
Competing Interests: OW has received research grants for clinical studies, speaker’s fees, honoraria and travel expenses from Amgen, Astellas, Bristol-Myers Squibb, Chiesi, Hexal, Janssen-Cilag, MSD, Novartis, Roche, Pfizer, and Sanofi. MN has received research grant for clinical studies, speaker´s fees, honoraria and travel expenses form Alexion, Roche, Otsuka and Sanofi. VK has received honoraria for lectures from Astellas, DaVita, and Chiesi. MZ and EW are employees of Roche Pharma AG. IAH served as advisory board member for the study and received speaker’s honoraria and travel grants from Astellas, Biotest, Novartis, Roche Pharma, and Sanofi during the last 36 months. There are no patents, products in development or marketed products to declare. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
¶ for the VIPP study group. The complete membership of the author group can be found in the Acknowledgments.
- Altmetric
Background
Cytomegalovirus (CMV) infection is amongst the most important factors complicating solid organ transplantation. In a large prospective randomized clinical trial, valganciclovir prophylaxis reduced the occurrence of CMV infection and disease compared with preemptive therapy in CMV-positive renal allograft recipients (VIPP study; NCT00372229). Here, we present a subanalysis of the VIPP study, investigating single nucleotide polymorphisms (SNPs) in immune-response-related genes and their association with active CMV infection, CMV disease, graft loss or death, rejection, infections, and leukopenia.
Methods
Based on literature research ten SNPs were analyzed for TLR4, three for IFN-γ, six for IL10, nine for IL37, and two for TNF-α. An asymptotic independence test (Cochran-Armitage trend test) was used to examine associations between SNPs and the occurrence of CMV infection or other negative outcomes. Statistical significance was defined as p<0.05 and Bonferroni correction for multiple testing was performed.
Results
SNPs were analyzed on 116 blood samples. No associations were found between the analyzed SNPs and the occurrence of CMV infection, rejection and leukopenia in all patients. For IL37 rs2723186, an association with CMV disease (p = 0.0499), for IL10 rs1800872, with graft loss or death (p = 0.0207) and for IL10 rs3024496, with infections (p = 0.0258) was observed in all patients, however did not hold true after correction for multiple testing.
Conclusion
The study did not reveal significant associations between the analyzed SNPs and the occurrence of negative outcomes in CMV-positive renal transplant recipients after correction for multiple testing. The results of this association analysis may be of use in guiding future research efforts.
Introduction
Cytomegalovirus (CMV) infection is the most important serious viral infection that complicates solid organ transplantation [1]. CMV infection can result in CMV disease, which, in severe cases, may lead to hospitalization, morbidity and death [2]. In addition, CMV infection has been associated with secondary effects on the immune system, predisposing patients to opportunistic infections [3, 4], diabetes mellitus, cardiovascular disease and graft loss [5, 6]. Renal transplant recipients without immunity against CMV (R−), who receive an organ from a CMV-positive donor (D+), are at the highest risk of developing CMV disease. Renal transplant recipients with immunity against CMV (R+) are at intermediate risk of active CMV infection [1]. Furthermore, severe organ-invasive CMV disease can also occur in R+ patients and be fatal [7].
Valganciclovir, a valyl ester prodrug of ganciclovir, is currently licensed for the prevention of CMV disease in high-risk (D+/R-) patients receiving a solid organ transplantation [8]. In the largest and longest valganciclovir randomized clinical trial reported to date (VIPP study), prophylaxis was compared with preemptive therapy in CMV R+ renal allograft recipients. The one-year and seven-year results showed that oral valganciclovir prophylaxis significantly reduced CMV infection and disease in intermediate-risk patients compared with preemptive therapy, particularly for CMV-positive donor and recipient (D+/R+) [9, 10]. However, incidences of long-term graft loss and death were similar in the valganciclovir prophylaxis and preemptive treatment groups [10].
Genetic variations in drug targets or in other points of targeted biosynthetic pathways may influence the drug response [11–14]. In renal transplant recipients, polymorphisms in genes whose products are implicated in modulating the human immune response, such as toll-like receptor 4 (TLR4) (Asp299Gly, Tyr399Ile) have been reported to be associated with more frequent serious infectious complications, including severe bacterial infections, CMV disease, and opportunistic infections [15]. Polymorphisms in interleukin (IL) 10 can contribute to CMV reactivation and disease after allogeneic stem cell transplantation [16]. Interferon (IFN)-γ plays an important role in the immune response. A statistically significant correlation was found between the IFN-γ +874 A>T polymorphism and the risk of CMV infection among 247 Hispanic renal transplant recipients [17]. On the other hand, a protective role against CMV infection has been reported for tumor necrosis factor (TNF)-α polymorphisms that are associated with a strong inflammatory response [18]. Anti-inflammatory effects of IL37, a newer member of the IL1 family, were described in a mouse model: Transgenic mice expressing IL37 were protected from colitis [19].
As several studies indicated an association with the occurrence of infection or reported contradictory information about such an association with polymorphisms in TLR4, IFN-γ, IL10, IL37 or TNF-α, we analyzed, as part of the prospective VIPP study mentioned above, single nucleotide polymorphisms (SNPs) in CMV-positive renal allograft recipients.
Methods
Design
This project was conducted as a retrospective analysis of the data of the VIPP study, a randomized multicenter trial comparing CMV prophylaxis vs. preemptive therapy with valganciclovir in CMV-positive renal allograft recipients compared in 22 centers in Germany and in 2 centers in Austria. The study included a recruitment period of 29 months, a study phase of 12 months after transplantation, and a follow-up period of 6 years.
(NCT00372229) [9, 10]. Ten SNPs were selected for TLR4, three SNPs for IFN-γ, six SNPs for IL10, nine SNPs for IL37 and two SNPs for TNF-α. Associations between these genetic variants and the occurrence of CMV infection, CMV disease, graft loss or death, rejection, infections, and leukopenia were analyzed.
According to the VIPP study protocol, CMV Disease was defined as either biopsy or clinically proven tissue invasive disease or CMV syndrome with viremia >400 copies/ml with at least one of the following signs: fever of ≥ 38°C, severe malaise (toxicity grading ≥ 3), leucopenia on 2 successive measurements separated by at least 24 hours defined as (1) a white blood cell (WBC) count of <3,500/μL or (2) a WBC decrease of >20% if the WBC count prior to development of viremia is <4,000/μL, atypical lymphocytosis of ≥5%, thrombocytopenia defined as (1) a platelet count of <100,000/μL or (2) a decrease of >20% if the platelet count prior to development of viremia is <115,000/μL, elevation of hepatic transaminases (alanine aminotransferase or aspartate aminotransferase to at least 2x ULN.
