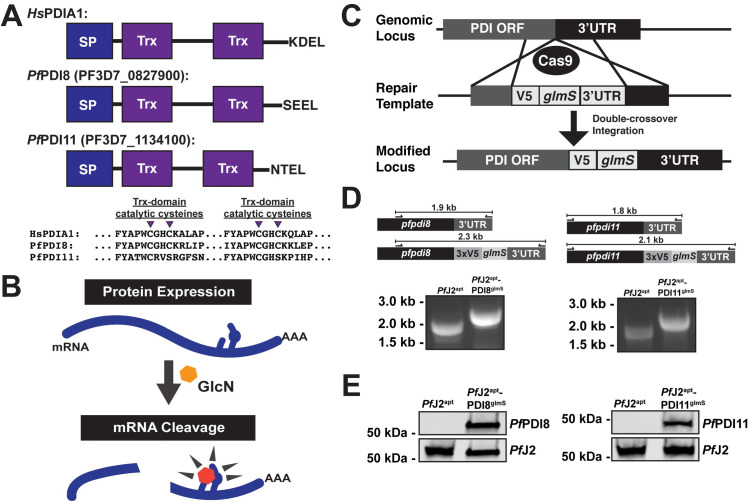
Generation of PfPDI8 (PF3D7_0827900) and PfPDI11 (PF3D7_1134100) conditional knockdown mutants using CRISPR/Cas9.
A) Predicted domain structure of PfPDI8, PfPDI11, and human PDIA1, showing signal peptide (SP), thioredoxin domains (Trx), and C-terminal ER retention signals. Essential, conserved cysteine residues are shown for each of the proteins’ Trx domains. B) Regulation of protein expression using the glmS ribozyme system. The mRNA of interest encodes the ribozyme in the 3’UTR. Upon addition of glucosamine (GlcN, orange hexagon), which is converted to glucosamine-6-phosphate (pink hexagon) by the parasite, the ribozyme is activated to cleave the mRNA, leading to transcript instability and degradation (Prommana et al., 2013) C) Schematic of CRISPR/Cas9 mediated introduction of the glmS knockdown system into the genome. A repair template was transfected, along with a plasmid to express Cas9 and a gRNA, to introduce sequences for a 3xV5 tag, ER retention signals, stop codon, and glmS ribozyme. D) PCR integration test confirming correct modification of pfpdi8 and pfpdi11. Correct integration results in increased amplicon size due to the V5 and glmS sequences. E) Western blots showing V5-tagged proteins in the PfJ2apt-PDI8glmS and PfJ2apt-PDI11glmS parasite lines at the predicted sizes for PfPDI8 and -11. For each western blot, one of two biological replicates shown.
The subcellular localizations of PfPDI8 and -11 were determined by IFA, and both proteins were found to co-localize with PfJ2 in the ER (Fig 6A). To test the functionality of the glmS ribozyme knockdown system, each parasite line was treated with glucosamine (GlcN), and samples were taken for western blot analysis over the course of the parasite lifecycle. Compared to -GlcN control samples, protein levels were found to be reduced during GlcN treatment (S3 Fig). In order to determine the effect that PDI knockdown had on parasite growth, each cell line was treated with GlcN and parasite growth was measured over the course of two life cycles. We observed dramatic inhibition of parasite growth during PfPDI8 knockdown, but no growth defects were observed when PfPDI11 was knocked down (Fig 6B). These results demonstrate that PfPDI8 is essential for the asexual lifecycle and suggest that PfPDI11 may be dispensable, though in the case of PfPDI11, the lack of a phenotype could be attributed to incomplete knockdown (S3 Fig). Both conclusions are supported by a genome-wide essentiality screen performed in P. falciparum [26].

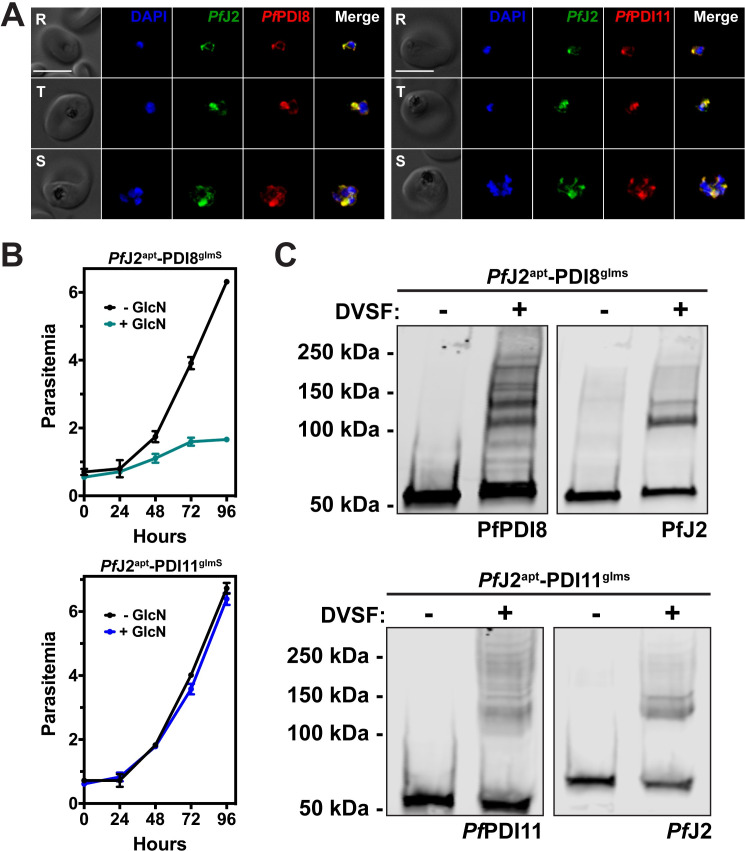
PfPDI8 and PfPDI11 are redox-active ER proteins.
A) PfJ2apt-PDI8glmS and PfJ2apt-PDI11glmS parasites were fixed and stained with DAPI (blue) and with antibodies against HA (green) and V5 (red). Ring (R), Trophozoite (T), and Schizont (S) stage parasites are shown. Z-stack Images were deconvoluted and shown as a single, maximum intensity projection. Scale bar represents 5 μm. B) Asynchronous PfJ2apt-PDI8glmS (top) and PfJ2apt-PDI11glmS (bottom) parasites were grown in normal (-GlcN) or knockdown (+ 5mM GlcN) conditions, and parasite growth was monitored daily for 96 hours via flow cytometry. Data points represent the mean of three technical replicates, with error bars representing standard deviation. For each growth curve, a representative experiment of two biological replicates is shown. C) PfJ2apt-PDI8glmS and PfJ2apt-PDI11glmS parasites were incubated with 3 mM DVSF in 1x PBS for 30 minutes at 37°C, then samples were taken for western blot analysis. Membranes were incubated with antibodies against HA (PfJ2) and V5 (PfPDI8 or -11). For each western blow, one of two biological replicates is shown.
