
Present address: LEAF- Linking Landscape, Environment, Agriculture and Food, Instituto Superior de Agronomia, Universidade de Lisboa, Portugal
- Altmetric
About 15,000 angiosperm species (∼6%) have separate sexes, a phenomenon known as dioecy. Why dioecious taxa are so rare is still an open question. Early work reported lower species richness in dioecious compared with nondioecious sister clades, raising the hypothesis that dioecy may be an evolutionary dead-end. This hypothesis has been recently challenged by macroevolutionary analyses that detected no or even positive effect of dioecy on diversification. However, the possible genetic consequences of dioecy at the population level, which could drive the long-term fate of dioecious lineages, have not been tested so far. Here, we used a population genomics approach in the Silene genus to look for possible effects of dioecy, especially for potential evidence of evolutionary handicaps of dioecy underlying the dead-end hypothesis. We collected individual-based RNA-seq data from several populations in 13 closely related species with different sexual systems: seven dioecious, three hermaphroditic, and three gynodioecious species. We show that dioecy is associated with increased genetic diversity, as well as higher selection efficacy both against deleterious mutations and for beneficial mutations. The results hold after controlling for phylogenetic inertia, differences in species census population sizes and geographic ranges. We conclude that dioecious Silene species neither show signs of increased mutational load nor genetic evidence for extinction risk. We discuss these observations in the light of the possible demographic differences between dioecious and self-compatible hermaphroditic species and how this could be related to alternatives to the dead-end hypothesis to explain the rarity of dioecy.
Introduction
Out of 261,750 angiosperm species, ∼15,600 (∼6%) are dioecious (Renner 2014). Dioecy is thus rare compared with the dominant sexual system in angiosperms, hermaphroditism, in which individuals carry bisexual flowers. Nonetheless, dioecy can be found in ∼40% of angiosperm families. The current view is that many independent (871 to ∼5,000) and mostly recent transitions occurred from hermaphroditism to dioecy in angiosperms (Renner 2014). These events probably followed two main evolutionary paths: one through a gynodioecious intermediate (with female and hermaphrodite individuals), called the gynodioecy pathway, and another one through a monoecious intermediate (with a single individual carrying separate female and male flowers) called the monoecy or paradioecy pathway (Barrett 2002). In few cases, dioecy might have evolved through alternative pathways, such as heterostyly (where individuals have different style and stamen lengths, Barrett 2002).
Species richness was originally found to be lower in dioecious compared with nondioecious sister clades, suggesting that dioecy might lead to an increased extinction rate in angiosperms and defining the evolutionary dead-end hypothesis (Heilbuth 2000). Theoretical work has explored this idea and shown that an increased extinction rate could result from two main evolutionary handicaps (reviewed in Muyle and Marais 2016). First, in dioecious species, only females contribute to seed production and dispersal, which could result in individuals being less dispersed spatially and competing more for local resources compared with populations with hermaphrodites (Heilbuth et al. 2001). This should cause dioecious species to have smaller geographic distributions and species abundance compared with nondioecious species. Second, in dioecious species with insect pollination, male intrasexual competition would result in males being more attractive than females for pollinators, making reproduction and population size sensitive to variation in pollinator density (Vamosi and Otto 2002). This should lead to fluctuating population sizes in dioecious species and in turn affect levels of genetic diversity. It has been proposed that these handicaps could be the reason why dioecy is associated with a specific suite of traits in angiosperms (fleshy fruits, wind-pollination, inconspicuous flowers, and woody growth) as well as ecological preferences because dioecious species are preferentially found in the tropics and on islands (Vamosi et al. 2003). Assuming that pollination mode evolves more slowly than dioecy, dioecy could be more frequent among wind-pollinated species because wind-pollinated species are not subject to the pollinator handicap. Similarly, assuming fruit type evolves more slowly than dioecy, dioecy could be more prevalent in species with fleshy fruits because seeds can be dispersed over longer distances by fruit-eating animals, thus limiting the seed-dispersal handicap.
The evolutionary dead-end hypothesis has recently been challenged (Kafer et al. 2017), in part because the sister clade analysis used in early work assumed that speciation events and trait transitions coincide (Heilbuth 2000). However, if dioecy evolved after the split between the two diverging lineages and was not coincident with the split, then species richness is not expected to be equal between sister clades even when diversification rates are similar (Kafer and Mousset 2014). Using a corrected version of the sister clade analysis on an updated data set, dioecious clades have been found to have actually higher species richness than nondioecious sister clades in angiosperms (Kafer et al. 2014). Using a phylogenetic method to study trait evolution and species diversification on several genera with dioecious species (Maddison et al. 2007), dioecy was not associated with an increased extinction rate (Sabath et al. 2016; Duan et al. 2018). Another hypothesis to explain the rareness of dioecy in angiosperms posits that reversion from dioecy to other sexual systems is frequent and that dioecy is more evolutionary labile than previously thought (Kafer et al. 2017). This hypothesis was supported by a study of 2,145 species from 40 genera in which the authors found evidence of frequent reversions from dioecy to other breeding systems (Goldberg et al. 2017). However, these works relied on macroevolutionary and comparative approaches and the population functioning of dioecious versus nondioecious species has so far been poorly explored. Investigating consequences both at the macroevolutionary and population scale have been fruitful in the study of the evolution of selfing from outcrossing, another frequent breeding system transition in angiosperms. Selfing was also proposed to be an evolutionary dead-end, which has been supported by several macroevolutionary analyses (Goldberg et al. 2010; de Vos et al. 2014; Zenil-Ferguson et al. 2019). In parallel, various negative genetic consequences of selfing (i.e., low genetic diversity, accumulation of deleterious mutations) have been reported in several species, even in those of recent origin (see Glémin et al. 2019 for a review). Interestingly, negative genetic consequences have been detected despite the short-term advantage of selfing provided by reproductive assurance, allowing higher colonization potential and larger species range (Randle et al. 2009; Grossenbacher et al. 2015).
If the seed-dispersal handicap of dioecy operates, it should reduce the ability of dioecious plants to spread and increase local competition, which should lead to smaller geographic distributions and lower species abundance in dioecious compared with nondioecious species. These expectations can be tested using estimates of geographic distributions and species abundance in dioecious and nondioecious species. If the pollinator handicap of dioecy exists, it should cause fluctuations in dioecious population sizes. The effective population size (Ne) is given by the harmonic mean of the varying population sizes over time (Wright 1938). The harmonic mean tends to be dominated by the smallest population sizes that the population goes through. Fluctuating population sizes are therefore expected to decrease the long-term Ne of a species. If the pollinator and the seed-dispersal handicap of dioecy exist, they should cause dioecious species to have smaller Ne compared with nondioecious species. This expectation can be tested by studying genetic diversity to estimate the long-term Ne of dioecious and nondioecious species. A reduced effective population size should then lead to a decrease in the efficacy of selection. This expectation can be tested by comparing closely related dioecious and nondioecious species. A study that used the ratio of nonsynonymous to synonymous substitution rate (dN/dS) as a proxy to selection efficacy found that one out of two dioecious Silene clade had a reduced efficacy of selection for some genes, but positive selection increased in other genes (Kafer et al. 2013). However, transitions to dioecy can happen after speciation, making dN/dS biased because it is computed on entire branches. We therefore decided to use instead a population genetics approach to confront predictions of the dead-end hypothesis in Silene. We relied on statistics that allow quantifying the strength of natural selection, such as rates of adaptive and deleterious nonsynonymous substitutions. If dioecy is not a dead-end, as suggested by macroevolutionary analyses, we expect no effect (or even a positive effect) of dioecy on genetic diversity and the efficacy of selection. Note that if extinction due to the handicaps of dioecy is too rapid to allow for populations to survive long enough to be sampled, our approach would not be able to detect the genetic footprints expected under the dead-end hypothesis. However, as mentioned earlier, similar approaches in selfing species have consistently detected the predicted effects, even in very recent species such as Capsella rubella that originated ∼20,000 years ago from the self-incompatible C. grandiflora (Brandvain et al. 2013; Slotte et al. 2013).