Population
All patients of the VIPP study were eligible for enrollment in this substudy. For inclusion, patients needed to fulfill the following criteria: written informed consent previously obtained for the VIPP study, enrollment in the VIPP main protocol, and written informed consent for the SNP analyses. There were no exclusion criteria. The protocol was approved by the ethics committee of the Hannover Medical School, Nr. 4116M (Hanover, Germany).
Selection of genetic variants
Various criteria for the selection of SNPs which we considered in the substudy, were used: allele frequencies (>1%) extracted from the Genome Aggregation Database [gnomAD v2.1, http://gnomad.broadinstitute.org/], the location in the coding regions of genes, their clinical relevance [Online Mendelian Inheritance in Man/OMIM, www.omim.org/; ClinVar Short Variants, www.ncbi.nlm.nih.gov/clinvar/; flagged by the SNP database, dbSNP, www.ncbi.nlm.nih.gov/projects/SNP/] as clinically associated), and PubMed records. S1 Table summarizes in detail the selection criteria for the 30 SNPs.
Blood sampling, DNA extraction, and genotyping
Blood samples could be obtained at any time during the VIPP study, including at scheduled blood sampling timepoints. The samples were frozen and stored at -20°C. The SNP analysis was performed in the Dr. Margarete Fischer-Bosch-Institut für Klinische Pharmakologie (Stuttgart, Germany). Genomic DNA was isolated from whole blood using the QIAamp DNA Blood BioRobot MDx Kit and the QIAamp DSP DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. DNA quality and concentration were assessed using the NanoDrop 1000 Spectrophotometer or the Qubit™ dsDNA BR Assay Kit and a Qubit Fluorometer (Thermo Fisher Scientific, Wilmington, DE, United States). Genetic variants were genotyped by TaqMan technology using predesigned SNP Genotyping assays (Thermo Fisher Scientific) and the Applied Biosystems 7900HT Fast Real-Time PCR System (Thermo Fisher Scientific) according to the manufacturers’ protocols. SNPs were evaluated with Sequence Detection Software version 2.4 (SDS 2.4).
Statistical analysis
The study was planned for 200 patients. Exploratory analysis was done for all patients and for subgroups of patients receiving valganciclovir prophylaxis or preemptive treatment. Statistical analyses were performed using R version 3.3.2 [20] or SAS 9.4. Tests were two-sided and statistical significance was defined as p<0.05, but results should be interpreted descriptively. Pearson’s Chi-squared test was used to analyze associations between valganciclovir prophylaxis or preemptive treatment and the occurrence of CMV infection, CMV disease, graft loss or death, rejection, infections (viral, bacterial, fungal and other non-CMV infections), and leukopenia. CMV infection was defined as CMV-PCR ≥400 CMV copies/mL from visit 2 (day 7) until follow-up visit 27, excluding unscheduled visits.
Associations between SNPs and the occurrence of CMV infection, CMV disease, graft loss or death, rejection, infections, and leukopenia were examined using an asymptotic independence test (Cochran-Armitage trend test). The trend tests were envisaged for n = 30 genetic variants and 6 endpoints in the overall cohort and two subgroups (prophylaxis and preemptive treatment). For two variants (IFN-γ rs2069723 and IL10 rs3024489), no nucleotide changes were observed in our cohort. Only two events occurred for CMV disease in the prophylaxis group. The effective number of tests was therefore determined as n = 476 (= 28*17), and the corresponding Bonferroni corrected significance level consequently as α* = 0.05/476 ≈ 0.00011. Odds-Ratios (OR) and 95% confidence intervals (95% CI) were calculated using a logistic regression model as an association between genotypes and endpoints. Patients with missing data were excluded. Lower OR correspond to lower odds of seeing an event in the largest genotype group.
Haplotype analyses were performed with R-library haplo.stats [21]. Haplotypes were estimated separately for each gene, using all studied variants. Associations between haplotypes and the occurrence of CMV infection, CMV disease, graft loss or death, rejection, infections, and leukopenia were investigated using logistic regression and an additive genetic model. The minimal haplotype frequency for a haplotype to be included as a separate term in the regression model was set to 5%. Results with unusually large coefficients from the logistic regression fit (occurring for dependent variables with less than 15 events or haplotype frequencies close to 5% in treatment subgroup analyses) were considered as unreliable and therefore disregarded.
Results
Patients
Blood samples were collected between March 2009 and January 2014 within the VIPP study. In total, 117 of 299 patients provided blood samples for the analysis. The SNP analysis could be performed on 116 of these samples (Fig 1).

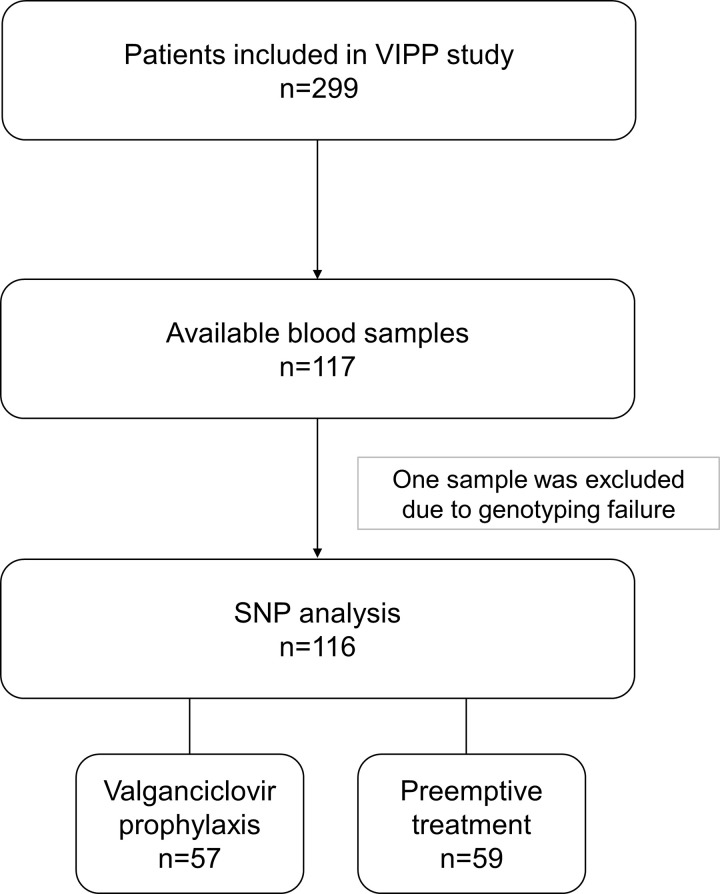
Disposition of patients and samples analyzed.
