
Competing Interests: The authors have declared that no competing interests exist.
- Altmetric
Treatment with hydrogen peroxide (H2O2) raises the hatching rate through the development and diapause termination of Artemia cysts. To comprehend the upstream genetic regulation of diapause termination activated by exterior H2O2 elements, an Illumina RNA-seq analysis was performed to recognize and assess comparative transcript amounts to explore the genetic regulation of H2O2 in starting the diapause termination of cysts in Artemia salina. We examined three groupings treated with no H2O2 (control), 180 μM H2O2 (low) and 1800 μM H2O2 (high). The results showed a total of 114,057 unigenes were identified, 41.22% of which were functionally annotated in at least one particular database. When compared to control group, 34 and 98 differentially expressed genes (DEGs) were upregulated in 180 μM and 1800 μM H2O2 treatments, respectively. On the other hand, 162 and 30 DEGs were downregulated in the 180 μM and 1800 μM H2O2 treatments, respectively. Cluster analysis of DEGs demonstrated significant patterns among these types of 3 groups. GO and KEGG enrichment analysis showed the DEGs involved in the regulation of blood coagulation (GO: 0030193; GO: 0050818), regulation of wound healing (GO:0061041), regulation of hemostasis (GO: 1900046), antigen processing and presentation (KO04612), the Hippo signaling pathway (KO04391), as well as the MAPK signaling pathway (KO04010). This research helped to define the diapause-related transcriptomes of Artemia cysts using RNA-seq technology, which might fill up a gap in the prevailing body of knowledge.
Introduction
During Artemia diapause, the metabolic process of egg cells reduces [1], thereby decreasing energy consumption; furthermore, molecular chaperones are accustomed to prevent or decrease protein denaturation. Brine shrimp possess a solid, semipermeable outer shell that protects their embryos while making sure embryo contact with external circumstances. Brine shrimp contain trehalose, which exhibits a fluid retention capacity that protects embryos from lethal harm due to dehydration [2, 3]. Diapause termination of Artemia cysts was proven to be regulated by environmental elements. Robbins et al. [4] indicated that the hatching rate of Artemia cysts improved by 50% when subjected to 180 μM hydrogen peroxide (H2O2). Based on the experimental outcomes obtained by these studies, chemical methods may be employed to terminate cyst diapause. However, the mechanism of diapause termination has not been elucidated and therefore is a subject that warrants in-depth investigation.
Several studies on cyst diapause have been around in arthropods utilizing a transcriptomic approach to elucidate the gene expression involved with diapause or diapause termination. For examples, transcriptomic profiling uncovered significant changes in gene expressions related to gonad development, lipid synthesis, and molt regulation, suggestive of a coupling of the processes in diapause initiation in copepod Calanus finmarchicus [5–7]. Tu et al. [8] used the proteomic and transcriptomic tools to investigate the distinctions between diapause and non-diapause eggs at both protein and mRNA expression levels. These differentially expressed proteins and genes were associated with metabolism and cryoprotection.
Researches on Artemia species have exposed to exterior biotic or abiotic elements to ensure that they provide a basic background data of Artemia transcriptomes, these researches demonstrate that the power of RNA-seq to recognize molecules in Artemia, however these studies were focusing on the adult individuals [9, 10]. In this research, we uncovered brine shrimp to H2O2 to induce their diapause termination. Subsequently, RNA-seq was used| to investigate the transcriptional profiles of H2O2-induced diapause termination of Artemia cysts to characterize the physiological regulation of the diapause termination process at the transcriptomic level.
Materials and methods
Artemia cyst samples
The modified procedure of the acquisition of Artemia salina diapause cysts was according to the method provided by Robbins et al. [4]. The cysts were incubated in artificial seawater in a 15 cm Petri dish for 7 days. The shell residuals and hatched individuals were removed each day. After 7-days incubation, the unhatched-cysts were used and harvested for diapause. The diapausing cysts were then incubated in a stationary condition in a Petri dish (15 cm) with H2O2 at 0, 180 and 1800 μM for 5 h.
RNA quantification and qualification
RNA qualification and quantification were determined by electrophoresis analysis at 1% agarose gels to monitor whether its degradation and contamination. RNA purity was measured by utilizing NanoPhotometer® spectrophotometer (IMPLEN, CA, USA). RNA concentration was determined using a Qubit® RNA Assay Kit in Qubit® 2.0 Fluorometer (Life Technologies, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
Library preparation for transcriptome sequencing
From the RNA sample to the ultimate data, each step, including sample testing, library collecting, and sequencing, influences the quality of the data which impacts the analysis consequences. To ensure the reliability of the data, quality control (QC) is conducted at each step in this research. The workflow is shown in S1A Fig.
A total amount of 1.5 μg RNA per sample was used for the RNA sample preparations. Sequencing libraries were produced using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer’s guidelines, and index codes were put into attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was completed using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). First strand cDNA was synthesized using random hexamer primers and M-MuLV Reverse Transcriptase. DNA polymerase I and RNase H were applied to synthesize the Second strand cDNA synthesis subsequently. The rest of the overhangs were changed into blunt ends via exonuclease/polymerase actions. After adenylation of the 3’ ends of DNA fragments, NEBNext Adaptors with hairpin loop structures were ligated for hybridization. To obtain the specific cDNA fragments of 150~200 bp in length, the library fragments were purified with the AMPure XP system (Beckman Coulter, Beverly, USA). Then, 3 μl USER Enzyme (NEB, USA) was used in combination with size-chosen, adaptor-ligated cDNA at 37°C for 15 min accompanied by 5 min at 95°C before PCR. After that, PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. Finally, PCR products were purified (AMPure XP system), and library quality was assessed on the Agilent Bioanalyzer 2100 system. Raw cDNA sequences have been deposited in Sequence Read Archive (SRA) at NCBI BioProject resources. The following accession numbers was PRJNA664008.
