
- Altmetric
A key challenge in understanding how organisms adapt to their environments is to identify the mutations and genes that make it possible. By comparing patterns of sequence variation to neutral predictions across genomes, the targets of positive selection can be located. We applied this logic to house mice that invaded Gough Island (GI), an unusual population that shows phenotypic and ecological hallmarks of selection. We used massively parallel short-read sequencing to survey the genomes of 14 GI mice. We computed a set of summary statistics to capture diverse aspects of variation across these genome sequences, used approximate Bayesian computation to reconstruct a null demographic model, and then applied machine learning to estimate the posterior probability of positive selection in each region of the genome. Using a conservative threshold, 1,463 5-kb windows show strong evidence for positive selection in GI mice but not in a mainland reference population of German mice. Disproportionate shares of these selection windows contain genes that harbor derived nonsynonymous mutations with large frequency differences. Over-represented gene ontologies in selection windows emphasize neurological themes. Inspection of genomic regions harboring many selection windows with high posterior probabilities pointed to genes with known effects on exploratory behavior and body size as potential targets. Some genes in these regions contain candidate adaptive variants, including missense mutations and/or putative regulatory mutations. Our results provide a genomic portrait of adaptation to island conditions and position GI mice as a powerful system for understanding the genetic component of natural selection.
Introduction
When members of a population display heritable differences in a trait that affects individual fitness, the trait distribution can change predictably from one generation to the next. This process of natural selection is the primary engine of evolutionary change responsible for the adaptation of populations to new environments. Elucidating the genetic component of natural selection—the identities and effects of the mutations that provide the basis for adaptive change—illuminates parts of the process, including whether different populations take the same paths to arrive at similar phenotypic optima. Theory and experimental evolution have usefully outlined the expected properties of genetic variants involved in selection (Orr 2005; Good et al. 2017). Field studies have characterized the intensity and form of natural selection on a variety of phenotypes (Endler 1986; Kingsolver et al. 2001). Nevertheless, the number of examples for which the specific genes and mutations targeted by natural selection are known is still small.
Recent colonizers of islands are compelling subjects for understanding natural selection. Colonizers often experience novel biotic and abiotic environments (Losos and Ricklefs 2009). Shifts in resource availability, predation risk, and competition compared with mainland environments can generate natural selection favoring larger bodies, reduced aggression, and higher population densities—characteristics seen in island populations of vertebrates (Foster 1964; Van Valen 1973; Lomolino 1985; Stamps and Buechner 1985; Adler and Levins 1994). Among the multitude of island colonizers, murid rodents have received considerable attention from evolutionary biologists and ecologists. Commensalism with humans has allowed mice and rats to invade islands around the world, exposing them to a wide range of new selective regimes on recent timescales. Skeletal phenotypes from island murids show some of the strongest evidence for rapid phenotypic change (Berry et al. 1978; Pergams and Ashley 2001; Millien 2006; Boell and Tautz 2011). Murid rodents usually evolve larger body sizes on islands (Adler and Levins 1994; Meiri et al. 2008), providing multiple compelling examples of the broader pattern of unusual size evolution in island populations, known as the island rule (Foster 1964; Van Valen 1973; Lomolino 1985).
House mice living on Gough Island (GI)—a remote island in the middle of the South Atlantic—offer a special opportunity to study natural selection in the context of the island rule. GI presents a novel habitat for house mice. There are no human-made shelters (other than a field station) for mice to reduce their exposure to inclement weather or find stored food. There are no predators or interspecific competitors for mice (Hill 1959). In response to these conditions and others, mice on GI have evolved unusual morphological, behavioral, and ecological traits. GI mice are the largest wild house mice on record (Rowe-Rowe and Crafford 1992; Jones et al. 2003; Gray et al. 2015), showing substantial, heritable differences from mainland relatives in growth trajectories (Gray et al. 2015) and skeletal dimensions (Parmenter et al. 2016). The mice routinely attack and eat endangered seabirds (Jones et al. 2003; Cuthbert and Hilton 2004)—wounding or killing millions of chicks per year by some estimates (Wanless et al. 2012; Caravaggi et al. 2019). The peak population density of GI mice is among the highest known for house mice on islands (Rowe-Rowe and Crafford 1992; Jones et al. 2003; Cuthbert et al. 2016). The colonization of a new environment and the accelerated evolution of striking phenotypic and ecological characteristics suggest that natural selection has influenced mice in significant ways since their arrival on GI.
A powerful strategy for understanding natural selection is to find the genomic regions, genes, and mutations that contribute to adaptive change. Positive selection distorts sequence variation linked to beneficial mutations, shifting the site frequency spectrum of polymorphisms (Tajima 1989; Braverman et al. 1995), linkage disequilibrium (Przeworski 2002; Kim and Nielsen 2004), and population differentiation (Charlesworth et al. 1997) in a localized manner. By comparing patterns of sequence variation across the genome to theoretical predictions under neutral evolution, the genetic substrates for positive selection can be located (Haasl and Payseur 2016). Although this population genetic framework lacks some of the ingredients necessary for fully demonstrating natural selection (Endler 1986), it offers the benefit that potential instances of selection can be discovered without biases imposed by focusing on certain phenotypes. It also enables the characterization of genomic properties associated with selection.
A key determinant of the success of genome-wide scans for selection is knowledge of the genome. In this regard, GI mice offer several advantages over most other island populations. Because these house mice belong to the same subspecies as laboratory mice (Mus musculus domesticus) (Gray et al. 2014), a high-quality reference genome sequence (Waterston et al. 2002), functional annotations for a large number of genes (Bult et al. 2019), and local recombination rate estimates across the genome (Cox et al. 2009) are already available. Genetic mapping in crosses between GI mice and a mainland strain has identified quantitative trait loci (QTL) that contributed to the evolution of body size (Gray et al. 2015), providing an opportunity to compare the genomic locations of targets of positive selection and alleles involved in phenotypic evolution.
In this article, we characterize positive selection in GI mice by examining genomic patterns of variation using a demography-aware approach. We identify genomic intervals with strong evidence for selection, including known genes and candidate variants that could have driven adaptation to the distinct environment on GI. Our findings provide a rare genomic portrait of natural selection on islands in the wild relatives of a model genetic organism.
Results
Genome Sequences and Variant Calls
High percentages of GI mouse sequencing reads mapped to the mouse reference genome sequence (ranging from 99.2% to 99.6% across individuals), covering high percentages of the genome (93.3–95.9%) (supplementary table 1, Supplementary Material online). Average fold-coverage ranged from 9.02× to 19.59× across mice (supplementary fig. 1, Supplementary Material online). Most variant calls had quality scores >100.
Demographic History
To formulate a reasonable null model for genome-wide scans for selection, we reconstructed major aspects of demographic history for GI mice. We focused our demographic inference on 8,248 5-kb windows on the autosomes and 364 5-kb windows on the X chromosome that were chosen to minimize effects of selection at linked sites (see Materials and Methods). Across this set of windows, GI mice show reduced nucleotide diversity (GI average π/site = 0.0022; Germany average π/site = 0.0026; P < 2.2 × 10−16, paired t-test), a stronger skew toward common alleles (GI average Tajima’s D = 1.23; Germany average Tajima’s D = 0.43; P < 2.2 × 10−16, paired t-test), and higher pairwise linkage disequilibrium (GI average R2 = 0.56; Germany average R2 = 0.48; P < 2.2 × 10−16, paired t-test), compared with German mice (fig. 1). These patterns are consistent with one or more recent reductions in population size since the ancestors of GI mice left western Europe and colonized the island (Gray et al. 2014).