In this study, we focused on 13 species of the large genus Silene (∼700 species) in the Caryophyllaceae family, in which dioecy has evolved independently at least twice (Desfeux et al. 1996). The Silene genus is considered a model system to study the evolution of sexual systems in angiosperms, thanks to its reasonably well-described sexual systems and phylogeny (Bernasconi et al. 2009). Dioecy in Silene is likely to have evolved through the gynodioecy pathway (Fruchard and Marais 2017). If dioecy is associated with the handicaps described above, dioecious Silene species should suffer from them because they do not have any of the traits named previously that likely compensate or prevent these handicaps. Indeed, Silene are short-lived herbs, many of them (but not all) are insect pollinated, they do not have fleshy fruits, and they live in temperate regions. The Silene genus is thus adequate to test for the dead-end hypothesis of dioecy.
We sampled dioecious species from two groups in which dioecy evolved independently (five species in the Melandrium section and two species in the Otites section) along with several hermaphroditic and gynodioecious close relatives for each group and one outgroup (Dianthus chinensis). Silene species can have very large genome sizes (Bernasconi et al. 2009) and no complete reference genome is available in the genus; only a fourth of the S. latifolia genome has been assembled to date (Papadopulos et al. 2015). We therefore used a population genomics approach relying on RNA-seq data, which has proven successful in organisms that lack a reference genome (Cahais et al. 2012; Gayral et al. 2013; Romiguier et al. 2014). Multiple individuals were sampled across the species’ geographic ranges and sequenced by RNA-seq. A pipeline specifically designed for such data was applied to perform SNP-calling (Tsagkogeorga et al. 2012; Gayral et al. 2013; Nabholz et al. 2014). The SNPs were then analyzed to estimate genetic diversity and the efficacy of selection in each species.
Results
Phylogeny, Geographic Range, Species Abundance, and Population Genetics Characterization of the Sampled Species
Our species sampling (13 species in total, see fig. 1) includes two hermaphroditic species (S. viscosa and S. paradoxa), three gynodioecious species (S. vulgaris, S. nutans W, and S. nutans E), and seven dioecious species (section Melandrium: S. heuffelii, S. dioica, S. marizii, S. diclinis, and S. latifolia and section Otites: S. pseudotites and S. otites) from subgenera Behenantha and Silene, the main subgenera in the Silene genus (Jenkins and Keller 2011). The criteria for selecting the nondioecious species are detailed in Materials and Methods. Briefly, we selected 1) close relatives of both dioecious sections based on existing phylogenies, 2) species for which the sexual system is well described, 3) species that had similar geographic ranges, and 4) hermaphroditic species not exhibiting high levels of selfing. The latter criterion was meant to perform a fair comparison between dioecious and hermaphroditic species, not confounded by strong differences in outcrossing rates.

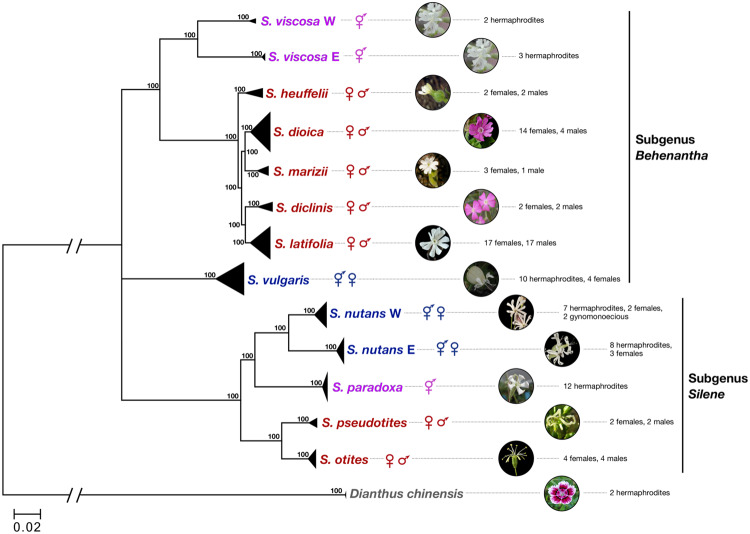
Sampled species and their phylogenetic relationships. The species breeding system is indicated by the corresponding set of greek symbols. Sample sizes and sex are indicated. Bootstrap support values were generated using 100 replicates. Pictures of Silene latifolia, S. dioica, S. nutans W, S. nutans E, and S. otites are from Maarten Strack van Schijndel. Pictures of S. marizii, S. vulgaris, S. paradoxa, and S. viscosa are from Oxelman et al. (2013). Picture of S. heuffelii is from Dr Daniel L. Nickrent.
For each species, between 4 and 34 individuals were sampled from different populations covering the geographic range of the species (see fig. 1 for the number of individuals sampled in every species and supplementary fig. S5, Supplementary Material online, for sample locations). Previous work has shown that reliable estimates of population genetics statistics can be obtained when individuals from populations spanning the whole geographic distribution of a species are sampled, even if the number of sampled individuals is small, that is, two or three (Cahais et al. 2012; Gayral et al. 2013; Romiguier et al. 2014).
Seeds were collected in the wild and plants were grown in controlled greenhouse conditions. Flower buds (and in a few cases, leaves) were sampled for 130 individuals in total and sequenced by RNA-seq (see supplementary table S1, Supplementary Material online, for details on samples). De novo reference transcriptomes were assembled separately for each species and ORFs and orthologs were annotated (see Materials and Methods and supplementary table S1, Supplementary Material online). All dioecious Silene species sampled here have sex chromosomes. Merging autosomal and sex-linked markers can be problematic in population genetics studies as they have different effective population sizes and selection dynamics (Gautier 2014). Therefore, only autosomal ORFs—as predicted by the SEX-DETector method (Muyle et al. 2016)—were kept for downstream analyses (see Materials and Methods).
