
The authors wish it to be known that, in their opinion, the first two authors should be regarded as Joint First Authors.
- Altmetric
Sulfuration of uridine 34 in the anticodon of tRNAs is conserved in the three domains of life, guaranteeing fidelity of protein translation. In eubacteria, it is catalyzed by MnmA-type enzymes, which were previously concluded not to depend on an iron–sulfur [Fe–S] cluster. However, we report here spectroscopic and iron/sulfur analysis, as well as in vitro catalytic assays and site-directed mutagenesis studies unambiguously showing that MnmA from Escherichia coli can bind a [4Fe–4S] cluster, which is essential for sulfuration of U34-tRNA. We propose that the cluster serves to bind and activate hydrosulfide for nucleophilic attack on the adenylated nucleoside. Intriguingly, we found that E. coli cells retain s2U34 biosynthesis in the ΔiscUA ΔsufABCDSE strain, lacking functional ISC and SUF [Fe–S] cluster assembly machineries, thus suggesting an original and yet undescribed way of maturation of MnmA. Moreover, we report genetic analysis showing the importance of MnmA for sustaining oxidative stress.
INTRODUCTION
Transfer RNAs (tRNAs) are essential components of the cellular translation machinery in the three domains of life. To achieve their function, these molecules feature a great variety of well-conserved post-transcriptional chemical modifications (1–3). In particular, sulfuration of uridine 34 (Figure 1A) at the wobble position of the anticodon in glutamate-, glutamine- and lysine-tRNA is conserved in bacteria, archaea and eukaryotes and guarantees fidelity of protein translation (4,5).

![s2U34-tRNA sulfuration and structure of Escherichia coli MnmA. (A) ATP-dependent reaction catalyzed by U34-tRNA thiolases through the formation of an adenylated intermediate. Ad stands for ‘adenylate’. (B) Structure of the active site of tRNA-bound MnmA (PDB code: 2DET) with the catalytic residues (green) and the U34 target (magenta) in stick representation. Cys102 and Cys199 form a disulfide bond. The Asp99 O2 atom is located 4.5 Å away from the Cys199 SG atom. (C) New mechanism proposed for [4Fe–4S]-dependent U34 sulfuration by E. coli MnmA.](/dataresources/secured/content-1766036516994-b225bb7d-35c8-4432-8636-fe22d62363ef/assets/gkab138fig1.jpg)
s2U34-tRNA sulfuration and structure of Escherichia coli MnmA. (A) ATP-dependent reaction catalyzed by U34-tRNA thiolases through the formation of an adenylated intermediate. Ad stands for ‘adenylate’. (B) Structure of the active site of tRNA-bound MnmA (PDB code: 2DET) with the catalytic residues (green) and the U34 target (magenta) in stick representation. Cys102 and Cys199 form a disulfide bond. The Asp99 O2 atom is located 4.5 Å away from the Cys199 SG atom. (C) New mechanism proposed for [4Fe–4S]-dependent U34 sulfuration by E. coli MnmA.
U34-tRNA sulfuration is catalyzed by MnmA-type enzymes in bacteria (6–8) and mitochondria (9), and by Ncs6-type enzymes in archaea and the eukaryotic cytosol (10–12). MnmA belongs to the minimal set of proteins that can sustain translation of the genetic code in Mollicutes (13), a lineage of the bacterial Firmicutes that has evolved by massive genome reduction. Mutations in the mnmA gene resulted in severe growth reduction in Escherichia coli (6,14) and Salmonella enterica serovar Typhimurium (15,16) and in nonviability in Bacillus subtilis (17). Altogether, these observations support both the key role of bacterial MnmA and its probable ancient origin.
MnmA from E. coli belongs to a sulfur relay pathway involving multiple proteins (18): IscS, a cysteine desulfurase that abstracts the sulfur atom from l-cysteine and transfers it via trans-persulfuration reactions onto a series of various sulfur carriers; TusA, a TusBCD complex; and TusE, which eventually interacts with the MnmA–tRNA complex (7). It was proposed that sulfur would be transferred from one conserved cysteine residue of TusE to one catalytic cysteine of MnmA although experimental evidence for the persulfide form of MnmA is lacking (6–8). MnmA, as the last element of the sulfur relay, introduces the sulfur atom into the tRNA substrate. Structural analysis of E. coli MnmA bound to tRNA in various states (initial tRNA binding, pre-reaction and adenylated intermediate) (Figure 1B) (8) strongly supported a key role for three neighboring residues in U34-tRNA sulfuration: Asp99, Cys102 and Cys199 (making up the DXXC + C motif in the catalytic domain; Supplementary Figure S1). In the currently accepted mechanism, Cys199 is proposed to carry the active persulfide, Cys102, to assist sulfur transfer via formation of a disulfide bond with Cys199, whereas Asp99 could act as an acid/base catalyst to facilitate proton transfer (Supplementary Figure S2) (8).
IscS acts as a pleiotropic sulfur donor in the cell. In particular, it forms, together with a series of protein partners IscU, IscA, HscAB and Fdx, the well-conserved ISC [Fe–S] cluster biogenesis machinery, which builds and delivers [Fe–S] clusters to most, if not all, [Fe–S] cellular proteins (19–21). Early work established the role of IscS in s2U34-tRNA modification in E. coli and S. enterica (6,16,22,23) but ruled out a role for the other ISC proteins in S. enterica (24), leading to the current belief that MnmA is not an [Fe–S] protein. This model was supported by the absence of such a cluster within the crystal structures of as-purified E. coli MnmA (8) and by the observation of a sulfuration activity by aerobically purified enzymes preparations under in vitro conditions, albeit very weak (6–8,25).