SNP, single nucleotide polymorphism; VIPP, ValgancIclovir Prophylaxis versus Preemptive therapy in cytomegalovirus-positive renal allograft recipients.
The patient characteristics are presented in Table 1. The samples were equally distributed between the prophylaxis and the preemptive treatment group (57 vs. 59 patients), with generally balanced demographic and clinical baseline characteristics. The mean age for all patients was 51.0 years, and more total male than female patients were included in the SNP analysis.

| Characteristic | All (n = 116) | Prophylaxis (n = 57) | Preemptive (n = 59) |
|---|---|---|---|
| Recipient age (years) | 51.0 ± 12.4 | 49.5 ± 13.2 | 52.5 ± 11.5 |
| Donor age (years)a | 52.7 ± 14.7 | 52.3 ± 15.6 | 53.1 ± 13.9 |
| Living donor, n (%) | 12 (10.3) | 8 (14.0) | 4 (6.8) |
| Male sex, n (%) | 76 (65.5) | 41 (71.9) | 35 (59.3) |
| Caucasian, n (%) | 110 (94.8) | 53 (93.0) | 57 (96.6) |
| Body weight (kg)b | 75.7 ± 14.6 | 77.0 ± 16.0 | 74.4 ± 13.2 |
| Sensitization of recipients against donor antigensc (%) | 1.0 ± 8.6 | 0.1 ± 0.6 | 1.9 ± 12.0 |
| Hypertensiond, n (%) | 87 (75.0) | 46 (80.7) | 41 (69.5) |
| Diabetes mellitusd, n (%) | 13 (11.2) | 7 (12.3) | 6 (10.2) |
| Stratification for ATG/ALG, n (%) | 3 (2.6) | 2 (3.5) | 1 (1.7) |
| Cold ischemia time (hours) | 12.2 ± 6.8 | 11.9 ± 7.3 | 12.5 ± 6.3 |
| HLA-A + B + DR mismatch | 2.6 ± 1.5 | 2.6 ± 1.5 | 2.6 ± 1.6 |
| Previous transplants, n (%) | |||
| 0 | 97 (83.6) | 49 (86.0) | 48 (81.4) |
| 1 | 19 (16.4) | 8 (14.0) | 11 (18.6) |
| Most frequent underlying kidney diseasese, n (%) | |||
| Glomerulonephritis | 24 (20.7) | 7 (12.3) | 17 (28.8) |
| Adult polycystic kidney disease | 14 (12.1) | 9 (15.8) | 5 (8.5) |
| IgA nephropathy | 19 (16.4) | 9 (15.8) | 10 (16.9) |
| Diabetic glomerulosclerosis/nephropathy | 6 (5.2) | 3 (5.3) | 3 (5.1) |
| Others | 33 (28.4) | 16 (28.1) | 17 (28.8) |
| CMV status donor/recipient, n (%) | |||
| D+/R+ | 65 (56.0) | 37 (64.9) | 28 (47.5) |
| D-/R+ | 51 (44.0) | 20 (35.1) | 31 (52.5) |
Values are means ± standard deviation unless otherwise indicated.
aAll: n = 115, prophylaxis: n = 56, preemptive: n = 59, due to missing value
bAll: n = 115, prophylaxis: n = 57, preemptive: n = 58, due to missing value.
cEstimated by the actual panel-reactive antibody test. All: n = 83, prophylaxis: n = 41, preemptive: n = 42.
dPrevious or concomitant disease.
eOccurrence in ≥3% of all patients.
ALG, antilymphocyte globulin; ATG, antithymocyte globulin; CMV, cytomegalovirus; D/R, donor/recipient; HLA, human leukocyte antigen; IgA, immunoglobulin A.
Occurrence of CMV infection, CMV disease, graft loss or death, rejection, infections, and leukopenia in the prophylaxis versus preemptive treatment group
The proportion of patients with CMV infection and CMV disease was higher in the preemptive treatment group than in the valganciclovir prophylaxis group (CMV infection: 35.6% vs. 14.0%, p = 0.014, 29 events in total; CMV disease: 7.8% vs. 1.7%, p = 0.031, 11 events in total). For occurrence of graft loss or death (15 events), rejection (93 events), infections (85 events), and leukopenia (32 events), no remarkable differences were observed between both groups.
Frequencies of genetic variants in all patients
Ten SNPs were analyzed for TLR4, three SNPs for IFN-γ, six SNPs for IL10, nine SNPs for IL37 and two SNPs for TNF-α. Selected SNPs, genotype and allele frequencies for all patients are provided in the S2 Table. Genotype frequencies of all SNPs are in Hardy-Weinberg equilibrium.
Association of genetic variants with CMV infection, CMV disease, graft loss or death, rejection, infections, and leukopenia in all patients
P-values were >0.05 for all analyzed SNPs and the occurrence of CMV infection, rejection and leukopenia (Tables 2 and 3). For IL37 2723186, an association with CMV disease was observed (p = 0.0499, OR 3.59, 95% CI 0.36–35.82). IL10 rs1800872 was associated with graft loss or death (p = 0.0207, OR 3.04, 95% CI 1.08–8.61). For IL10 rs3024496, an association with infections was observed (p = 0.0258, OR 1.63, 95% CI 0.725–3.64). All significant results did not hold true after Bonferroni correction.