Data quality control
Raw data (raw reads) in fastq file format were processed with in-house Perl scripts. Clean data (clean reads) were obtained by dethatching reads that contain adaptors, poly-N and low-quality reads from raw data in this step. Simultaneously, the Q20, Q30, GC content and sequence duplication level of the clean reads were calculated. All downstream analyses were based on clean data with high quality.
Transcriptome assembly
Due to the lack of a reference genome for Artemia samples, clean reads have to be assembled to obtain a reference sequence for the next analysis (S1B Fig). The software Trinity [11] was applied to complete the transcriptome reconstruction. The left files (read1 files) from all libraries/samples were pooled into one left.fq file, and another right files (read 2 files) were pooled into one right.fq file. Transcriptome assembly was achieved based on the left and right fq. using Trinity transcriptome assembler with min/kmer/cov set to 2 by default, and all the parameters were at default configurations.
The workflow of Trinity was described followed: Inchworm: Constructs a k-mer dictionary from all sequenced reads (k = 25), selects the most frequent seeding k-mer in the dictionary and extends the seed in both directions to create a contig assembly. Chrysalis: Chrysalis clusters minimally overlapping Inchworm contigs into sets of connected elements and constructs comprehensive de Bruijn graphs for each element. Each element defines an assortment of Inchworm contigs that will tend to be derived from alternative splice forms or closely related gene paralogs. Butterfly: Butterfly reconstructs full-length, plausible, linear transcripts by reconciling the individual de Bruijn graphs produced by Chrysalis with the original sequence reads and paired ends. Butterfly reconstructs unique transcripts for splice isoforms and paralogous genes and resolves ambiguities stemming from sequences >k bases length or from errors that are shared among transcripts. The final assembled result named as TRINITY.fasta.
Gene functional annotation
Gene function was annotated according to the information in following databases: Nt (NCBI non-redundant nucleotide sequences),Nr (NCBI non-redundant protein sequences), KOG/COG (Clusters of Orthologous Groups of proteins), Pfam (Protein family), Swiss-Prot (A manually annotated and reviewed protein sequence database), GO (Gene Ontology) and KO (KEGG Ortholog database).
The provided information and characteristics of the seven databases are specific and following described as: Nr (NCBI non-redundant protein sequences): the formal protein sequence database of NCBI, which include protein sequence information from such databases as GenBank, Swiss-Prot, PIR (Protein Information Resource), PRF (Protein Research Foundation), and PDB (Protein Data Bank).Pfam (Protein family) is the most comprehensive collection of protein families and domains, represented as multiple sequence alignments and profile by hidden Markov model. Pfam families are classified into two categories, Pfam-A and Pfam-B, based on the reliability of annotations. Both KOG (euKaryotic Orthologous Groups) and COG (Cluster of Orthologous Groups of proteins) are derive from NCBI’s gene orthologous relationships. COG is specific to prokaryotes, while KOG is definitely specific to eukaryotes. Based on the evolutionary relationships among species, COG/KOG defined the homologous genes from different species into different ortholog clusters. Swiss-Prot was an annotated and reviewed high-quality protein sequence database which combines computed features, experimental results, and scientific conclusions. And KEGG (Kyoto Encyclopedia of Genes and Genome) is a resource for understanding the high-level functions and utilities database of each biological system.GO (Gene Ontology) is the established standard for the functional annotation of gene products and is used to classify the three following functional attributes of gene: BP: biological process, MF: molecular function, and CC: cellular component.
Quantification of gene expression levels
The degree of gene expression was estimated by RSEM [12]; clean data had been mapped onto the assembled transcriptome, and the read count for each gene was calculated from the mapping consequences.
Differential expression analysis
The value of read count obtained from the gene expression analysis was used for the initial data to carry out further differential expression analysis. Group differential expression analysis was conducted by using the DESeq R package (1.10.1) [13]. DESeq provides statistical routines, in which the one of the models based on the negative binomial distribution was used to determining the differential expression with digital gene expression in this study. For controlling the false discovery rate obtained from digital gene expression analysis, the P value of DESeq analysis was then adjusted by using Benjamini and Hochberg’s approach. Genes with an adjusted p-value| <0.05 was considered as differentially expression.
Clustering and sequencing (Novogene Experimental Department)
Corset [14] functions by clustering contigs based on separates contigs and shared reads when different expression patterns observed between samples. Corset also filter out contigs with a low number of mapped reads with the information from reads (less than 10 reads). The clustering of the index-coded samples was performed on a cBot Cluster Generation System using the Illumina HiSeq PE Cluster Kit (cBot-HS) based on the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina HiSeq platform, and generated the paired-end reads.
Gene enrichment analysis
GO enrichment analysis of the DEGs was applied on the GOseq R package-based Wallenius noncentral hypergeometric distribution [15], which can adjust the bias of gene length in DEGs.