A phylogenetic tree was built in order to check for congruence with published phylogenies of the Silene genus (Rautenberg et al. 2010; Jenkins and Keller 2011). Orthologous ORFs were aligned among Silene species and D. chinensis, which were used to root the tree. A total of 55,337 variable positions, spread over 1,294 contigs, were used to build the tree (see Materials and Methods). All samples from a given species grouped together as expected (fig. 1). The two lineages of S. nutans formed two clearly distinct groups that should be treated as distinct species as shown in previous studies (Martin et al. 2016, 2017). Silene viscosa samples also formed two highly divergent lineages, even more divergent than the two lineages of S. nutans: a western lineage (S. viscosa W), which includes samples from Sweden and the Czech Republic, and an eastern lineage (S. viscosa E), which includes samples from Bulgaria, Russia, and Kyrgyzstan. We treated these lineages as two differentiated taxa and analyzed them separately hereafter. Unlike in previous work (Slancarova et al. 2013), S. vulgaris did not group with S. viscosa and the section Melandrium, but instead could not be reliably grouped with either subgenus (fig. 1).
A total of 1,088,269 variable nucleotides were called (table 1) using an RNA-seq pipeline (see Materials and Methods). Mean heterozygosity levels and Weir and Cockerham FIT statistics were computed for each species. FIT indicates deviations from Hardy–Weinberg equilibrium caused by processes such as selfing or population structure. FIT values were low to moderate (<0.2) for most sampled species. This indicates that neither population structure nor selfing is creating strong among-species differences in genetic diversity, as already observed (Moyle 2006). However, S. viscosa E exhibited a high FIT, a low heterozygosity level, and low genetic diversity, which could be due to recent demographic shifts or a high selfing rate (fig. 2).

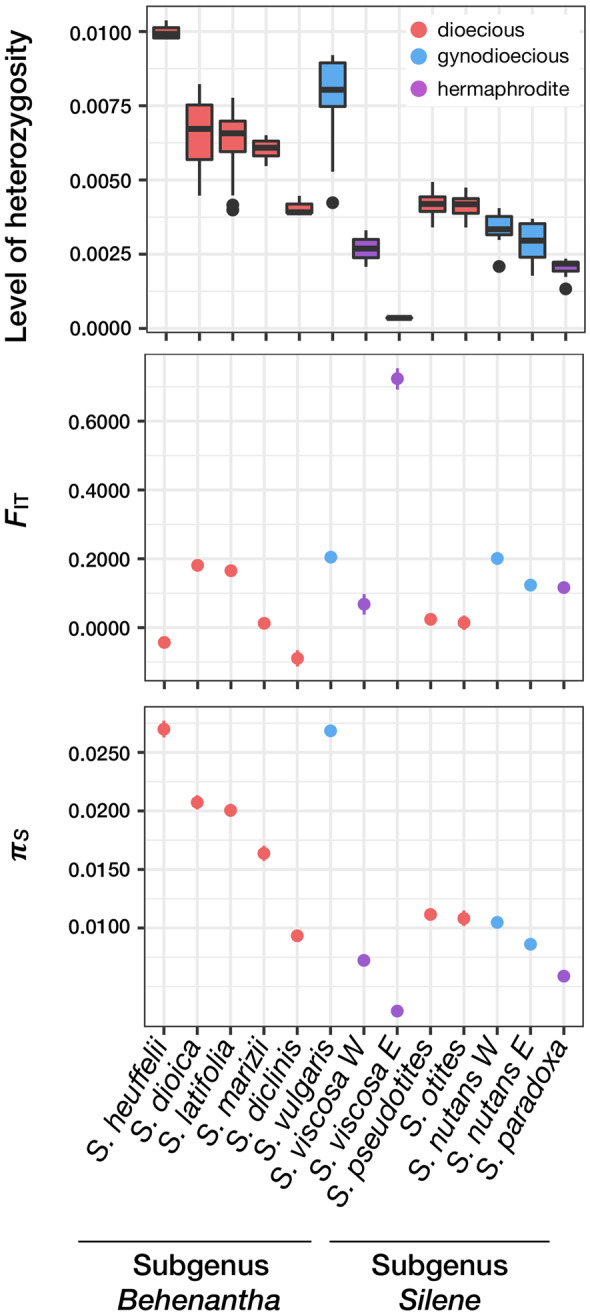
Levels of individual heterozygosity (proportion of heterozygote positions), Weir and Cockerham FIT statistic, and πS within each species. Dots show the mean value and vertical lines mark the 95% confidence interval (obtained by 1,000 bootstraps on contigs).

| Sexual System | Focal Species | Number of Samples | Number of Clean Contigs | Number of SNPs | Weir and Cockerham FIT | π S in Focal Species (%) | π N in Focal Species (%) | π N/πS |
|---|---|---|---|---|---|---|---|---|
| Section Behenantha | ||||||||
| Dioecious | Silene diclinis | 3 | 2,729 | 23,774 | −0.089 [−0.113; −0.065] | 0.934 [0.886; 0.982] | 0.179 [0.170; 0.187] | 0.191 [0.182; 0.200] |
| Silene dioica | 17 | 3,730 | 143,297 | 0.181 [0.176; 0.186] | 2.073 [2.013; 2.135] | 0.242 [0.234; 0.251] | 0.117 [0.113; 0.121] | |
| Silene heuffelli | 4 | 3,999 | 98,957 | −0.043 [−0.051; −0.035] | 2.698 [2.628; 2.771] | 0.342 [0.331; 0.353] | 0.127 [0.123; 0.131] | |
| Silene latifolia | 34 | 4,132 | 214,646 | 0.165 [0.161; 0.169] | 2.005 [1.950; 2.062] | 0.244 [0.236; 0.252] | 0.122 [0.118; 0.126] | |
| Silene marizii | 4 | 2,878 | 44,868 | 0.013 [−0.005; 0.030] | 1.637 [1.573; 1.703] | 0.246 [0.235; 0.256] | 0.150 [0.144; 0.156] | |
| Gynodioecious | S .vulgaris | 14 | 6,248 | 281,300 | 0.205 [0.200; 0.209] | 2.685 [2.639; 2.732] | 0.351 [0.343; 0.360] | 0.131 [0.128; 0.134] |
| Hermaphrodite | Silene viscosa E | 3 | 6,413 | 13,712 | 0.723 [0.691; 0.754] | 0.291 [0.277; 0.305] | 0.056 [0.053; 0.059] | 0.194 [0.184; 0.204] |
| Silene viscosa W | 2 | 5,959 | 24,305 | 0.068 [0.038; 0.098] | 0.723 [0.693; 0.757] | 0.134 [0.127; 0.142] | 0.186 [0.178; 0.193] | |
| Section Otites | ||||||||
| Dioecious | Silene pseudotites | 4 | 2,339 | 24,698 | 0.024 [0.006; 0.041] | 1.116 [1.074; 1.157] | 0.195 [0.185; 0.204] | 0.174 [0.166; 0.183] |
| Silene otites | 8 | 893 | 11,364 | 0.014 [−0.007; 0.035] | 1.081 [1.016; 1.150] | 0.180 [0.167; 0.194] | 0.167 [0.154; 0.180] | |
| Gynodioecious | Silene nutans E | 11 | 5,100 | 65,820 | 0.124 [0.106; 0.140] | 0.861 [0.829; 0.895] | 0.130 [0.125; 0.136] | 0.151 [0.146; 0.157] |
| Silene nutans W | 11 | 5,059 | 88,853 | 0.201 [0.182; 0.219] | 1.048 [1.011; 1.084] | 0.170 [0.165; 0.176] | 0.163 [0.157; 0.169] | |
| Hermaphrodite | Silene paradoxa | 12 | 5,955 | 52,675 | 0.116 [0.108; 0.124] | 0.589 [0.568; 0.611] | 0.102 [0.098; 0.105] | 0.172 [0.166; 0.179] |
Proxies for species abundance were calculated based on the number of occurrences of each species retrieved from the Global Biodiversity Information Facility (GBIF) database. The geographic range of each species was extracted from the Atlas Florae Europaeae (see Materials and Methods, supplementary fig. S6, Supplementary Material online, and fig. 3). Dioecious and nondioecious species had similar geographic ranges (622.6 vs. 784 Atlas dots on an average, Wilcoxon one-sided rank sum test P value = 0.52) and GBIF species abundance (30,777 vs. 23,968.5 on an average, Wilcoxon one-sided rank sum test P value = 0.43). This is not consistent with the dead-end hypothesis which predicts smaller geographic distributions and population sizes for dioecious species. However, some Silene dioecious species have very limited geographic ranges, making the study of their genetic diversity informative to test whether they are genetically endangered.