PfPDI8 contains two thioredoxin domains with classical CXXC active site cysteines, allowing the protein to function in oxidative folding as an oxidoreductase/isomerase [12,17]. PfPDI11 has two thioredoxin domains containing noncanonical CXXS active sites, but likely maintains the ability to form mixed disulfide bonds with client proteins through the conserved cysteine residues [32–34]. To determine whether we could trap redox interactions between these PDIs and their substrates, PfJ2apt-PDI8glmS and PfJ2apt-PDI11glmS cultures were treated with DVSF, and protein lysates were collected for western blot analysis. Several high molecular weight bands containing PfPDI8 appear following DVSF treatment, indicating that multiple substrates rely on the oxidoreductase activity of PfPDI8, in contrast to PfJ2, whose western blot shows a narrower set of redox substrates (Fig 6C). Similar results were observed for PfPDI11 (Fig 6C).
We next sought to determine if DVSF crosslinking occurs through Trx-domain cysteines. To do this, we attempted to generate parasites overexpressing either wild-type copies of PfJ2, PfPDI8, PfPDI11, or overexpressing these proteins with cysteine-to-alanine mutations in the Trx domain active sites. These types of mutations abolish DVSF-crosslinking in Trx proteins of model organisms [28,30]. We were unable to generate parasites overexpressing wild-type or mutant copies of PfJ2. We were also unable to generate parasites overexpressing a mutant version of PfPDI8, but were successful in creating parasites overexpressing the wild-type protein; characterization of that parasite line revealed mislocalization of the overexpressed PfPDI8 (S4 Fig). Given the essential nature of PfJ2 and PfPDI8, we concluded that the parasites may be sensitive to their overexpression and to mutations in their Trx domains.
In contrast, we were successfully able to generate both wild-type and cysteine-to-alanine PfPDI11 overexpression mutants (PfPDI11wt and PfPDI11mut, respectively) (S6 Fig). Both parasite lines displayed the expected ER co-localization with PfJ2 (S5 Fig). Importantly, treatment of these parasites with DVSF revealed extensive crosslinking between PfPDI11 and substrates in the wild-type parasites, but crosslinking is abolished in parasites with cysteine-to-alanine mutations in the Trx domain (S5 Fig). These data demonstrated the specificity of DVSF for trapping redox partnerships in P. falciparum.
Given the unusual nature of the PfPDI11 CXXS Trx-domain active site, we took advantage of these overexpression parasites to further investigate PfPDI11 function. Trx-domain-proteins with CXXS active sites are largely under-studied in all organisms [32–34]. It is possible that the remaining active-site cysteines in PfPDI11 forms mixed disulfides that may serve to retain proteins the in ER, prevent their aggregation, and/or block cysteines from non-productive bond formation as they fold [32,34]. Therefore, we hypothesized that we would be able to detect the mixed PfPDI11-substrate disulfide bonds by non-reducing SDS-PAGE and western blotting. Indeed, we were able to detect high-molecular-weight species of PfPDI11 when PfPDI11wt parasite lysates were used for western blotting under non-reducing conditions (S6 Fig). In contrast, these species were missing when PfPDI11mut parasite lysates were used (S6 Fig).
The PfBiP-PfJ2-PfPDI8 oxidative folding complex
Having shown that PfJ2 and PfPDI8 are redox partners and that both proteins are essential for the P. falciparum asexual lifecycle, we decided to focus on their interaction and what roles they may play together in the ER. To confirm the interaction between PfJ2 and PfPDI8, PfJ2 was immunoprecipitated from PfJ2apt-PDI8glmS parasites, and PfPDI8 was found to co-immunoprecipitate (Fig 7A). When performing the reciprocal co-IP, we were unable to detect PfJ2 pulling down with PfPDI8, perhaps due to inefficiency of the anti-V5 IP (S7 Fig). However, we were able to detect a band of overlapping PfJ2/PfPDI8 signal when the PfPDI8 IP was performed on cultures treated with DVSF, showing that the two proteins are interacting, redox partners (Fig 7B).

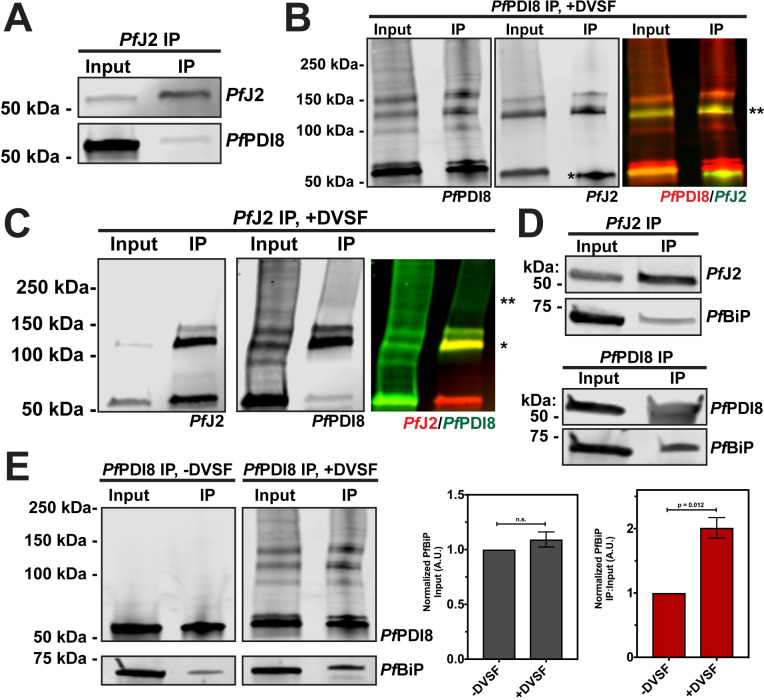
The PfBiP-PfJ2-PfPDI8 oxidative folding complex.