KEGG [16] is a database for understanding the high-level functions and utilities of genes in biological systems from molecular-level information. These genes could be collected from cells, organisms and ecosystems, especially large-scale molecular datasets generated by genome sequencing and high-throughput experimental technologies (http://www.genome.jp/kegg/) could provide more reliable evidence. Software KOBAS [17] was used to evaluated the statistical enrichment of differentially expressed genes in KEGG pathways. The software and parameters used in this study for data analysis are listed in S1 Table.
Results and discussion
Increase of hatching rates after H2O2 treatments
After 7-d incubation of Artemia cysts, the un-hatched ones were collected and cultured 12 hr for identification of H2O2 treatments on the hating rates. The results showed that the hatching rate was 3.1 ± 0.90% in control group (0 μM H2O2); 47.5 ± 5.31% in low-H2O2 group (180 μM), and 4.4 ± 1.30% in high-H2O2 group (1800 μM). Robbins et al. [4] and Hong et al. [9] have demonstrated that H2O2 increases the hatching rates of Artemia cysts and the hatching rates decreased in higher H2O2 than in lower H2O2 treatments. The pattern of H2O2 treatments on hatching rates is similar to our study.
Gene expression analysis
Transcriptomes of diapause cysts with or without H2O2-treated were sequenced individually, in which 26,252,846–49,563,734 raw reads were generated. The reads of adaptors and low quality were then removed by filtering process, so that there could have a clean reads for downstream analyses. The filtering process includes several steps: (1) to eliminate the reads with adaptor contamination. (2) to exclude the reads when uncertain nucleotides composition more than 10% of either reads (N > 10%). (3) to remove reads when low-quality nucleotides (base quality less than 20) constitute more than 50% of the total read. After filtering, 25,645,468–48,301,960 clean reads were obtained. The results of Kruskal-Wallis test revealed that the amounts of clean reads among the cysts with or without treating H2O2 weren’t considerably different (p = 0.2881). Therefore, the regulation of H2O2-induced diapause termination of Artemia cysts was related to the gene expression level rather than the amount of expressed genes. The de novo transcriptome filtered by Corset was used as a reference alignment. RSEM could map reads again to the transcriptome and quantify the expression level. S2 Table showed the mapping results. Both FPKM and RPKM used for gene expression level normalization. FPKM considered the fragment difference between these two reads of paired end reads, while RPKM considers reads: rather than using read counts, it approximates the relative abundance of transcripts regards to fragments observed from an RNA-seq experiment. That might not be represented by a single read, such as in paired-end RNA-seq experiments. In bioinformatic analysis without genome references, such as Artemia, the gene with FPKM>0.3 is considered to being expressed. This threshold is recommended and accepted by several prestigious journal and had been considered to be closely represent the high-level gene expression.
Clean reads were assembled by de novo method via Trinity to obtain the assembly transcriptome. To remove redundancies, the hierarchical clustering was carried out to the corset. Afterwards, the longest transcripts of each cluster were selected for unigenes. There were 114,153 transcripts and 114,057 unigenes were identified in the cyst samples (Table 1). With the priorities of genomic or transcriptomic techniques, recent researches had emphasized in-depth studies at the transcriptome level in Artemia organisms [9, 10, 18–20]. It was supposed that the genes involved with development or diapause termination might be correlated with the systematic changes| in each cell. Therefore, it could be the proof that there were more expressed genes in reactivated Artemia embryos in this study. For completeness of the Artemia transcriptome, the results provide valuable and supplementary information which could complete the presented library nowadays.

| Metric | w/o H2O2 | w/180μM H2O2 | w/1800μM H2O2 |
|---|---|---|---|
| Raw Reads | 41,470,557.33 ± 9,789,097.84 | 36,445,362.00 ± 11,441,445.00 | 28,630,529.33 ± 2,565,135.63 |
| Clean Reads | 40,518,019.33 ± 9,572,949.25 | 35,568,416.00 ± 11,117,664.18 | 27,965,944.67 ± 2,407,193.29 |
| Clean Bases | 6.07 ± 1.45 | 5.33 ± 1.63 | 4.20 ± 0.40 |
| Error (%) | 0.03 ± 0.00 | 0.03 ± 0.00 | 0.03 ± 0.00 |
| Q20 | 97.71 ± 0.21 | 97.70 ± 0.23 | 97.63 ± 0.12 |
| Q30 | 93.37 ± 0.47 | 93.32 ± 0.55 | 93.18 ± 0.32 |
| GC (%) | 40.66 ± 0.15 | 40.44 ± 0.03 | 40.28 ± 0.19 |
| Total transcripts | 114253 | ||
| Total unigenes | 114057 | ||
Data: mean ± sd
(1) Sample name: the names of samples
(2) Raw Reads: the original sequencing reads counts
(3) Clean Reads: number of reads after filtering
(4) Clean Bases: clean reads number multiply read length, saved in G unit
(5) Error Rate: average sequencing error rate, which is calculated by Qphred = -10log10(e)
(6) Q20: percentages of bases whose correct base recognition rates are greater than 99% in total bases
(7) Q30: percentages of bases whose correct base recognition rates are greater than 99.9% in total bases
(8) GC content: percentages of G and C in total bases
Gene functional annotation
To achieve comprehensive gene functional annotation, seven databases were applied by Novogene. These databases were utilized for functional annotation of identified unigenes, and the statistics of successfully annotated genes in this research by the database are shown in Fig 1 and S3 Table. Of the 114,057 unigenes, the number of genes successfully annotated in at least one database against NT, NR, KO, Swiss Prot, PFAM, KOG and GO was 47,020 (41.22%). The summary of gene annotations based on each database is listed in S3 Table. The functional annotation of these genes according to KOG, KEGG and GO classifications is demonstrated in Fig 2. In GO, the most top five annotated genes were linked to 1) cellular process, 2) binding, 3) metabolic process, 4) single-organism process, and 5) catalytic activity. Those in KOG were related linked to 1) general function mechanism only, 2) signal transduction mechanisms, 3) posttranslational modification, protein turnover, chaperones, 4) translation, and 5) ribosomal structure and biogenesis. In the KEGG pathway, these genes were involved with 1) signal transduction, 2) translation, 3) the endocrine system, 4) transport and catabolism, and 5) proteins folding, sorting and degradation.

