
- Altmetric
Uridine insertion/deletion editing of mitochondrial mRNAs is a characteristic feature of kinetoplastids, including Trypanosoma brucei. Editing is directed by trans-acting gRNAs and catalyzed by related RNA Editing Core Complexes (RECCs). The non-catalytic RNA Editing Substrate Binding Complex (RESC) coordinates interactions between RECC, gRNA and mRNA. RESC is a dynamic complex comprising GRBC (Guide RNA Binding Complex) and heterogeneous REMCs (RNA Editing Mediator Complexes). Here, we show that RESC10 is an essential, low abundance, RNA binding protein that exhibits RNase-sensitive and RNase-insensitive interactions with RESC proteins, albeit its minimal in vivo interaction with RESC13. RESC10 RNAi causes extensive RESC disorganization, including disruption of intra-GRBC protein–protein interactions, as well as mRNA depletion from GRBC and accumulation on REMCs. Analysis of mitochondrial RNAs at single nucleotide resolution reveals transcript-specific effects: RESC10 dramatically impacts editing progression in pan-edited RPS12 mRNA, but is critical for editing initiation in mRNAs with internally initiating gRNAs, pointing to distinct initiation mechanisms for these RNA classes. Correlations between sites at which editing pauses in RESC10 depleted cells and those in knockdowns of previously studied RESC proteins suggest that RESC10 acts upstream of these factors and that RESC is particularly important in promoting transitions between uridine insertion and deletion RECCs.
INTRODUCTION
Trypanosoma brucei, the causative agent of Human African Trypanosomiasis, belongs to the class Kinetoplastea, so named for the presence of their unique mitochondrial DNA, called the kinetoplast or kDNA (1,2). T. brucei kDNA has a bipartite organization, consisting of dozens of maxicircles and thousands of minicircles that are catenated together. Maxicircles contain 20 genes encoding ribosomal RNAs, ribosomal proteins, and numerous proteins important for the formation of respiratory complexes (1). However, primary transcripts from 12 of the 18 protein coding genes are incomplete, and must undergo substantial post-transcriptional modification to generate functional open reading frames. These transcripts require the precise insertion and less common deletion of uridine (U) residues through a process is known as U insertion/deletion (U-indel) RNA editing (reviewed in (3–6)). Editing is essential for the survival of both insect vector procyclic form (PF) and mammalian bloodstream form (BF) T. brucei and is conserved across the Kinetoplastea (7,8). Of the 12 edited transcripts in T. brucei, three need only a few to dozens of modifications, and these are known as moderately edited transcripts. However, nine of the 12 edited transcripts, referred to as pan-edited, undergo extensive editing involving hundreds of U insertions throughout their lengths, sometimes doubling the size of the transcript. Editing is directed by hundreds of small guide RNAs (gRNAs) that are primarily encoded in kDNA minicircles and specify U insertions and deletions through base pairing interactions (9,10). U-indel editing takes place in the 3′ to 5′ direction along an mRNA, beginning with the binding of the anchor region (10–15 nucleotides) of the first gRNA to pre-edited mRNA. Editing progresses throughout the length of a gRNA block, after which the first gRNA is removed by an unknown mechanism. The second gRNA then anchors to the previously created edited sequence, and the process is reiterated until editing is complete. Although editing proceeds generally 3′ to 5′, dictated by gRNA sequences, it does not proceed strictly 3′ to 5′ on a site-by-site basis. Rather, the majority of partially edited mRNAs contain junction sequences between the pre-edited and fully edited regions. Junctions contain edited sequence that matches neither the pre-edited sequence nor the canonical fully-edited sequence, and many junctions likely represent regions of active editing (5,11,12).
U-indel RNA editing is catalyzed by a holoenzyme comprising three dynamically interacting complexes: RECC (RNA Editing Core Complex), RESC (RNA Editing Substrate Binding Complex) and REH2C (RNA Editing Helicase 2 Complex), and most subunits of the holoenzyme are essential for cell viability (3). RECC contains the enzymes that catalyze endonuclease cleavage, U insertion/deletion and RNA ligation, and is present in three isoforms distinguished by their endonuclease modules. The Kinetoplast RNA Editing Endonuclease1 (KREN1) containing module functions in the deletion of U residues, whereas RECC variants containing KREN2 and KREN3 have somewhat overlapping function and are involved in U insertion (13–15). REH2C was recently reported to affect total editing and accuracy of editing in a site-specific and substrate-specific manner on RESC-associated transcripts (16). Interestingly, purified RECC lacks detectable RNA, while RESC and REH2C bind editing substrates, intermediates, and products, and interact with RECC through RNA linkers (6,17–19). RESC and REH2C are necessary for the 3′ to 5′ progression of editing, and are thought to coordinate interactions between mRNA, gRNA, and RECC by mechanisms that are only beginning to be unraveled (4,6).
The RESC component of the holoenzyme contains ∼20 proteins, most of which lack recognizable domains and have no homologs outside kinetoplastids. It is organized into subcomplexes termed the GRBC (Guide RNA Binding Complex) and REMCs (RNA Editing Mediator Complexes), and in some models the PAMC (Polyadenylation Mediator Complex) (3,4,6). The nomenclature of RESC components was recently revisited, and here we used terminology from (3); previous and current names of proteins discussed herein are listed in Table 1. Within RESC, the GRBC module appears relatively homogeneous and stable, although the RESC1/2 heterotetramer, which promotes gRNA stability and is often depicted as a dedicated GRBC component (19–22), is typically heterodisperse on glycerol gradients and is found in distinct small complexes with RESC14 and a subset of REMC proteins, with REH2C, and with other mitochondrial RNA binding proteins (19,22–27). In contrast to the GRBC module, REMCs describes a set of likely heterogenous complexes that interact with GRBC, and which were previously described to contain seven proteins (12,19). The functions and interactions of several REMC proteins have recently been studied using a combination of biochemical approaches and high throughput sequencing (HTS) to define editing defects in cells depleted of specific REMC factors. For example, interacting proteins RESC13 and RESC11A, which both bind RNA, facilitate progression through gRNA-defined blocks and promote the formation of mis-edited junctions, suggesting junctions are essential features of the editing process (12). The paralogous RNA binding proteins RESC12A and RESC12 also bind RESC13, and they function in editing initiation, likely facilitating recruitment of other RESC proteins, as well as to constrain the region of active editing (12,28). Recent evidence indicates that the ARM/HEAT repeat containing RESC8, previously denoted as a REMC protein (19), instead functions as a RESC organizer in the recruitment and sequential exchange of RESC factors and promotion of protein-protein and protein–RNA rearrangements during editing (29). Finally, RESC14, designated as neither a REMC nor GRBC component, plays a role in modulating RNA–protein interactions within RESC, and promotes editing progression through impacts on gRNA utilization. Sequence analysis of mRNAs in RESC14 depleted cells, as well as its presence in a small complex with gRNA stabilizing proteins RESC1/2, further suggests a role for this protein in gRNA exchange and gRNA discrimination (25).

| Old nomenclature | New nomenclature |
|---|---|
| GAP1/2 | RESC1/2 |
| MRB11870 | RESC5 |
| MRB3010 | RESC6 |
| MRB10130 | RESC8 |
| MRB800 | RESC10 |
| MRB8180 | RESC11A |
| MRB4150 | RESC11B |
| MRB4160 | RESC12 |
| MRB8170 | RESC12A |
| TbRGG2 | RESC13 |
| MRB7260 | RESC14 |
| MRB1590 | KRBP72 |
| TbRGG3 | KRGG3 |
| REH2 | KREH2 |
| RBP16 | KRBP16 |
| MRP1/2 | KMRP1/2 |
a (3).
In this study, we utilized biochemical and genomic approaches to characterize one of the less well-studied RESC proteins, RESC10 (Tb927.7.800). RESC10 is essential for PF T. brucei growth and for editing of all classes of mitochondrial mRNAs. It exhibits RNA binding activity in vitro, as well as RNase-sensitive and RNase-insensitive interactions with RESC proteins. RESC10 depletion lead to extensive RESC disorganization, including disruption of intra-GRBC protein-protein interactions, suggesting this protein is not a dedicated REMC factor as previously reported (19). Rather, its stoichiometry and interaction profile, as well as the analysis of edited mRNAs at the single nucleotide level in RESC10 knockdowns, are consistent with a model in which RESC10 interacts with other RESC proteins early in their assembly, and possibly transiently, to promote RESC integrity. Finally, the correlation between sites at which editing pauses in RESC10 depleted cells and those in previously reported RESC protein knockdowns suggests that RESC is particularly important in promoting transitions between U insertion and U deletion RECC variants.
MATERIALS AND METHODS
T. brucei growth conditions and cell line generation
Procyclic form (PF) T. brucei 29–13 strain was used for all the experiments. Cells were grown in standard conditions as described (30,31) at 27°C in SDM-79 medium supplemented with 10% FCS. To generate the RESC10 RNAi cell line, forward primer RESC10_5′-F and reverse primer RESC10_3′-R containing BamHI and HindIII sites, respectively, were used to amplify a 1021 bp amplicon and ligated into pJET1.2/Blunt (Thermo Scientific) plasmids. The resulting plasmids were digested with BamHI and HindIII and the resulting fallout fragment was ligated into p2T7–177 (32) between opposing tetracycline/doxycycline-regulated T7 RNA polymerase promoters. The plasmid was NotI digested, purified, and transfected into PF T. brucei stain 29–13. Cells were selected with 2.5 μg/ml phleomycin and clones obtained by limited dilution.
