
Competing Interests: The authors have declared that no competing interests exist.
- Altmetric
Multi-drug resistant (MDR) Acinetobacter baumannii (Ab) and Acinetobacter spp. present monumental global health challenges. These organisms represent model Gram-negative pathogens with known antibiotic resistance and biofilm-forming properties. Herein, a novel, nontoxic biocide, AB569, consisting of acidified nitrite (A-NO2-) and ethylenediaminetetraacetic acid (EDTA), demonstrated bactericidal activity against all Ab and Acinetobacter spp. strains, respectively. Average fractional inhibitory concentrations (FICs) of 0.25 mM EDTA plus 4 mM A-NO2- were observed across several clinical reference and multiple combat wound isolates from the Iraq/Afghanistan wars. Importantly, toxicity testing on human dermal fibroblasts (HDFa) revealed an upper toxicity limit of 3 mM EDTA plus 64 mM A-NO2-, and thus are in the therapeutic range for effective Ab and Acinetobacter spp. treatment. Following treatment of Ab strain ATCC 19606 with AB569, quantitative PCR analysis of selected genes products to be responsive to AB569 revealed up-regulation of iron regulated genes involved in siderophore production, siderophore biosynthesis non-ribosomal peptide synthetase module (SBNRPSM), and siderophore biosynthesis protein monooxygenase (SBPM) when compared to untreated organisms. Taken together, treating Ab infections with AB569 at inhibitory concentrations reveals the potential clinical application of preventing Ab from gaining an early growth advantage during infection followed by extensive bactericidal activity upon subsequent exposures.
Introduction
Acinetobacter baumannii (Ab) infections have historically been a major clinical challenge for both military and civilian health professionals, especially during the Iraq/Afghanistan conflicts. In the context of combat medical care and acquired wound, burn, blast and ventilator-associated pneumonia (VAP) infections, Ab represents a formidable multi-drug resistant (MDR-AB) pathogen and, as such, is a top 10 CDC priority organisms [1, 2]. Joint Program Committee 2, Military Infectious Disease Research Program (JPC-2/MIDRP) is a congressionally directed committee with a charter to research wound infection prevention as well as antimicrobial countermeasures. The focus research area for JPC-2/MIDRP includes poly-trauma and blast wound injuries [3, 4]. Ab, a common blast wound pathogen, is recognized as a nosocomial isolate that readily acquires resistance genes that especially plagues immunocompromised patients. In a retrospective study of wounded combat evacuees in 2003, Petersen et al. [5] found that of 56 patients that acquired infections, 84% were infections resulting from blast wound type injuries. Furthermore, 36% of the infections were caused by Ab, making it the predominant wound pathogen. Such organisms were also resistant to 80% of tested drugs [5], colloquially designating Ab as “Iraqibacter, [6]”. The 2003 and 2004 Acinetobacter baumannii-calcoaceticus complex (ABC) outbreak among U.S. military personnel treated in Military Treatment Facilities was investigated and discovered that 7 of 7 military field hospitals sampled recovered ABC strains [7]. This discovery further highlights the importance of Ab as a significant nosocomial pathogen as well as a top-priority, “serious threat” according to the CDC (https://www.cdc.gov/drugresistance/biggest_threats.html). In addition, mortality rates for nosocomial infections with multi-drug resistant Acinetobacter spp. were 26% higher than the mortality rate of uninfected patients [8]. Further compounding this problem is the fact that Ab possesses genomic resistance islands (Ab antibiotic resistance, AbaR) enabling the bacteria to rapidly acquire drug antibiotic resistance [9]. Furthermore, this bacterium is also naturally competent, thereby being highly proficient at the uptake of stable plasmids [10]. These characteristics make Ab and Acinetobacter spp. ideal model organisms and a huge challenge for the development of novel antibiotics or non-cytotoxic biocides, especially considering that the development of novel therapeutics has dramatically slowed in recent years.
AB569: A novel, two component formulation for the treatment of Ab infections
i. The EDTA component
AB569 is a non-toxic, bactericidal combination of EDTA and acidified nitrite (herein A-NO2-) buffered to pH of 5.5 to 6.5. The EDTA component of AB569 is widely known as a chelator of di/tri-valent cations with a binding preference prioritizing iron (Fe2/3+), calcium (Ca2+), and magnesium (Mg2+) [11]. EDTA also increases the permeability of the bacterial cell wall by binding Ca2+ and Mg2+ ions that bridge the vital lipopolysaccharide (LPS) component within the outer membrane of Gram-negative bacteria [12]. The ability of EDTA to also chelate metals, especially Fe, dramatically influences the ability of Ab to form complex, antibiotic-resistant, highly organized communities known as biofilms [13]. Biofilm formation by Ab is considered a significant virulence property, thereby enhancing its ability to cause disease [14]. Fe also influences the robustness of Ab biofilm formation [15] and we speculate that its sequestration may play a translational role in future clinical studies designed to treat highly problematic Ab and Acinetobacter spp. infections. Taken together, the role of EDTA in altering the permeability of bacterial cell walls, its ability to sequester Fe, and its effect on biofilm formation are just a few of the actions that contribute to the bactericidal status of EDTA that has been observed in multiple bacterial pathogens [16–18].
ii. The acidified nitrite (A-NO2-) component
Bacteria can also be exposed to potentially toxic doses of reactive nitrogen species (RNS) during the course of human infection [19]. The endogenous metabolic production of or exposure to exogenous RNS has been shown to enable bacteria to acquire resistance to some antibiotics [20]. Treatment of bacteria with A-NO2- has been shown to increase RNS formation in the form of nitric oxide (NO) production [21]. Given its ability to diffuse through bacterial cell membranes [22], NO can potentially combine with the one-electron reduction product of molecular oxygen, superoxide (O2-), to form peroxynitrite (OONO-), an exceedingly powerful oxidant that can react with virtually all known biomolecules at diffusion-limited rates (~1010 M-1s-1, [23]). Still, the predominant means by which OONO- causes cell death is thought to be predominantly due to DNA damage [24]. In addition, excess A-NO2- can lead to lipid peroxidation and increased cell permeability or tyrosine nitration that can alter cell function, structure or homeostasis [25]. NO also binds to cysteine residues on proteins, forming S-NO proteins (via S-nitrosylation/nitrosation), potentially inhibiting proper protein processing and overall cellular function [26].
Despite the fact that the genus Acinetobacter are strict aerobes, they still have the ability to reduce nitrate to nitrite using reductase enzymes [27, 28], further “recycling” A-NO2-. Ab also converts nitrate to ammonia and uses nitrogen as an energy source in the assimilatory nitrate reduction pathway. Ab is also capable of converting nitrite (NO2-) to ammonium hydroxide (NH3OH) via an NADH/NADPH nitrite reductase [29]. As there is likely a myriad of targets/processes adversely affected during the process of A-NO2- mediated bacterial killing, the direct mechanism of bactericidal action, as with any biocide (e.g., HOCl, H2O2, F-, etc.), is unknown. Finally, in a previous study, Yoon et al. [21] showed that at slightly acidic pH levels, A-NO2- can give rise to HNO2 that is unstable and spontaneously generates NO, N2O3, NO2. and ultimately NO3-.
Though many bacteria are sensitive to A-NO2-, a previous study by McDaniel et. al. [30] showed increased sensitivity of the ESKAPE pathogen Pseudomonas aeruginosa (Pa) to AB569 compared to using either EDTA or A-NO2- alone in a synergistic fashion. Pa differs from Ab in the metabolism of nitrate, due to its ability to grow under both aerobic and anaerobic conditions, the latter of which is by the process of denitrification. Moreover, current studies have also revealed sensitivity to AB569 in bacteria such as other ESKAPE pathogens including methicillin-resistant Staphylococcus aureus (MRSA) strain USA300, Enterococcus faecium, Klebsiella pneumoniae and Enterobacter sp., as well as Escherichia coli and many other Gram-positive and Gram-negative organisms [31].
The promise of AB569 as a therapeutic bacterial biocide is particularly relevant considering that bacteria are unable to develop resistance to it over time. In another combat wound/blast pathogen, Pa, AB569 significantly compromised >30 vital pathways including those involved in the biosynthesis of DNA, RNA, protein and ATP, leading to rapid killing of such bacteria [32]. Thus, given the interests of military and civilian practitioners in antibiotic resistance mechanisms and wound infection prevention and treatment, the goals of this study were to test AB569 against clinical isolates of Ab and Acinetobacter spp. with seven separate experimental challenges that are described in detail below. We found that all Iraq and Afghanistan battlefield clinical isolates as well as reference strains were sensitive to AB569, some of which were in a synergistic fashion. In addition, bactericidal concentrations of AB569 were non-toxic to primary adult human skin (dermal) fibroblasts. Regarding human use, both the NaNO2 and/or EDTA component(s) of AB569 have separately proven to be safe in studies related to the treatment of urinary tract infection [33], burn wounds [34], cystic fibrosis lung infection [35], chelation therapy [36], soaps [37] and cosmetics [38]. Thus, our results suggest that AB569 has great potential as a therapeutic agent form the treatment of battlefield wound/burn/blast infections as well as problematic ventilator-associated pneumonia (VAP) infections by Ab.