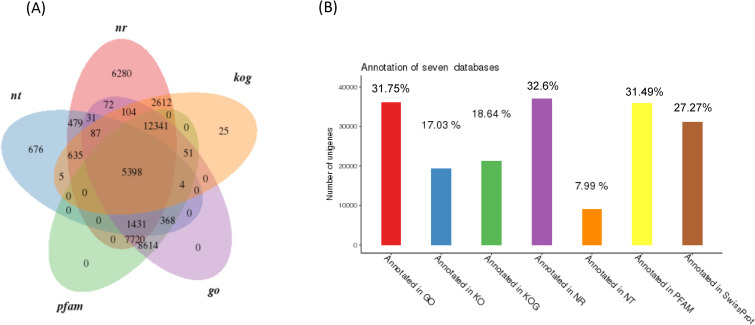
The number and percentage of identified unigenes based on the selected databases.
(A) the venn diagram is mapped with 5 selected database annotation result. (B) the number and percentage of identified unigenes through 7 databases.

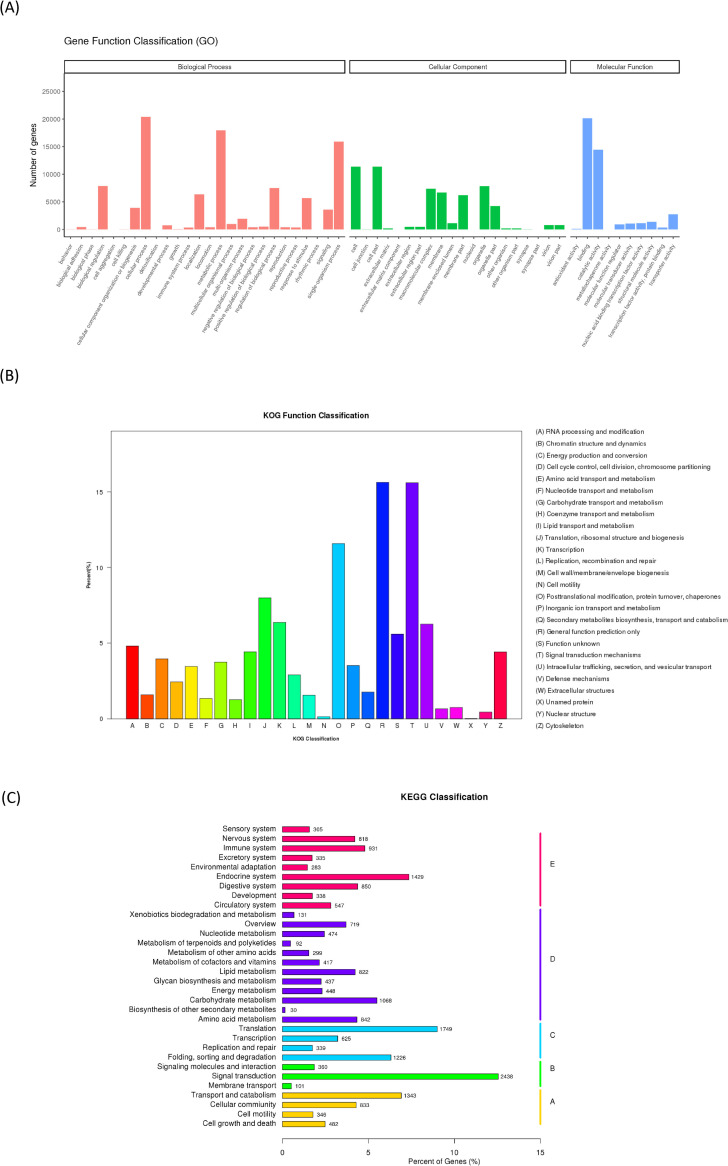
Functional annotation results of identified unigenes.
(A) Gene Ontology annotation that successfully annotated genes will group into three main GO domains: biological process (BP), cellular component (CC), molecular function (MF). (B) Karyotic Orthologous Groups (KOG) classification of putative proteins (C) Kyoto Encyclopedia of Genes and Genome (KEGG) pathway contains: metabolism, genetic information processing, environmental information processing, cellular processes, organismal systems and human diseases. Human diseases results were removed in the result of KOG and KEGG pathways.
The patterns of GO, KOG and KEGG, classifications were consistent with those of the A. sinica transcriptome [10], except for the amount of expressed genes. Thus, the additional genes recognized from this study provided a supplementary draft for Artemia transcriptomes.