To generate the cell lines harboring both PTP-tagged RESC6 and RESC10 RNAi or His-TEV-Myc(HTM)-tagged RESC13 and RESC10 RNAi (25), the same linearized DNA fragment was used for transfection into cells in which RESC6-PTP (33) and RESC13-HTM (25) were endogenously tagged, and the cells were selected with 2.5 μg/ml phleomycin and 1 μg/ml puromycin and cloned by limiting dilution. To generate a cell line harboring RESC5-MHT and RESC10 RNAi, the RESC10 RNAi cell line was transfected with a MHT-tagging cassette obtained by amplification of the pPOTv4-MHT-Puromycin vector with PCR primers (RESC5_SD-F and RESC5_SD-R) corresponding to the 3′ end of the RESC5 ORF without the stop codon and the 5′ end of 3′-UTR of RESC5 (34). Cells were selected with 2.5 μg/ml phleomycin and 1 μg/ml puromycin and clones obtained by limited dilution. All the RESC10 RNAi cell lines were confirmed by qRT-PCR and by using RESC10 specific antibodies. For monitoring cell growth, cells harboring the RESC10 RNAi construct were grown in the presence or absence of 4 μg/ml tetracycline and monitored in triplicate for 10 days. All primers used in this study are listed in Table 2.

| Primer Name | Sequences (5′→3′) |
|---|---|
| RESC10_5′-F | GGAAGCTTATGCGACGTCGGGTAGTTTTATG |
| RESC10_3′-R | GAGGATCCGGCTCAAGGTGAAATGGCGTAAG |
| pGEX_RESC10-F | CCCGGATCCATGCGACGTCGGGTAGTT |
| pGEX_RESC10-R | CCCGGAATTCGTTACCTGACGAAGCATCGCTTTAG |
| pMAL_RESC10-F | ATATATCTAGAATGCGACGTCGGGTAGTT |
| pMAL_RESC10-R | GGAAGCTTGTTACCTGACGAAGCATCGCTTTAG |
| RESC10_QPCR-F | GGAAAGTCGTCTTCGGAGTA |
| RESC10_QPCR-R | CATGCACACAAAACAACTGA |
| RESC5_SD-F | TCTTCAAGGCGAATGTTGGTGGCATGTTGTCGAGGAACAAGAGTCGTGGAGCCCGATGGCAGACGCATCAACTGCAGAAAGGTTCTGGTAGTGGTTCC |
| RESC5_SD-R | TGTTCCCTCTTGTGTACTAACCACAACCATGAGCACACACATACACGCACGCGCTAAACTACAAACAGTCTCCTCAGCCACCAATTTGAGAGACCTGTGC |
Anti-RESC10 antibody generation
The RESC10 gene was amplified from gDNA using the pGEX_RESC10-F and pGEX_RESC10-R primers having BamHI and EcoRI restriction overhangs in their 5′ends, respectively, and cloned into the pGEX-4TK expression vector (Novagen). After sequence confirmation, the plasmid was transformed in BL21 E. coli. Recombinant RESC10-GST(glutathione-S-transferase) was purified using glutathione resin (GE Health Care) as recommended by the manufacturer, and the protein was dialyzed in 1× PBS overnight. Antibodies were raised in rabbits at Bethyl Laboratories. For affinity purification of anti-RESC10 antibodies, GST-RESC10 (500 μg) was mixed with 2× SDS loading dye and transferred to nitrocellulose. The membrane was stained with Ponceau (Sigma) and the RESC10-GST band excised and incubated overnight with α-RESC10 containing rabbit serum at 4°C. Serum was removed, the membrane was washed with 1× PBST, and antibodies eluted with 0.1 mM glycine (pH 2.5) followed neutralization of the sample with 1mM Tris–HCl (pH 8.0). The specificity of this antibody was confirmed by probing an RNAi cell line targeting RESC10.
Determination of RESC10 protein abundance
The abundance of cellular RESC10 was estimated as previously described (12,35,36). To generate recombinant protein for these assays, the gene encoding RESC10 was amplified by PCR using Pfu DNA polymerase (Fermentas) and pMAL_RESC10-F and pMAL_RESC10-R (Table 2) primers having XbaI and HindIII restriction overhangs in their 5′ ends, respectively. The gel-purified PCR product was digested and ligated downstream of the IPTG inducible tac-lacUV5 promoter vector pMAL-C2 (NEB) to create plasmid for the expression of MBP (maltose binding protein)-RESC10 in Escherichia coli BL21. MBP-RESC10 was purified using amylose resin (NEB) as recommended by the manufacturer followed by anion exchange on Q Sepharose. Purified MBP-RESC10 was quantified on SDS-PAGE using a Bovine Serum Albumin (BSA) standard curve. Purified recombinant MBP-RESC10 was then titrated such that the bands were detectable by western blot, and known cell equivalents of whole cell and enriched mitochondrial extracts were also detectable, with the intensity of the latter signals falling within the standard curve of the recombinant protein. The intensities of Western blot bands were determined using ImageJ software (Bio Rad). A standard curve was created by linear regression analysis with Microsoft Excel and used to determine the amount of RESC10 in each whole cell and mitochondrial extract lane. This value was then converted to molecules per cell based on the number of cells loaded and the molecular mass of RESC10. Two biological replicates were performed to determine RESC10 protein abundance. In Figure 1B, we present a trimmed blot instead of the entire image, as any non-specific bands in this sample are irrelevant to the quantification. While there are indeed some bands above and below the region shown in the full image, for the reasons described above we believe showing the entire blot would only decrease the clarity of the figure without increasing its transparency.

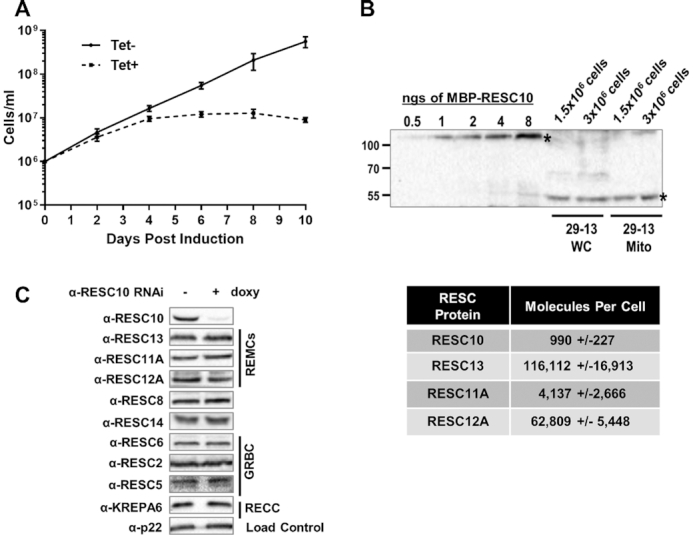
Effects of RESC10 knockdown on T. brucei growth and editing factor levels, and relative abundance compared to other RESC proteins. (A) Growth curve of PF T. brucei upon RESC10 RNAi induction. Cell growth was measured in triplicate for 10 days for uninduced (-tetracycline; Tet) and induced cells (+tetracycline). (B) Top, Determination of the number of molecules of RESC10 present per cell. Known amounts of recombinant RESC10 were probed with anti-RESC10 antibodies alongside known cell and mitochondrial equivalents. The experiment was performed twice, and shown is a representative experiment. Bottom, Estimated RESC10 molecules per cell compared to that of previously published RESC proteins (12). (C) Effect of doxycycline (doxy)-mediated RESC10 knockdown on the abundance of selected RESC proteins, including both GRBC and REMC components, and the KREPA6 RECC protein. p22 is a loading control.
Mitochondria were enriched by hypotonic lysis, DNase treatment, and centrifugation at 17 500 × g as described (37). Briefly cells were treated with hypotonic lysis buffer (1 mM Tris pH 8.0, 1mM EDTA, complete protease inhibitor cocktail (Roche), and RNase inhibitor) and Dounce homogenized on ice 10 times. Sucrose was then added to final concentration of 0.25 M. The sample was centrifuged at 15 800 × g for 15 min at 4°C. Pellets were resuspended in STM (250 mM sucrose, 20 mM Tris pH 8.0, 2 mM MgCl2 and 0.2 mM CaCl2) buffer along with 1 μg/ml DNase and incubated for 1 h on ice. An equal volume of STE (250 mM sucrose, 20 mM Tris pH 8.0 and 10 mM EDTA) buffer was then added, and the sample was mixed and centrifuged at 15 800 × g for 15 min at 4°C. The resulting pellet contains crude mitochondria that were used for determination of RESC10 abundance and RNA immunoprecipitations (see below).