A) PfJ2 and interacting proteins were immunoprecipitated from PfJ2apt-PDI8glmS parasite lysate using anti-HA antibodies. Input and eluted IP samples were used for western blot analysis. Membrane was probed with HA and V5 antibodies to detect PfJ2 and PfPDI8, respectively. B) PfJ2apt-PDI8glmS parasites were incubated with 3 mM DVSF as described above, then V5 antibodies were used to immunoprecipitate PfPDI8 and interacting proteins. Input and eluted IP samples were used for western blot analysis. Membrane was probed with V5 and HA antibodies to detect PfPDI8 and PfJ2, respectively. Antibody heavy chain is indicated by the single asterisk (*) in the PfJ2 panel. A merged image of the PfPDI8 (red) and PfJ2 (green) signal is shown, with the yellow overlap in signal indicated by a double asterisk (**). C) PfJ2apt-PDI8glmS parasites were incubated with 3 mM DVSF as described above, then HA antibodies were used to immunoprecipitate PfJ2 and interacting proteins. Input and eluted IP samples were used for western blot analysis. Membrane was probed with HA and V5 antibodies to detect PfJ2 and PfPDI8, respectively. A merged image of the PfJ2 (red) and PfPDI8 (green) signal is shown, with a single asterisk (*) indicating the yellow overlap in signal and a double asterisk (**) indicating PfPDI8+subtrates that co-immunoprecipitated with PfJ2. D) PfJ2 and PfPDI8 were immunoprecipitated from PfJ2apt and PfJ2apt-PDI8glmS parasite lysates, respectively. Input samples and eluted IP proteins were used for western blot analysis. Membrane was probed with HA and PfBiP antibodies (top) or V5 and PfBiP antibodies (bottom). Immunoprecipitations were performed in biological duplicate or triplicate, and representative results are shown in A-D. E) PfJ2apt-PDI8glmS parasite cultures were evenly split into two conditions: 3 mM DVSF or PBS only for 30 minutes at 37°C, after which parasite lysates were used for anti-V5 immunoprecipitation. Input and eluted IP proteins were analyzed by western blot using V5 and PfBiP antibodies. The PfBiP signal was measured for each lane and the ratio of IP-to-Input signal was determined. N = 3 biological replicates. Error bars represent standard deviation.
Our observations using the redox crosslinker DVSF showed that PfPDI8 is a major redox partner for PfJ2, whereas PfPDI8 has multiple other redox partnerships (Figs 4C and 6D). One explanation for this observation is that PfJ2 may work upstream to prime PfPDI8 for interaction with its substrates. Therefore, we asked whether we could detect PfPDI8+substrates co-immunoprecipitating with PfJ2. When PfJ2 was immunoprecipitated from PfJ2apt-PDI8glmS parasites treated with DVSF, we were able to detect PfPDI8 trapped to other substrates, indicated by smearing of the PfPDI8 signal above 150 kDa (Fig 7C). As a negative control, PfPMV was probed for in the proteins immunoprecipitating with PfJ2 and PfPDI8 and was not found to interact with either protein, confirming the specificity of the anti-HA and anti-V5 immunoprecipitations utilized (S8 Fig). Together, these results confirm the interaction between PfJ2 and PfPDI8 and suggest that PfJ2 may be part of a complex including PfPDI8 and its substrates.
Oxidative folding in the ER, mediated by proteins such as PfJ2 and PfPDI8, is only one aspect of protein folding in the ER, and likely works in conjunction with other folding determinants, such as the Hsp70 BiP. PfJ2 is an ER Hsp40—a class of co-chaperones that interact with the Hsp70 BiP—and BiP is likely involved in the folding of the same substrates that interact with PfPDI8. Such cooperation between BiP and mediators of oxidative folding has received limited investigation in most organisms, to our knowledge. Therefore, we next asked whether PfBiP interacts with PfJ2 and/or PfPDI8.
Western blot analysis of proteins co-immunoprecipitating with PfJ2 revealed that PfBiP does interact with PfJ2, consistent with our PfJ2 co-IP experiments and the proteins’ predicted chaperone/co-chaperone roles (Figs 3 and 7D, top). Lack of a suitable antibody precluded reciprocal PfBiP immunoprecipitation to probe for PfJ2. However, as a control we showed that PfBiP is not detected when wild-type parasites (lacking HA-tagged PfJ2) are subjected to anti-HA immunoprecipitation, ruling out nonspecific binding during the co-IP experiment (S9 Fig).
Next, we found that when PfPDI8 was immunoprecipitated, PfBiP was detected (Fig 7D, bottom). We further reasoned that because the same substrates may rely on both PfPDI8 and PfBiP to achieve their native state, trapping the PfPDI8-substrate interaction with DVSF may increase the amount of PfBiP co-immunoprecipitating with PfPDI8. To test this hypothesis, parasite cultures were equally split into +/- DVSF treated aliquots, PfPDI8 was immunoprecipitated, and lysates probed for PfPDI8 and PfBiP. Consistent with our hypothesis, we detected a two-fold increase in the amount of PfBiP that pulled down with PfPDI8 crosslinked to its substrates, with no significant difference in the starting amount of PfBiP detected in the sample prior to immunoprecipitation (Fig 6E). Together, these results suggest that PfJ2 and PfPDI8 work together with the major ER folding chaperone PfBiP to help substrates reach their native states.
ER redox interactions are druggable
Our data have shown that PfJ2 and PfPDI8, which participate in ER redox partnerships with each other as well as other substrates, are essential proteins in the P. falciparum asexual lifecycle. These proteins, through inhibition of their redox interactions, may represent unexploited targets for antimalarials. Further, our data show that DVSF can target these redox partners, suggesting that these proteins may be druggable. Indeed, consistent with the idea of targeting ER redox proteins in disease, high-throughput drug screens have identified potent inhibitors of human PDI in an effort to combat upregulation that is associated with some cancers and neurodegenerative diseases [35–38].
We tested four of these commercially available PDI inhibitors—16F16, LOC14, CCF642, and PACMA31—for activity against cultured asexual P. falciparum parasites. In contrast to their reported, highly potent activity against human cells, we observed a wide range of IC50 values for P. falciparum (S10 Fig). The compound with the best anti-Plasmodium activity was 16F16, with an IC50 value of approximately 4 μM (Fig 8A) [39].