Differential gene expression related to H2O2-induced diapause termination
Volcano plots were used to infer the overall distribution of differentially expressed genes (DEGs), and the number of DEGs for each paired comparison among the control (w/o H2O2) and H2O2 treatments (w/180- and 1,800-μM H2O2) are shown in Fig 3. When compared to control group, 34 and 98 DEGs were upregulated in the 180- and 1800-μM H2O2-treated groups, respectively. Furthermore, 40 and 162 DEGs were downregulated in the 180- and 1800-μM H2O2-treated groups, respectively. DEGs referring to H2O2-induced up- or downregulation compared to the control group were listed in Table 2.

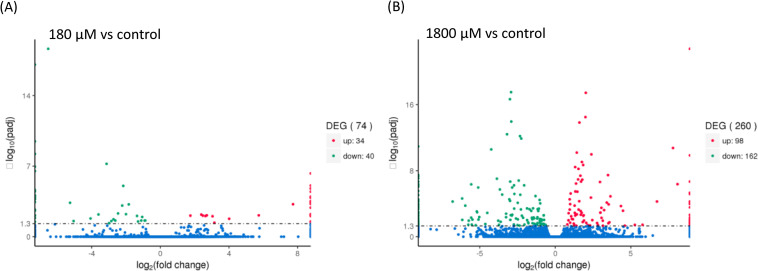
Volcano plots of differentially expressed genes associated with H2O2 treatments.
The X axis represents fold change, while the Y axis indicates the significance of differential expression. (A) 180 μM vs control. (B) 1800 μM vs control.

| Gene Name | 180μM | 1800μM | UniGenes | Term |
|---|---|---|---|---|
| VARS, valS FARSB | ↑ | Cluster2579.61530 | Aminoacyl-tRNA biosynthesis | |
| CYP15A1_C1 | ↑ | Cluster2579.21243 | Insect hormone biosynthesis | |
| MPDZ, MUPP1, Patj | ↑ | Cluster2579.55288 | Tight junction | |
| ATG12 | ↑ | Cluster2579.41553 | Regulation of autophagy/RIG-I-like receptor signaling pathway/FoxO signaling pathway | |
| E2F4_5 | ↑ | Cluster2579.24946 | TGF-beta signaling pathway/Cell cycle | |
| CTTN, EMS1 | ↑ | Cluster2579.53583 | Shigellosis/Pathogenic Escherichia coli infection/Bacterial invasion of epithelial cells/Proteoglycans in cancer | |
| GYS | ↑ | ↑ | Cluster2579.13656 | Starch and sucrose metabolism/AMPK signaling pathway/Insulin signaling pathway/PI3K-Akt signaling pathway |
| MLL1 | ↑ | Cluster2579.7161 | Lysine degradation/Transcriptional misregulation in cancer | |
| E1.14.11.2 | ↑ | Cluster2579.60582 | Arginine and proline metabolism | |
| MPDZ, MUPP1, Patj | ↑ | Cluster2579.55288 | Hippo signaling pathway—fly | |
| CACNA1H | ↑ | Cluster2579.6210 | Circadian entrainment/Calcium signaling pathway/Calcium signaling pathway/MAPK signaling pathway | |
| RASAL2 | ↑ | ↑ | Cluster2579.27404 | Ras signaling pathway |
| K16330 | ↑ | Cluster2579.24942 | Pyrimidine metabolism | |
| CTSO | ↑ | Cluster2579.28731 | Lysosome | |
| NDUFS3 | ↑ | Cluster2579.45089 | Non-alcoholic fatty liver disease (NAFLD)/Oxidative phosphorylation/Parkinson’s disease/Alzheimer’s disease/Huntington’s disease | |
| E3.5.4.3, guaD | ↓ | Cluster2579.11205 | Purine metabolism | |
| PDE1 | ↓ | Cluster2579.6962 | Morphine addiction/Calcium signaling pathway/Purine metabolism | |
| PDE4 | ↓ | Cluster2579.18105 | cAMP signaling pathway | |
| BMPR2 | ↓ | Cluster2579.35766 | Cytokinecytokine receptor interaction/TGF-beta signaling pathway/Signaling pathways regulating pluripotency of stem cells/MicroRNAs in cancer/Hippo signaling pathway | |
| TBK1 | ↓ | Cluster2579.19502 | RIG-I-like receptor signaling pathway/Cytosolic DNA-sensing pathway/Toll-like receptor signaling pathway/Hepatitis C/Measles/Hepatitis B/Ras signaling pathway/Herpes simplex infection/Influenza A/Epstein-Barr virus infection | |
| CACNA2DN CACNA1N | ↓ | Cluster2579.54638 | MAPK signaling pathway | |
| Cluster2579.67327 | ||||
| E3.2.1.24 | ↓ | Cluster2579.64238 | Other glycan degradation | |
| htpG, HSP90A | ↓ | Cluster25290.0 | NOD-like receptor signaling pathway/Plantpathogen interaction/Prostate cancer/Antigen processing and presentation/Progesteronemediated oocyte maturation/Estrogen signaling pathway/PI3K-Akt signaling pathway/Pathways in cancer/Protein processing in endoplasmic reticulum | |
| GIT1 | ↓ | Cluster2579.39583 | Epithelial cell signaling in Helicobacter pylori infection/Regulation of actin cytoskeleton | |
| SMC1 | ↓ | Cluster2579.34889 | Cell cycle yeast/Cell cycle | |
| EIF6 | ↓ | ↓ | Cluster2579.23978 | Ribosome biogenesis in eukaryotes |
| LSM4 | ↓ | Cluster2579.41833 | RNA degradation/Spliceosome | |