Materials and methods
Bacteria used in this study
All bacteria used in this study are listed in Table 1. The bacteria were maintained as frozen stocks in a 1:1 suspension of Luria-Bertani (L-broth) or Trypticase soy broth and 30% glycerol at -80°C.

| Bacterial Strain, Primer, or Cell Line | Short ID | Source | Description or DNA sequence (5′ to 3′) | Source or reference |
|---|---|---|---|---|
| Bacterial Strains: Acinetobacter baumannii (Ab) or Acinetobacter Spp. | ||||
| Ab | Ab-Epi | - | Clinical Isolate | USAFSAM PHE |
| A. iwoffi | AI-EPI | Urine | Clinical Isolate | USAFSAM PHE |
| Ab ACICU | ACICU | - | Clinical Isolate | Miami University |
| Ab AYE | AYE | - | Clinical Isolate | Miami University |
| Ab ATCC 17978 | 17978 | - | ATCC Strain | Miami University |
| A. iwoffi | WP#2 | - | Clinical Isolate | 88 DTS/ SGQC |
| Ab | WP#1 | - | Clinical Isolate | 88 DTS/ SGQC |
| Acinetobacter spp | GNR 3-9J | - | Clinical Isolate | CCHMC |
| Acinetobacter spp | GNR 3-IG | - | Clinical Isolate | CCHMC |
| Acinetobacter spp | GNR 3-2G | - | Clinical Isolate | CCHMC |
| Acinetobacter spp | GNR 3-10J | - | Clinical Isolate | CCHMC |
| Acinetobacter spp genomospecies 9 ATCC 9957 | 9957 | - | ATCC Strain | CCHMC |
| A. anitratus ATCC 49139 | 49139 | - | ATCC Strain | CCHMC |
| Ab ATCC 19606 | 19606-JM | - | ATCC Strain | CCHMC |
| Ab ATCC 747 | 747 | - | ATCC Strain | CCHMC |
| A. anitratus ATCC 49137 | 49137 | - | ATCC Strain | CCHMC |
| Acinetobacter spp | GNR 3-8E | - | Clinical Isolate | CCHMC |
| Acinetobacter spp | GNR 3-3C | - | Clinical Isolate | CCHMC |
| Ab | B2-MRO12 | - | Clinical Isolate | CCHMC |
| Ab | B1-MRO11 | - | Clinical Isolate | CCHMC |
| Ab | WR 1 | Bone | Clinical Isolate | WRNMMC |
| Ab | WR 2 | Urine | Clinical Isolate | WRNMMC |
| Ab | WR3 | Sputum | Clinical Isolate | WRNMMC |
| Ab | WR4 | Hip | Clinical Isolate | WRNMMC |
| Ab | WR5 | Wound | Clinical Isolate | WRNMMC |
| Ab | WR6 | Urine | Clinical Isolate | WRNMMC |
| Ab | WR7 | Groin | Clinical Isolate | WRNMMC |
| Ab | WR8 | Tissue | Clinical Isolate | WRNMMC |
| Ab | WR9 | Tissue | Clinical Isolate | WRNMMC |
| Ab | WR10 | Groin | Clinical Isolate | WRNMMC |
| Ab | WR11 | Hip | Clinical Isolate | WRNMMC |
| Ab | WR12 | Clinical Isolate | WRNMMC | |
| Acinetobacter spp | WR13 | Clinical Isolate | WRNMMC | |
Bacterial identification
Bacterial identification was confirmed using the Vitek MS (bioMerieux, Marcy-l-Etoile, France)—Matrix Assisted Laser Desorption Ionization–Time of Flight (MALDI-TOF) using and FDA cleared–IVD database following manufacturers guidelines. This database contains the following: Acinetobacter spp., Acinetobacter baumannii complex, Acinetobacter haemolyticus, Acinetobacter junii, and Acinetobacter lwoffii. Additional data is available for A. ursingii but is not sufficient for an IVD claim.
Minimum Inhibitory Concentration (MIC) testing
MIC testing was performed using the Vitek 2 system (bioMerieux, Marcy-l-Etoile, France) following manufacturer guidelines. Additional MICs were determined for colistin using the Etest system (bioMerieux, Marcy-l-Etoile, France) with 150 mm cation adjusted Mueller-Hinton agar plates (BD Diagnostics, Sparks, MD) incubated at 35°C for 18 to 24 hr in ambient air, following manufacturer guidelines.
Fractional Inhibitory Concentration (FIC) determinations
Microtiter plates (96-well) were prepared in a checkerboard scheme as previously described [39] (Fig 1). Stock EDTA and freshly prepared NaNO2 solutions were diluted in Luria-Bertani (LB) broth plus 100 mM potassium nitrate (KNO3) and 50 mM potassium phosphate buffer at pH 6.5 (Kpi, LBN 6.5) or Trypticase Soy Broth (TSB) plus 100 mM KNO3 and 50 mM Kpi at pH 6.5 (TSBN 6.5) at 2-fold increasing levels to final concentrations of 16 mM EDTA and 64 mM A-NO2- and also a combination of the two. The two media used was based upon the growth preferences of each strain, as some were difficult to grow. The nitrate was added to the medium since wounded/burned murine serum has been shown to possess higher nitrate/nitrite levels [40]. Most of the bacteria used in this study were grown in LB. However, if some bacteria could not grow or grew poorly in LB, TSB was used. Bacteria were incubated at 37°C overnight in LB or TSB broth and were centrifuged at 13,000 x g for 2 min at room temperature. The pellet was resuspended in PBS to a final optical density (O.D.600) of ~0.5. This suspension was then added to either LBN, 6.5 or TSBN, 6.5 at a dilution factor of 1:5000 (~5 x 105 CFU/ml). Subsequently, the diluted bacterial suspension was added to all wells except for the media control column. The plates were then incubated overnight at 37°C and analyzed on the O.D.630 channel of a Bio-Tek ELx 800 Universal Microplate Reader.

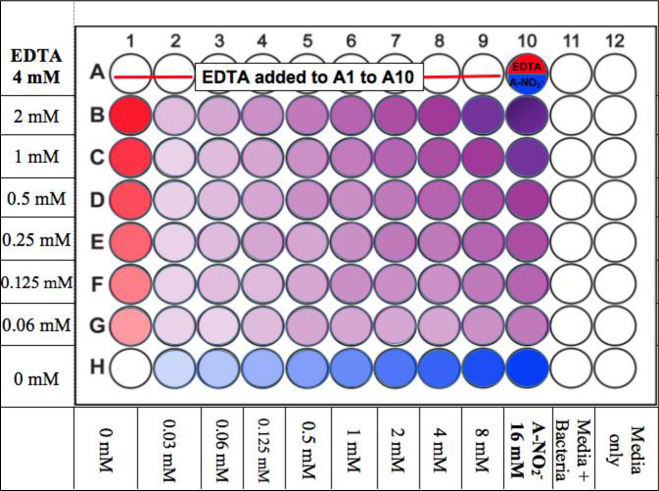
Illustration of the FIC checkerboard scheme, 96-well plate set up.
The red wells are EDTA, blue wells are A-NO2-, and purple wells are the AB569 combination at various concentration as indicated.
AB569 killing studies
Ab ATCC 19606 was grown from a -80°C frozen stock on LB agar at 37°C overnight. A single colony was inoculated in LB broth and grown aerobically overnight at 37°C with shaking at 200 rpm. The stationary phase culture was then diluted 100-fold in LBN 6.5 and grown under the same conditions for an additional 1 hr. Treatment with EDTA, A-NO2- or the AB569 combination at various concentrations were then added and viability of serially diluted samples evaluated over a period of 48 hr. Suspensions were diluted 10-fold in PBS and 50–100 μl aliquots were spread evenly on LB agar plates. After the plates were incubated at 37°C for 16–18 hr, bacterial colonies were enumerated and reported as Colony Forming Units (CFU) per ml.
Quantitative polymerase chain reaction (qPCR) of Ab gene expression after treatment with AB569
The primers used for these experiments were based on the DNA sequence in the 470.2129 version of the Ab ATCC 19606 genome in the Patric.org database [41]. All primers used are listed in Table 2. An overnight culture of Ab ATCC 19606 was diluted 100-fold with LBN 6.5. Diluted bacteria were incubated for 1 hr under the same conditions used in the aforementioned killing studies. Then, bacterial cultures were treated with AB569 or its individual components for 2.5 hr under the same conditions. Bacteria were pelleted and lysed following the Qiagen RNeasy Protect Bacteria Mini Kit (Qiagen) protocol for total RNA isolation. The lysate was then pipetted into RNeasy spin columns. DNase was introduced into the column to remove contaminating DNA from the RNA samples. After non-specific binding components were washed from the column, total RNA was then eluted using RNase-free water. Prior to qPCR analyses, total RNA samples were examined for contaminating DNA by PCR using ChoiceTaq Mastermix and total RNA as a template to amplify a known housekeeping gene, rpoD [42]. The concentration of DNA-free total RNA sample was next analyzed using the Thermo Scientific Nanodrop method. Total RNA samples were then reverse-transcribed to cDNA using the ImProm-IITM Reverse Transcription System by following the provided protocol. The cDNA was then used as a template to perform qPCR using a StepOnePlus System (Thermo Scientific, Applied Biosystems) analyzer with PowerUp SYBR Green Master Mix (Applied Biosystems) to amplify the genes of interest. To standardize the assay, 2-ΔΔCt values were calculated and normalized to rpoD [42], annotated in Table 2, using the raw cycle threshold (Ct) values (also known as cross point (Cp) values) generated from our analyses [43, 44].