| CMV infection | CMV disease | Infections | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Nucleotide change* | All p-value | Proph. p-value | Preem. p-value | All p-value | Proph. p-value‡ | Preem. p-value | All p-value | Proph. p-value | Preem. p-value |
| n/n† | n/n† | n/n† | n/n† | n/n† | n/n† | n/n† | n/n† | n/n† | ||
| TLR4_rs4986790 | g.13843A>G | 0.5118 | 0.3325 | 0.9157 | 0.2348 | 0.6330 | 0.2556 | 0.2838 | 0.8604 | 0.1022 |
| 29/114 | 8/55 | 21/59 | 11/114 | 2/55 | 9/59 | 84/114 | 41/55 | 43/59 | ||
| TLR4_rs4986791 | g.14143C>T | 0.2925 | 0.3325 | 0.4407 | 0.2479 | 0.6330 | 0.2729 | 0.3801 | 0.8604 | 0.1149 |
| 29/114 | 8/55 | 21/59 | 11/114 | 2/55 | 9/59 | 84/114 | 41/55 | 43/59 | ||
| TLR4_rs5030710 | g.13262T>C | 0.3102 | NA§ | 0.1901 | 0.5681 | NA§ | 0.4507 | 0.7721 | NA§ | 0.8037 |
| 29/115 | 8/56 | 21/59 | 11/115 | 2/56 | 9/59 | 85/115 | 42/56 | 43/59 | ||
| TLR4_rs7869402 | g.16573C>T | 0.2587 | 0.6799 | 0.2638 | 0.5037 | 0.8446 | 0.4459 | 0.2795 | 0.5554 | 0.1199 |
| 29/113 | 8/55 | 21/58 | 11/113 | 2/55 | 9/58 | 83/113 | 41/55 | 42/58 | ||
| TLR4_rs7873784 | g.17477G>C | 0.8143 | 0.5697 | 0.8880 | 0.7593 | 0.2799 | 0.7945 | 0.7328 | 0.7510 | 0.4875 |
| 28/112 | 8/55 | 20/57 | 11/112 | 2/55 | 9/57 | 83/112 | 42/55 | 41/57 | ||
| TLR4_rs11536871 | g.9039A>C | 0.1123 | 0.3958 | 0.1901 | 0.3721 | 0.6866 | 0.4507 | 0.0559 | 0.2419 | 0.1138 |
| 29/114 | 8/55 | 21/59 | 11/114 | 2/55 | 9/59 | 84/114 | 41/55 | 43/59 | ||
| TLR4_rs11536887 | g.16215A>G | 0.5545 | 0.6767 | NA§ | 0.7403 | 0.8431 | NA§ | 0.5527 | 0.5698 | NA§ |
| 29/112 | 8/54 | 21/58 | 11/112 | 2/54 | 9/58 | 83/112 | 41/54 | 42/58 | ||
| TLR4_rs11536889 | g.16672G>C | 0.3219 | 0.4372 | 0.8338 | 0.3320 | 0.3095 | 0.8106 | 0.0664 | 0.7817 | 0.0082¶ |
| 29/115 | 8/54 | 21/59 | 11/115 | 2/56 | 9/59 | 85/115 | 42/56 | 43/59 | ||
| TLR4_rs11536891 | g.17878T>C | 0.7480 | 0.4831 | 0.9590 | 0.8002 | 0.4043 | 0.8706 | 0.7038 | 0.2430 | 0.5593 |
| 29/113 | 8/56 | 21/57 | 11/113 | 2/56 | 9/57 | 85/113 | 42/56 | 43/57 | ||
| TLR4_rs11536892 | g.18023G>A | 0.5614 | NA§ | 0.4572 | 0.7439 | NA§ | 0.6687 | 0.0909 | NA§ | 0.0982 |
| 29/115 | 8/56 | 21/59 | 11/115 | 2/56 | 9/59 | 85/115 | 42/56 | 43/59 | ||
| IFN-γ_rs2069707 | g.4234C>G | 0.8586 | 0.4224 | 0.6716 | 0.1676 | 1.0000 | 0.2966 | 0.8769 | 1.0000 | 0.9266 |
| 29/114 | 8/55 | 21/59 | 11/114 | 2/55 | 9/59 | 85/114 | 42/55 | 43/59 | ||
| IFN-γ_rs2069723 | g.9928A>G | NA§ | NA§ | NA§ | NA§ | NA§ | NA§ | NA§ | NA§ | NA§ |
| 29/114 | 8/55 | 21/59 | 11/114 | 2/55 | 9/59 | 84/114 | 41/55 | 43/59 | ||
| IFN-γ_rs2430561 | g.6000A>T | 0.2821 | 0.1729 | 0.4540 | 0.2060 | 0.1610 | 0.3220 | 0.9635 | 0.2212 | 0.2839 |
| 29/114 | 8/57 | 21/57 | 11/114 | 2/57 | 9/57 | 83/114 | 42/57 | 41/57 | ||
| IL10_rs1800871 | g.4206T>C | 0.2093 | 0.5349 | 0.1975 | 0.1460 | 0.2333 | 0.3130 | 0.1584 | 0.0771 | 0.8477 |
| 28/114 | 7/55 | 21/59 | 11/114 | 2/55 | 9/59 | 84/114 | 41/55 | 43/59 | ||
| IL10_rs1800872 | g.4433A>C | 0.3458 | 0.6463 | 0.3710 | 0.2412 | 0.8784 | 0.2821 | 0.9756 | 0.9269 | 0.9377 |
| 29/115 | 8/56 | 21/59 | 11/115 | 2/56 | 9/59 | 85/115 | 42/56 | 43/59 | ||
| IL10_rs1800894 | g.4174G>A | 0.8808 | 0.5288 | 0.7218 | 0.9591 | 0.6896 | 0.9474 | 0.1604 | 1.0000 | 0.0981 |
| 29/115 | 8/56 | 21/59 | 11/115 | 2/56 | 9/59 | 85/115 | 42/56 | 43/59 | ||
| IL10_rs3024489 | g.4596G>T | NA§ | NA§ | NA§ | NA§ | NA§ | NA§ | NA§ | NA§ | NA§ |
| 29/113 | 8/54 | 21/59 | 11/113 | 2/54 | 9/59 | 84/113 | 41/54 | 43/59 | ||