UV crosslinking and guanylyltransferase assays
MBP-RESC10 was produced as described above, and RESC13-GST and p22 tagged protein controls were previously described (12,38). For in vitro UV crosslinking assays, guide RNA gA6 (39) and mRNA A6U5 pre-mRNA (79 nt) (40) were synthesized from plasmids using the T7 Maxiscript Kit (Ambion). Internally [α 32P]-UTP (800 Ci/mmol) labeled RNA was purified on 8% (w/v) acrylamide/7 M urea gel. UV cross-linking reaction containing 5 fmol of radiolabeled RNA and 2 μg of protein was performed as described previously (25). T. brucei RESC10 RNAi cell line was grown for three days in the presence or absence of 4 μg/ml tetracycline, RNA was purified from cells using TRIzol (Ambion), and guanylyltransferase reactions were performed as described previously (33).
qRT-PCR analysis
RESC10 RNAi cells were treated with 4 μg/ml tetracycline for 3 days to induce RNAi or left untreated. Equal number of cells were collected from both the samples, and RNA was purified with Trizol reagent (Ambion) and treated with DNase (Ambion) for 1 h. After treatment, RNA was purified using phenol/chloroform followed by ethanol precipitation. Purity of the RNA was measured by using Nanodrop 1000, and 260/280 ratio was ∼2. Quality of the RNA was assessed on a 1.0% of TBE agarose gel. cDNA was generated using random hexamer primers and the iScript reverse transcriptase kit (Biorad). To detect levels of RESC10 mRNA, qRT-PCR was performed using the RESC10 specific primers described in Table 2, and to detect levels of pre-edited and edited mitochondrial transcripts from T. brucei established primers were used, with normalization to 18S rRNA (40,41). Analysis of qRT-PCR results was performed using BioRad iQ5 software. All the results reflect two biological replicates, each with three technical replicates of the qRT-PCR reaction.
High-throughput sequence analysis
PF T. brucei RESC10 RNAi cell lines were grown in the presence or absence of 4 μg/ml tetracycline for 3 days. RNA was isolated using Trizol followed by phenol/chloroform extraction and ethanol precipitation. RNA samples were treated with DNase and phenol/chloroform extracted and ethanol precipitated. Two biological replicate experiments were performed, and qRT-PCR was used to validate the level of RESC10 knockdown (32–38% remaining). cDNA was generated from DNase-treated RNA using gene-specific primers (11). These cDNA samples were PCR amplified within the linear range of PCR to maintain the relative abundance of unique fragments. Paired-end Illumina Mi-Seq was used for high throughput sequencing of amplicons as described previously (11). All reads were aligned to the published pre-edited and fully edited mRNA sequences (42). Reads which had non-T SNPs or insertions/deletions relative to the published canonical sequence were excluded from the analysis. All remaining reads in each sample were normalized to 100 000 counts in order to compare relative abundances of specific sequences between samples. The sequencing data used in this study have been deposited in the Sequence Read Archive, accession numbers SRP271414 (RESC10 knockdown, RPS12 and ND7–5′), SRP185791 (RESC10 knockdown, CYb and MURF2) and SRP097727 (previously published uninduced samples (12) included in the uninduced pool).
Immunoprecipitation
RESC6-PTP, RESC5-MHT and RESC13-HTM tagged cells harboring RESC10 RNAi constructs were grown for 3 days following induction with 4 μg/ml doxycycline. Cells (1 × 1010) were collected and washed with 1X PBS. The cell pellet was lysed in N150 buffer (50 mM Tris–Cl pH 7.5, 150 mM NaCl,0.1% NP-40,1 mM MgCl2 and 5 mM β-ME) with 1% (v/v) Triton X-100 and 1 μg/ml DNase 1 by incubation for 30 min. The cell suspension was centrifuged at 18 000 rpm for 30 min. Supernatants (cleared cell lysates) from RESC6-PTP and RESC5-MHT cell lines were incubated in 20 ml BioRad columns with 200 μl of IgG beads (GE Healthcare), and RESC13-HTM supernatant was similarly incubated with 200 μl of anti-myc beads (ICL), for 2 h at 4°C. The beads were washed with 10 ml N150 buffer and followed by 10 ml tobacco etch virus (TEV) cleavage buffer (10 mM Tris–Cl (pH 7.5),150 mM NaCl, 0.5 mM EDTA) and incubated in 1 ml of TEV cleavage buffer with 10 μl of TEV protease (Thermo Scientific) overnight at 4°C. RESC10 was immunoprecipitated using a similar method, except using 2 × 1010 PF T. brucei 29–13 cells, lysate of which was incubated with α-RESC10 antibody pre-bound to Protein A fast flow beads (GE Healthcare). Prior to incubation, lysate was divided into two 20 ml fractions. One 20 ml fraction was incubated with 60 U of RNase inhibitor (Applied Biosystems) and DNase 1 (1 μg/ml), while the other 20 ml portion was treated with DNase1 and a nuclease cocktail containing 8 μg RNase A (Thermo Scientific), 2500 U RNase T1 (Ambion), 28 U RNase H (Invitrogen), and 2040 U micrococcal nuclease (Thermo Scientific) for 1 h on ice. After washing with N150 buffer, RESC10 complexes were eluted with 0.1 mM glycine pH 2.5 followed by neutralization with 1 mM Tris–HCl (pH 8.0). The levels of target proteins were normalized by western blot using either specific antibodies against the target or antibodies recognizing the tag, as indicated on each figure. Co-purifying proteins were then detected by western blotting with specific antibodies against RESC6 (23), RESC13 (40), RESC11A (12), RESC12A (43), RESC8 (29), RESC14 (25), RESC10 (this study) and RESC2 (33).
RNA immunoprecipitation (RIP)
PF T. brucei RESC6-PTP and RESC13-HTM cells harboring the RESC10 RNAi construct were grown in presence or absence of 4 μg/ml of doxycycline for 3 days. Cells (1 × 1010) were collected and mitochondria enriched as described above, and RIP was performed as described previously (25,29). Briefly, RESC6-PTP and RESC13-HTM cells were immunoprecipitated with IgG fast flow (GE Healthcare) and Myc (ICL) beads, respectively. Superdex 200 (GE Healthcare) and anti-HA (ICL) beads were used as negative controls for PTP and HTM tagged proteins, respectively. Five percent of the beads were taken from each sample, and western blot was performed to confirm the pulldown of specific protein and their levels. DNase 1 was then added to the beads, and the sample was incubated for 30 min at 30°C, followed by addition of 50 μl of proteinase K (Roche) for 30 minutes at 55°C and 20 μl of 20% SDS. The supernatant was removed, and RNA was extracted with phenol/chloroform. RNA was DNase-treated (Ambion DNA-free DNase Kit) and 500 ng of RNA converted to cDNA with T. brucei established gene specific primers were used, with normalization to 18S rRNA gene (25,29) using the iScript cDNA synthesis kit (BioRad). cDNA was amplified by using SsoAdvanced PreAmp Supermix (BioRad) and then used for qRT-PCR. 18S rRNA was used for normalization. Fold change was determined using ΔΔCt method as described previously (25,29).
RESULTS
RESC10 is essential for cell growth and present in relatively low abundance
A previous report suggested that RESC10 depletion had only a modest effect on PF T. brucei cell growth; however, this result may reflect the incomplete knockdown achieved in that study (decrease in RESC10 RNA levels to 60% of wild type) (19). To further explore the essentiality of this protein in trypanosomes, we created a PF cell line harboring a tetracycline/doxycycline-inducible RESC10 RNAi construct. We observed a growth defect in induced compared to uninduced cells beginning 3 days post-induction, and by day 8 post-induction we observed a decline in cumulative numbers (Figure 1A). The more dramatic effect of RESC10 shown here is likely due to the robust level of knockdown, as we observed a nearly complete ablation of RESC10 protein (97 ± 3%; n = 4) by western blot with RESC10 specific antibodies (Figure 1C). Thus, RESC10 is essential for PF T. brucei cell growth. Interestingly, in the RESC10 RNAi cell line, we noted that the level of RESC10 protein ablation exceeds that of RESC10 RNA ablation (compare Figures 1C and 5A). This finding suggests that both mRNA stability and translation are affected by RNAi in this case. Indeed, two RNAi studies in T. brucei reported a large decline in protein in the absence of any effect on RNA levels, consistent with RNAi effects on translation in this system (44,45).
To begin to understand the role of RESC10 within the RESC complex and the basis for its essentiality, we first evaluated its stoichiometry with respect to other RESC components for which this parameter has been measured (12). We measured the western blot signal for known cell equivalents of whole cell or mitochondrial extract from PF 29–13 cells, and compared these values to a standard curve of recombinant RESC10 (Figure 1B). RESC10 is of relatively low abundance, present at only about 1–2% the levels of RESC13 and RESC12A, and lower than that of RESC11A. These data suggest that RESC10 may be present only in a subset of RESC complexes. To determine whether RESC10 depletion impacts the abundance of specific RESC components, as might occur if it were present in a distinct subcomplex, we analyzed by western blot the abundances of those RESC components for which antibodies are available (Figure 1C). We observed no reproducible changes in the abundances of any RESC proteins, nor did we see any change in the level of the KREPA6, suggesting that RECC abundance is also unaffected by RESC10 knockdown. The low relative abundance of RESC10 suggests that this protein is only present in a subset of RESC complexes or that it interacts transiently and sub-stoichiometrically with RESC.