| CYP18A1 | ↑ | Cluster2579.33894 | Insect hormone biosynthesis | |
| SCD, desC GYS | ↑ | Cluster30892.0 | AMPK signaling pathway | |
| Cluster2579.13656 | ||||
| NR5A2, FTF | ↑ | Cluster2579.27421 | Maturity onset diabetes of the young | |
| A2M | ↑ | Cluster2579.38707 | Complement and coagulation cascades | |
| BRCA2, FANCD1 | ↑ | Cluster2579.56302 | Homologous recombination/Fanconi anemia pathway/Pancreatic cancer/Pathways in cancer | |
| EXTL3 | ↑ | Cluster2579.55901 | Glycosaminoglycan biosynthesis heparan sulfate/heparin | |
| SCD, desC | ↑ | Cluster30892.0 | Biosynthesis of unsaturated fatty acids/PPAR signaling pathway/Fatty acid metabolism | |
| deoC, DERA | ↑ | Cluster2579.69015 | Pentose phosphate pathway | |
| SNW1, SKIIP, SKIP | ↑ | Cluster2579.36513 | Notch signaling pathway/Viral carcinogenesis/Spliceosome/Epstein-Barr virus infection | |
| ELOVL7 | ↑ | Cluster2579.48704 | Fatty acid elongation | |
| NOS1 | ↑ | Cluster2579.34982 | Amyotrophic lateral sclerosis (ALS)/Long-term depression/Arginine and proline metabolism/Circadian entrainment/Salivary secretion/Phagosome/Alzheimer’s disease | |
| GPD1 | ↑ | Cluster2579.39411 | Glycerophospholipid metabolism | |
| nagA, AMDHD2 | ↑ | Cluster2579.16917 | Amino sugar and nucleotide sugar metabolism | |
| FARSB, pheT | ↑ | Cluster2579.35161 | Aminoacyl-tRNA biosynthesis | |
| EYA1 | ↑ | Cluster2579.43795 | Transcriptional misregulation in cancer | |
| HSPA HSPA1_8 | ↓ | Cluster19454.1 | Antigen processing and presentation/Estrogen signaling pathway | |
| Cluster2579.2811 | ||||
| Cluster2579.68593 | ||||
| Cluster15619.0 | ||||
| htpG, HSP90, AhtpG | ↓ | Cluster25290.0 | NOD-like receptor signaling pathway/Plant-pathogen interaction/Prostate cancer/Progesteronemediated oocyte maturation/Pathways in cancer | |
| Cluster24430.0 | ||||
| EEF1A | ↓ | Cluster28693.0 | Legionellosis/Toxoplasmosis/Measles/Spliceosome/Endocytosis/Influenza A | |
| Cluster30677.0 | ||||
| CACNB2,CACNA2DN MKNK, MNK MAP3K13, LZK | ↓ | Cluster2579.5710 | MAPK signaling pathway | |
| Cluster2579.54638 | ||||
| Cluster2579.41607 | ||||
| Cluster2579.34400 | ||||
| LIMD1,ZDHHC9_14_1 MPDZ, MUPP1 Patj AJUBA, WTIP | ↓ | Cluster2579.46774 | Hippo signaling pathway—fly | |
| Cluster2579.55157 | ||||
| Cluster2579.20389 | ||||
| HSPA5, PDIA6, TXNDC | ↓ | Cluster2579.13853 | Protein processing in endoplasmic reticulum | |
| Cluster2579.21006 | ||||
| Cluster35149.0 | ||||
| DIAPH1 DOCK | ↓ | Cluster2579.53371 | Shigellosis | |
| Cluster2579.57004 | ||||
| STIP1 | ↓ | Cluster2579.68691 | Prion diseases/Protein export/Thyroid hormone synthesis | |
| CACNB2 | ↓ | Cluster2579.5710 | Arrhythmogenic right ventricular cardiomyopathy (ARVC)/Hypertrophic cardiomyopathy (HCM)/Dilated cardiomyopathy | |
| TFIIE1, GTF2E1, TFA1 tfe | Cluster15619.0 | Epstein-Barr virus infection | ||
| Cluster2579.25410 | ||||
| glnA, GLUL E4.1.3.1, ace | ↓ | Cluster28996.0 | Glyoxylate and dicarboxylate metabolism | |
| Cluster2579.67175 | ||||
| Cluster14244.0 | ||||
| VAV, DIAPH1 | ↓ | Cluster2579.54232 | Focal adhesion, Leukocyte transendothelial migration | |
| Cluster2579.53371 | ||||
| clpX, CLPX | ↓ | Cluster2579.41888 | Cell cycle Caulobacter | |
| PPP2R3, pckA, PEPCK | ↓ | Cluster2579.46613 | PI3K-Akt signaling pathway/mRNA surveillance pathway/Dopaminergic synapse/AMPK signaling pathway | |
| Cluster2579.37834 | ||||
| DOCK1 | ↓ | Cluster2579.57004 | Bacterial invasion of epithelial cells | |
| CACNB2 RGS2 | ↓ | Cluster2579.5710 | Oxytocin signaling pathway | |
| Cluster2579.8158 | ||||
| MPDZ, MUPP1 Patj | ↓ | Cluster2579.55157 | Tight junctio | |
| EYA1 | ↓ | Cluster2579.41820 | Transcriptional misregulation in cancer | |
| SIN3A | ↓ | Cluster2579.35774 | Thyroid hormone signaling pathway/Viral myocarditis/Pathogenic Escherichia coli infection/Salmonella infection/Adherens junction/Phototransduction fly/Vibrio cholerae infection/Platelet activation | |