| IL10_rs3024496 | g.8976T>C | 0.2235 | 0.5353 | 0.3041 | 0.3293 | 0.7507 | 0.1778 | 0.0258ǁ | 0.0111** | 0.4730 |
| 28/112 | 7/53 | 21/59 | 11/112 | 2/53 | 9/59 | 84/112 | 53 | 43/59 | ||
| IL10_rs3024498 | g.9311A>G | 0.7548 | 0.5481 | 0.8859 | 0.1761 | 0.3247 | 0.2349 | 0.8437 | 0.2454 | 0.1842 |
| 28/112 | 8/54 | 20/58 | 11/112 | 2/54 | 9/58 | 82/112 | 40/54 | 42/58 | ||
| IL37_rs2708943 | g.9162C>G | 0.9678 | 0.5417 | 0.4388 | 0.6971 | 0.0178†† | 0.4969 | 0.4886 | 0.2249 | 0.8664 |
| 29/114 | 8/55 | 21/59 | 11/114 | 2/55 | 9/59 | 84/114 | 41/55 | 43/59 | ||
| IL37_rs2708947 | g.10672T>C | 0.8902 | 0.7045 | 0.6878 | 0.5946 | 0.0381‡‡ | 0.6121 | 0.9563 | 0.7867 | 0.8221 |
| 29/115 | 8/56 | 21/59 | 11/115 | 2/56 | 9/59 | 85/115 | 42/56 | 43/59 | ||
| IL37_rs2723171 | g.3256G>C | 0.9829 | 0.7045 | 0.5264 | 0.6671 | 0.0381§§ | 0.5285 | 0.9150 | 0.7867 | 0.9898 |
| 29/115 | 8/56 | 21/59 | 11/115 | 2/56 | 9/59 | 85/115 | 42/56 | 43/59 | ||
| IL37_rs2723183 | g.9174A>G | 0.9243 | 0.7192 | 0.6563 | 0.6136 | 0.0404¶¶ | 0.5963 | 0.9911 | 0.7690 | 0.8511 |
| 29/113 | 8/55 | 21/58 | 11/113 | 2/55 | 9/58 | 83/113 | 41/55 | 42/58 | ||
| IL37_rs2723186 | g.9533A>G | 0.5303 | 0.1003 | 0.4572 | 0.0499ǁǁ | 0.00004*** | 0.6687 | 0.5285 | 0.9711 | 0.0982 |
| 29/113 | 8/54 | 21/59 | 11/113 | 2/54 | 9/59 | 84/113 | 41/54 | 43/59 | ||
| IL37_rs2723187 | g.9722C>T | 0.8549 | 0.7192 | 0.4388 | 0.7640 | 0.0404††† | 0.4969 | 0.7909 | 0.7690 | 0.8664 |
| 29/114 | 8/55 | 21/49 | 11/114 | 2/55 | 9/59 | 84/114 | 41/55 | 43/59 | ||
| IL37_rs2723192 | g.10834G>A | 0.9071 | 0.7192 | 0.6878 | 0.6041 | 0.0404‡‡‡ | 0.6121 | 0.9736 | 0.7690 | 0.8221 |
| 29/114 | 8/55 | 21/59 | 11/114 | 2/55 | 9/59 | 84/114 | 41/55 | 43/59 | ||
| IL37_rs3811046 | g.5831G>T | 0.5652 | 0.0651 | 0.4636 | 0.7663 | 0.0573 | 0.4726 | 0.9074 | 1.0000 | 0.8627 |
| 29/114 | 8/56 | 21/58 | 11/114 | 2/56 | 9/58 | 84/114 | 42/56 | 42/58 | ||
| IL37_rs3811047 | g.5863A>G | 0.5859 | 0.1348 | 0.5817 | 0.6982 | 0.0431§§§ | 0.4726 | 0.9680 | 0.8275 | 0.8627 |
| 29/112 | 8/54 | 21/58 | 11/112 | 2/54 | 9/58 | 82/112 | 40/54 | 42/58 | ||
| TNF-α_rs1800629 | g.4682G>A | 0.3230 | 0.8424 | 0.2298 | 0.4078 | 0.6703 | 0.4270 | 0.4372 | 0.1185 | 0.6212 |
| 29/115 | 8/56 | 21/59 | 11/115 | 2/56 | 9/59 | 85/115 | 42/56 | 43/59 | ||
| TNF-α_rs3093665 | g.7042A>C | 0.4821 | 0.3910 | 0.8793 | 0.8220 | 0.7183 | 0.7869 | 0.3160 | 0.2721 | 0.6741 |
| 29/108 | 8/51 | 21/57 | 11/108 | 2/51 | 9/57 | 78/108 | 37/51 | 41/57 | ||
NA, not applicable; preem., preemptive; proph., prophylaxis; SNP, single nucleotide polymorphism, Odds ratio (OR), 95% Confidence interval (CI). Asymptotic Cochran-Armitage trend test was used.
P-values <0.05 in bold. All p-values were >0.00011 (Bonferroni correction for multiple testing).
*Nucleotide change refers to the coding reference sequence.
†Number of patients with an event and total number of patients included for the statistical analysis.
‡Due to the low numbers of CMV disease events in the prophylaxis group (only two events) Bonferroni correction was not performed.
§NA since all patients of this study-group carrying the same genotype.
¶OR 0.194, 95% CI 0.055–0.69.
ǁOR 1.625, 95% CI 0.725–3.64.
**OR 6.73, 95% CI 1.61–28.20.
††OR 16.67, 95% CI 0.824–337.01.
‡‡OR 12.50, 95% CI 0.652–239.54.
§§OR 12.50, 95% CI 0.652–239.54.
¶¶OR 12.25, 95% CI 0.639–234.82.
ǁǁOR 3.591, 95% CI 0.36–35.823.
***OR 50.99, 95% CI 2.40-Inf.
†††OR 12.25, 95% CI 0.639–234.82.
‡‡‡OR 12.25, 95% CI 0.639–234.82.
§§§OR 12.23, 95% CI 0.728–205.42.
¶¶¶OR 12.25, 95% CI 0.639–234.82.