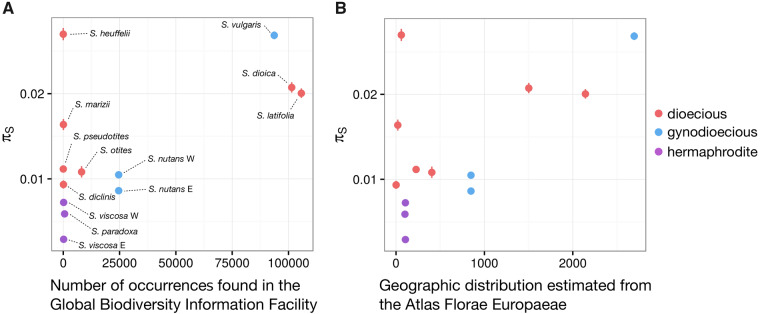
Relationship between synonymous polymorphism (πS) and (A) census population size estimated from the Global Biodiversity Information Facility (GBIF) and (B) geographic range estimated from the Atlas Florae Europaeae. For each species, dots represent the mean πS and vertical lines mark the 95% confidence interval (obtained by 1,000 bootstraps on contigs). Census population sizes estimated from GBIF refer to the number of occurrences, and geographic distributions from the Atlas Florae Europaeae refer to the number of dots (native occurrences) counted on the Atlas maps (see Materials and Methods).
Genetic Diversity Is Higher in Dioecious Silene Species, Regardless of Census Population Size Differences
In order to test whether dioecious and nondioecious Silene species differ in their effective population size (Ne), synonymous genetic diversity (πS) was used as a proxy for Ne with πS = 4Neμ (the mutation rate μ was assumed constant among species). Ne is a direct measure of the long-term mean intensity of drift in populations. Under the evolutionary dead-end hypothesis, dioecious species are expected to have lower Ne and therefore lower values of πS compared with nondioecious species (see Introduction). Table 1 and figure 2 showed that dioecious species have higher πS values than nondioecious species (Wilcoxon one-sided rank sum test W = 35, P value = 0.026), which does not support the dead-end predictions.
However, the 13 species studied here have very different census population sizes (N) with some species being cosmopolitan (e.g., S. latifolia, S. vulgaris), whereas others are rare or even endemic (e.g., S. diclinis). Genetic diversity can be affected by N, although the relationship can be complex such that N and Ne are quite different (Kalinowski and Waples 2002). In order to make a fair comparison of dioecious and nondioecious species, we accounted for the effect of N when looking at the effect of sexual systems on genetic diversity. Two different proxies for N were used: 1) the species abundance in the GBIF data set, and 2) the geographic range of each species according to the Atlas Florae Europaeae.
We used a linear model to jointly assess the effects of N and sexual systems on πS. GBIF data and Atlas geographic range (estimates of N) were significantly positively associated with πS (P values = 0.030 and 0.012, t values = 2.529 and 3.054, R2 = 0.390 and 0.483, respectively, and see fig. 3). This shows that genetic diversity is strongly influenced by N in our Silene sample set. This also suggests that, in the case of the Silene genus, GBIF abundance and Atlas geographic ranges are sensible proxies for N. After controlling for the effect of N on genetic diversity, dioecious species had a significantly higher πS than nondioecious species (linear model pairwise t-test P values = 0.142 and 0.048, t values = 1.595 and 2.247 using GBIF data and Atlas geographic range estimates, respectively). Adding FIT to the linear model correlating genetic diversity and N did not significantly improve our linear model (ANOVA P = 0.102 and P = 0.097 for GBIF and Atlas geographic range estimates, respectively, see Materials and Methods for details). This suggests that the increase in genetic diversity observed under dioecy is not attributable to inbreeding evasion.
These effects remained after correcting for phylogenetic relationships among species using generalized least square models in the R package ape (Paradis and Schliep 2019): N was significantly positively correlated to πS (P values = 0.012 and 0.03, t values = 3.05 and 2.53 for Atlas and GBIF, respectively) and dioecious species had a significantly higher πS compared with nondioecious species when using Atlas geographic range estimates (P value = 0.048, t value = 2.25), but not when using GBIF estimates (P value = 0.14, t value = 1.6). In this analysis, different sets of genes were used in different species. As this might affect the results, the analysis was repeated on two smaller sets of genes, one shared by every species in the subgenus Behenantha and the second shared by every species in the subgenus Silene. Similar results were found using these shared gene subsets, that is, GBIF data and Atlas geographic range have a significant positive effect on πS (P values = 0.021 and 0.012, t values = 2.725 and 3.074, R2 = 0.429 and 0.488, respectively). And, dioecious species had higher πS compared with other species after correcting for N (linear model pairwise t-test P values = 0.127 and 0.045, t values = 1.662 and 2.283 using GBIF data and Atlas geographic range, respectively, see supplementary fig. S1, Supplementary Material online).
Selection Is More Efficient in Dioecious Silene Species, Regardless of Differences in Ne
We then tested whether selection efficacy differs in dioecious and nondioecious Silene species. Selection should be more efficient in species with larger Ne. The ratio of nonsynonymous to synonymous substitution rate, dN/dS, could not be used here because the studied species are closely related and some shifts in sexual systems might have occurred too recently to allow for fixed differences to evolve (Desfeux et al. 1996; Bernasconi et al. 2009). Rather, the ratio of nonsynonymous over synonymous polymorphism, πN/πS, was used, because it is a more appropriate statistic to study the efficacy of selection on a short evolutionary timescale (Mugal et al. 2014). One expects increased πN/πS values when purifying selection is reduced and hence the dead-end hypothesis would predict elevated πN/πS in dioecious species. However, we found no statistical association between sexual systems and πN/πS (Wilcoxon rank sum test P = 0.295, table 1), perhaps reflecting the opposing effects that purifying and positive selection have on πN/πS, or perhaps reflecting the fact that breeding systems do not have an effect on πN/πS.