| Ab | S1 Mutant | Ab ATCC 19606 acinetobactin siderophore mutant | Miami University |
|---|---|---|---|
| Primers | |||
| RNA polymerase sigma factor rpoD (Housekeeper Gene) | rpoD | Forward CCGATCAGGCTCGTACAATTC Reverse CACGGCCCATTTCCTGTAATA | IDT |
| Pyruvate dehydrogenase E1 component | PDHcE1 | Forward GCATGTTCCAATTCAACGTCTC Reverse ACGAATACGGCGTTCCATATC | IDT |
| LSU ribosomal protein L4p (L1e) | L4p | Forward CTCTGCTGTTGAATTGTCTGAAG Reverse GACGACCACCTGCTAAGTAAG | IDT |
| SSU ribosomal protein S11p | S11p | Forward CGGCTTTGGATTACGGTTTG Reverse TATAACCCACTGCGCCTAATG | IDT |
| Siderophore biosynthesis non-ribosomal peptide synthetase modules | SBNRPSM | Forward CCGACGTCCGGCATATTATT Reverse TCGTCTGTTGTAGCGTGTTT | IDT |
| Siderophore biosynthesis protein, monooxygenase | SBPM | Forward TCGTTGGATCACACGTTCTG Reverse CTCTAGGCAGGCTTTGGAAATA | IDT |
| SSU ribosomal protein S2p | S2p | Forward GATGCTTTGAACTTCGCTAACC Reverse AGCTTGTTCACGGATGATGT | IDT |
| Translation elongation factor Ts | EFTs | Forward AGGCGAACAATTGGCTATCT Reverse ACGTGCATTGCAATACCTTTAC | IDT |
| SSU ribosomal protein S7p | S7p | Forward GTGAGATCCTTCCAGATCCTAAAT Reverse CCGTAAACGATACTTTCAGCAATAG | IDT |
| LSU ribosomal protein L28p zinc-independent | L28p | Forward TCTCACACGCCAACAACAA Reverse AGTGGTTAAACGAAGACGTACAA | IDT |
| ATP synthase delta chain | atpD | Forward AGCAAGGTGCAACAGACA Reverse GGAGTAAGTTCAGGGCGATTTA | IDT |
AB569 biofilm inhibition testing
Experiment were performed in 96-well plates as described for the FIC studies above. After an overnight incubation at 37°C, the bacteria biofilms were first washed twice with PBS and stained with filter-sterilized 0.1% crystal violet. After a 30 min incubation at room temperature, unattached bacteria were removed gently by rinsing with water. Ethanol (95%) was then added to each well to dissolve the crystal violet that was bound to the peptidoglycan layer of biofilm bacteria [45]. The quantity of solubilized crystal violet was then measured at O.D.595 using Bio-Tek ELx 800 Universal Microplate Reader.
AB569 biofilm dispersion testing
An overnight culture of Ab in LB or TSB broth was diluted 100-fold into the same media and 200 μl of diluted bacteria culture was transferred into each well in 96-well plates and incubated at 37°C for 24 hr without shaking to allow the bacteria biofilms to mature. After unattached, planktonic bacteria were removed and the biofilms washed with LBN or TSBN, pH 6.5, media containing 2-fold dilutions of EDTA, A-NO2- or AB569 was transferred into the plates. The plates were then incubated under the same conditions as above. After 24 h, the optical density of the 96-well plate suspensions was measured on the O.D.630 nm channel using a Bio-Tek ELx 800 Universal Microplate Reader. This was followed by the crystal violet biofilm staining assay as described above.
Cell culture
Adult Human Dermal Fibroblast (HDFa) cells (ThermoFisher Scientific, Waltham, MA, Cat# CO135C) were cultured in DMEM supplemented with 5% fetal bovine serum (FBS), 5 μg/ml insulin, 10 ng/ml epidermal growth factor (EGF), 0.5 μg/ml hydrocortisone, and 1% penicillin streptomycin. Cells were grown in 150 mm plates in 20 ml of supplemented media with a typical dilution of 1:5 or 1:10 to promote uniform cell growth.
SYTOX® orange dead cell staining
HDFa cells were plated at 5 x 104 or 1 x 105 cells per well in 12- or 24-well plates, respectively. After an overnight incubation at 37°C in an atmosphere of 5% CO2, medium was aspirated and the cells were treated with varying concentrations of EDTA and/or A-NO2- in supplemented DMEM media which was acidified with 50 mM KPi, pH 6.5 (supplemented DMEM 6.5). Control and treated HDFa cells were incubated under the same conditions for 24 hr. To quantify the dead cells, the media from each well was transferred to individual microcentrifuge tubes. Dulbecco’s Phosphate Buffered Saline (DPBS) was used to rinse the cells in each well followed by the addition of 0.05% trypsin. The plates were subsequently incubated for 30 min at 37°C in an atmosphere of 5% CO2. Next, the removed media was used to neutralize the trypsin and control and treated HDFa cells were centrifuged for 5 min at 500 x g. The cell pelleted was re-suspended in 250 or 500 μl of supplemented DMEM media containing the Sytox Orange dead cell stain at a ratio of 1:1000 at a final concentration of 250 nM. After incubation at room temperature for 30 min, the live/dead cell ratio was analyzed by flow cytometry (Becton Dickinson (BD) FACS Calibur analysis). Stained cells were excited at 488 nm and read on the orange FL2 channel at an emission wavelength of 570 nm. Results were reported as the percentage of dead cells.
Statistical analyses
Statisitical analyses were performed using GraphPad Prism 7 software. Statisitical significance was set at a p-value of <0.05.
Results
Bacterial identification and antibiotic minimum inhibitory concentration (MIC) determinations
Ab and Acinetobacter spp. are aerobic, oxidase-negative, Gram-negative bacilli that possess the ability to form highly problematic biofilms during disease [13]. As previously mentioned, Ab is known for easily acquiring multi-drug resistance during treatment of infected patients. The ensemble of bacteria used in this study included 25 Ab and 8 Acinetobacter spp. strains, including 13 MDR clinical strains provided by Walter Reed Army Medical Center (WRAMC) and 11 other clinical strains from other sources that are listed in Table 1. MALDI-TOF was used to identify each strain and our results showed 6/8 were A. ursingii, 1/8 was A. radioresistens, and 1/8 was A. iwoffii, respectively (Table 1). Antibiotic sensitivities as a collective result of minimum inhibitory concentration (MIC) testing and bacterial identification analysis were performed on each isolate. Our MIC results showed that all of the bacterial isolates used in this study were found to be resistant to cefazolin (CFZ), a first-generation cephalosporin (Table 3), consistent with Ab being a notorious β-lactamase producer [46] and also with previous studies [47, 48]. In addition, none of the bacteria tested were resistant to colistin (CST, polymyxin E) an antibiotic with high human toxicity and, as a result, a last choice for the treatment of MDR organisms [49]. According to current clinical standards of antibiotic selection by physicians for Ab treatment [50], meropenem (MEM) is typically the first choice in Ab antibiotic therapy. We found that the strains used in this study were 37% resistant to MEM (Table 3). Second-tier antibiotics used in this study revealed lower resistance with tobramycin (TOB, 23%) and gentamicin (GEN, 39%), respectively. It is worth noting that the 25 clinical isolates in this study meet the definition of Multi-Drug Resistant Organisms (MDRO) in that each strain was resistant to more than one class of antibiotics (Table 3). The percentage of antibiotic resistance for each organism was 33% for Aa, 0–43% for Au strains, 0–7% for Ai and ~14% for Ar, while ABC strains revealed far higher antibiotic resistance, reaching an average of 66% (range of 34–94%), respectively (Table 3).