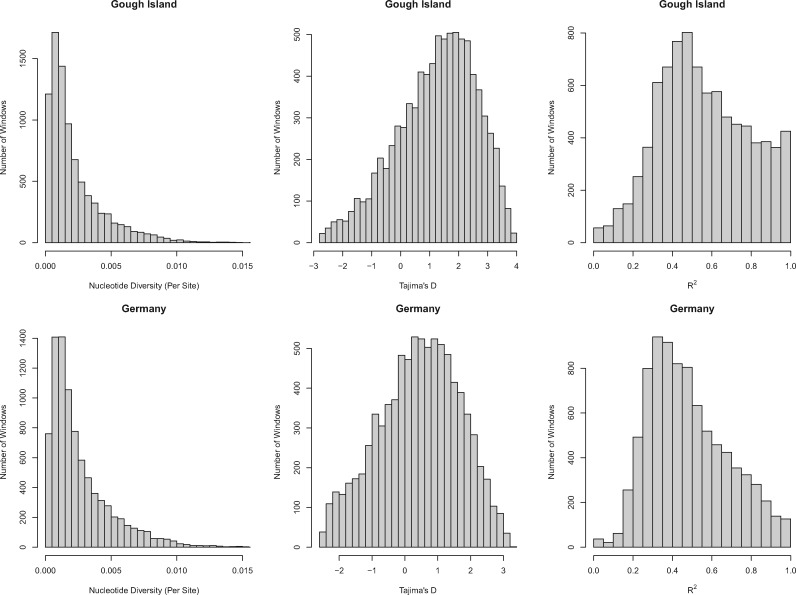
GI mice show reduced nucleotide diversity, a stronger skew toward intermediate frequency alleles, and higher linkage disequilibrium than German mice. Histograms display nucleotide diversity, Tajima’s D, and pairwise R2 in the two populations at the 8,248 5-kb autosomal windows and 364 5-kb X-linked windows used for demographic analyses.
Among the three primary demographic models we considered (supplementary fig. 2, Supplementary Material online), approximate Bayesian computation (ABC) analyses identified a single colonization event followed by a low rate of migration as the most likely scenario (supplementary table 2, Supplementary Material online). Under this model, posterior distributions are distinct from prior distributions for all parameters (supplementary fig. 3, Supplementary Material online) and the likelihood of the data exceeds the likelihood of retained simulations (P > 0.999), indicating patterns of variation were informative about demographic history. Estimates of posterior modes suggest mice first colonized GI close to 1,900 generations ago with an initial effective population size of 145 that later expanded into the thousands (table 1). We estimated a low rate of continuous migration to the island (0.00079 per generation) following initial colonization. Given the remoteness of GI, we interpret these findings as evidence that our sample of mice is descended from multiple colonization events. The exact geographic sources of these events are unclear, though they were probably in western Europe (Gray et al. 2014).

| Parameter | Priors | Posteriors (Arithmetic Scale) | ||||||
|---|---|---|---|---|---|---|---|---|
| Distribution | Minimum | Maximum | Mode | Mean | Median | 50% Lower Confidence Limit | 50% Upper Confidence Limit | |
| Colonization time (generations) | Log-Uniform | 1.6 | 3.7 | 1,896 | 1,248 | 744 | 204 | 1,832 |
| Colonization effective population size | Log-Uniform | 0.3 | 3.7 | 145 | 359 | 112 | 36 | 316 |
| GI effective population size | Log-Uniform | 3.0 | 5.0 | 3,181 | 18,648 | 7,529 | 3,050 | 23,903 |
| Migration rate following colonization | Uniform | 2.5e-6 | 1.3e-3 | 7.9e-4 | 6.9e-4 | 6.9e-4 | 4.1e-4 | 9.4e-4 |
| Mainland effective population size | Log-Uniform | 4.0 | 5.7 | 206,814 | 175,397 | 174,341 | 97,154 | 241,652 |
| Mainland bottleneck strength | Log-Uniform | −4.0 | −1.0 | 6.1e-4 | 1.9e-3 | 7.3e-4 | 4.9e-4 | 1.3e-3 |
| Mainland bottleneck duration (generations) | Log-Uniform | 3.0 | 4.2 | 5,672 | 5,484 | 5,006 | 2,943 | 7,581 |
| Mutation rate | Log-Uniform | −9.5 | −7.0 | 6.4e-9 | 1.4e-8 | 7.3e-9 | 5.5e-9 | 1.2e-8 |
Genomic Targets of Positive Selection—Broad Patterns
We used simulations assuming the best-fit demographic history to identify 5 kb windows that depart significantly from neutral expectations separately for each summary statistic. Although all tests yield low P values for many windows, using the false discovery rate to account for multiple testing across the genome leaves a small percentage of significant windows (using genome-wide q < 0.01 as the threshold) for most statistics (table 2). One exception is Fst: 8% of tested windows on the autosomes and 16.8% of X-chromosomal windows show higher population differentiation between GI mice and German mice than expected under neutrality (q < 0.01). This result probably reflects a higher false-positive rate for Fst; inter-locus variance in Fst fit our demographic model less well than other summary statistics used for ABC inference (data not shown). Fay and Wu’s H (on the autosomes) and SweeD (on the X chromosome) are the only other tests that detect appreciable percentages of windows as departing from neutrality after multiple test correction. The full list of summary statistics, P values, and genome-wide q-values for each individual test of neutrality (along with nucleotide diversity) for every window is accessible through Data Dryad (supplementary table 3, Supplementary Material online).

| Autosomes | X Chromosome | |||||||
|---|---|---|---|---|---|---|---|---|
| Gough | Germany | Gough-Specific | Gough vs. Germany | Gough | Germany | Gough-Specific | Gough vs. Germany | |
| Tajima’s D | 22a (460,141)b | 56 (469,515) | 22 0.005%c | — | 0 (23,509) | 0 (26,051) | 0 0% | — |
| Fay and Wu’s H | 1,275 (437,200) | 7,731 (454,385) | 1,152 2.63% | — | 0 (19,370) | 58 (20,823) | 0 0% | — |
| SweeD | 0 (477,679) | 0 (477,682) | 0 0% | — | 1,232 (33,562) | 0 (33,562) | 1,232 3.67% | — |
| H12 | 0 (478,477) | 0 (478,477) | 0 0% | — | 0 (32,968) | 0 (32,968) | 0 0% | — |
| iHS | 23 (300,057) | 5 (298,742) | 23 0.008% | — | 0 (14,271) | 0 (15,132) | 0 0% | — |
| nSL | 443 (447,414) | 1 (464,843) | 443 0.0004% | — | 0 (23,036) | 0 (24,644) | 0 0% | — |
| OmegaPlus | 0 (477,685) | 1 (477,685) | 0 0% | — | 0 (33,563) | 0 (33,563) | 0 0% | — |
| Fst | — | – | — | 38,287 (478,477) 8.00% | — | — | — | 5,539 (32,966) 16.80% |
| xpEHH | — | — | — | 582 (305,747) 0.19% | — | — | — | 34 (17,748) 0.19% |
| SWIF(r) | — | — | 1,375d (480,707) 0.29% | — | — | — | 88 (33,563) 0.26% | — |
a Counts are numbers of significant windows identified by conducting neutral simulations of the best-fit demographic history and accounting for multiple testing at a genome-wide q < 0.01.
b Parentheses indicate total windows that could be examined for each test.
c Percentages are percentages of significant windows found in GI mice but not in German mice.
d For SWIF(r), significant windows were identified as those with posterior probabilities ≥0.9.