We thus turned to a more sophisticated approach to quantify selection efficacy and used a modified version of the McDonald–Kreitman framework, Implemented in the method polyDFE (Tataru et al. 2017; Tataru and Bataillon 2020). This method estimates the distribution of fitness effects (DFE) of both deleterious and beneficial mutations using polymorphism data only, thus avoiding the problem of shifts in population sizes. Silene species underwent a postglacial age population expansion in Europe and for this reason, it would be biased to use methods that also rely on divergence (Keightley and Eyre-Walker 2007; Galtier 2016). Indeed, other methods to estimate the DFE use both polymorphism and divergence data and usually return estimates of proportion of adaptive substitution that are assuming that population size remained constant since divergence with the outgroup. Silene viscosa E was not included because its diversity was too low due to selfing and the polyDFE model did not apply. PolyDFE was used to estimate the DFE (supplementary fig. S2, Supplementary Material online) for each species with a gamma distribution for deleterious mutations and exponential distribution for beneficial mutations (see estimated parameter ranges in supplementary fig. S3, Supplementary Material online). From this, the predicted proportion of adaptive substitutions (α), the predicted rate of adaptive substitutions (ωA), and the predicted rate of deleterious substitutions (ωNA) were estimated. Under the dead-end hypothesis, dioecious species are expected to have a higher ωNA, a lower α, and a lower ωA because of a faster accumulation of deleterious mutations and a lower rate of adaptation due to genetic drift, when compared with nondioecious species. Because Ne is known to influence the efficacy of selection, we correct for differences in Ne (using πS as a proxy) between species in order to make a fair comparison of dioecious and nondioecious species.
A linear model was used to jointly assess the effects of Ne (using πS as a proxy) and of the sexual system on α. πS was significantly positively correlated to α (P value = 0.003, t value = 4.07, fig. 4). The same result has previously been observed in animal species (Galtier 2016) where the correlation between α and πS was solely due to the link between πS and ωNA. Because α = ωA/(ωA+ωNA), when selection is more efficient (higher Ne and higher πS), the rate of deleterious substitutions ωNA is reduced, which in turn increases α. However, in the present data set, the correlation between α and πS was both due to higher adaptive evolutionary rates and lower deleterious evolutionary rates when effective population size is high. Indeed, πS was significantly positively linked to ωA (linear model P value = 0.011, t value = 3.19) and significantly negatively correlated to ωNA (linear model P value = 0.00017, t value = −6.15, see fig. 4). These effects remained after correcting for phylogenetic relationships among species using gls models in the R package ape (Paradis and Schliep 2019): πS was significantly positively correlated to α and ωA (P values = 0.026 and 0.0054, t values = 4.11 and 3.64 for α and ωA, respectively) and πS was negatively correlated to ωNA (P value = 0.0015 and t value = −4.49).

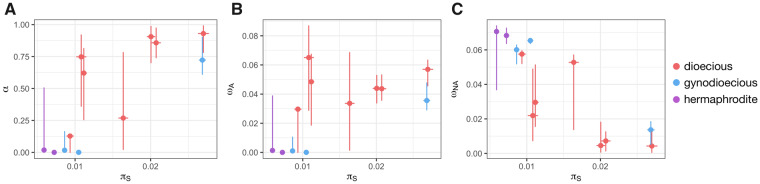
Relationship between synonymous polymorphisms (πS) and (A) the proportion of adaptive amino acid substitutions, α, (B) the rate of adaptive nonsynonymous substitutions (ωA), and (C) the rate of nonadaptive nonsynonymous substitutions (ωNA). Dots show the mean value, horizontal lines mark the 95% πS confidence interval (obtained by 1,000 bootstraps on contigs), and vertical lines show the y axis 95% confidence interval (obtained on 500 bootstraps on the SFS SNPs).
Estimates of α, ωA, and ωNA were then compared between dioecious and nondioecious species, after controlling for the effect of Ne (using πS as a proxy). Nondioecious species had a significantly lower α and ωA and a significantly higher ωNA compared with dioecious species (linear model pairwise t-test P values = 0.015, 0.00031, and 0.019, t values = −2.99, −5.65, and 2.87 for α, ωA, and ωNA, respectively). These effects remained significant after correcting for phylogenetic relationships among species using gls models in the R package ape (P values = 0.032, 0.0007, and 0.058, t values = −2.53, −5.03, and 2.17 for α, ωA, and ωNA, respectively). Dioecious species thus experience fewer fixations of slightly deleterious mutations due to genetic drift and more adaptive evolution, which is opposite to what we expect under the dead-end hypothesis. The analyses were repeated using orthologous genes shared within the subgenus Behenantha or within the subgenus Silene as different sets of genes in different species could affect the results. The conclusions remained unchanged; differences were still in the same direction although statistical power was reduced in some cases (see supplementary fig. S4, Supplementary Material online, legend for details).
Discussion
In the set of Silene species studied here, dioecy is associated with higher genetic diversity, more efficient purifying selection, and increased adaptive evolution. These results contradict the dead-end hypothesis and are consistent with macroevolutionary analysis that rejected the dead-end from different lines of evidence. High levels of genetic diversity in dioecious species had previously been observed in S. dioica (Giles and Goudet 1997), S. chlorantha (Lauterbach et al. 2011), S. otites (Lauterbach et al. 2012), Ficus (Nazareno et al. 2013), Antennaria dioica (Rosche et al. 2018), and a comparison of 12 monoecious and 11 dioecious plants (Nazareno et al. 2013). Some of these studies have shown that the sex ratio strongly influences genetic diversity through its influence on the outcrossing rate. We do not have access to sex ratios of the species sampled here but their study would be insightful to better understand patterns of diversity in Silene species in the future. Our results clearly show that dioecious Silene species are not at risk of extinction given their high genetic diversity. Observed rates of adaptive nonsynonymous substitution patterns suggest that dioecious species undergo more adaptation than nondioecious species, however, the underlying causes of this pattern remain to be determined.
The Silene genus is a good model system to test for the dead-end hypothesis because dioecious Silene species do not have any of the ecological traits thought to possibly compensate for the handicaps of dioecy (see Introduction). In other genera, in which dioecy is ancient, it is likely that enough time has passed for species to evolve compensating mechanisms to counteract the handicaps of dioecy, if these truly exist. This is less likely to be an issue in the Silene genus where dioecy has evolved fairly recently: ∼11 Ma old in the Melandrium section (Krasovec et al. 2018) and even more recently in section Otites (Slancarova et al. 2013).
Our approach, however, also has its limitations. First, our set of species represents only two independent events of dioecy evolution and a small sample of the whole Silene genus (∼700 species). However, we did not find a significant phylogenetic signal on genetic diversity in our species set (Pagel’s λ = 0.848, P = 0.133), suggesting that each species can be considered as independent even though some species share the same transition event to dioecy. Nonetheless, other genera should be studied in the future to assess whether our findings can be generalized to other angiosperm species, especially to groups in which dioecy evolved through different routes (see Introduction).
A second limitation is that sampling was limited by the number of Silene species for which the sexual system is well described and for which the geographic distribution is similar among species, especially for hermaphrodites (see Materials and Methods for detailed criteria used to select species). None of the Silene hermaphrodite species has geographic distributions as wide as the widest dioecious species range. However, dioecious species that have geographic ranges comparable with those of the hermaphroditic species we sampled still have higher diversity and selection efficacy compared with hermaphrodites (figs. 3 and 4), which is conservatively in disagreement with the dead-end hypothesis. For 2 species out of 13, namely S. otites (dioecious) and S. paradoxa (nondioecious), we were not able to sample individuals across the whole geographic range of the species and their genetic diversity might be underestimated compared with others.