RESC10 is an RNA binding protein with numerous RNase-sensitive and RNase-insensitive RESC interactions
At least seven RESC proteins exhibit RNA binding activity, despite five of these proteins harboring no known RNA binding domain (12,20,29,40,43). These findings suggest that although RESC10 also has no recognizable domains and no homology to proteins outside kinetoplastids, it might also possess RNA binding activity. To test this, we performed in vitro cross-linking experiments with MBP-tagged RESC10 and in vitro transcribed RNA comprising the gA6[14] gRNA and the 3′A6U fragment of unedited A6 mRNA (39,40). GST-RESC13 served as a positive control and his-p22 as a negative control, and we previously showed that the MBP tag does not bind these RNAs (29). Figure 2A shows that RESC10 binds to both gRNA and mRNA in vitro.

![RESC10 is an RNA binding protein that exhibits RNA dependent and independent interactions with other RESC proteins. (A) Recombinant MBP-RESC10, GST-RESC13, or his-p22 were incubated with radiolabeled gA6[14] gRNA or a 79-nt fragment of pre-edited A6 mRNA, UV-cross-linked, treated with RNase A, and separated on a 12% SDS-acrylamide gel. The left panel shows a Coomassie-stained gel image, and the right panel shows a phosphorimage. Asterisks indicate the positions of full length proteins. Molecular mass standards are on the left (kDa). (B) Top, Immunoprecipitation (IP) of RESC10 with α-RESC10 antibodies from cell lysates of T. brucei either treated with RNase inhibitor (−) or an RNase cocktail (+). Elutions were normalized for the levels of RESC10, and RESC proteins were probed with the respective antibodies shown on the left. Arrows indicate proteins whose RESC10 interaction is decreased following RNase treatment. Bottom, RESC13 was probed again, with both original loading (top two panels) and higher loading of the elution (3×) and lower loading of input (1/4X) (bottom panel). (C) Quantification of the experiment in B (top panels). The levels in the -RNase samples were set to 1.0. Bar graphs represent the average and standard deviation of two biological replicates, each with two technical replicates. ND; Not Detectable. (D) IP of RESC13-HTM from cell lysates treated as in B, probed with antibodies against RESC10. (E) IP of RESC14-MHT from cell lysates treated as in (B), and probed with antibodies against RESC10. In all panels, Input indicates cleared cell lysates.](/dataresources/secured/content-1766009954888-08f7e48c-c432-40c7-8528-3e99d41e5c8c/assets/gkab129fig2.jpg)
RESC10 is an RNA binding protein that exhibits RNA dependent and independent interactions with other RESC proteins. (A) Recombinant MBP-RESC10, GST-RESC13, or his-p22 were incubated with radiolabeled gA6[14] gRNA or a 79-nt fragment of pre-edited A6 mRNA, UV-cross-linked, treated with RNase A, and separated on a 12% SDS-acrylamide gel. The left panel shows a Coomassie-stained gel image, and the right panel shows a phosphorimage. Asterisks indicate the positions of full length proteins. Molecular mass standards are on the left (kDa). (B) Top, Immunoprecipitation (IP) of RESC10 with α-RESC10 antibodies from cell lysates of T. brucei either treated with RNase inhibitor (−) or an RNase cocktail (+). Elutions were normalized for the levels of RESC10, and RESC proteins were probed with the respective antibodies shown on the left. Arrows indicate proteins whose RESC10 interaction is decreased following RNase treatment. Bottom, RESC13 was probed again, with both original loading (top two panels) and higher loading of the elution (3×) and lower loading of input (1/4X) (bottom panel). (C) Quantification of the experiment in B (top panels). The levels in the -RNase samples were set to 1.0. Bar graphs represent the average and standard deviation of two biological replicates, each with two technical replicates. ND; Not Detectable. (D) IP of RESC13-HTM from cell lysates treated as in B, probed with antibodies against RESC10. (E) IP of RESC14-MHT from cell lysates treated as in (B), and probed with antibodies against RESC10. In all panels, Input indicates cleared cell lysates.
Since RESC10 is an RNA binding protein, we next asked whether its interaction with distinct RESC components is sensitive to RNase treatment. To this end, we immunoprecipitated RESC10 from cell extracts that were either treated with RNase inhibitor (-RNase) or with an RNase cocktail (+RNase), and performed western blots for RESC proteins (Figure 2B). We note that we generated specific antibodies against RESC10 for these studies because we observed that both PTP-tagged and myc/his/TAP (MHT)-tagged RESC10 were very inefficiently imported into mitochondria. Additionally, myc-tagged RESC10, while imported, associated with smaller complexes than did native RESC10 (data not shown). In RNA replete conditions, native RESC10 interacted with all seven RESC components tested. However, we were surprised by the reproducibly very minimal interaction with RESC13, as this protein is highly abundant and consistently associated with RESC complexes (12). Figure 2C shows quantification of the impacts of RNase treatment on RESC10 protein-protein interactions, and indicates that RESC10 association with RESC12A, RESC8, and RESC6 is somewhat RNase-sensitive, while its interaction with the other RESC factors is RNase-insensitive. Because of the faint RESC13 signal, we could not determine with certainty the RNase sensitivity of this interaction. To both determine if RESC10-RESC13 interact slightly and, if so, whether this interaction is impacted by RNase treatment, we repeated the pull down with native RESC10, loading less input and more eluate (Figure 2B, bottom). We also performed a reciprocal co-immunoprecipitation using our previously reported RESC13-his/TEV/myc (HTM) expressing cells (25) (Figure 2D). Figure 2B (bottom) and Figure 2D demonstrate that RESC10 and RESC13 do minimally interact and that this interaction is insensitive to RNase. Finally, we confirmed the RNase insensitivity of the RESC10-RESC14 interaction with a reciprocal co-immunoprecipitation using previously described RESC14-MHT expressing cells (Figure 2E) (25). Together, these data show that RESC10 is an RNA binding protein with both RNase-sensitive and RNase-insensitive interactions within the RESC complex. The absence of a robust interaction between RESC10 and RESC13 suggests that these two proteins may dynamically interact with RESC in a temporally distinct manner.
Depletion of RESC10 impacts both GRBC-REMC and intra-GRBC interactions
Because RESC10 interacts with numerous RESC components, we wanted to determine whether and how its depletion affects RESC integrity. To begin, we generated a series of cell lines harboring the regulatable RESC10 RNAi construct and constitutively tagged versions of either RESC13 (a factor of REMCs), RESC6, or RESC5 (the latter two are GRBC components). We then analyzed the effects of RESC10 knockdown on distinct RESC interactions by co-immunoprecipitation (Figure 3). When RESC13 was immunoprecipitated, we observed no effect of RESC10 knockdown on its association with its REMC partners, RESC11A or RESC12A, or with the gRNA binding protein, RESC2 (Figure 3A and B). In contrast, interaction between RESC13 and the organizer proteins, RESC14 and RESC8, as well as with GRBC component, RESC6, were dramatically decreased when RESC10 was depleted. To confirm the decrease in the RESC6-RESC13 interaction and probe additional interactions with GRBC, we immunoprecipitated RESC6-PTP from RESC10 replete and depleted cells (Figure 3C and D). Depletion of RESC10 decreased the interaction of RESC6 with all RESC proteins tested, with the exception of RESC2. Disruption of the RESC6–RESC5 interaction was particularly remarkable. These two GRBC components interact directly in yeast two-hybrid assays and associate strongly, and in an RNA-independent manner, in co-immunoprecipitations (19,23). If RESC10 is a REMC component as previous suggested (19), it would not be expected to affect the strong and direct RESC6–RESC5 interaction. To confirm this result, we precipitated RESC5-MHT and probed its RESC interactions in RESC10 replete and depleted conditions (Figure 3E and F). RESC10 depletion indeed disrupts the intra-GRBC interaction between RESC5 and RESC6, confirming the results of the RESC6 pulldown, but does not impact the RESC5-RESC2 interaction. Additionally, interactions between RESC5 and all other proteins tested, including components of REMCs and organizer proteins, are reduced by RESC10 depletion. From these data, we conclude that RESC10 does not impact intra-REMC interactions, but affects both GRBC-REMC and intra-GRBC associations. This suggests that RESC10 is not a REMC component, but rather plays an important role in the integrity of the GRBC module and that of RESC in general.