| Strain | ID by MALDI-TOF | AMP | SAM | TZP | CFZ | FOX | CAZ | CRO | FEP | MEM | GEN | TOB | CIP | NIT | SXT | CST |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacterial Strains: Acinetobacter baumannii (AB) or Acinetobacter Spp. | ||||||||||||||||
| Percent Resistance | 88.6% | 31.4% | 64.3% | 100.0% | 94.3% | 80.0% | 94.3% | 65.7% | 37.1% | 38.9% | 22.9% | 51.4% | 97.1% | 57.1% | 0.0% | |
| Ab-Epi | ABC | ≥32 | ≥32 | ≥128 | ≥64 | ≥64 | 4 | 16 | ≥64 | ≥16 | ≤1 | ≤1 | ≥4 | ≥512 | ≤20 | 0.38 |
| R | R | R | R | R | R | I | R | R | S | S | R | R | S | S | ||
| AI-EPI | AI | * | * | * | * | * | * | * | * | * | ≤1 | * | * | * | * | 0.094 |
| * | * | * | * | * | * | * | * | * | S | * | * | * | * | S | ||
| ACICU | ABC | ≥32 | 16 | ≥128 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | 4 | 8 | ≥4 | ≥512 | ≥320 | 0.38 |
| R | I | R | R | R | R | R | R | R | S | I | R | R | R | S | ||
| AYE | ABC | ≥32 | 4 | ≥128 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | 1 | ≥16 | ≥16 | ≥4 | ≥512 | ≥320 | 0.38 |
| R | S | R | R | R | R | R | R | S | R | R | R | R | R | S | ||
| 17978 | ABC | ≥32 | ≤2 | ≤4 | ≥64 | ≥64 | 4 | 16 | 2 | ≤0.25 | ≤1 | ≤1 | ≤0.25 | ≥512 | 160 | 0.19 |
| R | S | S | R | R | S | I | S | S | S | S | S | R | R | S | ||
| WP#2 | ABC | ≥32 | 4 | ≥128 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | 1 | 4 | ≤1 | ≥4 | ≥512 | ≤20 | 0.5 |
| R | S | R | R | R | R | R | R | S | S | S | R | R | S | S | ||
| WP#1 | ABC | ≥32 | 4 | ≥128 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | 1 | 4 | ≤1 | ≥4 | ≥512 | ≤20 | 0.38 |
| R | S | R | R | R | R | R | R | S | S | S | R | R | S | S | ||
| GNR 3-9J | AU | 16 | ≤2 | * | ≥64 | ≥64 | 32 | 32 | 8 | ≤0.25 | ≤1 | ≤1 | 0.5 | 256 | ≤20 | 0.125 |
| I | S | * | R | R | R | I | S | S | S | S | S | R | S | S | ||
| GNR 3-IG | AU | 16 | ≤2 | * | ≥64 | ≥64 | ≥64 | 32 | 8 | ≤0.25 | ≤1 | ≤1 | ≤0.25 | ≥512 | ≤20 | 0.125 |
| I | S | * | R | R | R | I | S | S | S | S | S | R | S | S | ||
| GNR3-2G | AU | 4 | ≤2 | * | 32 | 16 | 2 | 16 | ≤1 | ≤0.25 | ≤1 | ≤1 | ≤0.25 | ≥512 | ≤20 | 0.125 |
| S | S | * | R | I | S | I | S | S | S | S | S | R | S | S | ||
| GNR 3-10J | AU | 8 | ≤2 | * | ≥64 | ≥64 | 16 | 16 | 2 | ≤0.25 | ≤1 | ≤1 | ≤0.25 | ≥512 | ≤20 | 0.125 |
| S | S | * | R | R | I | I | S | S | S | S | S | R | S | S | ||
| 9957 | AI | ≤2 | ≤2 | * | 16 | ≤4 | ≤1 | 4 | ≤1 | ≤0.25 | ≤1 | ≤1 | ≤0.25 | 32 | ≤20 | 0.094 |
| S | S | * | I | S | S | S | S | S | S | S | S | S | S | S | ||
| 49139 | ABC | ≥32 | ≤2 | 8 | ≥64 | ≥64 | 16 | 16 | 32 | 1 | ≥16 | 2 | 1 | ≥512 | ≥320 | 0.25 |
| R | S | S | R | R | I | I | R | S | R | S | S | R | R | S | ||
| 19606 | ABC | ≥32 | ≤2 | ≤4 | ≥64 | ≥64 | 16 | 16 | 16 | 1 | 8 | ≤1 | 1 | 128 | ≥320 | 0.125 |
| R | S | S | R | R | I | I | I | S | I | S | S | R | R | S | ||
| 747 | ABC | 16* | ≤2 | ≤4 | ≥64 | ≥64 | 4 | 16 | 4 | ≤0.25 | ≤1 | ≤1 | ≤0.25 | ≥512 | ≤20 | 0.38 |
| R | S | S | R | R | S | I | S | S | S | S | S | R | S | S | ||
| 49137 | ABC | 16* | ≤2 | ≤4 | ≥64 | 32 | 8 | 16 | 4 | ≤0.25 | ≤1 | ≤1 | ≤0.25 | 128 | ≤20 | 0.38 |
| R | S | S | R | R | S | I | S | S | S | S | S | R | S | S | ||
| GNR 3-8E | AU | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 0.094 |
| * | * | * | * | * | * | * | * | * | * | * | * | * | * | S | ||
| GNR 3-3C | AU | 16 | ≤2 | * | ≥64 | ≥64 | 16 | 16 | 8 | ≤0.25 | ≤1 | ≤1 | ≤0.25 | 128 | ≤20 | 0.125 |
| I | S | * | R | R | I | I | S | S | S | S | S | R | S | S | ||
| B2-MRO12 | ABC | ≥32 | 8 | ≥128 | ≥64 | ≥64 | ≥64 | ≥64 | 32 | ≥16 | ≥16 | ≥16 | ≥4 | ≥512 | ≥320 | 0.38 |
| R | S | R | R | R | R | R | R | R | R | R | R | R | R | S | ||
| B1-MRO11 | ABC | ≥32 | 4 | ≥128 | ≥64 | ≥64 | ≥64 | ≥64 | 32 | 2 | ≤1 | ≤1 | ≥4 | ≥512 | ≥320 | 0.5 |
| R | S | R | R | R | R | R | R | S | S | S | R | R | R | S | ||
| WR 1 | ABC | ≥32 | 16 | ≥128 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | ≤1 | 2 | ≥4 | ≥512 | 160 | 0.5 |
| R | I | R | R | R | R | R | R | R | S | S | R | R | R | S | ||
| WR 2 | ABC | ≥32 | 16 | ≥128 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | ≥16 | ≥16 | ≥4 | ≥512 | 160 | 0.75 |
| R | I | R | R | R | R | R | R | R | R | R | R | R | R | S | ||
| WR3 | ABC | ≥32 | 16 | ≥128 | ≥64 | ≥64 | ≥64 | ≥64 | 32 | ≥16 | ≥16 | ≥16 | ≥4 | ≥512 | ≥320 | 0.5 |
| R | I | R | R | R | R | R | R | R | R | R | R | R | R | S | ||
| WR4 | ABC | ≥32 | ≤2 | 8 | ≥64 | ≥64 | 16 | 16 | 16 | 4 | ≤1 | ≤1 | ≥4 | ≥512 | ≤20 | 0.25 |
| R | S | S | R | R | I | I | I | S | S | S | R | R | S | S | ||
| WR5 | ABC | ≥32 | 16 | ≥128 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | 8 | ≥16 | ≥4 | ≥512 | ≥320 | 0.38 |
| R | I | R | R | R | R | R | R | R | I | R | R | R | R | S | ||
| WR6 | ABC | ≥32 | 4 | ≥128 | ≥64 | ≥64 | ≥64 | ≥64 | 8 | ≥16 | 4 | ≤1 | ≥4 | ≥512 | ≤20 | 0.38 |
| R | S | R | R | R | R | R | S | R | S | S | R | R | S | S | ||
| WR7 | ABC | ≥32 | ≥32 | ≥128 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | ≥16 | ≥16 | ≥4 | ≥512 | ≥320 | 0.5 |
| R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | ||
| WR8 | ABC | ≥32 | ≥32 | ≥128 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | ≥16 | ≤1 | ≥4 | ≥512 | ≥320 | 0.5 |
| R | R | R | R | R | R | R | R | R | R | S | R | R | R | S | ||
| WR9 | ABC | ≥32 | 4 | ≥128 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | 4 | 8 | ≥4 | ≥512 | ≥320 | 0.5 |
| R | S | R | R | R | R | R | R | R | S | I | R | R | R | S | ||
| WR10 | ABC | ≥32 | ≥32 | ≥128 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | ≥16 | ≤1 | ≥4 | ≥512 | ≥320 | 0.75 |
| R | R | R | R | R | R | R | R | R | R | S | R | R | R | S | ||
| WR11 | ABC | ≥32 | 16 | ≥128 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | 8 | ≤1 | ≥4 | ≥512 | ≥320 | 0.5 |
| R | I | R | R | R | R | R | R | R | I | S | R | R | R | S | ||
| WR12 | ABC | ≥32 | ≤2 | 16 | ≥64 | ≥64 | 8 | 16 | 8 | 0.5 | ≤1 | ≤1 | ≤0.25 | ≥512 | ≤20 | 0.38 |
| R | S | S | R | R | S | I | S | S | S | S | S | R | S | S | ||
| WR13 | AR | 4 | ≤2 | * | ≥64 | 8 | 2 | 8 | ≤1 | ≤0.25 | ≤1 | ≤1 | ≤0.25 | ≥512 | ≤20 | 0.125 |
| S | S | * | R | S | S | S | S | S | S | S | S | R | S | S | ||
| S1 Mutant | ABC | ≥32 | ≤2 | 8 | ≥64 | ≥64 | 16 | 32 | 16 | 1 | 8 | 2 | 1 | 256 | ≥320 | 0.19 |
| R | S | S | R | R | I | I | I | S | I | S | S | R | R | S | ||
NOTE; Antibiotic interpretation: Sensitive (S), Resistant (R), and Intermediate (I) Antibiotic abbreviations for MICs include: Ampicillin (AMP), ampicillin-sulbactam (SAM), piperacillin-tazobactam (TZP), cefazolin (CFZ), cefoxitin (FOX), ceftazidime (CAZ), ceftriaxone (CRO), cefepime (FEP), meropenem (MEM), gentamicin (GEN), tobramycin (TOB), ciprofloxacin (CIP), nitrofurantoin (NIT), trimethoprim-sulfamethoxazole (SXT), and E-test (ug/mL) for colistin (CST) Bacterial ID abbreviations: Acinetobacter baumannii-calcoaceticus Complex (ABC), Acinetobacter iwoffii (AI), Acinetobacter radioresistens (AR) Acinetobacter ursingii (AU) *Insufficient Growth or interpretation adjustment by software.
