
- Altmetric
G-quadruplexes are noncanonical nucleic acid structures formed by stacked guanine tetrads. They are capable of a range of functions and thought to play widespread biological roles. This diversity raises an important question: what determines the biochemical specificity of G-quadruplex structures? The answer is particularly important from the perspective of biological regulation because genomes can contain hundreds of thousands of G-quadruplexes with a range of functions. Here we analyze the specificity of each sequence in a 496-member library of variants of a reference G-quadruplex with respect to five functions. Our analysis shows that the sequence requirements of G-quadruplexes with these functions are different from one another, with some mutations altering biochemical specificity by orders of magnitude. Mutations in tetrads have larger effects than mutations in loops, and changes in specificity are correlated with changes in multimeric state. To complement our biochemical data we determined the solution structure of a monomeric G-quadruplex from the library. The stacked and accessible tetrads rationalize why monomers tend to promote a model peroxidase reaction and generate fluorescence. Our experiments support a model in which the sequence requirements of G-quadruplexes with different functions are overlapping but distinct. This has implications for biological regulation, bioinformatics, and drug design.
INTRODUCTION
In addition to the familiar double helix, nucleic acids can adopt a variety of noncanonical structures (1). One of the most well-studied examples is the G-quadruplex, which is a four-stranded structure typically formed by stacked guanine tetrads (2,3). A remarkable feature of G-quadruplexes is their functional diversity. These structures interact with more than 30 proteins (4,5) and bind a variety of small-molecule ligands (6), including some which are biologically important (7–10). They catalyze various peroxidase reactions in the presence of a hemin cofactor (11,12) and form structures that are intrinsically fluorescent (13–15). G-quadruplexes have also been proposed to play widespread biological roles, especially as regulatory elements that modulate processes such as transcription, DNA replication, telomere function, and translation (16–19). This functional diversity raises an important question: what factors determine the biochemical specificity of G-quadruplex structures? The answer to this question is particularly important from the perspective of biological regulation. The genomes of higher eukaryotes typically contain hundreds of thousands of G-quadruplexes with a range of functions (20–22), and the cellular machinery must have a way of distinguishing them. It is also relevant for bioinformatics. Algorithms currently used to identify G-quadruplexes in genomes (20–21) cannot in general distinguish structurally different classes of G-quadruplexes, but if such structures have distinct functional properties they should be analyzed separately. Understanding factors that modulate G-quadruplex biochemical specificity also has implications for drug design. Ligands used as lead compounds in G-quadruplex-based drug discovery efforts are typically generated by targeting a single G-quadruplex, such as the Pu22 sequence in the c-MYC promoter (23). Such ligands typically bind a range of G-quadruplexes in addition to the target sequence, and developing new approaches to generate more specific ligands is a significant challenge in the field.
In principle, the biological mechanism by which G-quadruplexes attain biochemical specificity could be unrelated to the sequence of the G-quadruplex. For example, specificity could be determined entirely by genomic context or the action of cellular proteins. However, the simplest mechanism is that at least some specificity is determined by the sequence of the G-quadruplex itself. This could be achieved by mutations that affect any structural feature which differs among G-quadruplexes. Examples include strand orientation (parallel, mixed, or antiparallel), guanine glycosidic angle (anti or syn), loop configuration (propeller, lateral or diagonal), groove width, and accessibility of terminal tetrads (24–28). Experimental studies have confirmed that in some cases such features can modulate G-quadruplex specificity. For example, ligands which bind to G-quadruplexes by stacking on terminal tetrads (6) can have significantly different affinities for the 5′ and 3′ tetrad due to differences in the structural context of these tetrads (29–31). Loops can also modulate G-quadruplex binding affinity and specificity (32–34). Diagonal loops can help to form binding sites for ligands that stack on terminal tetrads (34), while propeller loops can form smaller pockets into which the side chains of ligands can bind (33). In some cases, the basis for these effects can be rationalized from the perspective of high-resolution structures. For example, the loops of a telomeric G-quadruplex form water-filled channels that interact with the side chains of the ligand BRACO-19, and the affinity of the G-quadruplex for this ligand progressively decreases when these side chains are extended with hydrophobic groups (33).