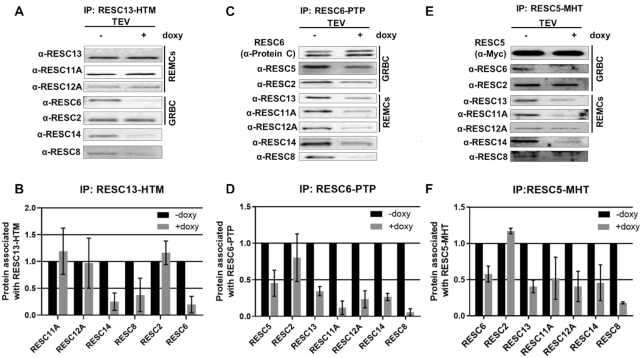
Effect of RESC10 depletion on RESC protein-protein interactions. (A) IP of RESC13-HTM was performed from lysates of RESC10 replete (–doxy) and depleted (+doxy) cells (three day induction). Bound proteins were released by TEV cleavage, and RESC13 was normalized in the elutions by western blot. Normalized elutions were analyzed by western blot with antibodies against RESC proteins, including GRBC and REMC components. (B) Quantification of A. Levels in -doxy samples were set to 1.0. Bar graphs represent the average and standard deviation of two biological replicates each with two technical replicates. (C) IP of RESC6-PTP was performed in lysates of RESC10 replete (–doxy) and depleted (+doxy) cells as in A. (D) Quantification of C. Levels in -doxy samples were set to 1.0. Bar graphs represent the average and standard deviation of two biological replicates each with two technical replicates. (E) IP of RESC5-MHT IP of RESC13-HTM was performed in lysates of RESC10 replete (-doxy) and depleted (+doxy) cells as in (A). The RESC12A panel contains three times the amount of eluate as do the other panels. (F) Quantification of (E). Levels in -doxy samples were set to 1.0. Bar graphs represent the average and standard deviation of two biological replicates each with two technical replicates.
RESC–mRNA interactions are disrupted by RESC10 depletion
Because RESC10 is essential for RESC integrity, its depletion is likely to impact distinct protein-RNA interactions as well. To identify changes in RESC-mRNA and RESC-gRNA associations that occur upon RESC10 knockdown, we performed RNA immunoprecipitation (RIP) experiments as described (25,29), using the RESC6-PTP/RESC10 RNAi and RESC13-HTM/RESC10 RNAi cell lines described above. To detect the largest pool of mRNA for a specific pan-edited transcript, we used primers that hybridize to the 5′ never-edited region and a portion of the 5′ pre-edited sequence of several mRNAs (Supplementary Figure S1A). These primers will detect the entire pool of pre-edited and partially edited versions of a given pan-edited transcript, apart from those that are edited in the extreme 5′-most region (25). CYb mRNA was detected using primers corresponding to never edited regions downstream of its small editing domain. Because this strategy measures the bulk of a given transcript population, regardless of editing status, RIP data are not confounded by large changes in the input levels of a given mRNA (Supplementary Figure S1B) (18,22,24). To begin, we confirmed that targeted mRNAs and gRNAs were enriched in both RESC6-PTP and RESC13-HTM pulldowns, compared to mock RIPs (Supplementary Figure S1C and S1D). We next analyzed protein-RNA interactions upon RESC10 depletion and found that three different pan-edited mRNAs exhibited decreased association with RESC6, by 1.5- to 2-fold (Figure 4A). At the same time, these mRNAs were approximately two-fold increased association with RESC13 (Figure 4B). The moderately edited CYb mRNA behaved differently from pan-edited mRNAs in response to RESC10 depletion, being increased in its association with both proteins. Remarkably, while CYb mRNA increased just under two-fold in its interaction with RESC6, the amount of CYb mRNA associated with RESC13 increased almost eight-fold in RESC10 depleted cells, despite the absence of any change in total CYb mRNA levels (Supplementary Figure S1B). gRNA-protein interactions were more heterogenous as measured in RIP assays. In general, most gRNAs had decreased association with RESC6 (Figure 4A), while RESC13-associated gRNAs exhibited no reproducible differences (Figure 4B). Together, RIP assays demonstrate the presence of substantially disrupted protein-RNA interactions within RESC when RESC10 is depleted, likely due in part caused by changes in protein-protein interactions (Figure 3).

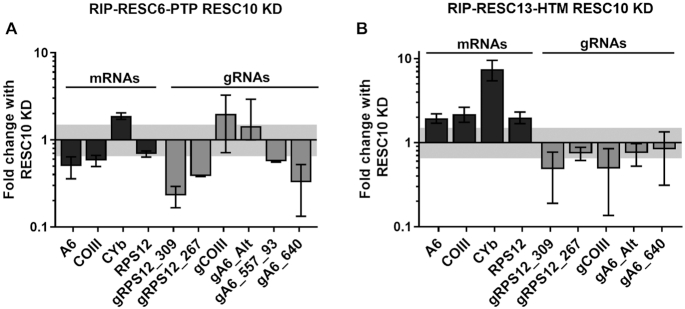
Effect of RESC10 depletion on RESC protein-RNA interactions. (A) Comparison of mRNAs and gRNAs immunoprecipitated with RESC6-PTP from cells grown with or without doxycycline to induce RESC10 knockdown for 3 days. Fold change with RESC10 knockdown (KD) represents the RNA levels detected in the RIP from doxycycline induced cells compared to the RIP from uninduced cells. RNA levels were standardized against 18S rRNA, and numbers represent the mean and standard deviation of two biological replicates, each with three technical replicates. (B) Comparison of mRNAs and gRNAs immunoprecipitated with RESC13-HTM from cells grown with or without doxycycline for three days to induce RESC10 knockdown. Quantification as in (A). Shaded area indicates 1.5-fold change.
RESC10 similarly affects levels of all classes of edited transcripts
The dramatic impacts of RESC10 depletion on RESC integrity implies that RNA editing will be disrupted. To examine potential editing defects, we analyzed mRNA levels by qRT-PCR using primer sets that specifically detect either edited or pre-edited versions of each transcript (41). We analyzed pan-edited (A6, RPS12 and COIII) and moderately edited (CYb, MURF2 and COII) transcripts. Interestingly, all edited mRNAs tested were decreased by 70–80%, while pre-edited levels changed modestly for a subset of mRNAs (Figure 5A). This is a rather atypical pattern, only previously observed in RESC8 knockdowns (29). It is much more commonly observed that depletion of RESC factors has a greater effect on pan-edited transcripts as compared to moderately edited transcripts (12,24,25,33,40,46,47). The dramatic decrease observed in edited COII mRNA, a transcript whose editing requires just one gRNA that is cis-acting, implies that the widespread impact of RESC10 knockdown is not due to a wholesale effect on the gRNA population. Indeed, a guanylyltransferase labelling experiment demonstrates no alterations in gRNA abundance when RESC10 is depleted (Figure 5B). The above results indicate that RESC10 knockdown impacts editing of all classes of mRNAs and that this effect is likely due to the disorganization of RESC and not to gRNA destabilization.

![Effect of RESC10 depletion on mitochondrial RNA levels. (A) RNA was quantified by qRT-PCR using primer sets that specifically detect the pre-edited (Pre) and edited (Edit) versions of the pan-edited and moderately edited RNAs indicated. Relative RNA abundance represents RNA levels in tetracycline-induced cells compared to levels in uninduced cells (three day induction). RNA levels were normalized to 18S rRNA levels, and numbers represent the mean and standard deviation of two biological replicates, each with three technical replicates. The level of RESC10 knockdown was quantified using RESC10 specific primers. (B) RNA was isolated from RESC10 RNAi cells either grown in the absence or presence of tetracycline for three days, and then labeled with [α32P]-GTP using guanylyltransferase to identify gRNAs and resolved on a denaturing gel. A cytoplasmic RNA that was used for normalization and the labeled gRNA are indicated.](/dataresources/secured/content-1766009954888-08f7e48c-c432-40c7-8528-3e99d41e5c8c/assets/gkab129fig5.jpg)
Effect of RESC10 depletion on mitochondrial RNA levels. (A) RNA was quantified by qRT-PCR using primer sets that specifically detect the pre-edited (Pre) and edited (Edit) versions of the pan-edited and moderately edited RNAs indicated. Relative RNA abundance represents RNA levels in tetracycline-induced cells compared to levels in uninduced cells (three day induction). RNA levels were normalized to 18S rRNA levels, and numbers represent the mean and standard deviation of two biological replicates, each with three technical replicates. The level of RESC10 knockdown was quantified using RESC10 specific primers. (B) RNA was isolated from RESC10 RNAi cells either grown in the absence or presence of tetracycline for three days, and then labeled with [α32P]-GTP using guanylyltransferase to identify gRNAs and resolved on a denaturing gel. A cytoplasmic RNA that was used for normalization and the labeled gRNA are indicated.