A third limitation comes from the comparison of dioecious and nondioecious species which implies comparing species with sex chromosomes to species with only autosomes. Sex-linked genes, which are located on the nonrecombining region of sex chromosomes, are known to suffer from lower Ne, which could bias comparisons of dioecious to nondioecious species. Because sex-linked genes in Silene represent <10% of genes (discussed in Muyle et al. 2012), we chose to subset our study to autosomal genes only. Our results therefore overlook the potential negative effects of sex chromosomes on selection efficacy in dioecious species.
A fourth limitation of our study is that, if the extinction dynamics of dioecious species is extremely fast, there might not be enough time for deleterious genetic footprints to accumulate in dioecious species before they go extinct. The microevolutionary approach taken here can only detect handicaps of dioecy that do not lead to immediate extinction. Such a study, as others testing evolutionary dead-end hypotheses, suffers from a “survivor bias” as only nonextinct species can be analyzed. However, some of the dioecious species studied here have small geographic distributions and one is even endemic. These species should be strongly affected by the handicaps of dioecy, if there are any. But even these species exhibited high levels of diversity and selection efficacy.
In the Silene genus, all hermaphrodites are self-compatible and therefore able to self-fertilize. Outcrossing promotes genetic diversity by increasing effective population size and minimizing the effects of linkage (Nordborg 2000; Glémin et al. 2019). Dioecy could be beneficial in Silene by preventing the negative effects of selfing that occur in hermaphroditic and gynodioecious species (Charlesworth and Charlesworth 1978). One selfing hermaphrodite subspecies (S. viscosa E) could not be included in the analysis of selection efficacy because its genetic diversity was too low. However, two hermaphrodite species that are not heavy selfers (fig. 2) were included in our comparison of dioecious and nondioecious species. However, the fact that significant differences in terms of selection efficacy between dioecious and nondioecious species remain, even without this selfing species, highlights that the advantages of dioecy that we observe do not result from comparing selfing and outcrossing species. What is more, the increased genetic diversity observed in dioecious species could not be explained by differences in inbreeding levels in our data set.
However, the main difference between dioecious species and hermaphrodites is the ability to “occasionally” self-fertilize, which can strongly affect the demography of a species. Indeed, in hermaphroditic self-compatible species, a single individual can start a new population (Schoen and Brown 1991). This reproductive assurance provides an ecological advantage to hermaphroditic self-compatible species over dioecious species. However, the other side of the coin is that hermaphroditic self-compatible species can experience recurrent bottlenecks, even if the mean selfing rate is close to zero. This reduces genetic diversity and Ne and makes selection less efficient. On the other hand, dioecious populations require both males and females to be present in sufficient density in a location in order to reproduce. Therefore, dioecious population sizes cannot decline below a given threshold at risk of declining to extinction, a phenomenon called the Allee effect (Courchamp et al. 2008). A counter-intuitive prediction is thus that, “conditioning on survival,” the Allee effect promotes genetic diversity in dioecious populations, especially during colonization (Roques et al. 2012; Romiguier et al. 2014). Silene species are known to have colonized Europe northward after the last ice age and have experienced population expansion. The Allee effect could have played a strong role during this expansion with self-compatible hermaphrodites colonizing new habitats fast with as few as a single individual and undergoing strong bottlenecks and diversity loss. Dioecious species that survived, on the other hand, would have expended slowly, maintaining sufficiently dense populations throughout the expansion, resulting in the maintenance of genetic diversity. In established dioecious species, the Allee effect could provide a long-term safeguard against genetic depletion.
Our results suggest that established dioecious Silene species are genetically healthy. Our work revisits the fascinating question of why dioecy is rare in angiosperms, since the dead-end hypothesis does not seem to apply, at least to our sample of species. However, our data set might suffer from a “survivor bias” and we cannot exclude that potential handicaps of dioecy could lead to very early extinction during the emergence of new dioecious populations from hermaphrodite lineages. At a macroevolutionary scale, this rapid dead-end would lead to an observed low rate of transition toward dioecy. However, current estimates do not show low transition rates to dioecy in angiosperms (Goldberg et al. 2017), which goes against the possibility of an early dead-end of dioecy.
An alternative hypothesis to explain the rarity of dioecy is that rates of transition to dioecy could be low in angiosperms and/or dioecy could be more evolutionary labile than previously thought and easily lost and reversed to nondioecious states depending on environmental conditions (Goldberg et al. 2017; Kafer et al. 2017). The combination of a low transition rate to dioecy and a high reversibility to nondioecy together would lead to a low probability to observe dioecious angiosperms in nature, as they would only be transiently existing. This would make dioecy rare without the need for it to be an evolutionary dead-end. Larger and fully resolved angiosperm phylogenies with annotated breeding systems will be required to precisely estimate more precisely transition and reversion rates to and from dioecy.
Materials and Methods
Sampling and Sequencing
We investigated patterns of polymorphism and divergence of three sexual systems (hermaphroditism, gynodioecy, and dioecy) in two subgenera of the Silene genus, subgenus Behenantha and subgenus Silene (sample sizes and sexes are indicated in fig. 1). For subgenus Behenantha, we sampled five dioecious species (S. latifolia, S. diclinis, S. dioica, S. marizii, and S. heuffelli), one gynodioecious species (S. vulgaris), and one hermaphroditic species (S. viscosa). For the subgenus Silene, we sampled two dioecious species (S. otites and S. pseudotites), two gynodioecious subspecies (S. nutans W—Western lineage, and S. nutans E—Eastern lineage, as defined in Martin et al. [2016]), and one hermaphroditic species (S. paradoxa). Two samples of D. chinensis were added to root the phylogeny. Some of the individuals were already used in previous studies.
The two known dioecious sections in the genus Silene were sampled: section Melandrium (all five species of the section were selected) and section Otites (2 closely related dioecious species were selected). The selection of the nondioecious species was done using different criteria. First, close relatives of the two dioecious sections were selected based on available phylogenies (Desfeux et al. 1996; Jenkins and Keller 2011; Slancarova et al. 2013). We focused on those for which the sexual system is well described. The Silene genus is a large genus comprising ∼700 species but only ∼60 have a well-described sexual system (dioecious species included, Jürgens et al. 2002). Highly selfing species were avoided by focusing on hermaphroditic species that are perennial, insect pollinated (or at least not autogamous/cleistogamous), and with a high pollen/ovule ratio (Jürgens et al. 2002). These criteria left us with only a handful of hermaphroditic species, of which S. viscosa and S. paradoxa were selected for seed availability.