| Graft loss or death | Rejection | Leukopenia | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Nucleotide change* | All p-value | Proph. p-value | Preem. p-value | All p-value | Proph. p-value | Preem. p-value | All p-value | Proph. p-value | Preem. p-value |
| n/n† | n/n† | n/n† | n/n† | n/n† | n/n† | n/n† | n/n† | n/n† | ||
| TLR4_rs4986790 | g.13843A>G | 0.1571 | 0.2769 | 0.3673 | 0.5827 | 0.3479 | 0.9792 | 0.5624 | 0.2009 | 0.6018 |
| 15/114 | 9/55 | 6/59 | 92/114 | 48/55 | 44/59 | 30/114 | 18/55 | 12/59 | ||
| TLR4_rs4986791 | g.14143C>T | 0.1858 | 0.3105 | 0.3845 | 0.7259 | 0.3479 | 0.6387 | 0.3705 | 0.2427 | 0.8136 |
| 14/114 | 8/55 | 6/59 | 92/114 | 48/55 | 44/59 | 31/114 | 19/55 | 12/59 | ||
| TLR4_rs5030710 | g.13262T>C | 0.4967 | NA§ | 0.5497 | 0.3933 | NA§ | 0.2993 | 0.2863 | NA§ | 0.3690 |
| 15/115 | 9/56 | 6/59 | 93/115 | 49/56 | 44/59 | 31/115 | 19/56 | 12/59 | ||
| TLR4_rs7869402 | g.16573C>T | 0.4816 | 0.0225¶ | 0.5457 | 0.3167 | 0.6999 | 0.2935 | 0.2315 | 0.4815 | 0.3895 |
| 15/113 | 9/55 | 6/58 | 91/113 | 48/55 | 43/58 | 29/113 | 18/55 | 11/58 | ||
| TLR4_rs7873784 | g.17477G>C | 0.8773 | 0.7021 | 0.6268 | 0.6470 | 0.8288 | 0.5792 | 0.1445 | 0.2019 | 0.4641 |
| 14/112 | 8/55 | 6/57 | 91/112 | 49/55 | 42/57 | 31/112 | 19/55 | 12/57 | ||
| TLR4_rs11536871 | g.9039A>C | 0.2130 | 0.6278 | 0.1730 | 0.5211 | 0.4444 | 0.7467 | 0.4556 | 0.7323 | 0.3690 |
| 15/114 | 9/55 | 6/59 | 92/114 | 48/55 | 44/59 | 30/114 | 18/55 | 12/59 | ||
| TLR4_rs11536887 | g.16215A>G | 0.6928 | 0.6517 | NA§ | 0.6194 | 0.6969 | NA | 0.5435 | 0.4754 | NA§ |
| 15/112 | 9/54 | 6/58 | 90/112 | 47/54 | 43/58 | 30/112 | 18/54 | 12/58 | ||
| TLR4_rs11536889 | g.16672G>C | 0.5181 | 0.7263 | 0.1447 | 0.6640 | 0.8560 | 0.4153 | 0.6681 | 0.4579 | 0.1477 |
| 15/115 | 9/56 | 6/59 | 93/115 | 49/56 | 44/59 | 31/115 | 19/56 | 12/59 | ||
| TLR4_rs11536891 | g.17878T>C | 0.3996 | 0.1329 | 0.6632 | 0.7048 | 0.2297 | 0.6611 | 0.1233 | 0.1413 | 0.5120 |
| 15/113 | 9/56 | 6/57 | 91/113 | 49/56 | 42/57 | 31/113 | 19/56 | 12/57 | ||
| TLR4_rs11536892 | g.18023G>A | 0.6973 | NA§ | 0.7343 | 0.6252 | NA§ | 0.5559 | 0.5418 | NA§ | 0.6103 |
| 15/115 | 9/56 | 6/59 | 93/115 | 49/56 | 44/59 | 31/115 | 19/56 | 12/59 | ||
| IFN-γ_rs2069707 | g.4234C>G | 0.9427 | 0.1765 | 0.0862 | 0.1987 | 0.1336 | 0.0401ǁ | 0.4485 | 1.0000 | 0.1149 |
| 15/114 | 9/55 | 6/59 | 92/114 | 48/55 | 44/59 | 31/114 | 19/55 | 12/59 | ||
| IFN-γ_rs2069723 | g.9928A>G | NA§ | NA§ | NA§ | NA§ | NA§ | NA§ | NA§ | NA§ | NA§ |
| 14/114 | 8/55 | 6/59 | 92/114 | 48/55 | 44/59 | 31/114 | 19/55 | 12/59 | ||
| IFN-γ_rs2430561 | g.6000A>T | 0.6879 | 0.8932 | 0.5563 | 0.4006 | 0.1729 | 0.8859 | 0.3261 | 0.6566 | 0.0764 |
| 15/114 | 9/57 | 6/57 | 93/114 | 49/57 | 44/57 | 32/114 | 20/57 | 12/57 | ||
| IL10_rs1800871 | g.4206T>C | 0.0767 | 0.1002 | 0.4236 | 0.3383 | 0.3043 | 0.5898 | 0.3098 | 0.5911 | 0.3769 |
| 15/114 | 9/55 | 6/59 | 93/114 | 49/55 | 44/59 | 31/114 | 19/55 | 12/59 | ||
| IL10_rs1800872 | g.4433A>C | 0.0207** | 0.0037†† | 0.5812 | 0.5464 | 0.9521 | 0.3547 | 0.1966 | 0.2831 | 0.5966 |
| 15/115 | 9/56 | 6/59 | 93/115 | 49/56 | 44/59 | 31/115 | 19/56 | 12/59 | ||
| IL10_rs1800894 | g.4174G>A | 0.6273 | 0.3638 | 0.1359 | 0.5100 | 0.4328 | 0.8595 | 0.4999 | 0.6955 | 0.1707 |
| 15/115 | 9/56 | 6/59 | 93/115 | 49/56 | 44/59 | 31/115 | 19/56 | 12/59 | ||
| IL10_rs3024489 | g.4596G>T | NA§ | NA§ | NA§ | NA§ | NA§ | NA§ | NA§ | NA§ | NA§ |
| 15/113 | 9/54 | 6/59 | 91/113 | 47/54 | 44/59 | 30/113 | 18/54 | 12/59 | ||
| IL10_rs3024496 | g.8976T>C | 0.3191 | 0.2406 | 0.9144 | 0.9153 | 0.3403 | 0.4156 | 0.1309 | 0.1489 | 0.4811 |
| 15/112 | 9/53 | 6/59 | 90/112 | 46/53 | 44/59 | 28/112 | 16/53 | 12/59 | ||
| IL10_rs3024498 | g.9311A>G | 0.1424 | 0.1244 | 0.6925 | 0.6220 | 0.3501 | 0.9062 | 0.9558 | 0.5622 | 0.5521 |
| 15/112 | 9/54 | 6/58 | 91/112 | 48/54 | 43/58 | 29/112 | 17/54 | 12/58 | ||
| IL37_rs2708943 | g.9162C>G | 0.4101 | 0.6278 | 0.2350 | 0.5603 | 0.4277 | 0.5214 | 0.6515 | 0.0613 | 0.7474 |
| 15/114 | 9/55 | 6/59 | 92/114 | 48/55 | 44/59 | 30/114 | 18/55 | 12/59 | ||
| IL37_rs2708947 | g.10672T>C | 0.4316 | 0.8021 | 0.2430 | 0.4262 | 0.3758 | 0.2454 | 0.5506 | 0.1970 | 0.9767 |
| 15/115 | 9/56 | 6/59 | 93/115 | 49/56 | 44/59 | 31/115 | 19/56 | 12/59 | ||
| IL37_rs2723171 | g.3256G>C | 0.3845 | 0.8021 | 0.2160 | 0.5200 | 0.3758 | 0.3556 | 0.6766 | 0.1970 | 0.8438 |
| 15/115 | 9/56 | 6/59 | 93/115 | 49/56 | 44/59 | 31/115 | 19/56 | 12/59 | ||
| IL37_rs2723183 | g.9174A>G | 0.4180 | 0.8177 | 0.2377 | 0.3875 | 0.3705 | 0.1976 | 0.5228 | 0.1728 | 0.9528 |
| 15/113 | 9/55 | 6/58 | 93/113 | 49/55 | 44/58 | 30/113 | 18/55 | 12/58 | ||
| IL37_rs2723186 | g.9533A>G | 0.4525 | 0.4538 | 0.7343 | 0.3450 | 0.5180 | 0.5559 | 0.5586 | 0.5537 | 0.6103 |
| 15/113 | 9/54 | 6/59 | 91/113 | 47/54 | 44/59 | 30/113 | 18/54 | 12/59 | ||
| IL37_rs2723187 | g.9722C>T | 0.3672 | 0.8177 | 0.2350 | 0.2830 | 0.3705 | 0.1880 | 0.8345 | 0.2093 | 0.7474 |
| 15/114 | 9/55 | 6/59 | 93/114 | 49/55 | 44/59 | 31/114 | 19/55 | 12/59 | ||
| IL37_rs2723192 | g.10834G>A | 0.4248 | 0.8177 | 0.2430 | 0.4378 | 0.3705 | 0.2454 | 0.5078 | 0.1728 | 0.9767 |
| 15/114 | 9/55 | 6/59 | 93/114 | 49/55 | 44/59 | 30/114 | 18/55 | 12/59 | ||
| IL37_rs3811046 | g.5831G>T | 0.2698 | 0.9411 | 0.0920 | 0.6565 | 0.5657 | 0.9233 | 0.3573 | 0.7309 | 0.3060 |