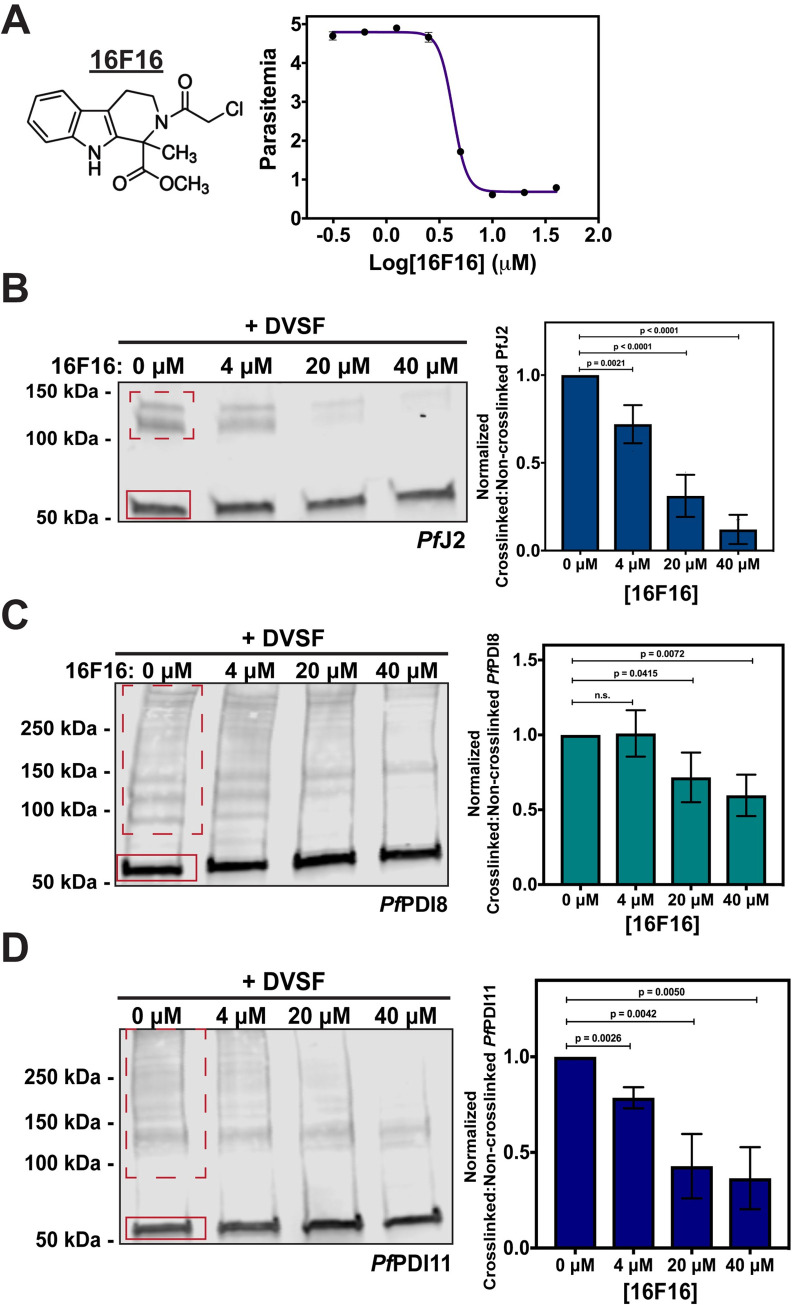
ER redox interactions are sensitive to interruption by a small molecule.
A) Asynchronous PfJ2apt parasites were incubated in various concentrations of the human PDI inhibitor 16F16. Parasite growth was determined via flow cytometry at 72 hours and the 16F16 IC50 was determined to be approximately 4 μM. Each data point in the curve represents the mean parasitemia at a given concentration, in technical triplicate. Error bars are not seen for data points in which they are smaller than the circle symbol, represent standard deviation from the mean. A representative IC50 curve is shown for one of three biological replicates. B) PfJ2apt, C) PfJ2apt-PDI8glmS, and D) PfJ2apt-PDI11glmS parasites cultures were equally split and incubated with three concentrations of 16F16 for 3 hours prior to removal of 16F16, then incubation with 3 mM DVSF as described above. Samples were taken for western blot analysis, loading equal parasite equivalents into each gel. Membranes were incubated with antibodies against HA or V5. Signal for non-crosslinked (the band at approximately 50 kDa, solid red box) and crosslinked proteins (dashed red box) was measured. Inhibition was measured by determining the ratio of crosslinked to non-crosslinked signal. N = 3 biological replicates for each parasite line. Error bars represent standard deviation.
16F16 inhibits human PDI function by covalently binding the cysteines of the Trx domain active sites, thereby blocking their ability to catalyze oxidative folding [35,36]. If 16F16 behaves similarly in P. falciparum, we reasoned that treatment of cultures with 16F16 prior to performing redox crosslinking with DVSF would prevent crosslinking from occurring, as both compounds rely on the same cysteine residues for their activity. Indeed, pre-treatment with increasing amounts of 16F16 significantly and reproducibly decreased the amount of crosslinked PfJ2 detected by western blot (Fig 8B). Our data indicate that DVSF treatment crosslinks PfJ2 to PfPDI8 and PfPDI11 (Fig 4). Therefore, the observed reduction in PfJ2 crosslinking likely occurs due to direct reaction of 16F16 with the PfJ2 Trx-domain active site and/or the PfPDI8 and -11 active sites. Similar experiments showed that pre-treatment of cultures with 16F16 also blocked crosslinking of PfPDI8 and -11 with their substrates, though to a lesser extent than what was observed for PfJ2 (Fig 8C and 8D). The quantification of the Western blot signals for all replicates is found in S3 Table. These data suggest that 16F16 inhibits the redox activity of PfJ2, PfPDI8, and PfPDI11 (Fig 8B, 8C and 8D) and it is likely that this compound inhibits other Trx-domain containing proteins in P. falciparum.
These data suggest that redox interactions within the P. falciparum ER, occurring between essential proteins like PfJ2 and PfPDI8 and their substrates, are sensitive to small molecule inhibition. Additionally, the disparity in activity observed for the PDI inhibitors against human and P. falciparum cell lines suggest that development of Plasmodium-specific inhibitors is likely possible (S10 Fig).
Discussion
The ability to conduct oxidative folding likely underlies the diverse functions of the P. falciparum ER. The oxidizing environment of the ER encourages disulfide bond formation, but only the correct bonds allow proteins to reach their native states. Therefore, organisms must maintain a way to reduce/isomerize nonproductive disulfides. We have used CRISPR/Cas9 genome editing and conditional knockdown to show here that a putative disulfide reductase in the P. falciparum ER—PfJ2—Is essential for the parasite asexual lifecycle (Figs 1 and 2).