Single nucleotide resolution analysis of RPS12 mRNA demonstrates that RESC10 impacts editing progression
While qRT-PCR measures those mRNAs that are nearly fully edited, it does not measure the levels of most partially edited mRNAs or the extent to which their editing has progressed from 3′ to 5′. To gain deeper insight into the step(s) in editing impacted by RESC10, we performed HTS analysis on the pan-edited RPS12 transcript using a previously described protocol that quantifies pre-edited, partially edited, and fully edited transcripts and, additionally, defines the regions in which the 3′ to 5′ progression of editing is paused upon depletion of essential editing factors (11,12). Sequences were obtained from two RESC10 RNAi-induced cDNA samples and ten uninduced control samples, two from this study and eight from a previous study (12). Libraries were aligned using the TREAT algorithm that defines editing sites (ES), editing stop sites, exacerbated pause sites (EPS), and junctions (11,12). An ES is defined as any space between two non-T nucleotides in the cDNA, numbered 3′ to 5′ corresponding to the direction in which editing progresses. An editing stop site is the 5′ most ES containing canonical edited sequence; all sequence 3′ of the editing stop site is fully edited. An EPS is defined as the editing stop site at which canonical editing pauses to a significant extent in a given knockdown cell line compared to uninduced controls (Padj < 0.05) (12,25,29,48). Junctions are stretches that contain sequence matching neither the pre-edited nor fully edited sequence. TREAT defines junctions as extending from the 3′ most ES that does not match the fully edited sequence to the 5′ most ES that shows any editing action. In this way, the TREAT tool permits analysis of junction lengths and sequence (11,12).
Sequences for each sample were normalized to 100 000, and the number of normalized pre-edited, partially edited, and fully edited transcripts for the uninduced and RESC10 RNAi-induced samples were averaged and plotted in Figure 6A (see also Supplementary Table S1). We measured no change in pre-edited transcript levels, consistent with qRT-PCR results (Figure 5A) suggesting that RESC10 does not play a role in the initiation of RPS12 mRNA editing. As typically observed (12), we obtained very few fully edited mRNAs (<10 normalized reads) in any sample, and the scatter in the uninduced samples obscured any significance in this population. We did observe a slight but significant increase in total partially edited transcripts, suggesting an impact of RESC10 knockdown on editing progression. To determine whether RESC10 affects editing progression, and if so whether specific defects could inform its function, we quantified the EPSs in RESC10 depleted and replete samples (Supplementary Table S2; Supplementary Figure S2A). EPSs are shown aligned with the edited RPS12 sequence and its cognate gRNAs from a published study (Figure 6B, black diamonds and boxes) (42). In RESC10 depleted cells, EPSs are present throughout the lengths of several gRNA directed blocks, consistent with an effect utilization of a given gRNA following its association with cognate mRNA. Additionally, the EPS at ES22 falls at the end of gRNA-1, which could implicate RESC10 in gRNA exchange at this point. Analysis of those partially edited sequences with the highest abundance in RESC10 depleted cells (>100 normalized counts in the knockdown) revealed that many of these exhibited a failure to correctly execute a U deletion (Supplementary Figure S2B). For example, mRNAs with editing stop site 19 typically lacked the U deletion at ES20, an abundant mRNA with editing stop site 28 failed to execute the single U deletion at ES29, mRNAs with editing stop site 30 often failed to delete the four U’s at ES31, and mRNAs with editing stop site 31 often failed to delete the three U’s at ES32. In total, 80% of the abundant sequence variants shown in Supplementary Figure S2B are those whose correct editing ends at an ES 3′ of a U deletion site. From these data, we conclude that RESC10 primarily impacts editing progression and may be especially critical at sites of U deletion in RPS12 mRNA.

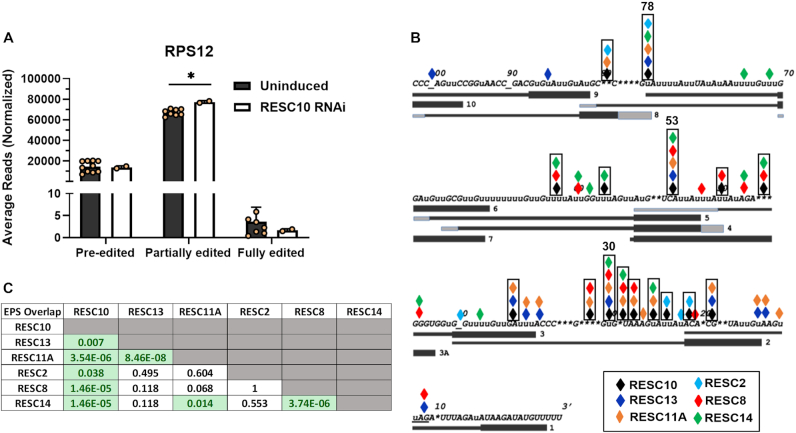
High-throughput sequence analysis of RPS12 mRNA in RESC10 depleted cells. (A) Average number of normalized pre-edited, partially edited, and fully edited RPS12 transcripts for ten uninduced (Avg UI) and two RESC10 RNAi induced samples. P-values (Student t-test): 0.002 for RPS12 partially edited (∗). (B) RPS12 mRNA 3′ edited sequence showing EPS arising upon RNAi of RESC10 (black diamonds and outlines), RESC13 (dark blue diamonds), RESC11A (orange diamonds), RESC2 (turquoise diamonds), RESC8 (red diamonds), and RESC14 (green diamonds). Exacerbated Pause Sites (EPSs) are defined as having padj <0.05 in both biological replicates (12). Those EPS found in five of the six knockdown cell lines are indicated by their ES number above the outline. Bars below the sequence indicate gRNAs, numbered 3′ to 5′ (42). gRNA anchor regions are depicted with bold lines. Gray regions at the 5′ and 3′ ends represent variation in gRNA lengths within a gRNA class. RPS12 stop codon is underlined; additional underscores are shown for clarity in stretches of unedited sequence to align numbers with the correct ES. (C) Significance of the overlaps in EPSs between six different RESC protein knockdowns (12,25,29).
Comparison of multiple RESC knockdowns suggests that RESC10 acts upstream of other RESC factors and that RESC is important for RECC transitions
We previously reported the effects of knockdown of several other RESC proteins on the editing of RPS12 mRNA at single nucleotide resolution, identifying common impacts of distinct protein pairs (RESC13 and RESC11A; RESC14 and RESC8) (12,25,29). To determine whether RESC10 function aligns with that of any other RESC factor, we asked whether the EPSs that arise upon RESC10 depletion significantly overlap those of other tested RESC proteins. We denote the EPSs arising upon depletion of each of these factors by different colored diamonds in Figure 6B and the significance of their overlap in Figure 6C (green boxes). Strikingly, RESC10 EPSs significantly overlap with those of all other RESC factors tested, with the exception RESC12A (not shown). This widespread overlap suggests that RESC10 depletion leads to secondary effects on other RESC proteins. These data are consistent with a model in which RESC10 acts upstream of the other RESC factors to perform its function, likely by assuring the integrity of RESC organization.
We noted that three EPSs (ES30, ES53 and ES78) are shared by five of the six factors tested, indicating that these sites are particularly sensitive to perturbations of RESC. Figure 6B shows that these three ES share a common characteristic: the last correctly edited ES is a U insertion site, while the next (incorrectly edited or unedited) site is a U deletion site. That is, the pauses in the progression of editing along RPS12 mRNA most often exacerbated by RESC disruption occur at sites where the editing machinery must exchange a U insertion RECC for a U deletion RECC. Examination of the most abundant sequences at ES30, ES53 and ES78 reveals that all five RESC knockdowns exhibit very similar sequences, in many cases struggling to delete the correct number of U’s or halting editing altogether (Figure 7). Together, these data suggest that RESC plays a critical role in coordinating RECC transitions.

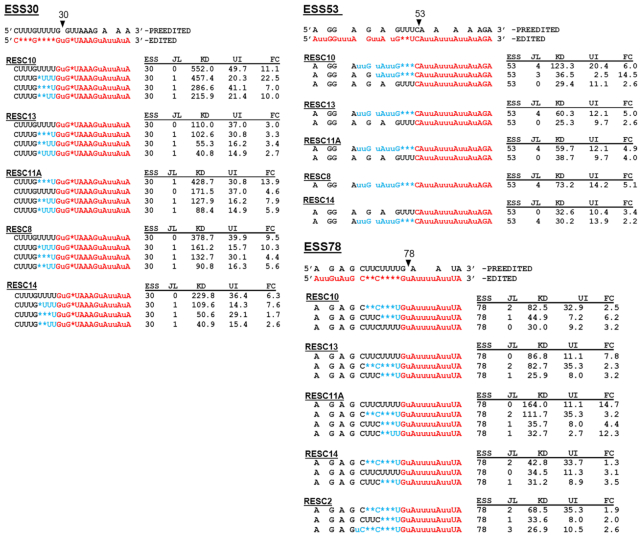
Comparison of RPS12 mRNA junction sequences at ES30, ES53 and ES78 in numerous RESC protein knockdown cell lines. Shown are those ES that were quantified as EPS in five of six cell lines tested. Abundant sequences that were present in >25 average normalized counts in knockdown cell lines are shown. The region matching fully edited is shown in red, pre-edited in black, and the junction region (containing mis-edited sequence) in blue. The editing stop site (ESS) number and the junction length (JL) are shown to the right of each sequence. To the right is indicated the average number of each sequence detected from 10 uninduced samples (UI) and two induced samples (KD). The fold change (FC) of each sequence compared to the average of uninduced is also indicated for all RESC proteins (12,25,29).