| 15/114 | 9/56 | 6/58 | 92/114 | 49/56 | 43/58 | 31/114 | 19/56 | 12/58 | ||
| IL37_rs3811047 | g.5863A>G | 0.1517 | 0.6574 | 0.0920 | 0.9456 | 0.8885 | 0.9233 | 0.4533 | 0.9559 | 0.3060 |
| 15/112 | 9/54 | 6/58 | 91/112 | 48/54 | 43/58 | 29/112 | 17/54 | 12/58 | ||
| TNF-α_rs1800629 | g.4682G>A | 0.2792 | 0.1927 | 0.9536 | 0.5649 | 0.3068 | 0.9196 | 0.3672 | 0.8167 | 0.2299 |
| 15/115 | 9/56 | 6/59 | 93/115 | 49/56 | 44/59 | 31/115 | 19/56 | 12/59 | ||
| TNF-α_rs3093665 | g.7042A>C | 0.2378 | 0.4085 | 0.4220 | 0.6799 | 0.5145 | 0.4669 | 0.9505 | 0.2274 | 0.2766 |
| 15/108 | 9/51 | 6/57 | 87/108 | 45/51 | 42/57 | 28/108 | 16/51 | 12/57 | ||
*Nucleotide change refers to the coding reference sequence.
†Number of patients with an event and total number of patients included for the statistical analysis.
‡Due to the low numbers of CMV disease events in the prophylaxis group (only two events) Bonferroni correction was not performed.
§NA since all patients of this study-group carrying the same genotype.
¶OR Inf, 95% CI 0-Inf.
ǁOR 0.201, 95% CI 0.039–1.035.
**OR 3.043, 95% CI 1.076–8.61.
††OR 5.342, 95% CI 1.331–21.45.
Association of genetic variants with the occurrence of events in the prophylaxis versus preemptive treatment group
Associations of SNPs with the occurrence of CMV infection, CMV disease, graft loss or death, rejection, infections, and leukopenia were also investigated in the prophylaxis and preemptive treatment group. In the prophylaxis group, TLR4 rs7869402 and IL10 rs1800872 were associated with the occurrence of graft loss or death (p = 0.0255, OR Inf., 95% CI 0-Inf. and p = 0.0037, OR 5.34, 95% CI 1.33–21.45), and IL10_rs3024496 was associated with infections (p = 0.0111, OR 6.73, 95% CI 1.61–28.2) (Tables 2 and 3). Additionally, in the prophylaxis group, associations with CMV disease were observed for nearly all analyzed IL37 SNPs. However, only two CMV disease events were observed in this subgroup. In the preemptive group, we found associations with infections for TLR4 rs11536889 (p = 0.0082, OR 0.194, 95% CI 0.055–0.693) and with rejection for IFN-γ rs2069707 (p = 0.201, 95% OR 0.039–1.035). Again, all significant results did not hold true after Bonferroni correction. For all other investigated SNPs and events, p-values were >0.05.
Association of haplotypes with the occurrence of events in all patients and the prophylaxis versus preemptive treatment group
In all patients, nominal significant differences between the most frequent haplotype and any of the other non-rare haplotypes (haplotype frequency ≥5%) identified were revealed for endpoint rejection and IFN-γ (p = 0.023) as well as infections and IL10 (p = 0.017). Moreover, the association between infections and TLR4 showed a borderline effect in all patients (p = 0.051) and was nominally significant in the preemptive treatment group (p = 0.038). However, in all three cases, the differing haplotype was determined by just one variant, which we already identified in our previous per-variant analysis (Tables 2 and 3, IFN-γ rs2069707, IL10 rs3024496, and TLR4 rs11536889, respectively). In addition, in the preemptive treatment group, a nominally significant association was identified between leukopenia and IFN-γ (p = 0.049). Here, the differing haplotype was solely determined by IFN-γ rs2430561. In the corresponding per-variant analysis, the association between IFN-γ rs2430561 and leukopenia showed a borderline effect (p = 0.0764, Table 3). Our haplotype analyses did not reveal any nominal significant associations for CMV infection, CMV disease as well as graft loss or death. All haplotype analyses results are not statistically significant after adjustment for multiple testing.
Discussion
This is the first association study of common genetic variants in patients after renal transplantation in a prospective randomized clinical trial.
The characteristics of the 116 patients, from whom blood samples were analyzed, were comparable to the overall population of the VIPP study, which comprised 299 patients [10]. The proportion of patients with CMV infection and CMV disease was higher in the preemptive treatment group than in the valganciclovir prophylaxis group (35.6% vs. 14.0% and 7.8% vs 1.7%). The percentages of CMV infection events in this analysis were comparable to the results seen in the overall population of the VIPP study after 84 months (incidence of CMV infection: 39.7% in the preemptive group vs. 11.5% in the prophylaxis group; p<0.0001) [10]. The percentages of CMV disease events were lower in this substudy than in the overall population (incidence of CMV disease in VIPP study after 84 months: 15.9% in the preemptive group vs. 4.7% in the prophylaxis group; p<0.01). Due to the low numbers of CMV disease events, analyses of CMV disease in the prophylaxis group (two events) were excluded from the Bonferroni correction.
The selection of the SNPs which were tested in this substudy was based on various criteria. One criterium considered the allele frequencies of variants >1%. Population frequency data were obtained from the Genome Aggregation Database (gnomAD v2.1) which comprises data from more than 140,000 individuals including their ethnic background. Of note total allele frequency data of the selected variants corresponds very well with allele frequency data of European (non-Finnish) individuals (> 60,000).