Fractional Inhibitory Concentration Index (FICI) determinations of AB569 against Ab isolates
The FICI is the lowest value that is optimal for two compounds to inhibit the growth of a single bacterial isolate. A FICI value can be calculated using the MIC and FIC values for the two components of AB569 (EDTA, A-NO2-) by the following equation (Eq 1):
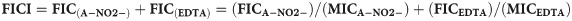
A FICI value of ≤ 0.5 indicates synergy between two compounds, whereas a FICI value of >0.5–4.0 indicates additive results and no interaction in combination. Finally, a FICI value >4 infers an antagonism by two test compounds [51]. The MIC and FICI results for all bacteria used in this study were summarized in Table 4 and Fig 1. Most of the FICI values of ABC strains used in this study were 0.56–2.25 which indicates that the majority are considered “additive” with the exception of two strains in this group (a clinical isolate from bone WR1 (WR1) and A. anitratus ATCC 49137 (49137)) which showed synergistic killing by AB569. The FICI values were 0.5 and 0.31, respectively, the same as A. iwoffii ATCC 9957 (9957), which had an FICI value of 0.375 (Table 4 and Fig 2). The FICI results listed in Table 4 revealed no trends with the percentage of antibiotic resistance and also no differences between Acinetobacter species.

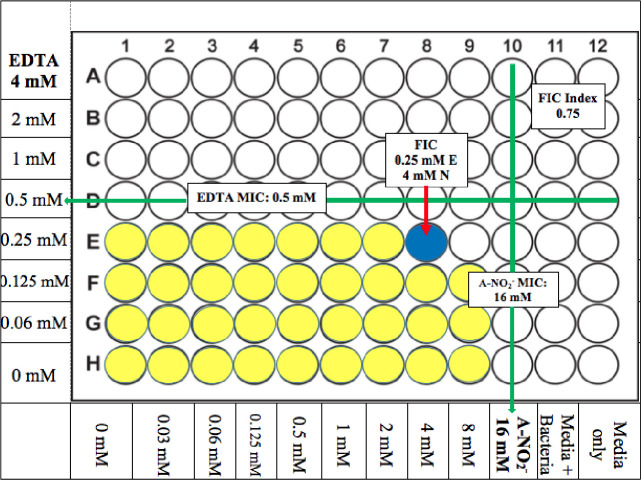
Acinetobacter spp. AB569 Average MIC/FIC.
Average FIC index results, average FIC, and MIC concentrations. Yellow wells indicate typical bacterial growth, blue indicates well that represents FIC with inhibited bacterial growth, and white wells indicated inhibited bacterial growth. EDTA concentrations are noted on the left, A-NO2- concentrations are annotated on the bottom, media only column 12 is listed on the bottom, and media plus bacteria column 11 is listed on the bottom. Each experiment was performed at least 3 times.

| Bacterial Strain | EDTA MIC (mM) | A-NO2- MIC (mM) | FIC index | EDTA FIC (mM) | A-NO2- FIC (mM) | % Antibiotic Resistance | |
|---|---|---|---|---|---|---|---|
| Acinetobacter baumannii calcoaceticus complex (ABC) | |||||||
| WP#2 | 0.25 | 4 | 2.25 | 0.06 | 8 | 60.00 | |
| WR 4 | 1 | 16 | 1 | 0.5 | 8 | 53.33 | |
| WR 6 | 0.5 | 16 | 1 | 0.25 | 8 | 60.00 | |
| S1 Mutant | 0.25 | 16 | 1 | 0.125 | 8 | 60.00 | |
| Ab-Epi | 0.5 | 16 | 0.75 | 0.125 | 8 | 0.00 | |
| AYE | 0.5 | 16 | 0.75 | 0.125 | 8 | 80.00 | |
| WP#1 | 1 | 16 | 0.75 | 0.5 | 4 | 60.00 | |
| 49139 | 0.5 | 16 | 0.75 | 0.125 | 8 | 60.00 | |
| WR 2 | 1 | 16 | 0.75 | 0.5 | 4 | 93.33 | |
| WR 3 | 0.5 | 16 | 0.75 | 0.125 | 8 | 93.33 | |
| WR 5 | 1 | 16 | 0.75 | 0.5 | 4 | 93.33 | |
| WR 7 | 1 | 16 | 0.75 | 0.5 | 4 | 93.33 | |
| WR 10 | 1 | 16 | 0.75 | 0.5 | 4 | 86.67 | |
| WR 11 | 1 | 16 | 0.75 | 0.5 | 4 | 86.67 | |
| 17978 | 1 | 16 | 0.75 | 0.25 | 8 | 40.00 | |
| 747 | 1 | 16 | 0.75 | 0.5 | 4 | 33.33 | |
| WR 12 | 1 | 8 | 0.75 | 0.5 | 2 | 33.33 | |
| 19606 | 0.5 | 16 | 0.625 | 0.06 | 8 | 60.00 | |
| B2-MRO12 | 1 | 16 | 0.625 | 0.125 | 8 | 86.67 | |
| B1-MRO11 | 0.5 | 16 | 0.625 | 0.06 | 8 | 66.67 | |
| WR 8 | 1 | 16 | 0.625 | 0.5 | 2 | 86.67 | |
| WR 9 | 1 | 16 | 0.625 | 0.5 | 2 | 80.00 | |
| ACICU | 1 | 16 | 0.56 | 0.06 | 8 | 86.67 | |
| WR 1 | 0.5 | 16 | 0.5 | 0.125 | 4 | 80.00 | |
| 49137 | 0.5 | 16 | 0.31 | 0.125 | 1 | 33.33 | |
| Acinetobacter iwoffii (Ai) | |||||||
| AI-EPI | 0.25 | 8 | 0.75 | 0.06 | 4 | 0 | |
| 9957 | 0.5 | 16 | 0.375 | 0.06 | 4 | 7.14 | |
| Acinetobacter radioresistens (Ar) | |||||||
| WR 13 | 0.25 | 16 | 0.625 | 0.125 | 2 | 14.29 | |
| Acinetobacter ursingii (Au) | |||||||
| GNR 3-IG | 0.125 | 8 | 1.5 | 0.06 | 8 | 42.86 | |
| GNR 3-3C | 0.25 | 8 | 1.25 | 0.06 | 8 | 42.86 | |
| GNR 3-2G | 0.25 | 8 | 1 | 0.125 | 4 | 28.57 | |
| GNR 3-8E | 0.25 | 8 | 1 | 0.125 | 4 | 0.00 | |
| GNR 3-10J | 0.25 | 16 | 0.75 | 0.06 | 8 | 35.71 | |
| GNR 3-9J | 0.125 | 8 | 0.56 | 0.06 | 0.5 | 42.86 | |
AB569 killing studies
Killing studies (or Time-kill kinetic tests (36)) were performed to plot a time course for Ab AB569 treatment and also to optimize the conditions for qPCR analysis of selected genes that may play a role in AB569 killing of Ab described below. After 8 hr of treating Ab ATCC 19606 with 1 mM EDTA or 32 mM A-NO2- or 1 mM EDTA plus 32 mM A-NO2-, there was no significant killing (Fig 3A). However, our results indicated a bacteriostatic effect on Ab growth by 32 mM A-NO2 or 1 mM EDTA + 32 mM A-NO2- treatments when compared to untreated bacteria (Fig 3A). The killing efficacy of either EDTA, A-NO2- or both was further investigation by extending the incubation time to 48 hr. Our CFU analyses revealed bacterial killing of ~2 log by 1.5 mM EDTA, ~5 log by 30 mM A-NO2- and ~9 log by 1.5 mM EDTA + 30 mM A-NO2-, respectively, when compared to untreated bacteria. These results clearly indicate that EDTA plus A-NO2- (AB569) has far greater bactericidal activity than either the EDTA or A-NO2- components alone (Fig 3B).