In contrast to the view presented above, we report here biochemical and spectroscopic results unambiguously showing that E. coli MnmA can assemble a [4Fe–4S] cluster, most likely chelated by Asp99, Cys102 and Cys199, and that this cluster is absolutely needed for in vitro activity. Intriguingly, we found that E. coli cells retain s2U34 biosynthesis in the ΔiscUA ΔsufABCDSE strain, lacking functional ISC and SUF [Fe–S] cluster assembly machineries, thus suggesting an original and yet undescribed way of maturation of MnmA.
MATERIALS AND METHODS
Media and growth conditions
Media used were Luria–Bertani (LB) rich medium or M9 minimal medium supplemented with glycerol (0.4%) and MgSO4 (1 mM). l-Arabinose (Ara) (0.2%), casamino acids (0.05%), thiamine (50 μg ml−1) and mevalonate ( MV), 1 mM were added when required. Solid media contained 1.5% agar. Antibiotics were used at the following concentrations: chloramphenicol (Cam), 30 μg ml−1; kanamycin (Kan), 30 μg ml−1; spectinomycin (Spc), 100 μg ml−1; and ampicillin (Amp), 100 μg ml−1. Note that Kan was used at 80 μg ml−1 for transduction experiments involving the ΔiscS mutants as recipients. For growth at pH 7.0 or 4.5, cultures grown in LB rich medium were diluted 1:100 into 200 μl of LB or LB–HCl (pH 4.5) in 96-well plates. Growth was monitored using a TECAN spectrophotometer by recording OD600 every 10 min over 16 h at 37°C.
Strains and plasmids
All strains used in this study are E. coli K-12 MG1655 derivatives. The mnmA mutant, FBE584, was obtained by in-frame deletion of mnmA and replaced with a kanamycin cassette (26). FBE597 was obtained from the E. coli KEIO Knockout Collection (27) by P1 transducing the mnmE::kan allele into MG1655 background. The mnmE::cam mutant was constructed by deleting and replacing the mnmE gene with a chloramphenicol resistance encoding cassette giving rise to the FBE707 strain (26). The conditional RExBADmnmA mutant, FBE583, was obtained by amplifying a fragment carrying the aadA7I (spectinomycin resistance gene) and araC genes by polymerase chain reaction (PCR) from TG1 spec RExBAD (FBE319) using the ML39 primers (28). The linear fragment was then inserted upstream of the mnmA gene in E. coli MG1655 carrying the λ red expression plasmid pKD46 (26). All mutants were P1-transduced into different E. coli K-12 backgrounds (Supplementary Table S1). The CF8310 strain, a MG1655 strain derivative carrying the T7 RNA polymerase encoding gene on a lambda prophage (DE3), was used as recipient for P1 transduction of ΔmnmA::kan, giving rise to the FBE598 strain. Strain FBE605 (ΔiscUA ΔsufABCDSE MVA+, kanR piscR::lacZ Δlac) is a MG1655 strain derivative, which can synthesize isopentenyl diphosphate from the introduced eukaryotic MVA-dependent pathway (29). Strain FBE605 was derived from BR404 (30) as follows. First, the CamR cassette was removed from the suf locus, yielding to BR412 that was used as a recipient for P1 transduction of the ΔiscUA::cat mutation, to generate FBE033. The Cat cassette was removed from the iscUA locus by using the pCP20 plasmid. PCR analysis was used to check the absence of the iscUA genes and the suf operon. DNA sequence analysis of the isc region was carried out and MVA-dependent viability was controlled. For all constructions, transductants were verified by PCR, using primers hybridizing upstream and downstream of the deleted gene. When necessary, antibiotic resistance cassettes were eliminated using plasmid pCP20 as described (31). Strains used in this study are listed in Supplementary Table S1.
Plasmid construction
To construct the pBADmnmA+ plasmid, the mnmA gene was first amplified from E. coli MG1655 chromosomal DNA using the ML40 primers, digested by EcoRI and XhoI and cloned into the EcoRI and SalI sites of pBAD24. The mnmA gene was also subcloned into the pET28a vector by PCR amplification from pBADmnmA+ to introduce the BamHI restriction site and the TEV nucleotide sequence in 5′ and the HindIII restriction site in 3′ using the ML52 primers. The PCR fragment was purified, digested by BamHI and HindIII and cloned into the BamHI and HindIII sites of pET28a. mnmA variants containing mutations were obtained by site-directed mutagenesis using the ML49–50-51 primers (NEB, E0554S). The sequence of the expression plasmids was confirmed by sequencing. Primers and plasmids used in this study are listed in Supplementary Tables S2 and S3, respectively.
Synthetic lethality test
Cells were cultivated in M9 minimal medium supplemented with 0.05% casamino acids and 0.4% glycerol overnight at 37°C. Saturated cultures were washed the next day twice with PBS and then diluted 1:50 into LB rich medium with or without 0.2% l-arabinose. Cultures were incubated at 37°C for 8 h. The optical density at 600 nm was measured over time.
Hydrogen peroxide sensitivity test
Overnight cultures were diluted using serial dilution in sterile PBS and 5 μl was directly spotted onto LB plates containing 1 mM H2O2. The plates were incubated overnight at 37°C before growth was recorded.
Overexpression of E. coli MnmA wild-type and the D99A-C102A and D99A-C102A-C199A variants
The mnmA gene was sub-cloned into the pET15b plasmid using a ligation independent cloning strategy by Eurofins to produce a 6His-MnmA protein construct whose 6His tag can be cleaved by the H3C protease. The mnmAD99A-C102A and mnmAD99A-C102A-C199A genes were sub-cloned into the pET28a plasmid. The plasmids were transformed into E. coli BL21(DE3) Star Codon Plus competent cells. One colony was used to inoculate 100–200 ml of LB medium supplemented with ampicillin or kanamycin (50 μg ml−1) for MnmA wild-type and variants, respectively. 60 ml (20 ml) of this culture grown overnight at 37°C was used to inoculate 6 L (2 L) of LB medium supplemented with the same antibiotic. Cultures were grown at 37°C to an OD600 0.6–0.8, and overexpression was induced with 0.5 mM Isopropyl β-D-1-thiogalactopyranoside. After 4 h incubation at 37°C, cells were collected by centrifugation and stored at −20°C.