For CMV infection, no association with the investigated SNPs in the genes TLR4, IL10, IFN-γ, IL37 and TNF-α was observed in this largely Caucasian population. Regarding IFN-γ, our results are in line with data by Aguado et al. [22], demonstrating no association between the +874 T/A (rs2430561) variant and CMV replication in R+ transplant patients. However, other studies demonstrated an association between SNPs in these genes and infection or CMV disease [15–18]. This can be partially explained by differences in the study populations, conditions and/or specific locations of investigated SNPs. Vu et al. observed an association of the IFN-γ polymorphism +874 A/T (rs2430561) with an increased risk of CMV infection in transplant recipients in a Hispanic population [18]. Mitsani et al. [23] also found a correlation of the IFN-γ +874 T/T genotype and CMV disease after lung transplantation, which was most striking among R+ patients. In another study, associations were found between other IL10 SNPs (rs1800896, rs3024492 and rs1878672) and the development of CMV disease [16].
For the further endpoints of interest, associations were observed in all patients for IL37 rs2723186 with CMV disease, for IL10 rs1800872 with the occurrence of graft loss or death, and for IL10 rs3024496 with the occurrence of infections. These results were confirmed in the prophylaxis group, however did not hold true after Bonferroni correction.
IL-37 is capable of reducing the activity of both innate and specific immune responses [24, 25]. IL37 rs2723186 is located in the intronic region of the gene. Previously, it was shown that the rs2723186 allele was significantly associated with a decreased risk of Graves’disease (an autoimmune thyroid disease) in female patients [26]. In addition, the AG genotype of rs2723186, compared to the GG genotype exhibited a decreased risk of hepatitis B virus infection [27]. In our analysis, ten patients with the GG genotype and one patient with the AA genotype developed CMV disease.
IL10 rs1800872 (-592C>A) is located in the promotor region of the gene and part of the A-T-A (-1082/-819/-592) haplotype, whose impact on the production of IL10 is discussed controversially [28–30]. In our analysis, patients with the GG genotype experienced fewer events of graft loss or death. IL10 rs3024496 is located in the 3’ untranslated region of the gene and it was previously shown to influence CD4+ T cell responses to antiretroviral therapy in patients with human immunodeficiency virus 1 infection [31].
The herein described study has the following limitations: The number of analyzed blood samples was smaller than the planned number of 200 patients, and no sample size calculation was performed in advance. The study population cannot be extended by additional patient samples since the VIPP study was a prospective randomised clinical trial. Furthermore, the study was not designed for the large number of statistical tests. Additionally, the number of events was low, particularly for CMV disease, and graft loss or death. Conversely, intensive immunosuppression may have hidden possible associations.
The results of this association analysis may guide future research efforts. Based on the complexity of the immune response, research on the combined use of biomarkers and gene profiles represents a better approach than the focus on single markers. Immunity against CMV is usually defined by serology. However, a cellular immune response with potent CMV-specific T cells seems to help prevent CMV disease in CMV R+ recipients. CMV-specific T-cell responses are not measured routinely in clinics, but their potential diagnostic value has been demonstrated [32–34]. T-cell response monitoring may be useful to guide prophylaxis and preemptive treatment. Thus, additional data regarding the use of biomarkers for risk stratification of renal transplant recipients should be explored [1].
Acknowledgements
The authors thank all patients who participated in the study. They further thank the study nurses Jasmin Hennig and Anne Heinrich, as well as Monika Elbl and Petra Metzner for excellent technical assistance. Stefan Winter (Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart) contributed to the statistical analysis. Medical writing assistance was provided by Physicians World Europe GmbH, Mannheim, Germany.
The complete membership of the author group for the SNP subanalysis within the VIPP study was as follows (principal investigators in brackets): Universitätsklinikum Aachen, Medizinische Klinik II, Nephrologie und klinische Immunologie (Anja Susanne Mühlfeld); Charité-Universitätsmedizin Berlin, Medizinische Klinik mit Schwerpunkt Nephrologie und Intensivmedizin, Campus Virchow (Petra Reinke); Universitätsklinikum Düsseldorf, Klinik für Nephrologie (Johannes Stegbauer); Universitätsklinikum Erlangen, Medizinische Klinik IV, Nephrologie (Katharina Heller); Universitätsklinikum Essen, Zentrum für Innere Medizin, Klinik für Nieren- und Hochdruckkrankheiten (Oliver Witzke); Klinikum der Johann-Wolfgang-Goethe Universität, Med. Klinik III/Nephrologie (Ingeborg A. Hauser); Medizinische Hochschule Hannover (Jürgen Klempnauer); Nephrologisches Zentrum Niedersachsen, Innere Medizin/Nephrologie (Volker Kliem); Universitätsklinikum Leipzig, Zentrum für Chirurgie, Klinik f. Viszeral-, Thorax- u. Gefäßchirurgie (Michael Bartels); Universitätsklinikum Schleswig-Holstein, Campus Lübeck/Medizinische Klinik I, Transplantationszentrum (Martin Nitschke); Universitätsklinikum Münster, Klinik und Poliklinik für Allgemeine Chirurgie (Heiner Wolters); Universitätsklinikum Köln, Medizinische Klinik IV (Tüely Kisner); Klinikum Bremen-Mitte GmbH, Innere Medizin III (Frans A. Zantvoort); Klinikum der Universität Regensburg, Klinik und Poliklinik für Innere Medizin II Nephrologie, Zentrum für klinische Studien (Bernhard Banas); Landeskrankenhaus-Universitätskliniken Innsbruck, Universitätsklinik für Chirurgie (Johann Pratschke); Universitätsklinikum Hamburg-Eppendorf, Zentrum für Innere Medizin, 3. Medizinische Klinik und Poliklinik (Rolf Stahl)
Abbreviations
| D/R | donor/recipient |
| CMV | cytomegalovirus |
| IFN | interferon |
| IL | interleukin |
| SNP | single nucleotide polymorphism |
| TLR | toll like receptor |
| TNF | tumor necrosis factor |
References
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
 No association of genetic variants in TLR4, TNF-α, IL10, IFN-γ, and IL37 in cytomegalovirus-positive renal allograft recipients with active CMV infection—Subanalysis of the prospective randomised VIPP study
No association of genetic variants in TLR4, TNF-α, IL10, IFN-γ, and IL37 in cytomegalovirus-positive renal allograft recipients with active CMV infection—Subanalysis of the prospective randomised VIPP study