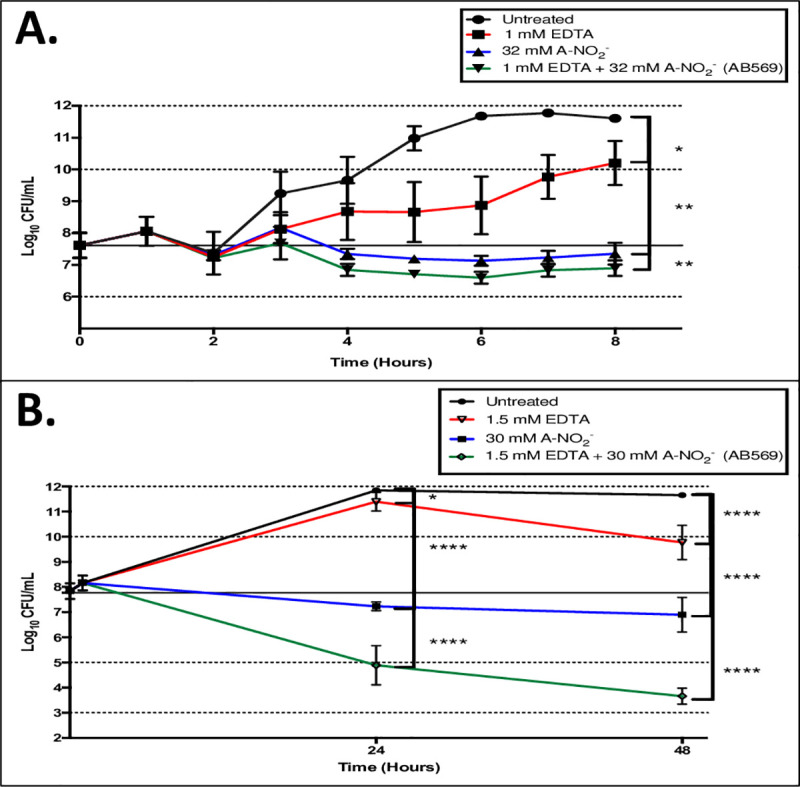
AB569 killing study Ab 19606.
CFU counts transformed to Log10 and plotted in semi-log scale. Black line represents untreated control values, red lines are EDTA only samples, blue lines are A-NO2- only, and green lines are EDTA + A-NO2- (AB569) combination. (A). Log10 CFU/ml counts with 8-hr exposure time and treated with 1 mM EDTA only, 32 mM A-NO2- only, and 1 mM EDTA + 32 mM A-NO2- AB569 combination, n = 3. (B). Log10 CFU/ml counts with 48-hr exposure time and treated with 1.5 mM EDTA, 30 mM A-NO2-, or 1.5 mM EDTA + 30 mM A-NO2- AB569. Each experiment was performed at least 3 times.
Quantitative PCR (qPCR) of Ab gene expression with AB569 treatment
We next delved into the preliminary mechanistic basis underlying AB569-mediated killing of Ab using qPCR after sublethal exposure to the drug. The genes that we predicted to be involved in this mechanism (refer to Table 1) were those that have orthologs in Pseudomonas aeruginosa (Pa), another combat wound/blast pathogen [1, 52], that we have recently shown are modulated by AB569 [32]. As both Pa and Ab are Gram-negative bacteria that cause highly problematic biofilm-based infections [53, 54], we hypothesized that the response to AB569 treatment potentially shared some common gene expression patterns. As above, the qPCR experiments were also performed using bacteria treated with EDTA, A-NO2- or AB569. Siderophore biosynthesis non-ribosomal peptide synthetase module (SBNRPSM) and siderophore biosynthesis protein monooxygenase (SBPM) genes were first tested to evaluate the Ab siderophore response to the aforementioned treatments as both the EDTA and A-NO2- components of AB569 have been recently shown to affect iron metabolism/uptake machinery in Pa [32]. Small subunit (SSU) ribosomal protein genes s11p, s2p and s7p as well as large subunit (LSU) ribosomal protein genes L19p, L4p, L28p, and translation elongation factor Ts (EFTs) genes were also selected to assess the response of these genes to the aforementioned compounds. Transcription of each of the aforementioned genes was dramatically down-regulated by EDTA, A-NO2-, and especially by AB569. The ATP synthase delta chain (atpD) and pyruvate dehydrogenase E1 component (PDHcE1) genes were selected to represent the ATP synthesis and TCA cycle transcriptional responses and these, too, showed a similar pattern of down-regulation. In contrast to the genes that were down-regulated by either treatment (Fig 4B and 4C), we observed a 22.2- and 17.6-fold up-regulation of the acinetobactin siderophore related genes SBNRPSM and SBPM gene transcription, respectively, in the EDTA sample only. SBNRPSM gene expression in the sample treated with AB569, however, was down-regulated 0.7-fold, a 0.3-fold lower expression level as compared to the untreated controls, but 0.63-fold higher than the A-NO2- sample. SBPM gene expression in the AB569 sample was down-regulated 0.23-fold, 0.77-fold lower compared to the untreated control but 0.17-fold higher than the A-NO2- sample. The EDTA samples were on average 0.16-fold higher than those of the A-NO2- samples and AB569 treated samples in expression of s11p, s2p, s7p, L4p, and L28p. There was an overall downregulation compared to the untreated control with the s11p, s2p and s7p as well as L4p, and L28p genes. The remainder of the genes screened demonstrated decreased expression compared to the untreated control. No differences in the expression of L19p, atpD, EFTs, and PDHcE1 genes were observed with either treatment.

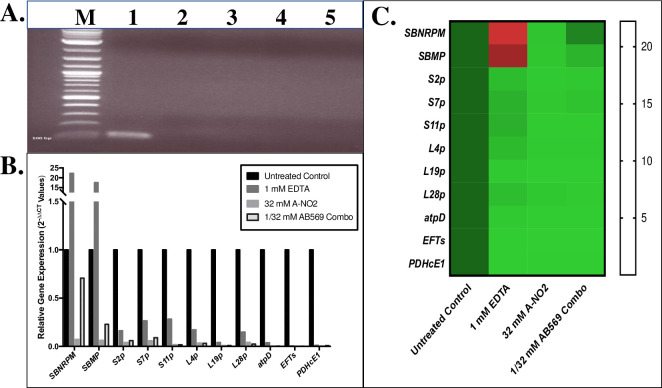
Quantitative PCR analysis of predicted genes affected by treatment of Ab with AB569.
Total RNA Isolation following treatment with EDTA, A-NO2-, or AB569 combination (n = 3). (A). Electrophoresis of purified RNA prior to RT-PCR, lane M is the ladder marker, lane 1 is genome control, lane 2 untreated control, lane 3 is sample treated with 1 mM EDTA, lane 4 is sample treated with 32 mM A-NO2-, and lane 5 is sample treated with 1 mM EDTA + 32 mM A-NO2- AB569 combination, note the absence of dsDNA in samples compared to the positive genome control in the first lane. (B). Ab relative gene expression (2-ΔΔCt Values). Black column represents untreated control, EDTA only is dark grey, A-NO2- only is light grey, and AB569 combination is the black outlined column. Genes tested are listed on the bottom and log scale 2-ΔΔCt values are listed on the left, n = 3. (C). Heat Map of qPCR 2-ΔΔCT values. The genes tested are listed on the left side and treatment conditions are noted on the bottom of the graph. Gene expression is indicated by green for down regulation, red for up regulation, and black for middle range of gene expression (adjusted for contrast, n = 3).
Effect of Ab569 on Ab biofilm formation and dispersion
Ab is an established biofilm-forming organism, a feature which also is attributed to its enhanced virulence properties [13]. The ability of Ab to form biofilms points to several phenotypic advantages for Ab pathogenesis. The production of extrapolymeric substance (EPS) is protective, as antibiotics have to penetrate the EPS to be effectively bactericidal [55]. In addition, the presence of EPS promotes increased horizontal gene transfer and antibiotic resistance in biofilm communities [56]. Therefore, the FICI of AB569 on Ab biofilm formation and also the effect of AB569 on biofilm dispersion was next investigated. The FIC of AB569 that inhibited Ab ATCC 19606 biofilm formation was 0.25 mM EDTA + 4 mM A-NO2- while the MIC of EDTA and A-NO2- alone to inhibit bacterial biofilm formation were 0.5 mM and 16 mM, respectively (Fig 5). In addition, the effect of AB569 on bacteria biofilm dispersion was also investigated. Our, results showed a 36% reduced bacterial biomass compared to untreated biofilms after treatment for 24 hr with AB569 at concentrations of 2 mM EDTA + 16 mM A-NO2-.