Despite considerable progress in understanding how model G-quadruplex structures interact with different types of ligands, little is known about how systematic changes in the sequence of a G-quadruplex can affect its biochemical specificity. To explore this question, we developed an experimental system to evaluate the effects of mutations on G-quadruplex biochemical specificity using a 496-member library of variants of a monomeric reference G-quadruplex. By screening this library for a series of biochemical functions associated with G-quadruplex structures, we reasoned that it would be possible to both assess the extent to which the specificity of a G-quadruplex can be modulated by changing its sequence and explore the mechanisms by which such specificity is achieved. Here we report the results of a comparative study of the ability of these library members to bind GTP (35–36), promote a model peroxidase reaction using hemin as a cofactor and ABTS as a substrate (35), generate fluorescence (15), form dimers (37), and form tetramers (37). Our results indicate that the sequence requirements of G-quadruplexes with these functions are different from one another. Mutations in the central tetrad of the monomeric reference G-quadruplex are more important than mutations in loops with respect to both biochemical function and specificity, probably because they are more likely to change the multimeric state of the G-quadruplex. Consistent with this idea, changes in biochemical specificity are correlated with changes in multimeric state. Fluorescence quenching experiments show that multimerization alters the functional properties of the 5′ and 3′ ends of these G-quadruplexes, highlighting the role of terminal nucleotides in modulating G-quadruplex specificity. To complement these biochemical studies, we determined the solution structure of a monomeric G-quadruplex from the library. The stacked and accessible tetrads in this structure help to explain why monomers in the library tend to promote the model peroxidase reaction and generate fluorescence efficiently. Taken together, these experiments provide a comprehensive view of the factors that modulate G-quadruplex biochemical specificity, and support a model in which the sequence requirements of G-quadruplexes with different functions are overlapping but distinct. This model has implications for biological regulation, bioinformatics, and drug design.
MATERIALS AND METHODS
Reagents
Desalted DNA oligonucleotides were purchased from Sigma. Oligonucleotides were typically resuspended in Milli-Q water at a concentration of 100–200 μM and used without additional purification in biochemical assays. A previous study showed that additional purification did not change the functional properties of these G-quadruplexes (15). Other reagents were purchased from Sigma. For more details see references 15, 35 and 37.
Biochemical assays
All experiments described in Figure 2 and analyzed in Figures 3–5 were performed at a DNA concentration of 10 μM. Since G-quadruplex multimerization is concentration-dependent, this ensured that the multimeric states of these G-quadruplexes was the same in assays for different biochemical functions. Assays for GTP binding activity, dimer formation, fluorescence, and tetramer formation were performed in a buffer containing 200 mM KCl, 1 mM MgCl2, 20 mM HEPES pH 7.1, and (for the GTP binding assay) 10 nM of 32P-γ-GTP. Assays for peroxidase activity were performed in a buffer containing 200 mM KCl, 1 mM MgCl2, 20 mM HEPES pH 8, 0.05% Triton X-100, 10% DMSO, 0.5 μM hemin, 5 mM ABTS and 600 μM hydrogen peroxide. For GTP-binding activity, the average standard deviation of a measurement was 40 ± 20% of the average value (35). For peroxidase activity, the average standard deviation of a measurement was 11 ± 15% of the average value (35). For fluorescence, the average standard deviation of a measurement was 6 ± 4% of the average value (15). For dimerization, the average standard deviation of a measurement was 1.3 ± 1.2% of the average value (this study). For tetramerization, the average standard deviation of a measurement was 16 ± 16% of the average value (this study). Protocols for these assays are described in more detail in (35) (GTP-binding and peroxidase assay), (37) (dimer and tetramer formation) and (15) (fluorescence assay).