RESC10 impacts initiation and progression of transcripts with internal initiating gRNAs
Having analyzed the impact of RESC10 knockdown on a pan-edited mRNA, we next asked if similar effects were observed in those transcripts whose editing initiates with gRNAs that anchor internally in the mRNA, rather than at the 3′ end. This includes moderately edited mRNAs, CYb and MURF2, as well as the 5′ domain of ND7, whose editing initiates in a never-edited region approximately 200 nt from its 5′ end (42). Analysis of the levels of pre-edited, partially edited, and fully edited mRNAs revealed a significant 50–100% increase in the levels of all three pre-edited mRNAs upon RESC10 depletion, thereby indicating an effect on editing initiation in all cases (Figure 8A–C; Supplementary Tables S1-S3). As previously reported, fully edited mRNAs constitute 25–30% of CYb and MURF2 populations (48), and a significant decrease in fully edited mRNAs was evident in these populations when RESC10 is depleted (Figure 8B and C). Calculation of EPSs revealed exacerbated pauses in both CYb (Figure 8D; Supplementary Figure S3) and MURF2 (Figure 8E; Supplementary Figure S4), indicating that editing progression is also affected. Of these, EPSs at CYb ES565 also occur in cells depleted of RESC13 and RESC2, and the EPS at ES572 is shared with RESC13 knockdowns (48). The MURF2 EPS at ES 446 is shared with knockdowns of RESC13 and RESC2, as well as the editing accessory factors KRBP16 and KMRP1/2, while that at ES455 is also found in the RESC13 knockdown (48). For ND7–5′, the very low number of fully edited reads prohibited any measure of significant change in this parameter. We did, however, observe a significant decrease in partially edited mRNAs, although no EPSs were observed indicating that the primary effect of RESC10 depletion on this region is at editing initiation. Thus, RESC10 depletion leads to transcript specific effects, impacting initiation of editing on those mRNAs with internal initiating gRNAs while primarily impacting editing progression along pan-edited RPS12.

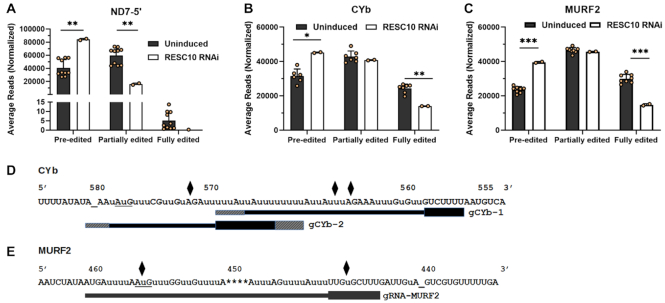
High-throughput sequence analysis of ND7–5′, CYb and MURF2 mRNAs in RESC10 depleted cells. (A) Average number of normalized pre-edited, partially edited, and fully edited ND7–5′ transcripts for ten uninduced and two RESC10 RNAi induced samples. P-values (Student t-test): 0.00087 for ND7–5′ pre-edited (∗∗) and 0.00088 for ND7–5′ partially-edited (∗∗). (B) Average number of normalized pre-edited, partially edited, and fully edited CYb transcripts for ten uninduced and two RESC10 RNAi induced samples. P-values (Student t-test): 0.0062 for CYb pre-edited (∗) and 0.00035 for CYb fully-edited (∗∗). (C) Average number of normalized pre-edited, partially edited, and fully edited MURF2 transcripts for ten uninduced and two RESC10 RNAi induced samples. P-values (Student's t-test): 0.000002 for MURF2 pre-edited (∗∗∗) and 0.000041 for MURF2 fully-edited (∗∗∗). (D) CYb edited mRNA sequence with exacerbated pause sites (EPS) arising upon RESC10 RNAi in two biological replicates indicated with black diamonds. EPSs are defined as having padj <0.05 in both biological replicates (12). (E) MURF2 edited mRNA sequence with exacerbated pause sites (EPS) arising upon RESC10 RNAi in two biological replicates indicated with black diamonds. Bars below the sequence indicate gRNAs, numbered 3′ to 5′ (42). gRNA anchor regions are depicted with bold lines. Hatched regions at the 5′ and 3′ ends represent variation in gRNA lengths within a gRNA class. Start codons are underlined; additional underscores are shown for clarity in stretches of unedited sequence to align numbers with the correct ES.
DISCUSSION
RESC is a non-enzymatic multi-protein apparatus that provides a platform for U-indel RNA editing in kinetoplastid mitochondria. It is responsible for coordinating dynamic protein-protein and RNA-protein interactions during editing, and it associates with RECC and REH2C components of the editing holoenzyme, editing accessory factors, and 5′ and 3′ RNA modification complexes (3,4,6,19,23,27,28,43,49–51). In this study, we identified RESC10 as a protein that plays a critical role in RESC integrity. The impacts of RESC10 depletion on RESC are more extensive than those of any other RESC protein reported to date, affecting both protein-protein interactions and protein-RNA interactions within RESC. Like RESC14 and RESC8 (25,29), RESC10 facilitates interactions between REMCs and GRBC. However, RESC10 is the only factor examined so far whose depletion also disrupts intra-GRBC protein interactions, apart from the apparently integral GRBC protein, RESC3 (24). In addition, while the knockdown of RESC14 and RESC8 has little or no impact on the amount of mRNA associated with the GRBC module, RESC10 knockdown leads to a consistent decrease in the amounts of pan-edited mRNAs associated with GRBC and a concomitant increase in those associated with RESC13 (a factor of REMCs). These protein-RNA interactions may reflect the alterations in protein-protein interactions. That is, prolonged loss of intra-GRBC integrity causes dissociation of RNA from the GRBC module, while REMC interactions remain intact, leading to accumulation of mRNAs on REMC variants containing RESC13. It is the RESC disorder caused by RESC10 depletion that likely leads to the observed dramatic and widespread effects on editing, with comparable impacts on both pan-edited and moderately edited mRNAs.
At what point during editing does RESC10 exert its effects on RESC macromolecular interactions? RESC is a dynamic and heterogeneous complex that appears to partially disassemble and reassemble, sometimes with different partners, during the editing process (3,4,25,29). RESC must accommodate constantly changing mRNA–gRNA base pairing and RNA structure as editing proceeds through a gRNA-directed block, and it must permit RECC interactions with a wide array of mRNA structures to promote editing catalysis. Moreover, most editing blocks contain both U insertions and U deletions, necessitating exchange of RECC isoforms. RESC must also allow the exchange of gRNAs upon completion of a gRNA-directed block. Finally, the REH2C interacts in an RNA-dependent manner with a subset of RESC proteins. The precise point(s) of entry of RECC variants and REH2C to the editing holoenzyme are not known. Nevertheless, each of these transitions conceivably requires changes in RESC organization and potentially release and acquisition of distinct proteins. Several lines of evidence suggest that RESC10 acts early, and potentially transiently, during RESC assembly at one or more of the above transitions (see Figure 9). First, RESC10 is of low abundance compared with other RESC proteins for which this parameter has been measured (12), suggesting that it is not a stoichiometric component of all RESC complexes. Second, co-immunoprecipitation experiments reveal minimal interaction between RESC10 and RESC13. Thus, one can envision a scenario in which RESC10 interacts with RESC components early in their assembly and then is released prior to association of the majority of RESC13. Third, the GRBC module is thought to be a relatively stable and RNA independent entity (19,23), so the impact of RESC10 on GRBC module integrity indicates a fundamental effect on RESC organization. Finally, single nucleotide resolution analysis of editing progression reveals that the sites at which RPS12 mRNA editing pauses in RESC10 knockdowns, as well as the editing effects at these sites, overlap those of five of the six RESC factors tested to date. This finding strongly suggests that RESC10 acts upstream of other RESC factors, orchestrating their proper organization and function as illustrated Figure 9.