Purification of E. coli wild-type MnmA and variants
Cells were resuspended in 50 mM Tris-HCl (pH 7.5), 500 mM NaCl, 10% glycerol, containing RNase A (2 μg ml−1), benzonase (1.6 U ml−1, Sigma Aldrich), lysozyme (0.3 mg ml−1), 1 mM PMSF PhenylMethylSulfonyl Fluoride, 1 mM β-mercaptoethanol and disrupted by sonication. Cells debris were removed by centrifugation at 25 000 rpm for 1 h at 4°C. The supernatant was then loaded on an immobilized metal affinity Ni-NTA column (HisTrap 5 ml, GE Healthcare) equilibrated in 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 1 mM PMSF and eluted with a linear gradient of 0–1 M imidazole. The proteins were collected, dialyzed twice against 1 L of 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 1 mM β-mercaptoethanol in the presence of the PreScission Protease (25 μg per mg wild-type MnmA) or TEV protease (43 μg/mg MnmA variants). Wild-type MnmA was further purified at 1 ml min−1 onto a gel filtration column (Hiload 26/60 Superdex 200, GE Healthcare) equilibrated with 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, using an ÅKTA system. The as-purified proteins were concentrated to 24 mg ml−1 (wild-type) with an Amicon Ultra filter device (30 kDa cutoff, Millipore) or 4–5 mg.ml−1 (variant) with an Amicon Ultra filter device (10 kDa cutoff, Millipore), frozen in liquid nitrogen and stored at −80°C.
The GST–3C-protease (PreScission, a gift from S. Mouilleron) was expressed using pGEX-2T recombinant plasmids. After induction at 25°C with 0.1 mM IPTG for 20 h, the protein was purified using glutathione–Sepharose chromatography.
[Fe–S] cluster reconstitution and purification of holo-MnmA wild-type and variants
The reconstitution of the [4Fe–4S] cluster and purification of holo-MnmA were performed in a glove box containing <0.5 ppm O2. After incubation of 100 μM as-purified MnmA with 10 mM dithiothreitol for 10 min, a 5.5-fold molar excess of ferrous ammonium sulfate and l-cysteine, as well as 2 μM E. coli cysteine desulfurase CsdA, were added, and incubation was extended overnight at room temperature. After centrifugation for 20 min at 12 300 g, holo-MnmA was loaded onto a Superdex 200 increase 10/300 GL column (GE Sciences) equilibrated in 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 5 mM dithiothreitol (DTT). The peak containing the holo-MnmA was then concentrated to 10–19 mg/ml on a Vivaspin concentrator (10 kDa cutoff). The same protocol was used for analyzing the capacity of the variants to host [Fe–S] clusters.
Quantification methods
The Bradford method was used to quantify the protein (32). The Fish and Beinert methods were routinely used after cluster reconstitution to quantify iron and sulfide, respectively (33,34).
SEC-MALS
Size exclusion chromatography coupled with multi-angle light scattering (SEC-MALS) experiments were performed using an HPLC-MALS system (Shimadzu) equipped with light scattering detector (mini DAWN TREOS, Wyatt Technology), refractive index detector (Optilab T-rEX, Wyatt Technology) and UV detector (SPD-20A, Shimadzu). Holo-MnmA (100 μl at 2 mg ml−1) was injected on a Superdex 200 10/300 GL increase column (GE Healthcare) equilibrated in 50 mM Tris-HCl (pH 7.5), 200 mM NaCl buffer, in the presence or absence of DTT at a flow rate of 0.5 ml.min−1. Molar masses of proteins were calculated using the ASTRA 6.1 software (Wyatt Technology) using a refractive index increment (dn/dc) value of 0.183 ml g−1.
CD analysis
Circular dichroism (CD) spectra were recorded on a Chirascan-plus CD Spectrometer (Applied Photophysics). The far ultraviolet spectra (195−260 nm) were measured at 20°C in quartz cells of 0.5 mm optical path length. The final concentration of MnmA proteins was 2 μM in 25 mM Tris-HCl (pH 7.5), 100 mM NaF. Spectra were acquired at a resolution of 1 nm, with time per points set at 1 s and a bandwidth of 1 nm. All spectra were corrected from the contribution of the buffer and are an average of ten accumulations.
Preparation of bulk tRNA and in vitro transcribed Ec-tRNAGlu
Bulk tRNA from various E. coli strains was purified as reported (35). The E. coli tRNAGluUUC (Ec-tRNAGlu) was synthesized in vitro by T7-RNA polymerase transcription as described in (36). Before use, the tRNA transcript was refolded by heating at 65°C for 15 min then 45°C for 15 min, and finally cooling at 4°C for 30 min.
In vitro enzyme assay
Holo-MnmA (1 or 10 μM) and Ec-tRNAGluUUC (15 μM) were incubated at 37°C in 100 μl of 50 mM Tris-HCl (pH 7.5), 200 mM NaCl in the presence of 0.25 mM ATP, 2.5 mM MgCl2 and 1 mM Na2S under anaerobic conditions for 60 min. The reaction was stopped by adding 1 μl of 3 M formic acid, followed by the addition of 3 μl of 1 M Tris (pH 8.5) to adjust the pH of the solution to 6.5.