Fluorescence quenching assays
In a typical binding assay, a 100 μM stock solution (stored at -20°C) containing DNA labeled with Cy5 via a phosphate group at its 5′ or 3′ end (Sigma) (Supplementary Figure S1) and a 100 μM stock solution (stored at –20°C) containing unlabeled DNA (Sigma) were thawed at room temperature. Dilutions were prepared by mixing 2.5 μl of 100 μM Cy5 labeled DNA with 250 μl of either 0 or 100 μM unlabeled DNA and 1.3725 ml of Milli-Q water. To fold the oligo, the solution was split into 65 μl aliquots. These were heated at 65°C for 5 min and cooled at room temperature for 5 min. Aliquots were then mixed with 35 μl of a mix containing 25 μl of 4× peroxidase buffer (4 mM MgCl2, 800 mM KCl, 80 mM HEPES pH 8, 0.2% Triton X-100) and 10 μl of hemin stock solutions at various concentrations in DMSO. After incubating for 30 min at room temperature in the dark, the solution was transferred to a standard black 96-well plate with a flat bottom (Corning; Sigma catalog number CLS3916). Fluorescence was measured at 670 nm using a plate reader (Tecan Spark). Final concentrations were 100 nM labeled DNA and either 0 μM or 10 μM unlabeled DNA in a buffer containing 200 mM KCl, 1 mM MgCl2, 20 mM HEPES pH 8, 0.05% Triton X-100, 10% DMSO and various concentrations of hemin. Optical emission spectrometry with inductively coupled plasma (ICP-OES) experiments confirmed that hemin was soluble at the highest concentrations used in these titrations. Binding activity was expressed relative to that of the no-hemin (DMSO) control, which was measured in every experiment. Data were fit with the program Gnuplot using a one-site binding model:
where Kd is the dissociation constant, [S] is the hemin concentration and Fmax is the maximum percent of bound DNA. For fitting, Fmax was considered to be between 90% and 110% to reflect experimental errors in determining maximal fluorescence. If there is no standard deviation for Fmax it means that the fit reached the bound. Samples were measured in duplicate, and reported values are averages of three independent experimentsNMR experiments
Sample preparation
Unlabeled DNA was purchased from Sigma and site-specifically labeled DNA was purchased from IDT. DNA was resuspended in Milli-Q water, heated at 65°C for 5 min, cooled at room temperature for 5 min, and folded by adding buffer. Final concentrations were 10 μM DNA, 20 mM Tris, pH 7.5, 200 mM KCl and 1 mM MgCl2. Labeled samples were then concentrated using Amicon Centrifugal filter units (cutoff 3 kDa) to a final concentration of 100 μM. Unlabeled sample was prepared as 70 ml of a 10 μM DNA solution which was further purified using ion-exchange chromatography MonoQ (1 ml volume, GE Healthcare) using a linear gradient from 0.2 to 1 M KCl. Eluted fractions were pooled, diluted to restore the KCl concentration to 200 mM and concentrated using Amicon Centrifugal filter units (cutoff 3 kDa). The buffer was also changed to d-Tris during the concentration. The final DNA concentration was 1.7 mM in a volume of 350 μl.
NMR measurements
NMR experiments were performed on a Bruker Avance III HD 850 MHz system equipped with an inverse triple resonance cryo-probe. Samples contained either 90% H2O and 10% D2O or 100% D2O. A trace amount of DSS was added as a frequency standard. Assignment of the imino protons of guanine residues was obtained using 1D SOFAST experiments (38) (8% 15N labeling), which filter out proton signals not coupled to 15N. Assignment of H8 protons was partially obtained from an HMBC spectrum correlating H1 and H8 resonances (39). Spectral assignments were made using NOESY and TOCSY spectra at various temperatures and mixing times. Spectral analyses were performed using TOPSPIN (Bruker) and Sparky (40–41).
Structure calculations
NOE distance restraints were obtained from a NOESY spectrum acquired in H2O at 200 ms. For non-exchangeable protons, the peaks were classified as strong, medium, or weak corresponding to distance restraints of 2.7 ± 0.8, 3.8 ± 0.9, or 5.5 ± 1.7 Å, respectively. Distances from exchangeable protons were classified as strong, medium, or weak corresponding to distance restraints of 3.6 ± 0.9, 4.8 ± 1.2 or 5.5 ± 1.7 Å, respectively.