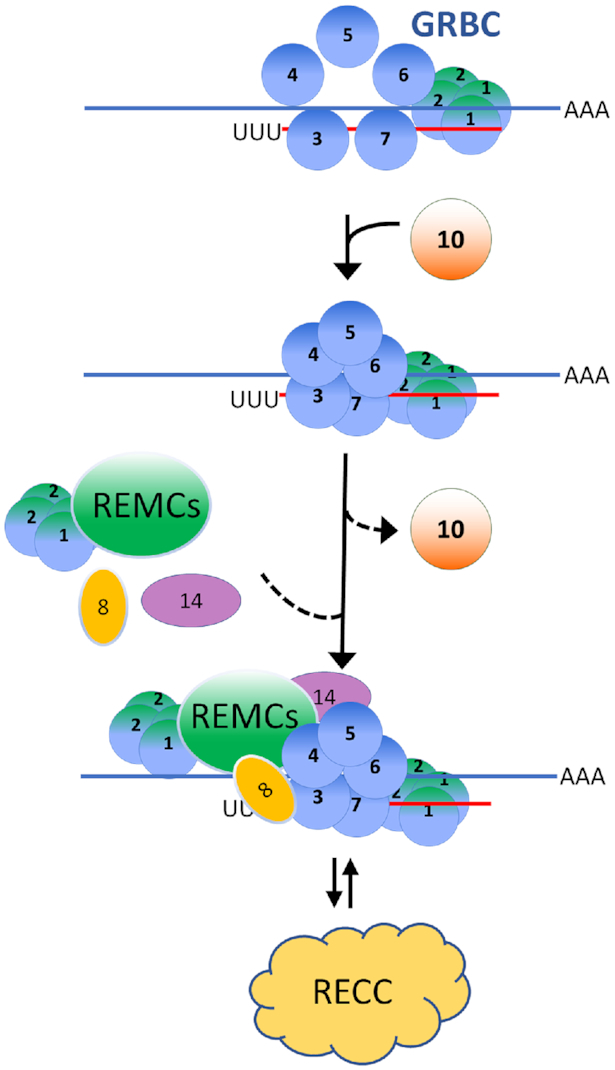
Working model of RESC10 action. RESC10 associates with GRBC to promote an editing-competent conformation of the GRBC module. Once GRBC is in the proper conformation, REMCs and organizer proteins can assemble into RESC. Dashed lines indicate that the precise timing and extent of RESC10 dissociation and association of distinct REMC and organizer proteins is not fully established. However, RESC10 appears to minimally interact with REMC factor, RESC13. The absence of a robust RESC10-RESC13 interaction, and the low abundance of RESC10 are consistent with RESC10 dissociation from the assembling RESC complex. Once RESC is assembled, RECC can associate with RESC/mRNA/gRNA and catalyze editing. This cycle may occur at many or most editing sites, and it appears to be especially critical when a deletion RECC needs to be substituted for an insertion RECC. Proteins are indicated by their RESC numbers, and ‘REMCs’ indicates heterogeneous REMC complexes. Blue line ending in AAA indicates mRNA shown 5′ to 3′; red line starting with UUU indicates gRNA shown 3′ to 5′. See text for additional details.
One possibility is that RESC partially disassembles after each editing site, and then reassembles in a RESC10-dependent manner (Figure 9). Alternatively, the holoenzyme may exhibit differing levels of processivity depending on sequence context and/or RNA structure. Sequence analysis of RESC knockdowns, including RESC10, suggests that the effect of RESC10 on RESC assembly is especially important during transitions between insertion and deletion RECCs. We demonstrated that cells depleted of RESC10 accumulate abundant partially edited RPS12 mRNAs that struggle to perform U deletions. Moreover, the most highly shared RPS12 EPSs among RESC factors are at sites that require a switch from a U insertion RECC to a U deletion RECC. Interestingly, recent proximity labeling studies suggested an increased interaction of RESC with U deletion RECC compared to those that perform U insertion (51). Specifically, three of the four RESC proteins tested by BioID interacted with the KREN1 deletion endonuclease and KREX1 U-specific exoribonuclease, while none of the four interacted with the KREN2 major insertion endonuclease. These findings support a critical function of RESC in recruitment and/or positioning of U deletion RECCs during editing progression.
A second important function for RESC10 was revealed through analysis of those mRNAs with internal initiating gRNAs. We observed no effect of RESC10 depletion on the initiation of editing for the pan-edited RPS12 mRNA, whose editing initiates at its 3′ end. In contrast, all three internal initiating mRNAs examined (ND7–5′, CYb, and MURF2) displayed significantly increased pre-edited mRNA upon RESC10 knockdown by HTS, and by qRT-PCR for those tested, indicating that their entry into the editing pathway is disrupted in the absence of RESC10. This distinction is reminiscent of our previous findings with RESC12A/RESC12, which exhibit the opposite phenotype, being essential for initiation of RPS12, but not for ND7–5′ (12). Together, these data suggest that different mechanisms exist for the initiation of editing at the 3′ end of a transcript versus internally. Several RNA binding proteins and processing factors associate with the 3′ ends of pre-edited mRNAs (3,51). Specifically, KPAF3 binds the 3′ end of pre-edited mRNAs to stimulate poly(A) tail addition by KPAP1; KPAF4/5 binds the poly(A) tail. Protein-protein interaction studies reveal that KPAP1 and KPAP4/5 make contacts with several RESC proteins, suggesting these factors may recruit RESC for editing initiation at the 3′ end in a manner that is not reliant on RESC10. However, in the absence of these 3′ end binding factors, RESC10 becomes essential for recruitment and/or assembly of RESC, such that editing can initiate internally. Again, this scenario is consistent with RESC10 acting early in a RESC assembly pathway.
The dissociation of RESC that occurs upon RESC10 knockdown raises questions regarding the abundance and interactions of the gRNA stabilizing protein, RESC1/2. The RESC1/2 heterotetramer interacts with the GRBC module in an RNA independent manner (19,23), and so is usually considered to be a GRBC component. However, RESC1/2 is also reportedly present in RESC variants with either RESC14 or KRGG3 that appear to contain little RESC6. Also, the REH2C complex co-purifies with a RESC variant with little or no RESC6. Together, those observations suggest RESC1/2 interactions outside of the canonical GRBC (4,18,25,26). Here, we observed that when RESC13 dissociates from GRBC, or when RESC5 dissociates from other GRBC proteins, the levels of RESC2 with each of the dissociated entities remains essentially the same (Figure 3). We previously reported similar results in RESC14 and RESC8 knockdowns, where RESC13 and RESC6 associated RESC2 remained unchanged despite dissociation of these two proteins (25,29). One possibility is that multiple RESC1/2 heterotetramers are present in a given RESC complex, and that these partition with distinct complex components when they separate from each other (Figure 9). Alternatively, there may be free RESC1/2 present in mitochondria, which becomes associated with multiple RESC components when they are disassociated due to knockdown of a RESC protein. RESC1/2 may also be transferred from one of the other RESC1/2 containing complexes described above during RESC disorganization. Future studies will be necessary to fully understand the interactions of RESC1/2 and which subsets of this heterotetramer are gRNA loaded.
A recent study suggested that RESC protects the 3′ ends of mRNAs that have entered the editing pathway from degradation by bridging the KPAF4/5 and PPsome complexes associated with the 3′ and 5′ ends of trypanosome mitochondrial mRNAs, respectively (51). The authors showed that depletion of RESC10 resulted in loss of edited RPS12 mRNA, as assayed by northern blot with a probe corresponding to 5′ edited sequence, and this finding was interpreted to mean that RESC10 is necessary for stabilization of edited RPS12 mRNA. However, our data provide an alternative interpretation of these results. The RPS12 probe used by Aphasizheva et al. (51) corresponds exactly to the region covered by our qRT-PCR assay (Figure 5), and as such we agree that RPS12 mRNA edited in this region is decreased over 90% by RESC10 knockdown. However, these authors failed to measure in any way mRNA that has entered the editing pathway but whose editing has not progressed to the 5′ region of RPS12 analyzed. We show here that RESC10 knockdown leads to extensive pausing in the 3′ to 5′ progression of editing at multiple ES that are 3′ of the region being measured by northern blot or qRT-PCR. Thus, while we cannot rule out a contribution of edited mRNA decay to the decrease in 5′ edited RPS12 mRNA, we conclude that a majority of the observed decrease is accounted for by an increase in partially edited mRNAs whose editing has not progressed into the 5′ third of the RPS12 transcript.
In summary, we show here that RESC integrity is dependent on RESC10, which acts early in RESC assembly. RESC disruptions caused by RESC10 depletion lead to widespread editing defects, manifested in both pauses in the progression of editing of pan-edited mRNAs and failure to initiate editing by internal initiating gRNAs. Moreover, the impacts of RESC disorganization appear especially prominent at sites of U deletion, implicating RESC in the orchestration of transitions between U insertion and U deletion RECCs.
DATA AVAILABILITY
The sequencing data have been deposited in the Sequence Read Archive, accession numbers SRP271414 (RESC10 knockdown, RPS12 and ND7–5′), SRP185791 (RESC10 knockdown, CYb and MURF2) and SRP097727 (previously published uninduced samples (12) included in the uninduced pool).
ACKNOWLEDGEMENTS
We thank Yijun Sun (Department of Microbiology and Immunology, University at Buffalo) and Runpu Chen (Department of Computer Science and Engineering, University at Buffalo) for assistance with statistical analysis of high throughput sequence data. We are also grateful to the University at Buffalo Center for Computational Research, especially Andrew Bruno, and the University at Buffalo Genomics and Bioinformatics Core, especially Jonathan Bard, for their support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [R01 GM129041 to L.K.R.]. Funding for open access charge: NIH [R01 GM129041].
Conflict of interest statement. None declared.
Notes
Present address: Natalie M. McAdams, Thermo Fisher Scientific, Buffalo, NY.
REFERENCES
1.
2.
3.
4.
5.
6.
7.
8.
9.
10.
11.
12.
13.
14.
15.
16.
17.
18.
19.
20.
21.
22.
23.
24.
25.
26.
27.
28.
29.
30.
31.
32.
33.
34.
35.
36.
37.
38.
39.
40.
41.
42.
43.
44.
45.
46.
47.
48.
49.
50.
 Trypanosome RNAEditing Substrate Binding Complex integrity and function depends on the upstream action of RESC10
Trypanosome RNAEditing Substrate Binding Complex integrity and function depends on the upstream action of RESC10