To boost power and combine evidence for selection across different aspects of variation, we used the machine learning approach SWIF(r) (Sugden et al. 2018) to estimate posterior probabilities of positive selection for 5 kb windows under the hypothesis of a complete selective sweep. We focus on findings from this approach for the remainder of the Results. We subsequently refer to genomic intervals with strong evidence for positive selection based on SWIF(r) analyses as “selection windows.” 97.9% of windows across the genome have posterior probabilities ≤0.01. A quantile–quantile plot of posterior probabilities from the data versus those from neutral simulations revealed a modest inflation in the data (supplementary fig. 4, Supplementary Material online), suggesting that posterior probabilities should be interpreted with caution. Posterior probabilities for every window can be found in Data Dryad (supplementary table 3, Supplementary Material online).
To identify broad genomic patterns of positive selection in GI mice, we first consider the set of 1,375 autosomal windows and 88 X-chromosome windows with SWIF(r) posterior probabilities ≥0.9. Neutral simulations suggested that using this conservative significance threshold leads to a low rate of false positives at the genome-wide level (Materials and Methods). Selection windows are more likely to overlap with genes on the autosomes (P = 0.0003; Fisher’s exact test). Among all autosomal windows that overlap with genes, selection windows are enriched for derived, nonsynonymous variants with frequencies ≥0.5 greater in GI mice than in German mice (P = 0.0005). Autosomal selection windows have higher recombination rates than nonselection windows (P = 3.7 × 10−5 Wilcoxon signed-rank test). Selection windows on the X chromosome are not enriched for genes (P = 1), are not enriched for nonsynonymous variants with large frequency differences (P = 1), and recombine at a slightly lower rate (P = 0.02). The proportion of selection windows on the autosomes and the X chromosomes is not significantly different (P = 0.46). To determine whether selection windows are nonrandomly distributed in the genome with regard to QTL for body size, we randomized the genomic locations of selection windows and computed the proportion of them located ±2 Mb from estimated QTL peaks for body weight and growth rate (Gray et al. 2015). This permutation test suggested that fewer selection windows are found near QTL than expected by chance (P = 0.0002; 10,000 permutations).