A co-IP/mass spectroscopy approach with stringent parameters for identifying interacting partners places PfJ2 in the broader context of ER biology, revealing that PfJ2 interacts with other folding determinants, such as BiP and Endoplasmin, as well as other members of the Thioredoxin superfamily, namely PDIs. The remaining proteins that were identified, most with unknown localization throughout the secretory pathway and many with no known function, may represent substrates that rely on PfJ2 and these other chaperones for their folding and/or trafficking. Among the proteins identified were large, complex proteins such as PfMSP1 and PfRhopH3 (Fig 3). Both proteins have numerous cysteine residues that must navigate oxidative folding when they are synthesized into the ER, likely relying on PfJ2 and PfPDIs to do so correctly. Consistent with this hypothesis, a recent study identified PfJ2, PfPDI11, and PfEndoplasmin as potential contributors to folding and trafficking of PfEMP1, a cysteine-rich transmembrane protein that serves as the major P. falciparum virulence factor [40]. Our analysis of the proteins identified by mass spectroscopy predicted interactions between PfJ2 and PfBiP, and between PfJ2 and PfPDI8. We confirmed these interactions via co-immunoprecipitation and western blot experiments. Notably, we also demonstrated an interaction between PfJ2 and PfPDI11, a protein which was identified by mass spectroscopy (S1 Table) but did not meet our 5-fold enrichment criteria, suggesting that our parameters err on the side of caution to reduce false-positives.
One major PfJ2 redox substrate—PfPDI8—was identified using a chemical biology approach. DVSF is a redox-specific crosslinker that has been used to identify redox partnerships between Thioredoxin proteins in the cytoplasm of model organisms [28–30]. To our knowledge, this compound had not yet been used to trap and define redox partnerships in Plasmodium, nor in the ER of any organism. We demonstrate its utility in the Plasmodium ER, using it to identify the redox partnership between PfJ2, PfPDI8 and PfPDI11 (Fig 4). Double conditional P. falciparum mutants showed that PfPDI8 is also an essential, ER-resident protein and allowed us to probe more deeply the relationship between PfJ2 and PfPDI8 (Figs 5,6 and 7). PfJ2 likely acts as a reductase, and previous in vitro characterization of PfPDI8 revealed that it behaves like a classical PDI, capable of both forming and reducing disulfide bonds [12,13,15–17].The propensity for PfPDI8 to use its Trx domains either for oxidation of cysteines or reduction of disulfides likely depends on the oxidation state of its own active site (i.e. reduced PfPDI8 can act as a reductase). One explanation for the redox partnership between PfJ2 and PfPDI8 is that PfJ2 primes PfPDI8 to act as a reductase for some or all of the numerous substrates we visualized using DVSF (Fig 6D). Consistent with this hypothesis, immunoprecipitation experiments showed that PfPDI8+substrates pull down with PfJ2 (Fig 7C). We also found that PfJ2 and PfPDI8 both interact with the Hsp70 PfBiP, and detection of that interaction increases for PfPDI8 when it is trapped to its substrates (Fig 7D and 7E). These data suggest a model in which PfJ2, PfPDI8, and PfBiP cooperate to ensure substrates in the ER correctly navigate the oxidative folding process to achieve their native states (Fig 9). As a predicted Hsp40 with an Hsp70-interacting J-domain, one of PfJ2’s roles in this complex may be to recruit PfBiP to into a possible folding complex.

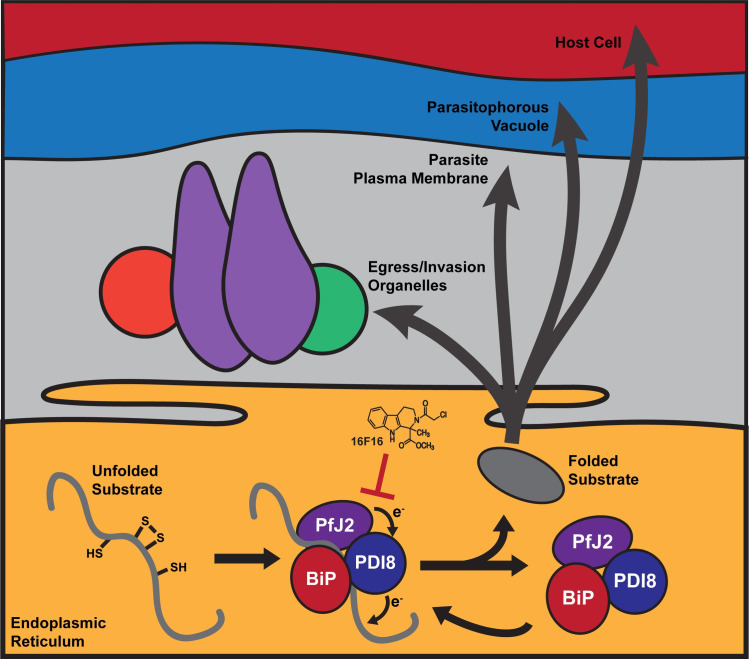
Oxidative folding in the P. falciparum ER.
We propose that Trx-domain proteins like PfJ2 and PfPDI8 work with PfBiP to help nascent proteins, which perform essential functions within the ER and throughout the parasite secretory pathway, achieve their native states. The redox interactions between PfJ2, PfPDI8, and their substrates are sensitive to inhibition by small molecules like 16F16, which could be expected to disrupt oxidative folding and impair the parasite’s ability to perform functions essential for survival and replication.
We also identified PfPDI11 as a redox substrate of PfJ2 (Fig 4). Our data demonstrate that PfPDI11 retains the ability to form mixed disulfides with client proteins despite the unusual CXXS Trx-domain active site (Figs 6, S5 and S6). Typically, the second cysteine of the Trx-domain active site is used to resolve enzyme-substrate mixed disulfides [10]. Therefore, the mechanisms used to resolve mixed disulfides between CXXS active sites and their substrates remains unclear, both in P. falciparum and other organisms. We propose that an ER-resident reductase such as PfJ2 helps resolve mixed disulfides, which would explain why PfJ2 and PfPDI11 were found to be redox partners.
Collectively, our data also demonstrate the power and specificity of using DVSF for trapping redox interactions between proteins with Trx domains and their substrates in P. falciparum. We showed that DVSF does not crosslink PfPMV or PfBiP to other proteins, despite the presence of numerous cysteine residues in the former, and that using NEM to block sulfhydryl groups in the parasite prevents crosslinking between PfJ2 and its substrates (Figs 4 and S2). Furthermore, we showed that DVSF traps PfPDI11 to its substrates, but cysteine-to-alanine mutations in the PfPDI11 Trx domain active sites abolish this crosslinking even though this mutant has two cysteine residues that are not in the active site (S5 Fig). These data are consistent with observations in yeast and mammalian cells, in which DVSF has been validated to specifically trap Trx-domain proteins to the substrates with which they exchange disulfide bonds [28–30].