Dihedral, hydrogen bond and planarity restraints
Dihedral angle restraints were imposed to the dihedral angle formed by O4′–C1′–N9–C4 of guanine residues, which was restricted to an angle of 240 ± 70°. Hoogsteen hydrogen bonds between guanines were restrained using H21–N7, N2–N7, H1–O6 and N1–O6 distances, which were set to 2.0 ± 0.2, 2.9 ± 0.3, 2.0 ± 0.2 and 2.9 ± 0.3 Å, respectively. Planarity restraints were used for the G1–G5–G10–G14, G2–G6–G11–G15 and G3–G7–G12–G16 tetrads.
Distance geometry simulated annealing
An initial extended conformation was generated using the XPLOR-NIH program (42). The system was then subjected to distance geometry simulated annealing by incorporating distance, dihedral, hydrogen bond, planarity and repulsion restraints. One hundred structures were generated and subjected to further refinement.
Distance-restrained molecular dynamics refinement
The 100 structures obtained from the simulated annealing step were refined with a distance-restrained molecular dynamics protocol incorporating all distance restraints. The system was heated from 300 to 1000 K in 14 ps and allowed to equilibrate for 6 ps, during which force constants for the distance restraints were kept at 2 kcal mol−1 Å−2. The force constants for non-exchangeable proton and exchangeable proton restraints were then increased to 16 and 8 kcal mol−1 Å−2 respectively in 20 ps before another equilibration at 1000 K for 50 ps. Next, the system was cooled down to 300 K in 42 ps, after which an equilibration was performed for 18 ps. Coordinates of the molecule were saved every 0.5 ps during the last 10.0 ps and averaged. The average structure obtained was then subjected to minimization until the energy gradient was less than 0.1 kcal mol−1. Dihedral (50 kcal mol−1 rad−2) and planarity (1 kcal mol−1 Å−2 for tetrads) restraints were maintained throughout the course of the refinement. The twenty lowest-energy structures were generated. See Supplementary Table S1 for a summary of NMR statistics.
1D 1H spectra of monomeric library members
For the experiments described in Supplementary Figure S17, 80 μl of a 200 μM solution of the G-quadruplex in Milli-Q water was heated at 65°C for 5 min and cooled at room temperature for 5 min. A solution containing 40 μl of 4× buffer (800 mM KCl, 4 mM MgCl2, 80 mM d-Tris, pH 7.1), 24 μl of Milli-Q water, and 16 μl of deuterated water was then added. Final concentrations were 100 μM DNA, 200 mM KCl, 1 mM MgCl2, 20 mM d-Tris, pH 7.1, and 10% D2O in a volume of 160 μl. After incubating for 30 min at room temperature, samples were heated in a thermal cycler at 97°C for 30 min, 95°C for 1 min and cooled to 25°C at a rate of 1°C/min. 1D 1H spectra were measured on a Bruker Avance III HD 850 MHz spectrometer. This folding protocol somewhat changed the NMR spectrum of A1, but significantly improved the spectra of some of the other monomeric G-quadruplexes shown in Supplementary Figure S17.
RESULTS
Identification of mutations that alter G-quadruplex biochemical specificity
In several recent studies we characterized the biochemical properties of a 496-member library of variants of a reference G-quadruplex (15,35–37) (Figure 1). This library contains all possible variants of the central tetrad in a monomeric reference G-quadruplex (note that these mutants do not necessarily form monomers or even G-quadruplexes themselves). It also contains all possible loop variants (A, C, or T but not G) in three different tetrad sequence backgrounds, each of which has a different multimeric state. Each variant has now been tested for five functions associated with G-quadruplex structures: the ability to bind GTP, to promote a model peroxidase reaction, to generate fluorescence, to form dimers, and to form tetramers (15, 35–37 and this work). This functional analysis was complemented by