Gene ontology analysis revealed that the genes found in selection windows are biased toward certain categories of biological components, processes, and functions (supplementary table 4, Supplementary Material online). A neurological theme is visible among enriched terms, which include “synapse,” “synapse part,” “neuron part,” “postsynaptic density,” “integral component of postsynaptic density,” “neuronal cell body,” “postsynaptic specialization,” “apical dendrite,” and “intrinsic component of postsynaptic specialization” (supplementary table 4, Supplementary Material online). Some of the highest enrichment values are associated with these neurological terms. Other themes include protein binding, membranes, and organelles.
Genomic Targets of Positive Selection—Candidate Genes and Variants
To gain deeper biological insights into the targets of selection in GI mice, we examined a subset of genomic regions in detail. Plots of the SWIF(r) probability of selection along chromosomes point to regions of interest (fig. 2), several of which we highlight here.

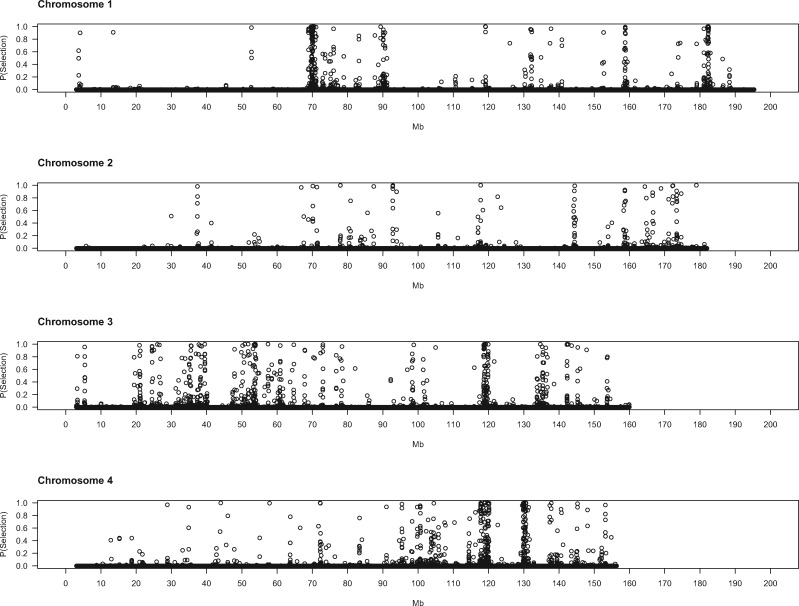
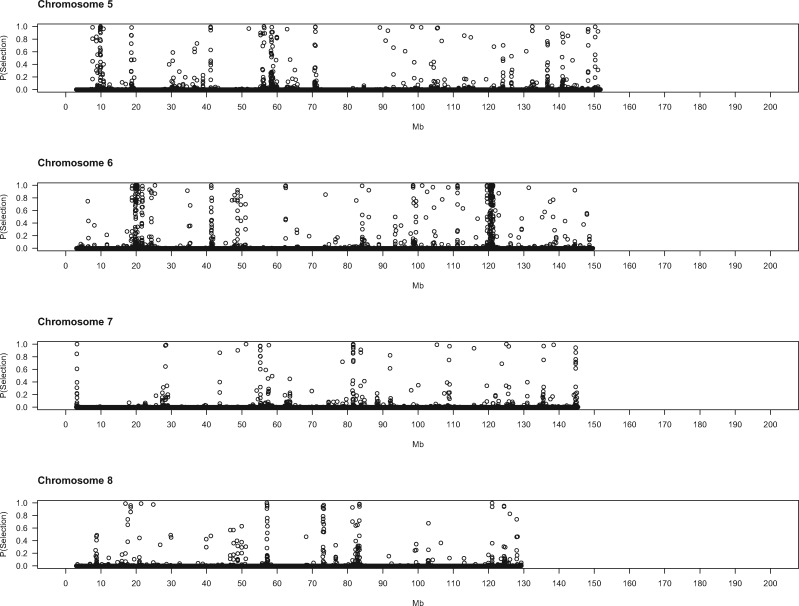
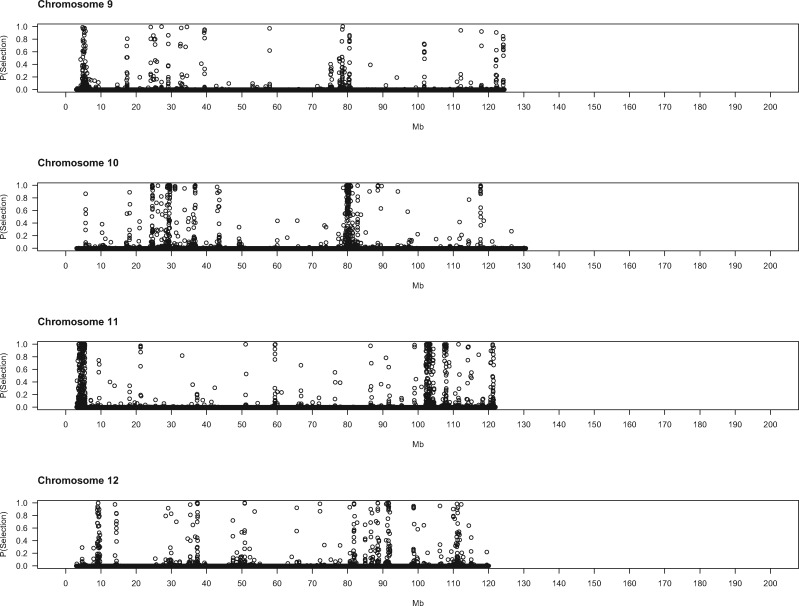
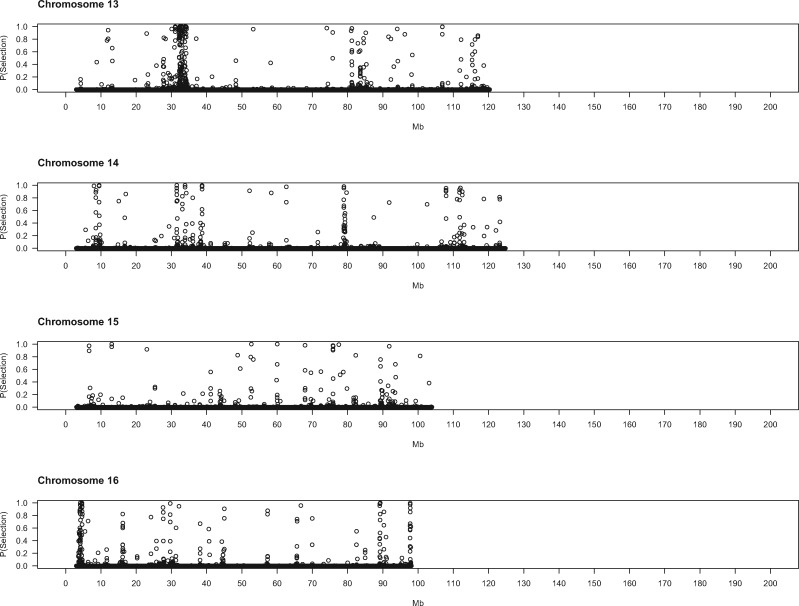
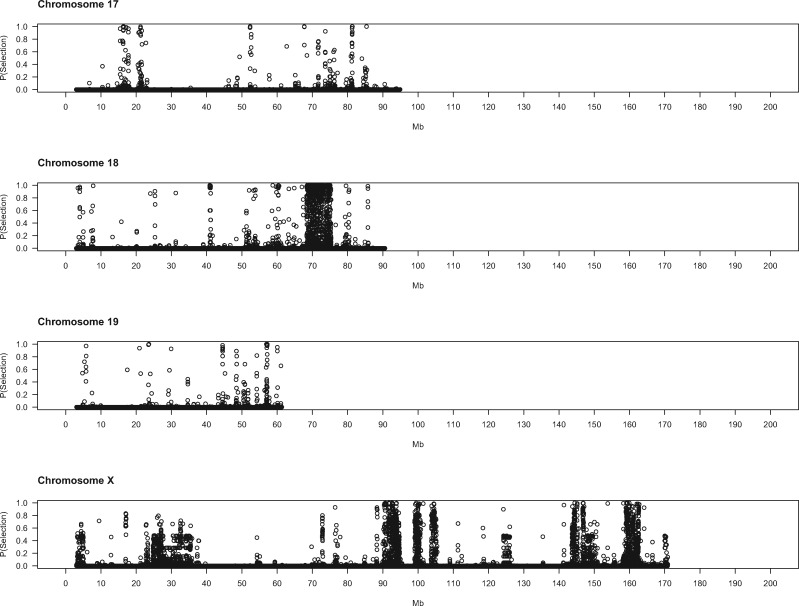
Evidence for positive selection across the genome. Each dot represents a 5-kb window. X-axis denotes the chromosomal start position of each window in megabytes. Y-axis shows the probability of selection estimated by SWIF(r).
Some genomic regions are notable for the spatial extent of their selection signatures along the chromosome. One region on chromosome 18 shows particularly striking patterns, with high SWIF(r) probabilities across approximately 5 Mb (68.5–73.5 Mb) (fig. 2). Deviations in multiple summary statistics distinguish this genomic interval (fig. 3). Many windows contain little to no nucleotide diversity in GI mice (but retain typical levels in German mice). Overall, the strongest signal is found from approximately 71.5–73.2 Mb, an interval overlapping with only one known gene: Dcc. There are no nonsynonymous variants at this gene in our sample. Potential regulatory variants include one mutation predicted to affect splicing and several other intron mutations that show large frequency differences of the derived allele between GI mice and German/French mice and are found in sequence elements significantly conserved across placental mammals (supplementary table 5, Supplementary Material online). The DCC protein is an axon guidance receptor that interacts with translational machinery in neurons (Russell and Bashaw 2018). Mice with disruptions at Dcc exhibit a range of nervous system defects, including ataxia and corticospinal lesions (Finger et al. 2002).

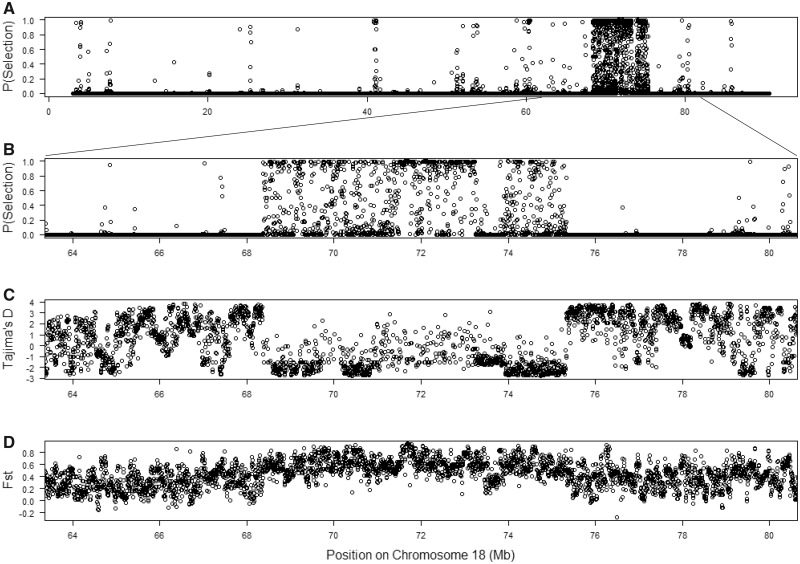
A region on chromosome 18 shows strong evidence for positive selection. The first panel plots the SWIF(r) probability of selection for the whole chromosome. Panels 2–4 display the SWIF(r) probability of selection, Tajima’s D, and Fst across the interval.