Importantly, given the recent stagnation observed in malaria elimination efforts, which is coincident with increasing cases of antimalarial resistance, we not only identified two proteins with essential functions; we further demonstrated that the redox partnerships of these proteins are sensitive to disruption by small molecule inhibition (Fig 8). 16F16 is a covalent inhibitor that blocks Trx-domain cysteines [35,36]. Such a compound, if specific for P. falciparum, could be expected to cripple oxidative folding in the ER and kill the parasite. Recently, interest in covalent inhibitors for treatment of human disease has renewed, with several covalent inhibitors approved for use by the United States Food and Drug Administration [41]. One particular concern with covalent inhibitors is the fact that mutagenesis of the target residue would result in resistance, but mutagenesis of Trx-domain cysteines would lead to loss of function in and of itself, presumably making this type of resistance harder to evolve.
Another concern is whether proteins with such conserved active sites and roles in biology would make appropriate drug targets. In reality, conserved proteins have given the field many of its validated and proposed drug targets. For example, the widely-used anti-malarial drug atovaquone targets Complex III of the mitochondrial electron transport chain [42]. Some other conserved proteins/complexes that have been proposed as anti-malarial drug targets include Cytochrome B, the TCP-1 Ring Complex chaperone, and the proteasome [43–46]. The precedence for targeting proteins that participate in conserved biology exists. Interest in targeting the P. falciparum proteasome began with observations that mammalian proteasome inhibitors have anti-malarial activity, which subsequently spurred development of Plasmodium-specific inhibitors [44,47,48]. Similarly, 16F16 likely targets many Trx-domains within the parasite, but we have used it to show that the redox interactions between PfJ2, PfPDI8, and their substrates can be disrupted with a small molecule, and development of a Plasmodium-specific inhibitor may be possible. In fact, given the disparity in activity observed for the PDI inhibitors against human cell lines and P. falciparum, enough diversity likely exists between these conserved proteins that Plasmodium-specific inhibitors could be developed (S10 Fig). Therefore, essential Trx-domain proteins in the parasite ER—like PfJ2 and PfPDI8—represent a class of proteins and a pathway in the ER that is apt for antimalarial drug development.
Materials and methods
Construction of plasmids
Parasite genomic DNA was isolated from 3D7 parasites using QIAamp DNA blood kit (QIAGEN). All constructs utilized in this study were confirmed by sequencing. Plasmids were constructed using the Sequence and Ligation Independent Cloning (SLIC) method. Plasmids to express Cas9 and gRNAs were constructed using pUF1-Cas9 as previously described [49–51]. All primers used in this study are listed in S4 Table. pfpdi8 cDNA was prepared using TRIzol-extracted mRNA and reverse transcription with primer P20 (SuperScript III, Invitrogen). All restriction enzymes used in plasmid construction were purchased from New England Biolabs.
To generate pMG74-PfJ2, approximately 500 bp of the sequence encoding the PfJ2 C-terminus was amplified using primers P1 and P2, and approximately 500 bp from the pfj2 3’UTR were amplified using P3 and P4. The two amplicons were joined together via PCR sewing using P1 and P4, then inserted into pMG74 [19] digested with AflII and AatII. For expression of a PfJ2 gRNA, oligos P31 and P32 were inserted into pUF1-Cas9.
To generate pV5-glmS-PDI8, approximately 500 bp of the sequence encoding the PfPDI8 C-terminus was amplified using primers P5 and P6. The 3x V5 tag was added to this amplicon via PCR sewing using a linearized plasmid encoding the 3xV5 sequence and primers P5 and P7. The glmS ribozyme sequence was amplified from pHA-glmS [31] using P8 and P9, then added to the PfPDI8 C-terminus+V5 amplicon via PCR sewing using P5 and P9. The resulting amplicon was inserted into pHA-glmS that had been digested with AfeI and NheI, creating pPDI8-Cterm. Approximately 500 bp of the pfpdi8 3’UTR was amplified using P10 and P11, then inserted into pPDI8-Cterm that had been digested with HindIII and NotI, creating pV5-glmS-PDI8. For expression of a PfPDI8 gRNA, oligos P33 and P34 were inserted into pUF1-Cas9.
To generate pV5-glmS-PDI11, approximately 500 bp of the sequence encoding the PfPDI11 C-terminus was amplified using primers P12 and P13. The 3x V5 tag was added to this amplicon via PCR sewing using a linearized plasmid encoding the 3xV5 sequence and primers P12 and P14. The glmS ribozyme sequence was amplified from pHA-glmS [31] using 15 and P16, then added to the PfPDI8 C-terminus+V5 amplicon via PCR sewing using P12 and P16. The resulting amplicon was inserted into pHA-glmS that had been digested with AfeI and NheI, creating pPDI11-Cterm. Approximately 500 bp of the pfpdi11 3’UTR was amplified using P17 and P18, then inserted into pPDI11-Cterm that had been digested with HindIII and NotI, creating pV5-glmS-PDI11. For expression of a PfPDI11 gRNA, oligos P35 and P36 were inserted into pUF1-Cas9.
PfPDI8 and PfPDI11 overexpression was carried out by using CRISPR/Cas9 to insert the open reading frame (ORF) of the tagged genes into the pfhsp110c locus. pUC57-Hsp110, the repair plasmid targeting pfhsp110, includes the last 429 bp encoding the PfHsp110c (PF3D7_0708800) C-terminus, a 2A skip peptide sequence, sequences for various peptide tags, then the first 400 bp from the pfhsp110c 3’UTR. This plasmid was synthesized by GeneScript. For expression of a PfHsp110c gRNA, oligos P37 and P38 were inserted into pUF1-Cas9.
To generate pUC57-Hsp110-PDI8wt, the PfPDI8 ORF was amplified from cDNA using P19 and P20. A sequence encoding the 3xV5 was attached to this amplicon via PCR sewing using a linearized plasmid encoding the tag and primers P19 and P21. The resulting amplicon was inserted into pUC57-Hsp110 digested with MfeI and SpeI.
To generate pUC57-Hsp110-PDI11wt, the PfPDI11 ORF was amplified using P22 and P23. A sequence encoding the 3xV5 was attached to this amplicon via PCR sewing using a linearized plasmid encoding the tag and primers P22 and P24. The resulting amplicon was inserted into pUC57-Hsp110 digested with MfeI and SpeI.