structural characterization of some variants, which showed that at least 41 out of 42 members of the library with at least one function we tested form G-quadruplexes (Supplementary Table S2). To determine the extent to which mutations in this library modulate G-quadruplex biochemical specificity, we compared the ability of each of the 496-members in our library to promote these five functions side-by-side. Each activity profile was distinct (Figure 2, Supplementary Figures S2 and S3, and Supplementary Tables S3 and S4), providing support for the idea that G-quadruplexes with different functions also have distinct sequence requirements. To compare the specificities of these G-quadruplexes in a quantitative way, the five functions described here were grouped into each of the ten possible pairwise combinations (GTP–tetramer, GTP–fluorescence, GTP–peroxidase, GTP–dimer, tetramer–fluorescence, tetramer–peroxidase, tetramer–dimer, fluorescence–peroxidase, fluorescence–dimer and peroxidase–dimer). For each pair, the ability of each sequence to promote the first function being analyzed was plotted on the x axis of a graph, and its ability to promote the second function was plotted on the y axis. When graphed in this way, sequences active for the first function but inactive for the second function will appear as a horizontal line with a y intercept of 1, while those active for the second function but inactive for the first one will appear as a vertical line with an x intercept of 1 (Supplementary Figure S4). Sequences with both activities should appear as a cluster of points with both x and y intercepts greater than one, while those with the same sequence requirements for both functions should exhibit a linear relationship. For most pairs of functions, sequences specific for one of the two functions could be readily identified (Supplementary Figure S4). Furthermore, in ∼15/20 cases, variants active for only one of the two functions being compared were present in the library (Supplementary Figure S4). A surprising feature of these graphs is that they were similar to controls in which the fold activity above background of each sequence was randomly scrambled (Supplementary Figures S4 and S5). This highlights the extent to which the sequence requirements of these five functions differ from one another. However, sequence requirements of different pairs of functions were not unrelated: for nine out of ten pairs analyzed, absolute values of correlation coefficients were at least two standard deviations higher for experimental datasets than for randomly scrambled controls, and for seven pairs they were at least five standard deviations higher (Supplementary Table S5).

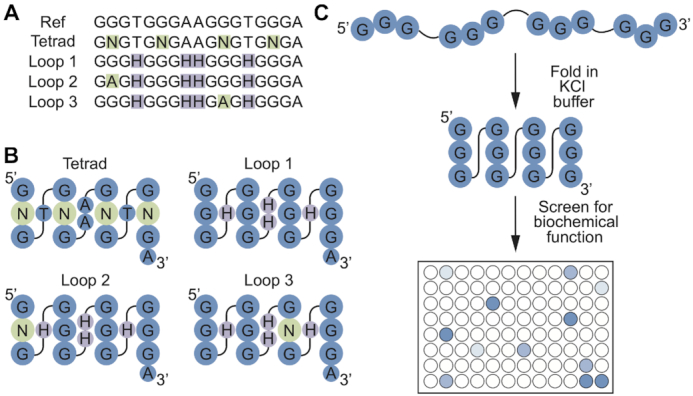
Identification of mutations which alter G-quadruplex biochemical specificity. (A) Library design. Ref = the reference G-quadruplex. Tetrad = library containing all possible variants of the central tetrad in the monomeric reference G-quadruplex. Note that these variants do not necessarily form monomers or G-quadruplexes themselves. Loop 1 = library containing all possible combinations of loop nucleotides (A, C or T but not G) in the reference G-quadruplex. Loop 2 = same as loop 1, but in a variant of the reference G-quadruplex containing a G to A mutation in position 2 of the central tetrad. Loop 3 = same as loop 1, but in a variant of the reference G-quadruplex containing a G to A mutation in position 11 of the central tetrad. (B) Positions of mutated nucleotides in the tetrad, loop 1, loop 2, and loop 3 libraries mapped onto the secondary structure of the reference G-quadruplex. Note that in some cases (especially in the loop 2 and loop 3 libraries) these mutations induce formation of structures that are different from that of the reference G-quadruplex. (C) Overview of the screening method.