On the proximal end of chromosome 11, near the centromere, is another expansive region (3.8–5.5 Mb) containing many windows with high probabilities of selection (fig. 2). Variants in or near several genes in this interval show substantial differences in derived allele frequency and fall within noncoding sequences that are conserved across mammals (supplementary table 5, Supplementary Material online). Disrupting Mtmr3 changes body size (Bush et al. 2012), making this gene a good candidate for selection in this interval of chromosome 11. Toward the other end of chromosome 11 lies another large area (102–104 Mb) displaying strong signatures of selection. Three genes in this region each harbor a single derived amino acid change with large allele frequency differences between GI mice and German/French mice (table 3): Hexim2, Map3k14, and Lrrc37a. Little information about the phenotypic consequences of disrupting these genes is available. Other genes in this region contain derived mutations with large allele frequency differences in conserved noncoding sequence elements (supplementary table 5, Supplementary Material online), including variants in splice regions (Atxn7l3, Ubtf), nearby intergenic variants (Tmub2, Ubtf, Slc25a39, Fzd2, Dcakd, Nmt1), and UTR sequences (Rundc3a, Fzd2).

| Chromosome | Selection Candidate Region (Mb) | Gene | Derived Allele Frequency, GI | Derived Allele Frequency, Germany | Derived Allele Frequency, France | Amino Acid in Outgroup (Mus spretus) | Derived Amino Acid | Amino Acid Position in Protein | Blosum Score | SIFT Prediction | PolyPhen-2 Prediction |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 83.3–83.4 | Mgat4d | 1 | 0.125 | 0.125 | Thr | Met | 222 | −1 | Tolerated | Possibly damaging |
| 9 | 5.0–6.0 | Casp12 | 0.8571 | 0 | 0 | Ile | Leu | 15 | 2 | Tolerated | Benign |
| 9 | 5.0–6.0 | Casp12 | 0.8929 | 0 | 0.0625 | Pro | Leu | 105 | −3 | Affect function | Benign |
| 10 | 79.6–80.5 | Madcam1 | 1 | 0.4375 | 0.3125 | Thr | Ala | 350 | 0 | Affect function (low confidence) | Benign |
| 10 | 79.6–80.5 | Fgf22 | 0.8929 | 0.0625 | 0.25 | Arg | Trp | 145 | −3 | Affect function | Probably damaging |
| 10 | 79.6–80.5 | Prss57 | 0.9286 | 0.0625 | 0.25 | Arg | Pro | 148 | −2 | Tolerated | Benign |
| 10 | 79.6–80.5 | Gpx4 | 0.8929 | 0.125 | 0.1875 | Ala | Thr | 51 | 0 | Affect function (low confidence) | Unknown |
| 10 | 79.6–80.5 | Sbno2 | 0.9643 | 0.125 | 0.1875 | Ser | Pro | 42 | −1 | Tolerated | Benign |
| 11 | 102.2–103.5 | Hexim2 | 0.8661 | 0.0625 | 0.1875 | Val | Gly | 294 | −3 | Affect function (low confidence) | Benign |
| 11 | 102.2–103.5 | Map3k14 | 0.8125 | 0.1875 | 0.375 | Val | Ala | 440 | 0 | Tolerated | Benign |
| 11 | 102.2–103.5 | Lrrc37a | 0.8214 | 0 | 0 | Arg | Gln | 250 | 1 | Tolerated | Possibly damaging |
| 12 | 91.6–91.7 | Ston2 | 1 | 0.3125 | 0.1875 | Met | Leu | 281 | 2 | Affect function (low confidence) | Unknown |
| 12 | 91.6–91.7 | Ston2 | 1 | 0 | 0 | Ala | Asp | 218 | −2 | Affect function (low confidence) | Benign |
| 14 | 78.9–79.0 | Vwa8 | 0.8929 | 0 | 0 | Lys | Arg | 555 | 2 | Tolerated | Benign |
| 14 | 78.9–79.0 | Vwa8 | 0.8929 | 0 | 0 | Glu | Lys | 724 | 1 | Tolerated | Benign |
| 16 | 3.9–4.2 | Nlrc3 | 0.9286 | 0 | 0.1875 | Pro | Ser | 155 | −1 | Affect function (low confidence) | Benign |
| 16 | 3.9–4.2 | Slx4 | 1 | 0.5 | 0.5 | His | Arg | 1225 | 0 | No results | Benign |
| 16 | 3.9–4.2 | Slx4 | 1 | 0.375 | 0.1875 | Gln | Pro | 98 | −1 | No results | Probably damaging |
| 19 | 56.9–57.3 | Vwa2 | 1 | 0.375 | 0.375 | Ala | Glu | 225 | −1 | Tolerated | No results |
| 19 | 56.9–57.3 | Vwa2 | 1 | 0.3125 | 0.125 | Asn | Asp | 713 | 1 | Tolerated | No results |
The stretch of chromosome 10 from 79.6 to 80.5 Mb is also of interest (fig. 2). This 900-kb region spans several genes with established connections to body size and/or behavior. Mice deficient in Sbno2 have higher bone mass caused by impaired osteoclast fusion (Maruyama et al. 2013). Disrupting Gpx4—a gene with a potential downstream regulatory variant in our sample (supplementary table 5, Supplementary Material online)—in mice produces a variety of phenotypes, including loss of neurons in the hippocampus, seizures, and decreased body weight (Seiler et al. 2008; Yoo et al. 2012). Hcn2 plays roles in locomotion, balance, and weight accumulation (Chung et al. 2009). Mice without Efna2 have abnormal neuron differentiation (Holmberg et al. 2005). Fgf22 facilitates differentiation of excitatory nerve terminals in hippocampal neurons (Terauchi et al. 2010). Five genes in this region of chromosome 10 harbor derived missense mutations with large frequency differences between GI mice and mice from Germany and France (table 3). One of these variants, located in Fgf22, is predicted to damage protein function (table 3).
Some genomic regions show spatially narrower signals of selection that point to single genes. On chromosome 13, within a broad region displaying evidence for selection that stretches from approximately 32–34 Mb (fig. 2), lies a more localized signal (32.2–32.6 Mb). Gmds is the only gene annotated in this interval. Mice with disrupted Gmds have altered numbers of immune cells and craniofacial morphology (Bush et al. 2012). Potential regulatory variants with large differences in derived allele frequency are found in the introns of Gmds and nearby intergenic sequence elements (supplementary table 5, Supplementary Material online).
One annotated gene is located within the 100-kb interval with the strongest signal (91.6–91.7 Mb) (fig. 2) on chromosome 12: Ston2. This gene functions in synaptic transmission; synapses from Ston2 knockout mice show impaired short-term plasticity (Kononenko et al. 2013). Consistent with this defect, Ston2 knockouts exhibit increased exploration in an open field test, including faster approach to a novel object (Kononenko et al. 2013). Two derived, missense variants in Ston2 are fixed in our sample of GI mice; one variant is absent from the Germany and France samples, and the other is polymorphic in both (table 3).