| AJUBA, LIMD1 WTIP | ↓ | Cluster2579.20389 | Hippo signaling pathway | |
| TOP3 | ↓ | Cluster2579.31547 | Homologous recombination/Fanconi anemia pathway | |
| SETDB1, OTX1 | ↓ | Cluster2579.30770 | Signaling pathways regulating pluripotency of stem cells | |
| Cluster2579.52125 | ||||
| MKNK, MNK | ↓ | Cluster2579.41607 | HIF-1 signaling pathway | |
| glnA, GLUL | ↓ | Cluster2579.67175 | Nitrogen metabolism/Arginine and proline metabolism/Glutamatergic synapse/Biosynthesis of amino acids/Two-component system | |
| Cl | ↓ | Cluster2579.62889 | Hedgehog signaling pathway | |
| dnaK, DCP1B | ↓ | Cluster2579.61706 | RNA degradation | |
| Cluster2579.42146 | ||||
| VAV | ↓ | Cluster2579.54232 | B cell receptor signaling pathway/Fc epsilon RI signaling pathway/Natural killer cell mediated cytotoxicity/Chemokine signaling pathway/cAMP signaling pathway | |
| mutY | ↓ | Cluster2579.58355 | Base excision repair | |
| EEF1A, XPOT, EEF1A | ↓ | Cluster28693.0 | RNA transport | |
| Cluster2579.30436 Cluster30677.0 | ||||
| PPT | ↓ | Cluster2579.1265 | Fatty acid elongation/Fatty acid metabolism | |
| PTGDS | ↓ | Cluster21276.0 | Arachidonic acid metabolism | |
| DNM1L | ↓ | Cluster2579.40879 | TNF signaling pathway | |
| PPP2R3 | ↓ | Cluster2579.46613 | Adrenergic signaling in cardiomyocytes | |
| RP-S6e, RPS6 | ↓ | Cluster14365.0 | Proteoglycans in cancer, Ribosome | |
| ABCA3 | ↓ | Cluster2579.40956 | ABC transporters | |
| UBE1, UBA1, ATPeF1B, ATP5B, ATP2, SIN3A, | ↓ | Cluster2579.42277 | Parkinson’s disease, Huntington’s disease | |
| Cluster31495.0 | ||||
| KCNMA1 | ↓ | Cluster2579.9471 | Insulin secretion/Phosphatidylinositol signaling system/Vascular smooth muscle contraction/Pancreatic secretion | |
| KIDINS220, ARMS | ↓ | Cluster2579.42798 | Neurotrophin signaling pathway | |
| dnaK | ↓ | Cluster2579.61706 | Tuberculosis | |
| CACYBP, SIP | ↓ | Cluster16736.0 | Wnt signaling pathway | |
| TFIIE1, GTF2E1, TFA1 | ↓ | Cluster2579.25410 | Viral carcinogenesis | |
| ATPeF1B, ATP5B, ATP2 | ↓ | Cluster31495.0 | Oxidative phosphorylation/Alzheimer’s disease |
Cluster analysis was used to discover genes with similar expression patterns under our experimental conditions (S2 Fig). By clustering genes with comparable expression patterns, it may be possible to discern unknown functions of previously characterized genes or the features of unknown genes. In hierarchical clustering, areas of different colors denote different groups (clusters) of genes, and each group may have similar functions or take part in the same biological process. It showed different patterns among these three groups significantly. GO and KEGG enrichment analyses were further utilized to gather genes into biological functions or pathways through bioinformatic databases and statistical tools. The GO enrichment analysis helped to elucidate the gene function of differentially expressed genes, while the KEGG enrichment analysis also helped to elucidate the metabolic pathways where the genes were involved.
Compared to the control group, the annotations of up- and downregulated DEGs in H2O2 treatments by GO and KEGG enrichment analysis are listed in Tables 3 and 4.

| GO accession | Description | Term type | Over represented p-Value | DEG item | DEG list |
|---|---|---|---|---|---|
| GO:0030193 | regulation of blood coagulation | biological process | 0.0010239 | 2 | 62 |
| GO:0050818 | regulation of coagulation | biological process | 0.0010239 | 2 | 62 |
| GO:0061041 | regulation of wound healing | biological process | 0.0010239 | 2 | 62 |
| GO:1900046 | regulation of hemostasis | biological process | 0.0010239 | 2 | 62 |
(1) Gene Ontology entry.
(2) Detailed description of Gene Ontology.
(3) GO types, including cellular component, biological process and molecular function.
(4) p-value in hypergenometric test.
(5) Number of DEGs with GO annotation.
(6) Number of all reference genes with GO annotation.