tRNA digestion and analysis of modified nucleosides
tRNA (15 μM) was digested overnight in 100 μl of 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 0.1 mM ZnSO4 at 37°C by nuclease P1 (two units, Sigma) followed by the addition of alkaline phosphatase for 2 h at 37°C (1 unit, Sigma). HPLC-tandem mass spectrometry analyses were performed with an ExionLC chromatographic system coupled with a QTRAP6500+ mass spectrometer (AB SCIEX INSTRUMENTS) equipped with a Turbo Spray IonDrive source used in the positive ionization mode. HPLC separation was carried out with a 2 × 150 mm, 2.7 μm Poroshell HPH-C18 column (Agilent, France) at 0.4 ml min−1 and at 35°C. A linear gradient of 0–15% acetonitrile in 0.1% formic acid over 7 min was used as the mobile phase. Mass spectrometry detection was carried out in the multiple reactions monitoring mode to obtain high sensitivity and specificity. The transitions used to quantify s2U (or s4U that has a similar fragmentation pattern) were 261→129 and 261→112, corresponding to the loss of ribose. Under the HPLC conditions used, s2U is eluted faster (4.3 min) than s4U (4.6 min). Quantification was performed by external calibration. The 5-methylaminomethyl-2-thiouridine (mnm5s2U) and 5-carboxymethylaminomethyl-2-thiouridine (cmnm5s2U) standards were synthesized via nucleophilic substitution of 5-pivaloyloxymethyl-2-thiouridine with methylamine or tetrabutylammonium salt of glycine, respectively, according to the previously described procedures (37). The structure and homogeneity of both nucleosides were confirmed by 1H and 13C NMR, mass spectrometry and reversed-phase HPLC analysis.
Characterization of the [4Fe–4S] cluster by UV-Visible and EPR spectroscopies
UV-visible absorption spectra were recorded in quartz cuvettes (1 cm optic path) under anaerobic conditions in a glove box on a Cary 100 UV-visible spectrophotometer equipped with optical fibers.
The EPR continuous wave measurements were performed on a spectrometer Bruker ELEXSYS-E500 operating at 9.38 GHZ, equipped with SHQE cavity cooled by a helium flow cryostat ESR 900 Oxford Instruments. The EPR spectra of frozen solution of 220 μM E. coli MmnA with reconstituted cluster, without treatment and reduced with 5.5 mM dithionite in 50 mM Tris-HCl (pH 7.5), 200 mM NaCl was recorded at 10, 15 and 20 K, under nonsaturating conditions and using the following parameters: a microwave power of 2–10 mW, a modulation amplitude of 0.4 mT, a modulation frequency of 100 kHz and an accumulation of 4 scans. For the quantification of unpaired spins, a Cu-EDTA standard sample (200 μM) was used. The simulation of the EPR spectrum was performed with the Easyspin software (http://www.easyspin.org/).
RESULTS
Escherichia coli MnmA binds a [4Fe–4S] cluster
Escherichia coli MnmA with an N-terminal histidine tag was purified under aerobic conditions using Ni-affinity chromatography. The faint brownish color and the weak absorbance at 410 nm suggested the presence of an [Fe–S] cluster (Supplementary Figure S3A). The tag was then removed using the H3C protease and the protein was further purified by size-exclusion chromatography, which led to a mixture of monomeric and dimeric species, as shown by SEC-MALS analysis (Supplementary Figure S3B and S3C). Because the monomer/dimer ratio increases with DTT concentration (Supplementary Figure S3B), the dimer is formed by disulfide bonds between two monomers. As-purified MnmA retained a faint brownish color and its UV-visible spectrum exhibited a band at around 410 nm (Supplementary Figure S3D). These unexpected features, which disappeared upon further aerobic purification, strongly suggested the presence of an [Fe–S] cluster within the as-purified protein, which would be destroyed by air during purification, and prompted us to evaluate the potential of MnmA to assemble a well-defined cluster. For that purpose, in vitro reconstitution of the cluster was carried out by anaerobically treating the protein with ferrous iron and l-cysteine in the presence of a cysteine desulfurase, as a source of sulfur. The protein was then purified by size-exclusion chromatography, also under anaerobic conditions, leading to a homogeneous brownish protein, indicative of the presence of an [Fe–S] cluster in the protein that was subsequently called holo-MnmA. SEC-MALS analysis (Supplementary Figure S3E) indicated that holo-MnmA is almost exclusively in the monomeric form in solution, the molar mass of the protein being 40.0 ± 2.6 kDa, close to the theoretical molar mass of the monomer (41 kDa).
Quantification of the protein-bound iron and sulfur content gave 3.02 ± 0.2 Fe and 3.9 ± 0.3 S per MnmA monomer, consistent with the presence of one [4Fe–4S] cluster per monomer. Accordingly, the UV-visible spectrum of purified holo-MnmA displays a broad absorption band at around 410 nm that is specifically characteristic for the presence of a [4Fe–4S] cluster (Figure 2A). Anaerobic addition of dithionite to holo-MnmA led to a rapid decrease in the intensity of the 410 nm absorption band, suggesting a fast reduction of the cluster (Figure 2A). The frozen solutions of holo-MnmA, before and after reduction, were analyzed by continuous wave EPR spectroscopy at different temperatures (Figure 2B and Supplementary Figure S3F). No EPR signal was observed for holo-MnmA solution, suggesting an S = 0 [4Fe–4S]2+ state, and excluding S = 1/2 paramagnetic clusters such as [4Fe–4S]+ or [3Fe-4S]+. Upon reduction, the EPR spectrum of the protein recorded at 15 K exhibited a signal with a rhombic g tensor (gx = 1.88, gy = 1.92, gz = 2.045) characteristic of the S = 1/2 [4Fe–4S]+ state (Figure 2B). Temperature variations allowed to discriminate between a [2Fe-2S]+ cluster and a [4Fe–4S]+ cluster because of their different spin relaxation behaviors (38). At 40 K, the EPR lines of the reduced holo-MnmA sample broadened beyond recognition (Supplementary Figure S3F), in agreement with the presence of a [4Fe–4S]+ cluster, which usually cannot be detected above 35 K, in contrast to [2Fe–2S]+ clusters. Quantification with Cu-EDTA standard solution indicated the presence of around 0.5 spin per holo-MnmA monomer.