A few additional regions showing strong evidence for selection are notable because of their proximity to QTL for body size evolution in GI mice (Gray et al. 2015). A selection interval on chromosome 6 (20.0–20.5 Mb) (fig. 2) is located approximately 2 Mb distal from a QTL peak (within the QTL confidence interval). This region features many contiguous windows with SWIF(r) probabilities of 1. This interval contains no SNPs that satisfy our criteria for regulatory variants, nor does it contain annotated genes. Another selection interval is found on chromosome 8 (83.3–83.4 Mb) (fig. 2), roughly 4 Mb distal from a QTL peak. Functional information is available for two genes in this region. Elmod2 affects body fat, cholesterol, bone mineral content, and gait in mice (Bush et al. 2012). Mouse mutants for Mgat4d exhibit abnormal morphology of the esophagus and spleen (Bush et al. 2012). Potential regulatory variants in this interval are found in UTR sequences (Elmod2), downstream intergenic elements (Elmod2), introns (Mgat4d), and splice sites (Mgat4d). Selection patterns in these genomic regions and others could help to narrow candidate intervals for QTL responsible for the evolution of extreme body size.
Discussion
Mice from GI provide a compelling example of the island rule (Gray et al. 2015). Compared with their mainland counterparts, these mice evolved enormous body sizes, as well as novel behaviors (Rowe-Rowe and Crafford 1992; Jones et al. 2003; Gray et al. 2015). In previous research, we used genetic mapping to identify genomic regions involved in morphological evolution in GI mice (Gray et al. 2015; Parmenter et al. 2016). In this article, we present evolutionary inferences from a complementary approach that is unbiased with respect to phenotype.
Our study is one of the first genome-wide scans for selection to be applied to an instance of the island rule. Genomic regions associated with changes in body size in selected lines of laboratory mice showed reduced variation on an SNP array in big house mice from the Faroe Islands and St. Kilda (Chan et al. 2012). In contrast to this work and other population genomic characterizations of selection in island populations that focus on SNP genotyping, exome sequencing, or other reduced representation methods, our approach considers full patterns of sequence variation from throughout the genome.
Application of a machine learning approach that integrates patterns of variation while accounting for demographic history located 1,463 genomic windows showing strong evidence for positive selection, suggesting that adaptive evolution has been frequent since mice colonized GI. The nonrandom genomic distribution of selection windows further supports the notion that positive selection has left footprints in the genome. We observed stronger signatures of selection in windows containing genes. Among genic windows, selection windows are enriched for nonsynonymous variants that are derived and at high frequency (or fixed) in GI mice. A subset of these variants likely contributed to island adaptation.
The instances of selection we uncovered probably reflect responses to diverse aspects of the novel environment faced by GI mice. The enrichment of genes with neurological functions in selection windows suggests the evolution of new behaviors. Some of the intervals with the most striking selection patterns contain genes with established effects on behavior, including the willingness and ability to explore novel environments. For a subset of these genes, the molecular mechanisms that mediate behavior have already been investigated. For example, Ston2 is an endocytic adaptor that sorts proteins in synaptic vesicles in the hippocampus (Kononenko et al. 2013). The enhanced boldness observed in Ston2 knockout mice resembles impulsivity seen in schizophrenia and Tourette’s syndrome, two disorders that may be associated with Ston2 variants in humans (Breedveld et al. 2010; Luan et al. 2011). Our sample of GI mice is fixed for two derived missense mutations in Ston2; these variants deserve functional characterization. In fact, many genomic regions with strong evidence for positive selection contain a single gene, making them good targets for genome editing. More broadly, the enrichment of neurological genes in selection windows should motivate the examination of behavioral evolution in GI mice. We have established inbred lines of GI mice that we are using for these purposes.
The evolution of extreme body size raises the possibility that selection stimulated the spread of mutations that increase size in GI mice. We considered this scenario by comparing the locations of selection windows with QTL for body size evolution (Gray et al. 2015). Although we found strong evidence for selection near a few QTL peaks, selection windows as a group were not enriched for QTL overlap. Perhaps the relatively low genomic resolution of body size QTL, which was mapped to Mb-scale intervals (Gray et al. 2015), obscures a true relationship between QTL and selection windows. Many QTL contain selection windows within their confidence intervals, but uncertainty about the positions of causative mutations makes it difficult to draw connections across disparate genomic scales. In addition, genetic drift could be responsible for the evolution of larger bodies, facilitated by a reduction in the effective population size that accompanied colonization of the island. This scenario seems less likely because the body size of GI mice has continued to increase over time (Rowe-Rowe and Crafford 1992) and the distribution of QTL effects supports the action of selection on size or traits correlated with it (Gray et al. 2015). Another possibility is that the evolution of large body size was caused by selection on standing genetic variation. Most of the signatures we searched for—including skews in site frequency spectra, unusual haplotype structure, and elevated population differentiation—are predicted when selection targets new beneficial alleles. In contrast, when selection acts on pre-existing variants that persisted in the population, recombination before the onset of selection can decouple beneficial and neutral mutations, substantially weakening detectable patterns in linked variation (Hermisson and Pennings 2005; Przeworski et al. 2005). If the evolution of large bodies was driven by selection on standing variation, we would not expect strong signatures of selection near most QTL for size. Low power to find selection targeting standing variation is a limitation of our study that extends beyond consideration of body size.
Our findings offer additional lessons about genomic scans for selection. Although analytical approaches that jointly consider multiple measures of sequence variation are becoming more popular (Grossman et al. 2010; Schrider and Kern 2018; Sugden et al. 2018), it is still common practice to focus on one or a few summary statistics. This is true despite the reasonable expectation that tests of neutrality vary in their statistical properties, including robustness to neutral departures from equilibrium and power to detect selection on different timescales. The disparate numbers of selection windows we detected using different tests (table 2) indicate that we would have reached divergent conclusions about the genomic determinants of selection in GI mice if we had relied on one or a few tests. This empirical result should help motivate comparisons of method performance on common simulated data sets generated under a diverse range of scenarios (Adrion et al. 2020), as well as the application of approaches that integrate different signals of selection (Grossman et al. 2010; Schrider and Kern 2018; Sugden et al. 2018).
Other caveats accompany our interpretations. Our genomic scans used Germany as a reference population. Although we were motivated to find targets of selection in GI mice, selection windows we identified could reflect instances of adaptive evolution that preceded colonization of the island (but followed the split between GI and German populations). Future research with larger samples from the island and additional mainland populations could enable estimation of the timescale of selection. The ability of demographic changes to produce genomic patterns that resemble selection signatures raises another caveat. We reconstructed major aspects of demographic history to control false-positive rates in selection scans. The simplified nature of the demographic models we considered likely missed important characteristics of the true colonization history. For example, because we lacked specific information about the source population for GI mice, we treated the colonization of GI and the split of GI and German populations as simultaneous events. If the GI-German split happened much earlier than island colonization, this modeling assumption could have led to overestimation of colonization time, which in turn could have reduced the accuracy of neutral predictions for selection scans. Accurate reconstruction of demographic history remains a challenge, even with genomic data.