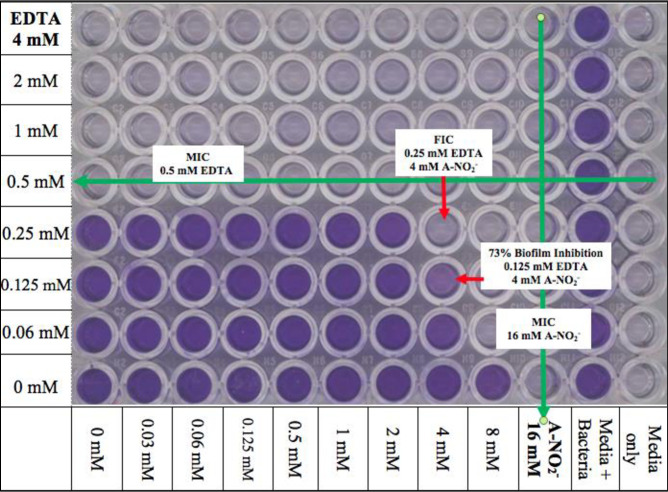
Biofilm inhibition.
Scanned 96-well plate following biofilm inhibition assay. EDTA row values are indicated on the left and A-NO2- column values are indicated on the bottom. Column 12 was the media only or background control and column 11 was media + bacteria only or the positive control. MIC values are marked with a green arrow. FIC concentrations and percent biofilm inhibition are annotated with a red arrow indicating the FIC well (n = 3).
Sytox Orange dead cell staining after AB569 treatment of HDFa cells
To address the potential utility of using AB569 for the treatment of Ab infections in humans, we felt strongly that it was first critical to perform some fundamental cytotoxicity studies on skin cells. Toward this end, human dermal fibroblasts (HDFa) were used to assess the potential utility of AB569 for future wound/burn/blast treatment regiments in planned human clinical trials. Individually, it is well established that both NO2- and EDTA have a long history of clinical use [57, 58]. A-NO2- is used topically to treat skin infections [59], as a lung treatment for asthma [57], vasodilator [60], and ingested for its gastric anti-microbial benefits [57]. EDTA is used in chelation therapy to treat coronary artery disease [61] and in heavy metal toxicity (e.g., lead) or iron chelation treatments [11]. Predetermined bactericidal concentrations of EDTA, A-NO2- and AB569 were used to treat HDFa cells and viability was assessed with the Sytox Orange dead cell stain and analyzed by FACS analysis. EDTA, A-NO2- and AB569 treatment was applied at individual and combined concentrations of 1 mM to 5 mM EDTA and 64, 128, or 256 mM A-NO2-. Note that in Fig 6, each bar represents an individual treatment condition listed on the x-axis with the % cell death indicated on the y-axis. Media only and media (pH 6.5) controls were used to compare results and set the toxicity cut off at 32%, indicated by the black line in Fig 6. One Way ANOVA with Dunnett’s multiple comparisons of Sytox Orange dead cell FACs analysis revealed statistically significant treatment conditions when compared to the media only control. The combined concentrations of 3 mM EDTA and 64 mM A-NO2- represents the highest combined concentration with insignificant cell death at a mean of ~32%. DMEM 1x supplemented media acidified to a pH of 6.5 control alone resulted in ~29% cell death.

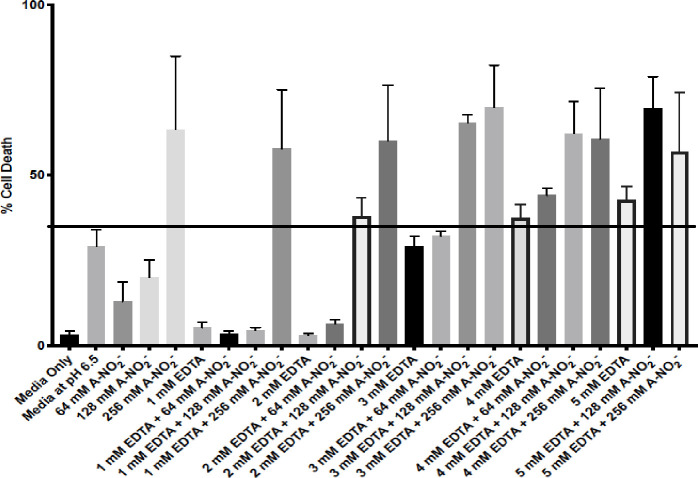
HDFa Sytox orange dead cell stain.
HDFa cells treated with various concentrations of EDTA, A-NO2- and AB569 combinations listed on the bottom axis. Each column represents a unique treatment condition. Percent cell death is listed on the left. Line indicates significant cell death at 37% compared to media only control (One way ANOVA F = 6.55, P Value = <0.0001).
Discussion
This study was initiated to determine the potential utility of using a novel two-component biocide, AB569, consisting of A-NO2- and EDTA at a pH of 6.5 to 5.5. Our primary analysis used the classical checkerboard technique [62] to measure the fractional inhibitory concentration (FIC) testing results for EDTA, A-NO2-, and AB569. The average FIC index of 0.75 revealed a non-synergistic or additive effect on Ab growth. Two of the reference strains demonstrated FIC values less than 0.5, an indication of synergy. These included Acinetobacter spp genomospecies 9, ATCC 9957, A. anitratus ATCC 49137 and Ab 19606T, respectively. All but one of the clinical strains demonstrated a lack of synergy; WR-1 revealed a FIC value of 0.5 with 80% antibiotic resistance. This lack of synergy differentiates Ab from the AB569 response in other Gram-negative bacteria including Pa, E. coli, K. pneumoniae and P. mirabilis [32]. Acinetobacter spp genomospecies 9, ATCC 9957 and A. anitratus ATCC 49137 were only 7.14% and 33.33% resistant to tested antibiotics when compared to the pronounced antibiotic resistance in the other isolates used in this study (Table 2).
Killing studies with AB569 demonstrated bacteriostatic activity with A-NO2- only, while EDTA only treated samples showing little to no effect on planktonic growth with an 8 hr exposure time. The drop-in growth between the 3 and 4 hr time points in the A-NO2- and AB569 combination samples, Fig 3A, demonstrates growth inhibition at shorter treatment exposure time. The inability of EDTA to kill planktonic cells was demonstrated by Lee et.al. [13] in a study that demonstrated that it played a role in Ab biofilm reduction. Inhibition of Ab growth with exposure to NO2- was also demonstrated in a study by Weon et. al. [63] that showed the effect of NO2- on the phosphate removal capacity of Ab in wastewater treatment. Specifically, inhibition is observed in less than 8 h of exposure. This study revealed that AB569 bactericidal activity in Ab treatment requires extended treatment times at concentrations that are not toxic to cells, as there was a greater than 2 log reduction in bacterial growth after 24 h of treatment.
Biofilm formation by Ab clearly demonstrated inhibition upon treatment with AB569. Our results demonstrated that AB569 prevented biofilm formation at treatment levels consistent with FIC results, 0.125 mM EDTA and 4 mM A-NO2- levels for biofilm inhibition compared to the FIC results at 0.06 mM EDTA and 8 mM A-NO2-. Biofilm dispersion was at an average of 34% reduction, indicating that the tested concentrations reduced pre-formed biofilm in vitro. These two results together indicate that once doses are optimized for in vivo use (requiring costly animal toxicology studies first), AB569 may be useful as a preventative therapy for Ab biofilm infections or to coat surfaces used in treatment, such as indwelling catheters, tubing, ventilators or prosthetic devices.
To attempt to address the preliminary mechanism of AB569 bactericidal activity, we found significant upregulation of the siderophore-related biosynthetic genes SBNRPSM and SBPM genes in the presence of EDTA. This event may not play a direct role in killing Ab as the EDTA only samples did not demonstrate phenotypic killing of planktonic bacteria during AB569 killing studies. AB569, however, did demonstrate an average of 75% biofilm inhibition. Lee et.al. [13] validated the role of EDTA in reducing Ab biofilm formation along with McConnell et.al. [15] indicating that iron influences the amount of biofilm formed by Ab. This may explain the role of EDTA in the treatment of Ab with AB569, as increased siderophore activity indicates lower than normal iron levels during growth [14]. There is a 14-fold decrease in A-NO2- only SBNRPSM gene transcription compared to the SBNRPSM gene transcription in the AB569 combination. Although, both were down-regulated compared to the untreated control. SBPM gene activity showed a similar trend to SBNRPSM transcription. In addition, the loss of response for siderophore production in AB569 (which still includes EDTA) could indicate a potentially important point for mechanistic studies as to why the combination is able to kill bacteria, even when not synergistic. Overall there was down-regulation with the other genes tested compared to untreated controls indicating lower ribosomal transcription levels.
The genes selected for qPCR analysis in this study were based on previous studies with AB569 and up- and down-regulated genes in another prominent wound/blast pathogen, Pa. After reviewing the expression patterns in both organisms with one exception, all the genes tested in Pa and Ab showed a predicted decreased gene expression when related to untreated control with no significant difference in gene expression between individually treated EDTA and A-NO2- compared to the AB569 combination (Fig 7 [32]). With the exception of poxB in Pa, gene expression upon exposure to AB569 in Pa poxB was at ~0.9-fold while EDTA was at ~1.0-fold and A-NO2- was at 0.39-fold expression. Pa poxB expression was decreased compared to untreated controls and only increased in the EDTA sample by 0.09-fold. The Pa gene poxB is pyruvate dehydrogenase (cytochrome b) compared to PDHcE1 pyruvate dehydrogenase E1 component in Ab that enzymatically catalyzes a mechanism of activation of the required co-factor, thiamine pyrophosphate coenzyme.