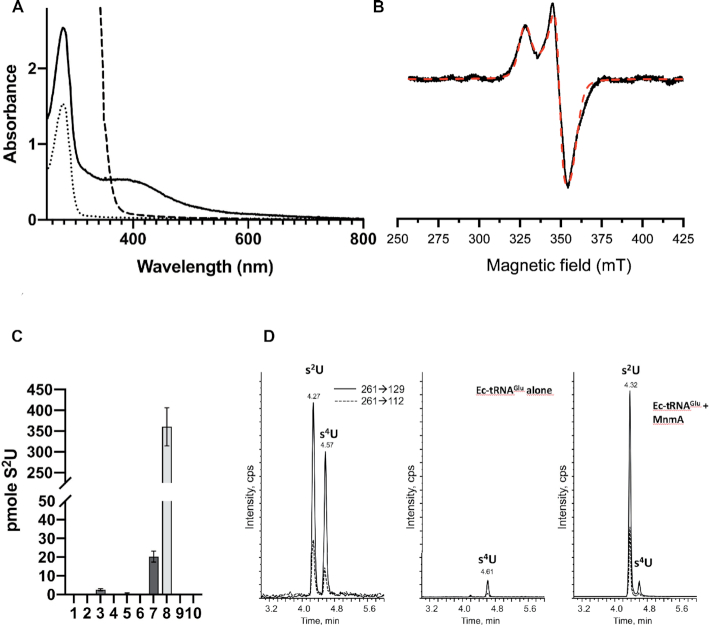
Spectroscopic and enzymatic characterizations of E. coli MnmA. (A) UV-visible spectra of 40 μM apo-MnmA (dotted line), 40 μM holo-MnmA (thick line) and reduced holo-MnmA after 20 min of incubation with 1 mM dithionite (dashed line). The spectra were recorded with 40 μM protein in 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 5 mM DTT in 1 cm (apo-MnmA and reduced holo-MnmA) or 1 mm (holo-MnmA) pathlength cuvettes and normalized. (B) X-band EPR spectrum (10 mW microwave power; modulation amplitude of 0.4 mT) of 220 μM holo-MnmA reduced with 5.5 mM dithionite at 15 K. The experimental (black solid line) and simulated (dashed line) spectra are superimposed. The cluster was simulated with the following values of the g-tensor: gx = 1.880, gy = 1.920, gz = 2.045 and Gaussian distribution deviations σ(gx) = 0.04, σ(gy) = 0.02, σ(gz) = 0.04. (C) In vitro tRNA sulfuration activity tests of MnmA under anaerobic conditions. After tRNA digestion, s2U was separated by HPLC-MS/MS and quantified using a synthetic s2U standard. The data shown are mean values based on three different experiments, with the standard error of the mean indicated as a bar. In vitro transcribed Ec-tRNAGlu (15 μM) was incubated for 1 h at 37°C in 50 mM Tris (pH 7.5), 200 mM NaCl with apo or holo-MnmA (wild-type or mutant) in the presence or absence of 1 mM Na2S, 2.5 mM MgCl2, 0.25 mM ATP. Ec-tRNAGlu alone 15 μM (1), apo-MnmA 1 μM (2) or 10 μM (3), 1 μM holo-MnmA and no Na2S (4), no MgCl2 (5) or no ATP (6), holo-MnmA 1 μM (7) or 10 μM (8), 1 μM holo-MnmAD99A-C102A mutant (9) or 1 μM holo-MnmAD99A-C102A-C199A mutant (10). (D) HPLC MS/MS detection of s2U and s4U (m/z 261). Samples were analyzed using the two transitions 261→129 and 261→112; the most intense one corresponds to the loss of the ribose moiety. Left: mixture of s2U and s4U synthetic standards (0.5 pmole injected); middle: Ec-tRNAGlu alone after hydrolysis; right: Ec-tRNAGlu (15 μM) after incubation with holo-MnmA (10 μM) in the presence of Na2S, ATP, MgCl2 and hydrolysis.
The [4Fe–4S] cluster is required for the tRNA sulfuration activity of E. coli MnmA
The sulfuration activity of E. coli MnmA was tested using in vitro transcribed Ec-tRNAGlu as a substrate in the presence of ATP, Mg2+ and inorganic sulfide, a sulfur donor often used in enzyme sulfuration assays (39–41). The thiolated nucleosides were quantified after digestion of the tRNA products by nuclease P1 and alkaline phosphatase. The s2U product was first identified by its elution position after HPLC-coupled mass spectrometry (MS), then quantified by MS/MS using a synthetic s2U standard (Figure 2C). The fragment spectrum corresponding to the loss of sugar (-132) was monitored (Figure 2D). Holo-MnmA (1 μM) was able to catalyze sulfuration of tRNA (15 μM) in contrast to apo-MnmA (1 or 10 μM) (Figure 2C). Moreover, the sulfur atom that was inserted into the nucleoside is derived exclusively from the inorganic sulfide salt, and not from the [4Fe–4S] cluster, since no sulfuration could be observed in the absence of sulfide. These control experiments indicated that the cluster and inorganic sulfide were required for the tRNA sulfuration activity of MnmA. Moreover, the amount of product formed increased with enzyme concentration (1 μM versus 10 μM in Figure 2C). Mg-ATP was also required, in agreement with its role in activating the substrate via adenylation of the C2 oxygen atom (Figure 1A and Supplementary Figure S2) (8).