Despite these issues, our results provide an initial portrait of the genomic landscape of positive selection in an island population exhibiting substantial phenotypic evolution. The repeated colonization of islands by house mice and other murid rodents will make it possible to determine whether the targets of selection we discovered have been used to adapt to novel conditions on other islands. This comparative population genetic approach is appealing, especially given the common characteristics of phenotypic evolution that characterize adaptation to islands (Foster 1964; Van Valen 1973; Lomolino 1985; Adler and Levins 1994).
Materials and Methods
Mice
Fifty mice from GI were collected by Henk Louw and Paul Visser, supervised by Richard Cuthbert and Peter Ryan. Livers were extracted in the field, stored in 70–100% ethanol, and transported to the University of Wisconsin – Madison. Genotypes at 21 dinucleotide microsatellites (Gray et al. 2014) were used to choose a subset of 14 mice that were not closely related for genome sequencing. These mice were sampled near a field station on GI. We considered publicly available genome sequences for eight house mice from Germany (Harr et al. 2016) as a reference sample for comparison in all analyses. Although the source population(s) for GI mice is (are) not known, mitochondrial DNA sequences and microsatellite genotypes indicate the mice likely originated in western Europe (Gray et al. 2014). Additional genome sequences for eight house mice from France (Harr et al. 2016) were used solely in analyses of selection candidate variants to improve confidence. All mice used in population genetic analyses belonged to the Mus musculus domesticus subspecies (Gray et al. 2014; Harr et al. 2016).
Sequencing
Genomic DNA was extracted from livers of GI mice using a Qiagen DNeasy blood and tissue kit. DNAs were further cleaned by phenol and chloroform to meet quality requirements for genome sequencing. Genome sequencing was completed in the University of Wisconsin – Madison Biotechnology Center. DNA concentration and sizing were verified using the Qubit dsDNA HS Assay Kit (Life Technologies, Carlsbad, CA, USA) and the Agilent DNA 1000 chip (Agilent Technologies, Inc., Santa Clara, CA, USA), respectively. Samples were then prepared using the TruSeq PCR Free Sample Preparation kit (Illumina Inc., San Diego, CA, USA), with minor modifications. Libraries were size-selected for an average insert size of 550 bp using SPRI-based bead selection. Quality of the finished libraries was assessed using an Agilent DNA 1000 chip and qPCR quantification was performed using the Kapa Illumina NGS Library Quantification Kit (KAPA Biosystems, Wilmington, MA). Libraries were standardized to a concentration of 2 nM. Cluster generation was performed using the Illumina Rapid PE Cluster Kits v2 and the Illumina cBot. Paired-end, 100-bp sequences were generated using Rapid v2 SBS chemistry on an Illumina HiSeq2500 sequencer. Images were analyzed using the standard Illumina Pipeline, version 1.8.2.
Mapping Sequencing Reads and Calling Variants
Sequencing reads were mapped to the mouse mm10 genome reference sequence (downloaded from http://genome.ucsc.edu/index.html on 12/11/2014) using bwa-mem (Li and Durbin 2009) with default settings. Samtools was used to sort and merge bam files, Picard tools software suite (http://broadinstitute.github.io/picard/) was used for marking and removing duplicates, and GATK (McKenna et al. 2010) IndelRealigner was used for indel realignment. Raw SNP and indel calls were obtained from the alignment files using GATK (McKenna et al. 2010) HaplotypeCaller with the following setting: –genotyping_mode DISCOVERY,–output_mode EMIT_VARIANTS_ONLY, -stand_call_conf 30. Variants with QUAL <100 were removed. To improve consistency among samples, we re-mapped reads to the same reference sequence and called variants for the German mice and French mice (Harr et al. 2016) following the same procedures.
Inference of Demographic History
To facilitate genome-wide scans for selection, we first used patterns of sequence variation to reconstruct the demographic history of our sample of GI mice. Our primary goal was to minimize the risk of false positives in genome-wide selection scans by formulating an improved null (neutral) model that incorporated major aspects of demographic history.
We summarized patterns of sequence variation for demographic inference using statistics designed to capture information about levels of variation, the site frequency spectrum, population differentiation, and haplotypes and linkage disequilibrium. Nucleotide diversity (Nei and Li 1979), Watterson’s theta (Watterson 1975), Tajima’s D (Tajima 1989), numbers of unique and shared SNPs (in GI mice vs. German mice) (Wakeley and Hey 1997), Fst (in GI mice vs. German mice) (Hudson et al. 1992), and average R2 (across SNP pairs) (Weir et al. 2004) were computed from unphased genotypes. Summary statistics computed from unphased genotypes used the same sample size (n = 28 for autosomal loci), regardless of whether some genotypes were missing. Haplotype number, haplotype heterozygosity, and frequency of the most frequent haplotype were computed from haplotypes phased using Beagle version 4.1 (Browning and Browning 2007) over 20 iterations and assuming cM recombination distances estimated from the genetic map for the laboratory mouse (Cox et al. 2009) (downloaded from http://cgd.jax.org/mousemapconverter/help/data_resource). For phased data, missing genotypes were imputed by Beagle. All summary statistics were computed in nonoverlapping 5-kb genomic windows. To minimize the effects of selection at linked sites, windows for analyses of demographic history were chosen based on the following criteria: at least 100 kb away from any annotated gene, at least 20 kb away from each other, local recombination rate at least 0.5 cM/Mb (estimated from the mouse genetic map) (Cox et al. 2009), and two or more SNPs so that all summary statistics could be calculated. This filtering resulted in 8,248 windows on the autosomes and 364 windows on the X chromosome. We used averages and variances of each summary statistic across these windows (treating the autosomes and the X chromosome separately) for demographic inference.
We used ABC (Beaumont et al. 2002) facilitated by ABCtoolbox (Wegmann et al. 2010) to evaluate the fit of three models (supplementary fig. 2, Supplementary Material online) to the data: a single colonization of GI modeled as an instantaneous population split (with colonization time, colonization effective population size, island current population size, mainland current population size, mainland bottleneck time, and mainland bottleneck size as parameters) (Model 1), a single colonization of GI followed by continuous migration to the island (parameters for Model 1 plus migration rate) (Model 2), and two colonization events (parameters for Model 1 plus second colonization time and colonization size) (Model 3). Prior distributions for all parameters were bounded to include likely scenarios (Gray et al. 2014). Prior distributions were log-uniform, except for migration rate, which followed a uniform distribution. X-linked loci were treated the same as autosomal loci, but with ¾ the population size. Simulations were performed using ms (Hudson 2002). To match the data set, we simulated 8,248 autosomal windows (made up of 10 recombination rates: 0.67, 1.42, 2.38, 3.38, 4.43, 5.47, 6.49, 7.55, 8.52, and 9.16 cM/Mb) and 364 X-linked windows (made up of three recombination rates: 0.69, 1.19, and 3.17 cM/Mb) for each parameter combination.
Summary statistics were transformed via partial least squares in ABCtoolbox (Wegmann et al. 2009). A subset of PLS components was used by the rejection method implemented in ABCestimator (ABCtoolbox) to infer posterior distributions for demographic parameters. We assessed model fit by comparing the likelihood of the observed data with likelihoods of 1,000 retained simulations in ABCtoolbox. We compared the fits of the three primary models by computing Bayes factors.