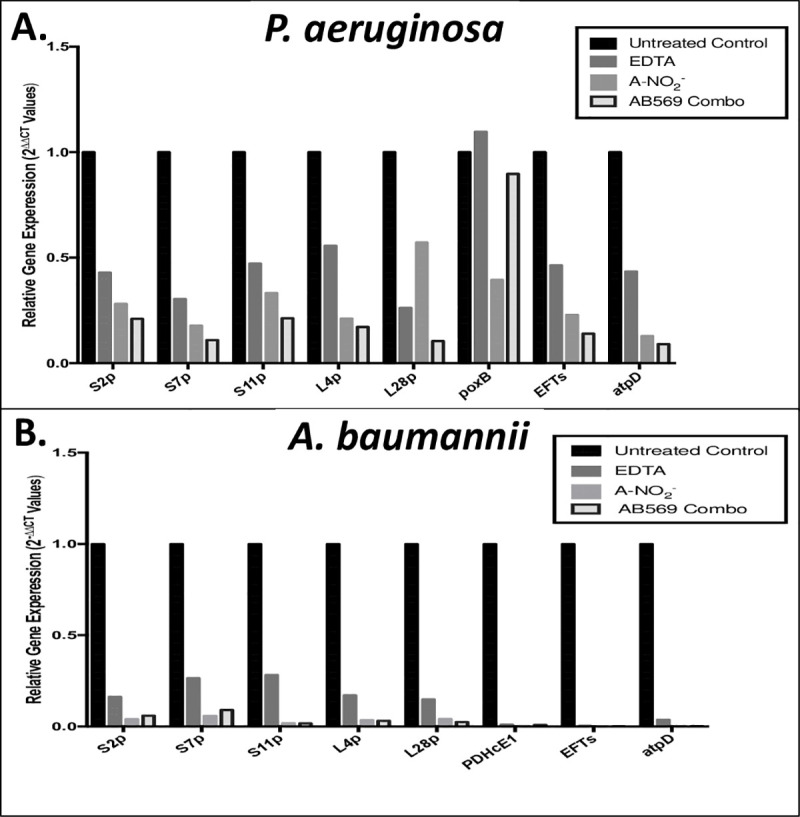
Comparison of Pa and Ab qPCR, gene expression following treatment with EDTA, A-NO2- and AB569 combination.
The black column represents the untreated control, EDTA is in dark grey, A-NO2- is in light grey, and the combination is the black outlined column 2-ΔΔCt values. Genes tested are listed on the x-axis and log scale values are listed on the y-axis. (A). Pa relative gene expression (2-ΔΔCt Values). (B). Ab relative gene expression (2-ΔΔCt Values).
Toxicity studies with HDFa cells established a workable upper limit for cell toxicity at 3 mM EDTA and 64 mM A-NO2-. This upper range is within the average FIC values for 29 Ab and Acinetobacter spp. isolates tested at 0.25 mM EDTA and 4 mM A-NO2-. However, the media at pH 6.5 control demonstrated ~29% cell death while the media only control demonstrated 3.3% cell death. This notable difference in % cell death by acidifying the cell culture media to a pH of 6.5 (necessary for optimal bactericidal activity) may be due to the use of phosphate buffer in the treatment, bicarbonate buffers in the DMEM 1x media, or the presence of 5% CO2 during incubation periods, as cell culture conditions are tailored to maintain an optimal pH of 7.4 (blood pH). However, in previous studies with A-NO2-, pH levels of 6.5 were tolerated in mice using a chronic lung infection model [21]. Treatment with AB569 in wound, burns, blast or ventilator-related infections and other applicable tissues is a viable therapeutic option as the treatment ranges are below the toxicity ranges for HDFa cells. However, it should be clarified that a topical agent, whether in solution or cream form, should be buffered to a pH of 6.5 or 5.5 for optimal antimicrobial activity [32].
Overall combination therapy, such as AB569, typically can decrease antibiotic resistance and combining bactericidal agents with bacteriostatic agents can often be antagonistic [64]. In this study samples treated with EDTA only failed to demonstrate killing of planktonic bacteria. However, significant growth inhibition or killing was demonstrated with A-NO2- and AB569 combination, respectively, suggesting that, in the case of Ab treatment AB569, is not antagonistic. The definition of bactericidal versus bacteriostatic in some cases is dependent on multiple factors to include bacterial load, test duration, bacterial targets and reduction in bacteria. From a clinical perspective, antibiotics are known to possess both bactericidal and bacteriostatic properties [65]. The AB569 combination has been recently shown to act synergistically with many Gram-negative pathogens [32] including Pa, Salmonella typhimurium, E. coli, Klebsiella pneumoniae and Proteus mirabilis [32]. As such, the potential benefit to the early growth inhibition with Ab is that in mixed infections it could prevent Ab from gaining a growth advantage while killing other pathogens.
A final and very important issue that is critical to the understanding of the mechanism underlying the bactericidal activity of AB569 against Ab are through extensive and costly mechanistic studies that are beyond the scope of this work. This would include those involving sophisticated (i) chemistry (biophysical/biochemical), (ii) RNA-seq/Tn-seq bioinformatics analyses, and (iii) potential but highly doubtful mechanisms of resistance. We offer some insight as to the possible mechanism based upon only recent discoveries in P. aeruginosa grown under strict anaerobic conditions, reminiscent of late-stage, chronic CF airway disease [32]. Although the mechanism of action of AB569 remains a mystery, it is clear that negative pleiotropic effects are in play. AB569 caused an overwhelming transcriptional dysregulation in PA, leading to a loss of vital cellular functions, events that are likely similar in Ab. Our current knowledge of the chemistry of A-NO2- plus EDTA points to increasing the amount of NO, via both generation and protection of dinitrosyliron complexes (DNICs) and S-nitrosylated proteins. RNA-Seq analyses of P. aeruginosa with AB569 resulted in a cataclysmic loss of essential core pathways that include those involved in the biosynthesis of DNA, RNA, protein, and ATP biosynthesis, as well as Type III secretion and iron metabolism.
Future considerations include expanding killing studies to test higher treatment levels, as toxicity levels were found 3 mM EDTA and 64 mM A-NO2- in HDFa cells. Additional human cell lines as well as toxicology studies in animals is warranted. Optimizing treatment conditions and ultimately performing RNA-seq/proteomic analyses would aid in the determination of the mechanisms of the bactericidal action of AB569. The aforementioned data, coupled with an analysis of downregulated genes that are deemed “essential” in PA [32], would increase dramatically the overall effectiveness of AB569 in clinical settings. Given the potential role of EDTA in this study with biofilm inhibition and dispersion of mature biofilms, the Ab siderophore acinetobactin [66] was predicted to greatly add clues to one critical component of fundamental bacterial metabolism, iron. Additional toxicity studies with keratinocytes, given the frequency of blast wounds infections associated with Ab, would support the data from this study and would. In addition, investigating the cell culture conditions to improve the percentage of cell death in the media at pH 6.5 control may provide more consistent results in future cytotoxicity assays. Finally, animal studies to test AB569 in a possible wound treatment or flush solution model will likely be considered a positive for novel treatments against naturally antibiotic-resistant bacteria that include Ab.
Acknowledgements
We thank the Laboratory Officers at Travis Air Force Base for assistance with the first author’s package selection into AFIT. Dr. Luis Actis (Miami University provided several Acinetobacter spp. strains [67]). Jessica Johnson and Michael Roper (88 DTS/SGQC, Wright Patterson AFB, Clinical Microbiology Department) provide several clinical Acinetobacter spp. strains. Terri Kerivan and Major Lisa Edwards at the US Air Force School of Aerospace Medicine/Public Health Epidemiology Laboratory (USAFSAM/PHE) Wright Patterson AFB also provided additional Acinetobacter spp. strains. Dr. Joel Mortensen (Division of Pathology and Laboratory Medicine at Cincinnati Children’s Hospital Medical Center (CCHMC)) shared Acinetobacter spp. strains and Ashley Marshall in his lab performed sensitivity and bacterial identification testing on more than 30 isolates. Major Edwin Kamau and the team at Walter Reed National Military Medical Center (WRNMMC) submitted several key clinical strains of Acinetobacter spp. isolated from Iraq/Afghanistan battle wounds, pending patient history confirmation, for this study.
Disclaimer
The views expressed in this article are those of the author and do not reflect the official policy or position of the United States Air Force, Department of Defense, or the U.S. Government.
References
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
 AB569, a non-toxic combination of acidified nitrite and EDTA, is effective at killing the notorious Iraq/Afghanistan combat wound pathogens, multi-drug resistant Acinetobacter baumannii and Acinetobacter spp.
AB569, a non-toxic combination of acidified nitrite and EDTA, is effective at killing the notorious Iraq/Afghanistan combat wound pathogens, multi-drug resistant Acinetobacter baumannii and Acinetobacter spp.