MnmA is necessary for E. coli to resist to stress
It was previously shown that mnmA mutation in E. coli leads to reduced growth rate (14). We reproduced this observation (Figure 3A) and also noticed that the mutant exhibited a small colony phenotype when plated on rich medium (Figure 3C). We constructed the ΔmnmA strain and checked by HPLC-MS that the bulk tRNA from this strain did not contain mnm5s2U nor cmnm5s2U (Table 1). By monitoring growth in liquid culture, we found that the ΔmnmA strain showed slower growth in the presence of a mild acid stress (i.e. growth medium at pH 4.5) (Figure 3B) and that it exhibited hypersensitivity to H2O2 (Figure 3C). Note that survival plate assays run in parallel showed that the viability of the mnmA strain was not impaired at pH 4.5.

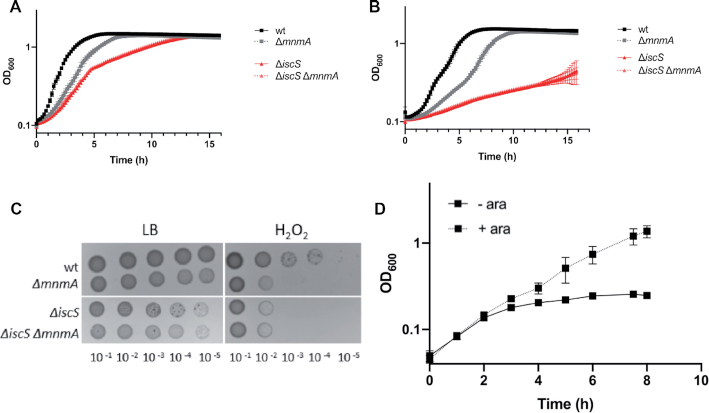
Phenotypes of ΔmnmA and derivative strains. (A) Growth in LB pH 7.0 medium. Strains studied are wild-type (wt) (FBE051), ΔmnmA (FBE584), ΔiscS (FBE653) and ΔiscS ΔmnmA (FBE703). Data are representative of three independent experiments (n = 3). (B) Growth in LB–HCl (pH 4.5). Strains studied are wild-type (wt) (FBE051), ΔmnmA (FBE584), ΔiscS (FBE653) and ΔiscS ΔmnmA (FBE703), (n=3). (C) Hypersensitivity of ΔmnmA to H2O2 (1 mM). Strains tested are wt, ΔmnmA, ΔiscS and ΔiscS ΔmnmA. Each spot represents a 10-fold serial dilution (n = 3). (D) The ΔmnmE and ΔmnmA mutations are lethal synthetic in E. coli. The RExBADmnmA ΔmnmE strain (FBE601) was grown in LB rich medium without (- ara) or with (+ ara) 0.2% l-arabinose, (n = 3).

| s2C | cmnm5s2U | mnm5s2U | |
|---|---|---|---|
| Wild-type | 9.5 ± 1.7 | 107.6 ± 39.8 | 1080.7 ± 647.0 |
| ΔmnmA | 14.8 ± 2.2 | 0.0 ± 0.1 | 1.9 ± 1.0 |
| ΔiscUA ΔsufABCDSE | 0.0 ± 0.0 | 64.5 ± 20.9 | 1421.9 ± 601.7 |
| ΔttcA | 0.4 ± 0.1 | 68.0± 44.4 | 1756.1 ± 1003.5 |
U34-tRNA modification pathway in E. coli
Biochemical studies together with a genetic approach coupled with mass spectrometry (ribonucleome analysis) have shown that MnmA receives sulfur from the IscS cysteine desulfurase (6,22) and the TusABCDE sulfur relay pathway (7). Thus, the prediction is that the ΔiscS strain should exhibit defects similar to those of the ΔmnmA strain, as shown in S. enterica (16). The ΔiscS mutation was found to cause extreme sensitivity to pH 4.5, even to a larger extent than that caused by the ΔmnmA mutation (Figure 3B). Such a feature has never been described for IscS so far. Introducing the ΔmnmA mutation in a ΔiscS background did not worsen the defect (Figure 3B), in line with the idea that MnmA depends upon IscS for getting sulfur. Similarly, when challenging the strains with H2O2 (Figure 3C), the ΔiscS strain displayed hypersensitivity comparable to that of the ΔmnmA strain, and the combination of both deletions failed to enhance the defect.
Besides hypersensitivity to H2O2, we wished to set-up another MnmA-associated phenotypic test. For this, we took advantage of the synthetic lethality between mnmA and mnmE mutations. MnmE, along with MnmG, adds the aminomethyl and carboxymethylaminomethyl groups to the C5 position of U34 (42). We constructed a strain named RExBADmnmA, in which expression of the mnmA gene was under the control of an arabinose inducible promoter and tested whether it could accommodate a ΔmnmE mutation. We observed that the ΔmnmE mutation could be introduced by transduction only when arabinose was added to the medium (Figure 3D). This confirmed that a strain lacking both mnmA and mnmE is not viable, as previously shown by using a strain, in which the mnmA gene was deleted and the mnmE gene put under the control of an arabinose inducible promoter (42).
Then, we reasoned that if iscS was required for MnmA to function, then a strain carrying both ΔiscS and ΔmnmE should not be viable. Therefore, we asked whether we could introduce a ΔiscS mutation into a ΔmnmE mutant. The result was negative, suggesting that a ΔiscS mutation recapitulated the ΔmnmA mutation, in agreement with IscS providing the sulfur atom to MnmA.
Taken together, these genetic analyses showed that the mnmA gene is crucial for E. coli to grow under both balanced and stress conditions, and established its close functional interaction with iscS, in agreement with early studies in S. enterica (23).