Seeds were sampled across the geographic distribution of each species (supplementary fig. S5, Supplementary Material online) and were sown and grown in controlled greenhouse conditions. In case plants did not flower, young leaves were sampled instead (details on individual names, sex, origin, and sampled tissue can be found in supplementary table S1, Supplementary Material online). Total RNA was extracted through the Spectrum Plant Total RNA kit (Sigma, Inc.) following the manufacturer’s protocol and treated with DNAse. Libraries were prepared with the TruSeq RNA sample Preparation v2 kit (Illumina Inc.). Each 2-nM cDNA library was sequenced using a paired-end protocol on an Illumina HiSeq2000 sequencer, producing 100-bp reads. Demultiplexing was performed using CASAVA 1.8.1 (Illumina) to produce paired-sequence files containing reads for each sample in Illumina FASTQ format. RNA extraction and sequencing were done by the sequencing platform in the AGAP laboratory, Montpellier, France (http://umr-agap.cirad.fr/). The Melandrium and Dianthus samples were sequenced as described in Zemp et al. (2016, p. 2016) at the Quantitative Genomics Facility (QGF, ETH Zürich, Switzerland) and by GATC Biotech AG (Konstanz, Germany).
Reference Transcriptome Assemblies
Reference transcriptomes were assembled de novo for all species (except for S. marizii, S. heuffelli, S. diclinis, and S. dioica). As assemblies were carried in different labs, they differ slightly for the two Silene taxa. For the subgenus Behenantha, reads from all individuals were pooled for each species. Steps in the assembly are represented in supplementary figure S3, Supplementary Material online. About 100% identical reads, assumed to be PCR duplicates, were filtered using the ConDeTri trimming software (Smeds and Künstner 2011). Illumina reads were then filtered for sequencing adapters and low read quality using the ea-utils FASTQ processing utilities (Aronesty 2013). Filtered reads were assembled using trinity with default settings (Haas et al. 2013). Poly-A tails were removed using PRINSEQ (Schmieder and Edwards 2011) with parameters -trim_tail_left 5 -trim_tail_right 5. rRNA-like sequences were removed using riboPicker version 0.4.3 (Schmieder et al. 2012) with parameters -i 90 -c 50 -l 50 and the following databases: SILVA Large subunit reference database, SILVA Small subunit reference database, the GreenGenes database and the Rfam database. Obtained transcripts were further assembled, inside of trinity components, using the program Cap3 (Huang and Madan 1999) with parameter -p 90 and custom perl scripts.
Raw data of the five taxa of subgenus Silene were filtered for sequencing adapters using Cutadapt (Martin 2011), low read quality, and poly-A tails with PRINSEQ (Schmieder and Edwards 2011). For each focal species of the subgenus, reference transcriptomes were assembled de novo by combining filtered reads from two individuals from the same population (the two individuals with the highest number of reads) using default settings of Trinity (Haas et al. 2013) and an additional Cap3 (Huang and Madan 1999) run. Finally, in both subgenera, coding sequences were predicted using TransDecoder (Haas et al. 2013) with PFAM domain searches as ORF retention criteria (see website: http://transdecoder.github.io, last accessed October 13, 2020). Assembly statistics are presented in supplementary table S2, Supplementary Material online.
Mapping, Genotyping, and Alignment
Reads from each species were mapped onto their respective reference transcriptome using BWA (Li and Durbin 2009) version 0.7.12 with parameter -n 5 for the subgenus Behenantha and Bowtie2 (Langmead and Salzberg 2012) for the subgenus Silene. Mapping statistics can be found in supplementary table S3, Supplementary Material online. For the Melandrium section (S. latifolia, S. marizii, S. heuffelli, S. diclinis, and S. dioica), all reads were mapped to the S. latifolia reference transcriptome. SAM files were compressed and sorted using SAMtools (Li et al. 2009). Individuals were genotyped using reads2snp_2.0 (Tsagkogeorga et al. 2012; Gayral et al. 2013) with parameters -par 1 -min 3 (ten for the subgenus Silene) -aeb -bqt 20 -rqt 10 that is to say: stringent filtering of paralogous positions, minimum coverage of three (or ten) reads for calling a genotype, accounting for allelic expression bias, minimum read mapping quality threshold of 10 and minimum base quality threshold of 20. The output obtained was two unphased sequences for each individual and each ORF.
Orthologous ORFs were identified using OrthoMCL (Li et al. 2003) Basic Protocol 2 with default parameters. Best reciprocal BLAST hits were considered orthologs among species and orthologous ORFs were aligned for all individuals using MACSE (Ranwez et al. 2011). Species from the subgenus Behenantha were aligned separately with S. paradoxa, considered as an outgroup for this section and species from the subgenus Silene were aligned with S. viscosa considered as an outgroup for this section.
Phylogenetic Reconstruction and Phylogenetic Signal
Orthologous ORFs among species were obtained using best reciprocal BLAST hits among the 14 species (all species from the Silene genus and D. chinensis). For each orthologous ORF, the nine species were added one by one into the alignment using MACSE (Ranwez et al. 2011). The data were filtered so that a maximum of 20% missing data per site and per species was allowed, for a total of 55,337 variable nucleotides across 1,294 ORFs. We used the maximum number of ORFs available and did not restrict our analysis to autosomal ORFs identified in dioecious species. The phylogenetic tree was reconstructed using the GTRGAMMA model in RaxML (Stamatakis 2006). Bootstrap support values were generated using 100 replications.
Phylogenetic signal and its significance were computed using the R function phylosig in the phytools R package (Revell 2012).
Data Filtering
Analyses were carried out on autosomal genes only, in order to avoid potential confounding effects of sex chromosomes and sexual systems on estimates of selection efficiency. Autosomal genes were predicted for each dioecious species separately by SEX-DETector (Muyle et al. 2016) thanks to the study of a cross (parents and progeny) sequenced by RNA-seq (Martin et al. 2019; A. Muyle, N. Zemp, A. Widmer, G.A.B Marais, in preparation). About 11,951 contigs were inferred as autosomal in S. latifolia, 7,695 for S. diclinis, 11,675 for S. heuffelli, 8,997 for S. marizii, 10,698 for S. dioica, 2,035 in S. otites, and 6,234 in S. pseudotites.
The data were further filtered in order to remove ORFs with frameshifts. Only biallelic sites were kept for analyses (sites with two or less alleles). Only ORFs longer than 100 codons and with a maximum of 20% of missing individuals of the focal species in the alignment were retained, after removing incomplete codons and gaps. We obtained final data sets ranging from 893 retained ORFs for S. otites up to 6,413 retained ORFs for S. viscosa E (see table 1). Finally, to verify that observed trends are not due to differences in studied ORFs among species, we performed the same analyses on a smaller set of orthologous ORFs present in all species within a subgenus, so that parameters were estimated on the same set of ORFs. For these tests, 188 orthologous ORFs were analyzed for the subgenus Silene, and 298 for the subgenus Behenantha.
Polymorphism, Heterozygosity, and FIT
Population statistics were computed on the filtered data set for each ORF and averaged using the PopPhyl tool dNdSpiNpiS_1.0 (Gayral et al. 2013) with parameter kappa = 2. The following statistics were computed for each contig: the mean fixation index FIT (Weir and Cockerham 1984), per-individual heterozygosity (proportion of heterozygous positions), per-site synonymous polymorphism (πS), per-site nonsynonymous polymorphism (πN), and the ratio of nonsynonymous to synonymous polymorphism (πN/πS). Note that, we could not compute FIS or FST as we do not have the required two individuals per population. Confidence intervals were obtained by 1,000 bootstraps on contigs.