| Term | Pathway ID | Sample number | Background number |
|---|---|---|---|
| Antigen processing and presentation | ko04612 | 6 | 148 |
| Hippo signaling pathway | ko04391 | 5 | 196 |
| MAPK signaling pathway | ko04010 | 7 | 366 |
(1) Term: Description of KEGG pathways
(2) Pathway ID: KEGG ID.
(3) Sample Number: Number of DGEs with pathway anntation.
(4) Background Number: Number of all reference genes with pathway annotation.
GO enrichment analysis of DEGs showed that the biological functions of DEGs were involved in the regulation of blood coagulation, coagulation, wound healing and hemostasis (Table 3). The interactions of multiple genes could be involved in certain biological functions. As KEGG is a collection of manually curated databases regarding genomes, biological pathways, diseases, drugs, and chemical substances, KEGG was a powerful tool in utilized for pathway enrichment analysis that identified significantly enriched metabolic pathways or signal transduction pathways associated with differentially expressed genes compared with the control group (Table 4 and Fig 4). The degree of KEGG enrichment was measured by the Rich factor that refers to the ratio of the amount of DEGs in the pathway and the amount of all genes annotated in the pathway. The Q-value is the p-value after normalization, and its range is [0,1]. The smaller the q-value is definitely representing the significance of the enrichment analysis. The top 20 significantly upregulated DEGs enriched pathways in H2O2 treatments compared with the control group are shown in Fig 4.

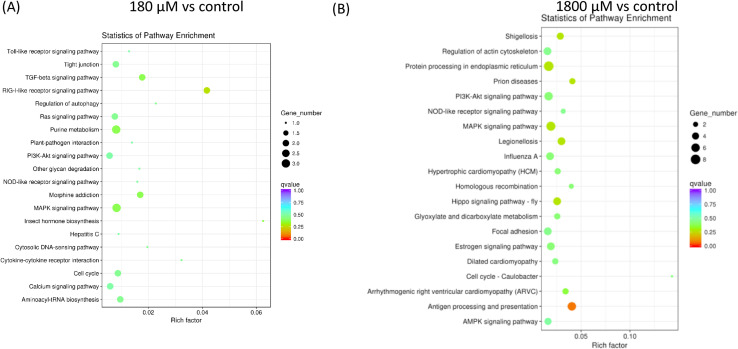
Scatter diagram of top 20 pathway enrichments for DEGs.
The X axis corresponds to the Rich factor that is the ratio of DEGs to all the genes in this pathway. The Y axis represents the different pathway. (A) 180 μM vs control. (B) 1800 μM vs control.
In previous studies, most of them on genes related to the regulation of diapause termination in Artemia cysts focused on genes related to adversity resistance [9, 21–24]. For examples, these reported genes or proteins are involved in the regulation of diapause termination have been elucidated including HSP70 [25], HSP p26 [26, 27], ArHSPs [28–32] and heat shock factor 1 (Hsf1) [33, 34]. In addition, There are also many recent studies on late embryogenesis abundant (LEA) proteins in their roles on Artemia [35–42]. The late embryogenesis abundant (LEA) proteins in plants, bacteria and invertebrates can prevent organisms from protein aggregation caused by low temperature-related dryness or osmotic pressure, and then LEA proteins prevent damage caused by drying, cold or high salinity of a variety of organisms. Thus, diapause termination and adversity resistance have many related mechanisms in regulation.
These previous studies provide us with a protective mechanism against adversity to explore the regulation of diapause termination. NGS was used to study the gene expressions of Artemia under high salinity, and it was found that high salinity would increase the influence of cell cycle arrest through La-related protein and many genes related to lipid transporters [9]. The DEGs involved in binding and catalytic activity were observed to be upregulated during initial development in Artemia cysts [43]. This finding is similar to the results obtained in this research in reactivated diapaused cysts, in which the physiological process for development or diapause termination is close to binding and catalysis. In our previous study, with a metabolomic approach to elucidate the metabolic regulation of H2O2-induced diapause termination. MetaboAnalyst pathway analysis showed the pathway impact of metabolites in groups displaying amino acid degradation and biosynthesis and aminoacyl-tRNA biosynthesis [44]. Among them, aminoacyl-tRNA biosynthesis that was also identified and up-regulated in H2O2 treatments in this study at transcriptional level. Additionally, we also discovered the other regulation pathways involved in H2O2 treatments which were the first findings in diapause termination of Artmeia cysts.
In this study, we analyzed global gene expressions in Artemia cysts in the use of H2O2 to induce diapause termination. These findings were different from the previous studies that focused on genes or proteins involved in the regulation or responses against environmental stress. We found that these DEGs are involved in coagulation regulation and wounds. healing, hemostasis, antigen processing and presentation, Hippo signaling pathway and MAPK signaling pathway. Therefore, this work shows that diapause termination of Artemia cysts is regulated by other pathways which are important on known or novel mechanisms in diapause termination.
Conclusions
In previous studies, most of the diapause termination focused on the regulation related to anti-stress responses, however, in this study, apply with a RNA-seq approach to discover upstream genetic regulation mechanisms that have not been described in Artemia cysts. The novel findings show a lot of genes and pathways which provide a new view to diapause termination of Artemia cysts.
References
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
 Transcriptomic analysis elucidates the molecular processes associated with hydrogen peroxide-induced diapause termination in Artemia-encysted embryos
Transcriptomic analysis elucidates the molecular processes associated with hydrogen peroxide-induced diapause termination in Artemia-encysted embryos