The ligands of the [4Fe–4S] cluster are most likely Asp99, Cys102 and Cys199
The D99A, C102S and C199A mutants of E. coli MnmA were reported to lack in vitro U34-tRNA sulfuration activity using cysteine as the sulfur donor, and the IscS and Tus proteins as sulfur carriers (8). We produced the double D99A-C102A and the triple D99A-C102A-C199A mutants (Supplementary Figure S4). Both mutants were correctly folded, as shown by their circular dichroism spectrum, which did not differ from that of the wild-type protein (Supplementary Figure S4F). After cluster reconstitution, they both almost completely lost the absorption band at 410 nm (Supplementary Figure S4G) and were shown to contain very little Fe, namely 0.46 ± 0.03 and 0.25 ± 0.05 iron per MnmA monomer, respectively. Finally, they did not exhibit any catalytic activity (Figure 2C, columns 9 and 10) in standard in vitro assays with sulfide used as a sulfur donor. Therefore, the mutagenesis results point out Asp99, Cys102 and Cys109 as being involved in cluster binding and catalysis (Figure 1B) (8). But one cannot unambiguously conclude on the in vitro analysis of only the double and triple mutant variants that all three residues contribute to ligation. Yet, we did not analyze the single variants because, in many instances, a single mutation of a cluster ligand was shown to be not sufficient to disrupt cluster ligation (43,44). Therefore, in addition, we tested the functional importance of the Asp99, Cys102 and Cys109 residues by changing each of them to alanine and testing the activity of the resulting single mutants in vivo using the series of phenotypic tests described above (Supplementary Figure S5). When expressed in trans from plasmids, all mutated variants, namely D99A, C102A and C109A failed to (i) suppress growth rate defects of the ΔmnmA recipient in rich medium (Supplementary Figure S5A, top), (ii) suppress hypersensitivity of the ΔmnmA strain to acid (Supplementary Figure S5A, bottom) and oxidative stress (Supplementary Figure S5B) and (iii) allow growth in the absence of arabinose in the RExBADmnmA ΔmnmE containing background (Supplementary Figure S5C). Note that expression of all three alleles gave rise to large amounts of soluble protein so that the possibility of a destabilizing effect of the introduced mutations was ruled out (Supplementary Figure S5D). Hence, based upon their inability to complement different defects of the ΔmnmA mutant, we concluded that substitution of Asp99, Cys102 or Cys109 by alanine yielded a nonfunctional MnmA protein. Altogether, the simplest interpretation of the in vitro characterization of the mutants and the in vivo complementation assays is that Asp99, Cys102 and Cys109 are the ligands of the [4Fe–4S] cluster.
Biosynthesis of mnm5s2U and cmnm5s2U in tRNAs occurs in the absence of the ISC and SUF [Fe–S] cluster biogenesis systems
Since maturation of [Fe–S] proteins in E. coli relies on the two ISC and SUF machineries, we analyzed the s2U content in bulk tRNA from the ΔiscUA ΔsufABCDSE strain (29) after tRNA digestion and HPLC-MS analysis (Table 1) to check whether MnmA maturation also uses these pathways. The ΔiscUA ΔsufABCDSE strain is not viable but can be grown by introducing the eukaryotic [Fe–S]-independent mevalonate-dependent isoprenoid biosynthesis pathway (29) to supply isopentenyl diphosphate, which is essential and normally synthesized by the [Fe–S] dependent IspG/IspH proteins. First, we observed that s2C was absent in bulk tRNA from the mutated strain (Table 1), in full agreement with TtcA, the unique enzyme responsible for s2C formation, requiring a [4Fe–4S] cluster (39) assembled by the ISC system (24). Then, we compared the content in mnm5s2U and cmnm5s2U in bulk tRNA from a wild-type and the ΔiscUA ΔsufABCDSE strains. Unexpectedly, we found that the amounts of mnm5s2U and cmnm5s2U from bulk tRNA in the mutant strain were comparable to those in the wild-type strain. This suggests that [Fe–S] cluster biogenesis of the E. coli MnmA protein can occur in the absence of the ISC and SUF systems.
DISCUSSION
We show here that MnmA, the only E. coli enzyme responsible for the sulfuration of U34 at C2 position in tRNAs (Figure 1A), assembles a [Fe–S] cluster. Moreover, the presence of this cluster proved essential for activity, under standard sulfuration assays previously used for other tRNA-sulfurating [Fe–S] enzymes such as TtcA (39) and TtuA (41). As a further confirmation of the functionality of the cluster, no in vitro activity could be obtained using MmnA variants, in which the residues that presumably bind the cluster (Asp99, Cys102 and Cys199) were changed into alanine. The occurrence of a [4Fe–4S] cluster was clearly established by Fe and S quantification and from UV-visible and EPR spectroscopic characteristic features, after cluster reconstitution under anaerobic conditions. Interestingly, residual cluster was also unambiguously observed within the as-purified protein before any chemical treatment, indicating that MnmA carried a cluster within the cell as well.
Shigi et al. recently reported that MnmA from Thermus thermophilus also contains an active [4Fe–4S] cluster (43). However, the authors proposed that MnmA proteins should be subdivided into [Fe–S]-containing and [Fe–S]-independent types (43). In the first C-type class, the three cysteines from the CXXC + C motif would ligate the cluster, such as in the T. thermophilus enzyme. The second D-type class would harbor a DXXC + C motif instead, like in the E. coli enzyme, which would be unable to bind a cluster. However, our results are strongly in agreement with Asp99, Cys102 and Cys199 of the DXXC + C motif being the ligands of the cluster of E. coli MnmA. Indeed, both the D99A-C102A and D99A-C102A-C199A mutants were unable to assemble a [4Fe–4S] cluster in vitro, in agreement with the observation that the D99A, C102A and C199A mutants were not able to rescue the growth phenotype of the ΔmnmA strain in vivo. It is interesting to note that Asp99, Cys102 and Cys199 are in close proximity to each other in the crystal structure of apo-MnmA (8), the only 3D structure available so far, and with the appropriate configuration for binding a [Fe–S] cluster (Figure 1B). In fact, amino acids other than cysteine can act as [Fe–S] ligands, such as histidine, glutamate, arginine and threonine (45). There are also precedents for aspartate as a ligand of a [4Fe–4S] or a [2Fe-2S] cluster, such as in ferrodoxin from hyperthermophilic archaea (46,47), in a nitrogenase-like enzyme named protochlorophyllide reductase (48,49), in a transcriptional regulator (50) and in IscA, involved in [Fe–S] cluster assembly (51).