To generate pUC57-Hsp110-PDI11mut, which required mutagenesis of the 2 Trx-domain active site, the PfPDI11 ORF was amplified in 3 parts. Part 1 was amplified using P25 and P26. Part 2 was amplified using P27 and P28. Part 3 was amplified using P29 and P30. Parts 1+2 were joined together using PCR sewing and primers P25 and P28. The resulting amplicon was attached to Part 3 using PCR sewing and primers P25 and P30. A sequence encoding the 3xV5 was attached to this amplicon via PCR sewing using a linearized plasmid encoding the tag and primers P25 and P24. The resulting amplicon was inserted into pUC57-Hsp110 digested with MfeI and SpeI.
Parasite culture and transfection
P. falciparum asexual parasites were cultured in RPMI 1640 medium supplemented with AlbuMAX I (Gibco) and transfected as described earlier [52,53].
To generate the PfJ2apt parasite line, uninfected RBCs were transfected with 20 μg pMG74-PfJ2 (linearized prior to transfection using EcoRV) and 50 μg pUF1-Cas9-PfJ2, then fed to 3D7 parasites. Drug pressure was applied 48 hours after transfection, selecting for integration using 0.5 μM aTc and 2.5 μg/mL Blasticidin. After parasites grew back up from transfection and were cloned using limiting dilution, clones were maintained in medium containing 10 nM aTc and 2.5 μg/mL Blasticidin. Unless started otherwise, all +/- aTc growth experiments were conducted in medium containing 10 nM aTc and 2.5 μg/mL Blasticidin or medium containing only 2.5 μg/mL Blasticidin.
To generate the PfJ2apt-PDI8glms parasite line, RBCs were transfected with 50 μg pV5-glmS-PDI8 and 50 μg pUF1-Cas9-PDI8, then fed to PfJ2apt parasites. Drug pressure was applied 48 hours after transfection, selecting with 0.5 μM aTc, 2.5 μg/mL Blasticidin, and 1 μM 5-Methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)naphthalen-2-ylamine (DSM1) [50]. After parasites grew back up from transfection and were cloned using limiting dilution, clones were maintained in medium containing 50 nM aTc and 2.5 μg/mL Blasticidin. PfJ2apt-PDI11glms parasites were generated in the same manner, using 50 μg pV5-glmS-PDI11 and 50 μg pUF1-Cas9-PDI11.
To generate the PfPDI8wt overexpression parasite line, RBCs were transfected with 50 μg pUC57-Hsp110-PDI8wt and 50 μg pUF1-Cas9-Hsp110, then fed to PfJ2apt parasites. Drug pressure was applied 48 hours after transfection, selecting with 0.5 μM aTc, 2.5 μg/mL Blasticidin, and 1 μM Drug Selectable Marker 1 (DSM1) [50]. After parasites grew back up from transfection and were cloned using limiting dilution, clones were maintained in medium containing 10 nM aTc and 2.5 μg/mL Blasticidin. PfPDI11wt and PfPDI11mut parasites were generated in the same manner, using pUC57-Hsp110-PDI11wt and pUC57-Hsp110-PD11mut, respectively.
Parasite synchronization was carried out as described [54].
Western blotting
Western blots were performed as previously described [55]. Briefly, ice-cold 0.04% saponin in 1x PBS was used to isolate parasites from host cells. Parasite pellets were subsequently solubilized in protein loading dye to which Beta-mercaptoethanol had been added (LI-COR Biosciences) and used for SDS-PAGE. Primary antibodies used in this study were rat-anti-HA 3F10 (Roche, 1:3000), mouse-anti-HA 6E2 (Cell Signaling Technology, 1:1000), rabbit-anti-HA 715500 (Thermofisher, 1:100), mouse-anti-V5 TCM5 (eBioscence, 1:1000), rabbit-anti-V5 D3H8Q (Cell Signaling Technology, 1:1000), rabbit anti-PfBiP MRA-1246 (BEI resources, 1:500), rabbit-anti-PfEF1α (from D. Goldberg, 1:2000), and mouse-anti-PfPMV (from D. Goldberg 1:400). Secondary antibodies used were IRDye 680CW goat-anti-rabbit IgG and IRDye 800CW goat-anti-mouse IgG (Li-COR Biosciences, 1:20,000). Membranes were imaged using the Odyssey Clx Li-COR infrared imaging system (Li-COR Biosciences). Images of membranes were processed using ImageStudio, the Odyssey Clx Li-COR infrared imaging system software (Li-COR Biosciences). Densitometry analysis of western blot signal was also performed using ImageStudio (Li-COR Biosciences).
Microscopy and image analysis
Parasites were fixed for IFA using 4% Paraformaldehyde and 0.03% glutaraldehyde, then permeabilized with 0.1% Triton-X100. Primary antibodies used were rat-anti-HA 3F10 (Roche, 1:100), mouse-anti-HA 6E2 (Cell Signaling Technology, 1:100), mouse-anti-V5 TCM5 (eBioscence, 1:100), rabbit-anti-V5 D3H8Q (Cell Signaling Technology, 1:100), and mouse-anti-PfPMV (from D. Goldberg 1:1). Secondary antibodies used were Alexa Fluor 488 and Alexa Fluor 546 (Life Technologies, 1:100). Cells were mounted to slides using ProLong Diamond with DAPI (Invitrogen). Fixed and stained cells were imaged using a DeltaVision II microscope system with an Olympus IX-71 inverted microscope. Images were collected as a Z-stack and deconvolved using SoftWorx, then displayed as a maximum intensity projection. Images were processed using Adobe Photoshop, with adjustments made to brightness and contrast for display purposes.
For imaging of parasite cultures using light microscopy, aliquots of culture were smeared onto glass slides and field-stained using Hema3 Fixative and Solutions (Fisher Healthcare), which is comparable to Wright-Giemsa staining. Slides were imaged using a Nikon Eclipse E400 microscope with a Nikon DS-L1-5M imaging camera. To measure parasite size, images were taken and parasites measured using ImageJ (NIH).
Growth assays using flow cytometry
Aliquots of parasites culture were incubated in 8 μM Hoescht 33342 (Thermofisher Scientific) for 20 minutes at room temperature, then fluorescence was measured using a CytoFlex S (Beckman Coulter, Hialeah, Florida). Flow cytometry data were analyzed using FlowJo software(Treestar, Inc., Ashland, Oregon). For IC50 experiments, data were analyzed using the 4-parameter dose-response-curve function of Prism (GraphPad Software, Inc.).