Genome-Wide Scans for Selection
We conducted a wide variety of well-established tests of neutrality in all nonoverlapping 5-kb windows in the genome to which sequences mapped reliably to the reference and which featured high-quality SNP calls. Tests focused on the site frequency spectrum included Tajima’s D (Tajima 1989), a normalized version (Zeng et al. 2006) of Fay and Wu’s H (Fay and Wu 2000), and SweeD (Pavlidis et al. 2013), each computed using unphased data. Tests emphasizing haplotype structure and linkage disequilibrium included OmegaPlus (Alachiotis et al. 2012), nSL (Ferrer-Admetlla et al. 2014), iHS (Voight et al. 2006), and H12 (Garud et al. 2015). Tests of population differentiation included xpEHH (computed from phased data) (Sabeti et al. 2007) and Fst (computed from unphased data) (Hudson et al. 1992). We used software sweed (https://github.com/alachins/sweed) for SweeD and omegaplus (https://github.com/alachins/omegaplus) for OmegaPlus, setting the minimum window size to 100, the maximum window size to 100,000, and the number of windows equal to chromosome length divided by 5,000. We used selscan (Szpiech and Hernandez 2014) with default settings for iHS, nSL, and xpEHH tests. For tests that required polarization of SNP alleles as derived or ancestral, we assumed that alleles present in an available genome sequence from Mus spretus (Keane et al. 2011) were ancestral.
The expected distribution of each statistic under neutrality was approximated by conducting 1,000,000 simulations in ms (Hudson 2002). These simulations assumed parameter values drawn from the modes (with 50% confidence intervals) of posterior distributions inferred for the best-fitting demographic model, with six recombination rates for autosomal windows (0, 1, 2, 4, 8, and 20 cM/Mb) and three recombination rates for X-linked windows (0, 1, and 3 cM/Mb). For each window and test, a P value was computed as the proportion of simulated windows with more extreme values than the observed value. Tests were one-tailed in the direction expected for a selective sweep. To account for multiple testing, a q-value for each window and test was calculated from the genome-wide distribution of P values (treating autosomal windows and X-linked windows separately) using the q-value package in R. We called significant windows with q < 0.01.
To jointly consider evidence for selection across tests, we applied SWIF(r) (Sugden et al. 2018) to each 5-kb window. SWIF(r) is a probabilistic method that computes the posterior probability of a selective sweep by learning the distributions of multiple summary statistics under contrasting evolutionary scenarios. Using cosi2 (Shlyakhter et al. 2014) (https://software.broadinstitute.org/mpg/cosi2/), we simulated 1-Mb genomic regions, assuming either neutrality or selection targeting a single beneficial mutation at the center of the interval. All simulations used demographic parameter combinations drawn from the modes (including 50% confidence intervals) of posterior distributions from the best-fitting demographic model. The recombination rate was set to 1 × 10−8/bp (for autosomal loci) or 0.75 × 10−8/bp (for X-linked loci). Selection coefficients were calculated according to equation (4) in Sugden et al. (2018) using estimates of the current effective population size and the time of colonization drawn from 50% confidence intervals of posterior distributions. The resulting selection coefficients ranged from 0.0069 to 0.037. Selection simulations assumed fixation of the beneficial mutation. Summary statistics from simulations were used to train SWIF(r) to discriminate between neutral and selective scenarios. To be conservative, we set the prior probability of selection in a window at 0.0001 (prior probability of neutrality = 0.9999). To examine the performance of SWIF(r) in a genome-wide scan like the one we conducted, we applied SWIF(r) to 480,707 windows simulated using cosi2 and assuming the neutral, best-fit demographic history. In neutral simulations, 99.7% of windows had posterior probabilities ≤0.01, and the proportion of windows that had posterior probabilities ≥0.9 was 0.00025 (121/480,707). For the identification of genomic patterns, we conservatively set a posterior probability threshold at ≥0.9.
Properties of Selection Windows
To identify broad patterns that characterize genomic targets of selection, we examined windows with SWIF(r) probability ≥0.9. We identified all REFSeq genes (https://ncbi.nlm.nih.gov) for which at least part of the protein-coding region is located within a selection window. We used Fisher’s exact tests to determine whether selection windows are enriched for genes and to compare the proportions of selection windows on the X chromosome and the autosomes. We used a t-test to compare recombination rates (Cox et al. 2009) between selection and nonselection windows. We used gene ontology analyses implemented in GOrilla (Eden et al. 2009) to identify biological processes, functions, and components enriched among genes found in selection windows. We compared ontology terms for genes in selection windows with terms for all genes in the genome. We focused on ontology terms with q-values less than 0.05.
Candidate Genes and Variants
To better understand the nature of selection, we examined candidate genes and variants in genomic regions with especially strong evidence for selection. We used plots of the SWIF(r) probability to visually locate regions containing a series of windows with high selection probabilities (usually higher than 0.9) and subsequently inspected patterns of diversity in these windows. Then we found genes with functional annotations in these windows using the UCSC Genome Browser (https://genome.ucsc.edu) and the Mouse Genome Informatics website (https://informatics.jax.org). We focused on genes with known connections to specific organismal phenotypes.
To pinpoint candidate variants, we used the UCSC Genome Browser to create a custom track containing all SNPs from our data set for which GI mice carry the derived allele and the frequency difference of the derived allele is at least 0.5 in two comparisons: GI mice versus German mice and GI mice versus French mice (considering French mice for added confidence). We then used the Variant Annotation Tool in the UCSC Genome Browser to identify 1) the subset of these variants that are missense mutations and 2) the subset of these variants that are noncoding with annotated locations and significant PhastCons conservation across eutherian mammals (using default settings). We subsequently used the Blosum score (https://www.ncbi.nlm.nih.gov/Class/FieldGuide/BLOSUM62.txt) (Henikoff and Henikoff 1992), SIFT (https://sift.bii.a-star.edu.sg) (Sim et al. 2012), and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2) (Adzhubei et al. 2010) to predict whether each missense variant affects protein function.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Data Availability
Summary statistics, P values, q-values, and SWIF(r) posterior probabilities for all analyzed genomic windows (supplementary table 3, Supplementary Material online) are accessible through Data Dryad. Sequencing reads for Gough Island mice are available via the NCBI Sequence Read Archive (BioProject accession PRJNA587779). Variant call format (VCF) files for Gough Island Mice are available through Data Dryad. Scripts for computational analyses are available through GitHub (https://github.com/pjing999/GoughSelectionScan).
Acknowledgments
We thank Peter Ryan, Richard Cuthbert, Henk Louw, and Paul Visser for collecting mice from Gough Island and sharing their tissues. We thank Bettina Harr and Diethard Tautz for early access to genome sequences from mice from Germany and France. We thank Lauren Sugden and Sohini Ramachandran for advice on using SWIF(r). We thank Jeff Jensen for advice on analyses. We thank members of the Payseur lab for useful discussions. This work was supported by National Institutes of Health grants GM100426 and GM120051 to B.A.P.
 Genomic Targets of Positive Selection in Giant Mice from Gough Island
Genomic Targets of Positive Selection in Giant Mice from Gough Island