Given that Ncs6-type proteins also use a cluster for s2U34 formation in tRNAs (52) (PDB code: 6SCY), the biochemical findings presented here lead us to propose that this sulfuration reaction depends on a [4Fe–4S] enzyme in all organisms and in mitochondria (9) (Supplementary Figure S1). More specifically, we propose that C-type and D-type MnmA are all [Fe–S] enzymes.
In Figure 1C, we propose a tentative mechanism for the holo-MnmA form, in which the cluster, adjacent to the substrate, serves to bind and activate a hydrosulfide nucleophilic substrate for subsequent attack on C2 from U34 to displace the AMP leaving group and form the final C–S bond in the product. A cluster with only three protein-bound ligands is appropriate for such a function since it provides a free coordination site on the fourth Fe atom where hydrosulfide can bind (53). We and others previously proposed a similar mechanism with the involvement of a [4Fe-5S] intermediate for TtuA, another thiouridine synthetase that targets position 54 in tRNA (41,54,55), and for TtcA responsible for the formation of s2C32-tRNA (39). While more studies are required to firmly establish such a mechanism for tRNA thiolation enzymes, we have recently structurally characterized a relevant [4Fe-5S] catalytic intermediate during the desulfuration of 4-thiouracil by thiouracil desulfidase TudS (56), which thus provides a unique precedent for this class of intermediate clusters. Obviously, this mechanism excludes persulfides as key reaction intermediates, as proposed previously, since the cysteines of the active site proposed to carry the persulfide function are engaged as ligands of the [4Fe–4S] cluster.
As a consequence of the results reported here, we need to examine why an [Fe–S] cluster in E. coli MnmA was previously excluded, especially considering its essential catalytic role. First, the conclusion that MnmA could not be an [Fe–S] enzyme came from an early study examining the s4U and mnm5s2U levels in ISC defective backgrounds; i.e. the ΔiscU, ΔhscA, Δfdx and ΔiscA strains showed wild-type like levels, while both s2C and ms2io6A levels were found altered (24). It was concluded that downstream IscS, which is the shared sulfur source for the four reactions, two distinct routes were likely to occur for biosynthesis of thiolated nucleotides in tRNA: the [Fe–S]-dependent route, responsible for the formation of s2C and ms2io6A, and the [Fe–S]-independent route, leading to s4U and mnm5s2U. Therefore, previous MnmA preparations, purified exclusively under aerobic conditions, have not been analyzed for the presence of small amounts of protein-bound clusters (6–8), as we did here. It is well known that most clusters, and more specifically those with a labile coordination site, like in aconitase (57) or radical-SAM enzymes (58), are very sensitive to air and degrade during purification, generally leading to colorless protein solutions. Second, while these seemingly ‘cluster-free’ enzyme preparations were active, the reported activities were very weak, despite very large, far from catalytic, amounts of protein used (MnmA:tRNA ratios of 1:1 (6), 2.1:1 (7), 4:1 (8) or 6.4:1 (25). It is tempting to suggest that any measured activity might have been due to the presence of a small fraction of holo form in the as-purified MnmA, as we observed it here.
To date, maturation of all tested [Fe–S] proteins in E. coli has been found to depend upon ISC, SUF or both systems. Therefore, after having shown that MnmA contains an [Fe–S] cluster, we reinvestigated the contribution of ISC and/or SUF to mature MnmA. Surprisingly, the mnm5s2U/cmnm5s2U content in tRNAs of the ΔiscUA ΔsufABCDSE strain was comparable to that in wild-type cells, arguing against the involvement of either machinery for maturation of MnmA. This result is consistent with previous work by Björk’s (24) and Leimkühler’s groups (59), who showed that ISC and SUF were dispensable for s2U34 biosynthesis. Thus, to explain the s2U content of tRNAs in the mutated strain, we are forced to entertain the possibility for MnmA to be targeted by an as yet unknown [Fe–S] cluster biogenesis pathway, which our current studies are aiming to identify.
DATA AVAILABILITY
All data are available in the manuscript; strains and constructs are available on request.
ACKNOWLEDGEMENTS
We thank the French EPR CNRS Facility, Infrastructure de Recherche Renard (IR 3443); the Macromolecular Interaction Platform of I2BC for use of its facilities and expertise; Ludovic Pecqueur for maintenance of the glove boxes; Bruno Faivre for advice in the purification experiments; Djemel Hamdane for assistance in CD experiments, Simon Arragain and Ornella Bimai for fruitful discussions that initiated this project; and the SAMe Unit members for discussion, technical help and suggestions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la recherche Scientifique and French State Program ‘Investissements d’Avenir’ [LABEX DYNAMO, ANR-11-LABX-0011, IBEID ANR-10-LABX-62]; Institut Pasteur. Funding for open access charge: LABEX DYNAMO.
Conflict of interest statement. None declared.
REFERENCES
1.
2.
3.
4.
5.
6.
7.
8.
9.
10.
11.
12.
13.
14.
15.
16.
17.
18.
19.
20.
21.
22.
23.
24.
25.
26.
27.
28.
29.
30.
31.
32.
33.
34.
35.
36.
37.
38.
39.
40.
41.
42.
43.
44.
45.
46.
47.
48.
49.
50.
51.
52.
53.
54.
55.
56.
57.
58.
 Iron–sulfur biology invades tRNA modification: the case of U34 sulfuration
Iron–sulfur biology invades tRNA modification: the case of U34 sulfuration