Estimation of Geographic Distribution and Population Size
Geographic distributions of the Silene species were estimated by counting the black dots (native occurrence) on the Atlas maps by Jalas and Suominen (1986). This was partly achieved using the Dot Count program (http://reuter.mit.edu/software/dotcount, last accessed October 13, 2020). As no map was available for S. pseudotites, dots were counted for Italy and adjacent regions of France and the Balkans using the S. vulgaris map (the Atlas indicates these are the regions where this plant is abundant).
Census population sizes of the Silene species were estimated using raw species occurrence numbers from GBIF (http://www.gbif.org January 17, 2017 Download of S. dioica: http://doi.org/10.15468/dl.fim8f2, S. marizii: http://doi.org/10.15468/dl.yywbrk, S. diclinis: http://doi.org/10.15468/dl.vizuiv and December 1, 2016 Download of S. pseudotites: http://doi.org/10.15468/dl.yrvenq, S. otites: http://doi.org/10.15468/dl.fxoocf, S. paradoxa: http://doi.org/10.15468/dl.wizsb6, S. nutans: http://doi.org/10.15468/dl.poqxc1, S. viscosa: http://doi.org/10.15468/dl.bwdu9z, S. vulgaris: http://doi.org/10.15468/dl.vmvebr, S. latifolia: http://doi.org/10.15468/dl.mfvyds, all previous links were last accessed October 13, 2020). Some countries contribute poorly to this database, however, we assumed that this affected all Silene species in the same way. The GBIF database was filtered for showing occurrence based on human observation and literature. Only data from European countries and Russia, which represent the native range of those species, were kept. For each species, all existing names of the species were included.
Both estimates (geographic range and population size) were divided by two when two subspecies were identified with our phylogeny (i.e., for S. viscosa and S. nutans). Atlas geographic ranges and GBIF species abundance were compared among dioecious and nondioecious species using a one-sided Wilcoxon rank sum test in R (R Core Team 2016). Geographic range and GBIF data were correlated to πS and sexual systems or FIT using R linear model function lm as follows:
The significancy of the effect of sexual system and FIT were tested thanks to a comparison to a nested model using the ANOVA function in R. The nested models are:
The same models were used and corrected for phylogenetic relationships among species using gls models with Martins and Hansen’s correlation structure (corMartins) in the R package ape (Paradis and Schliep 2019).
Selection Efficacy
The unfolded synonymous and nonsynonymous site frequency spectra (SFS, the distribution of derived allele counts across SNPs) were computed for each species with the sfs program provided by Nicolas Galtier (Galtier 2016). These SFS were used to estimate α (the proportion of adaptive amino acid substitutions) in each species. We estimated α according to a modified version of the Eyre-Walker and Keightley method (Keightley and Eyre-Walker 2007), which relies on polymorphism data only to estimate α (Tataru et al. 2017). polyDFE (https://github.com/paula-tataru/polyDFE, last accessed October 13, 2020) was used to estimate the distribution of fitness effects of mutations (DFEM), α, the (per synonymous substitution) rate of adaptive nonsynonymous substitution ωA=α dN/dS, and the rate of nonadaptive nonsynonymous substitutions ωNA = (1−α) dN/dS. For each species separately, four different models were fitted to the SFS:
Model 1: full DFEM with adaptive mutations (p_b>0), estimation of an orientation error rate for ancestral alleles in the SFS (eps_an), estimation of demography (r_i≠1) for a total of six parameters plus the number of demography parameters r_i which depends on the sample size of each species.
Model 2: same as Model 1 but without ancestral allele orientation error rate (eps_an = 0), for a total of five parameters plus the number of demography parameters r_i.
Model 3: same as Model 1 but without adaptive mutations (p_b = 0), for a total of five parameters plus the number of demography parameters r_i.
Model 4: same as Model 1 but without adaptive mutations (p_b = 0) and without ancestral allele orientation error rate (eps_an = 0), for a total of four parameters plus the number of demography parameters r_i.
Each model provided estimates for α, ωA, and ωNA. These values were averaged using Akaike weights as follows:
For each species, 500 bootstrapped data sets were randomly generated by random sampling SNPs with replacement from the original SFS. Average values among models were computed as explained previously for each bootstrap. The distribution of values for the 500 bootstraps was used to compute approximate 95% confidence intervals for α, ωA, and ωNA. For this, 2.5% of values were removed from each side of the bootstrap distribution (Efron and Tibshirani 1993). α, ωA, and ωNA values were analyzed in a linear model using R lm function (R Core Team 2016). The model tested the effect of πS and sexual system (dioecious, gynodioecious, hermaphroditic) as fixed effect, whereas taking into account the variance of the estimations with weights computed as follow:
The same models (without weights) were used and corrected for phylogenetic relationships among species using gls models with Martins and Hansen’s correlation structure (corMartins) in the R package ape (Paradis and Schliep 2019).
Acknowledgments
We would like to thank Jos Kafer for his help with fieldwork and a number of colleagues for sending us seeds: J.J. Wieringa, Deborah Charlesworth, Bohuslav Janousek, Tatiana Giraud, Honor C. Prentice, Salvatore Cozzolino, Delphine Griveau, Henk Schat, Cristina Gonnelli, Alessia Guggisberger, Sophie Karrenberg, Adrien Favre, Maria Domenica Moccia, Aria Minder, Carmen Rothenbuehler, Massimiliano Gnesotto, and Kerstin Kustas. A.M., R.T., and G.A.B.M. thank Elise Lacroix technician of the University of Lyon 1 greenhouse and H.M., S.Ga., and P.T. thank Eric Schmitt for taking care of the Silene plants. We are grateful to Sylvain Santoni and the AGAP platform staff for the RNA extraction and RNA-seq. The numerical results presented in this article were carried out using the computing cluster of Laboratoire Biométrie et Biologie-Pôle Rhone-Alpin de Bioinformatique (LBBE-PRABI) at University of Lyon 1 and the HPC service of the Centre de Ressources Informatiques (CRI) of University of Lille 1, for which, we thank Stéphane Delmotte and Bruno Spataro. We thank Nicolas Galtier for help with his program sfs. H.M., S.Ga., and P.T. are grateful to Jonathan Aceituno for technical support in R and to Céline Poux for discussion on phylogenetic inference. We are thankful to Brandon Gaut for commenting on the article. This work was supported by the Agence Nationale de la Recherche (ANR-11-BSV7-013-03, TRANS) to S.Gl., G.A.B.M., and P.T., the Swiss National Science Foundation (31003A-182675) to A.W., a PhD fellowship from the French Research Ministry to H.M and two Postdoctoral fellowships to A.M (EMBO ALTF 775-2017 and HFSPO LT000496/2018-L).
All the RNA-seq reads generated for this study can be downloaded from the European Nucleotide Archive accession PRJEB39526.
These authors contributed equally as co-first authors to this work.
These authors contributed equally as co-senior authors to this work.
Data availability
All the RNA-seq reads generated for this study can be downloaded from the European Nucleotide Archive Accession PRJEB39526.
 Dioecy Is Associated with High Genetic Diversity and Adaptation Rates in the Plant Genus Silene
Dioecy Is Associated with High Genetic Diversity and Adaptation Rates in the Plant Genus Silene