Immunoprecipitation assays
Anti-HA immunoprecipitation (IP) assays were performed as previously described, using anti-HA magnetic beads (Pierce) [56]. Anti-V5 IP assays were performed in the same manner as with anti-HA, but anti-V5 magnetic beads were used according to manufacturer instructions (MBL International Corporation).
Mass spectrometry and data analysis
CoIP samples were sent to Emory University Integrated Proteomics Core and analyzed using a Fusion Orbitrap mass spectrometer, or to the proteomics core at the Fred Hutchinson Cancer Research Center, where samples were analyzed using an OrbiTrap Elite. Data were searched using Proteome Discoverer 2.2 with UP000001450 Plasmodium falciparum (Uniprot Nov 2018) as the background database. The validation also included Sequest HT and Percolator to search for common contaminants. Results consisted of high confidence data with a 1% false discovery rate. Protein abundance was calculated by summing the total intensities (MS1 values) of all matched peptides for each selected protein, and normalizing by the total summed intensity of all matched peptides in the sample, as previously described [20]. The resulted calculated value was then averaged between replicates. To filter for high confidence hits, we calculated the abundance ratio between PfJ2 IP and the parental control, and set a threshold of fivefold enrichment over the parental control. This list was further curated to exclude proteins without a signal peptide or a transmembrane domain to identify biologically relevant interactors, because proteins with an signal peptide and/or transmembrane domain potentially traverse through the ER and can come in contact with PfJ2 in this cellular context.
Identification of PfJ2 redox partners
PfJ2apt parasites were incubated with 3 mM divinyl sulfone (DVSF, Fisher Scientific) in 1x PBS for 30 minutes at 37°C, then used for anti-HA immunoprecipitation as described above. Immunoprecipitated proteins were separated by SDS-PAGE. Polyacrylamide gel slices corresponding to the protein molecular weights of interest were excised and the peptides extracted by in-gel enzymatic digestion. The gel slices were dehydrated in 100% acetonitrile and dried using a speed vac. The proteins were then reduced by rehydrating the gel slices in 10mM dithiothreitol in 100mM ammonium bicarbonate solution, and alkylated in 50mM iodoacetamide in 100 mM ammonium bicarbonate solution. The gel slices were then washed in 50% acetonitrile in 50mM ammonium bicarbonate solution before being dehydrated and dried again using 100% acetonitrile and a speed vac. Proteins were then digested in-gel by rehydrating the gel slices in trypsin enzyme solution consisting of 6ng/μl trypsin (Promega) in 50mM ammonium bicarbonate solution. Digestion was performed at 37°C overnight. Peptides were extracted through stepwise incubations with 2% acetonitrile and 1% formic acid solution, 60% acetonitrile and 0.5% formic acid solution, and 100% acetonitrile solution. Supernatants were combined and dried in a speed vac before resuspension in 20 μl water with 0.1% formic acid.
LC-MS/MS analysis was performed using a 50 cm fused silica capillary (75 μm ID) packed with C18 (2 μm, Dr. Maisch GmbH), and heated to 50°C. Prior to loading the column, sample was loaded onto a 2 cm Acclaim PepMap 100 (Thermo Fisher Scientific) trap (75 μm ID, C18 3 μm). For each sample injection, 5 μl of sample was loaded onto the trap using an Easy nLC-1000 (Thermo Fisher Scientific). Each sample was separated using the Easy nLC-1000 with a binary mobile phase gradient to elute the peptides. Mobile phase A consisted of 0.1% formic acid in water, and mobile phase B consisted of 0.1% formic acid in acetonitrile. The gradient program consisted of three steps at a flow rate of 0.3 μL/min: (1) a linear gradient from 5% to 40% mobile phase B over two hours, (2) a 10 minute column wash at 80% mobile phase B, and (3) column re-equilibration for 20 minutes at 5% mobile phase B.
Mass spectra were acquired on a Fusion Lumos Tribrid (Thermo Fisher Scientific) mass spectrometer operated by data dependent acquisition (DDA) using a top 15 selection count. Precursor ion scans were performed at 120,000 resolution over a range from 375 to 1375 m/z. DDA was performed with charge exclusion of 1 and greater than 8, with isotope exclusion, and dynamic exclusion set to 10 seconds. MS/MS was performed using an isolation window of 1.6 m/z for selection, normalized collision energy (NCE) of 28, and higher energy collision induced dissociation (HCD). MS/MS spectra were acquired at 15000 resolution with an automatic gain control (AGC) target of 70,000 and maximum injection time of 50 ms.
Mass spectra (.raw files) were converted to mzML format using MSConvert (version 3.0.1908) [57] and peptide sequences were identified using database searching with Comet [58] (version 2016.01 rev 2). Spectra were searched against a subset of P. falciparum secretory proteins, common laboratory contaminants, and an equal number of randomized decoy sequences (4386 total protein sequence). Comet parameters included variable modifications of +57.021464 Da or +118.0089 Da on cysteine and a variable modification of +15.994915 Da on methionine or tryptophan. Precursor mass tolerance was set to 25 ppm and a fragment_bin_tolerance of 0.02 and fragment_bin_offset of 0 were used. Full-tryptic enzymatic cleavage was set, allowing for up to 3 missed cleavages. Peptide spectrum matches (PSM) were analyzed using the Trans-Proteomic Pipeline [59] (TPP, version 5.0.0 Typhoon), to assign peptide and protein probabilities using PeptideProphet [60] and iProphet [61], respectively. Spectral counts and precursor ion intensities were exported for each non-redundant PSM at a 1% false discovery rate (FDR). Protein inference was performed with ProteinProphet [62], using a 1% FDR.
Acknowledgements
We thank Dan Goldberg for antibodies against PfPMV and PfEF1α; Julie Nelson at the CTEGD Cytometry Shared Resource Laboratory for help with flow cytometry and analysis; and Muthugapatti Kandasamy at the Biomedical Microscopy Core at the University of Georgia for help with microscopy. We acknowledge assistance of the Proteomics Resource at Fred Hutchinson Cancer Research Center and the Emory University Integrated Proteomics Core for mass spectrometry and data analysis.
References
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
 A redox-active crosslinker reveals an essential and inhibitable oxidative folding network in the endoplasmic reticulum of malaria parasites
A redox-active crosslinker reveals an essential and inhibitable oxidative folding network in the endoplasmic reticulum of malaria parasites