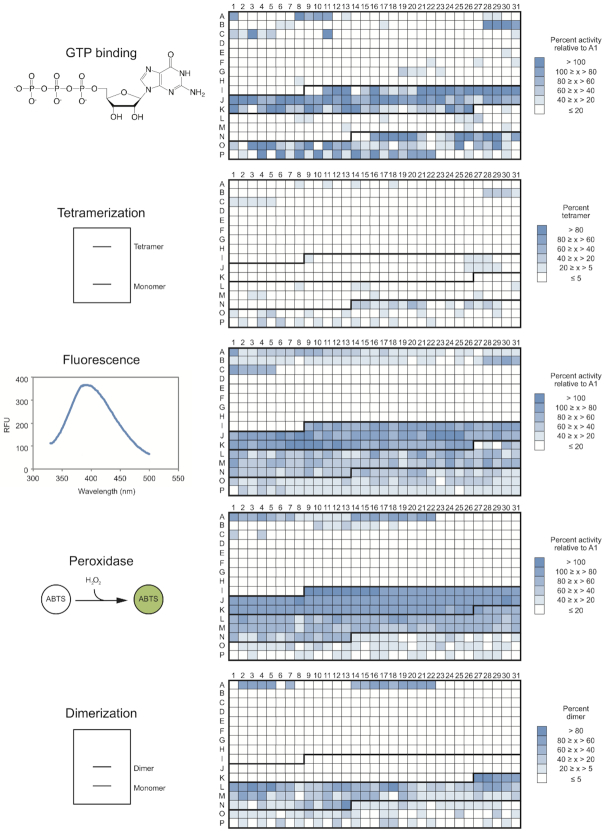
Distinct sequence requirements of G-quadruplexes with different biochemical functions. Heat maps showing the ability of the 496 G-quadruplex variants in the library to bind GTP, form tetramers, generate fluorescence, promote a model peroxidase reaction, and form dimers. Sequences A1-I8 are in the tetrad library, sequences I9-K26 are in the loop 1 library, sequences K27-N13 are in the loop 2 library, and sequences N14-P31 are in the loop 3 library. The boundaries of each library are indicated with dark lines. In each case, darker blue indicates stronger biochemical function.
To facilitate analysis of sequences active for both functions but with a preference for one of them, a specificity score was calculated for each member of the library by dividing the ability of the sequence to promote the first function being compared by its ability to promote the second function being compared. Specificity scores were normalized such that a score of 1 corresponds to the specificity of the monomeric reference sequence (sequence A1 in Figure 2) for any pair of functions being compared. The range of specificity scores varied from >90-fold (for the fluorescence-peroxidase pair) to >50 000-fold (for the GTP-dimer pair) (Figure 3). To determine whether ranges of specificity scores are affected by sequences in the library that do not form G-quadruplexes but are nonetheless active in these assays, we repeated this analysis using a dataset of 41 library members that had been confirmed to form G-quadruplexes using CD or NMR. This dataset yielded almost identical results: specificity scores varied from more than 57-fold (for the fluorescence-peroxidase pair) to >47 000-fold (for the GTP-dimer pair) (Supplementary Figure S6). In addition, ranges of specificity scores for each pair of functions were strongly correlated in these two datasets (R = 0.96). Although these ranges should be thought of as lower limits due to difficulties in accurately determining the background in some assays, they highlight the differences in biochemical specificities of the G-quadruplexes in this library. Taken together, these experiments support the idea that the sequence requirements of G-quadruplexes with different functions are overlapping but distinct.

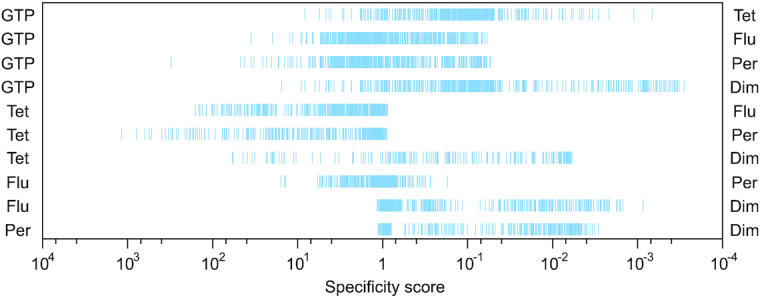
Range of specificity scores for each of the 10 possible pairs of biochemical functions analyzed in this study. For each pair, the specificity score of each of the 496 sequences in the library (represented by vertical blue lines) was determined by dividing the ability of the sequence to promote the first function being analyzed (shown on the left) by the ability to promote the second function being analyzed (shown on the right). Specificity scores were normalized such that a value of 1 corresponds to the specificity of the reference G-quadruplex. Due to difficulties in accurately measuring background assays for some assays, these ranges should be thought of as lower limits. GTP = GTP-binding activity. Tet = ability to form tetramers. Flu = ability to generate fluorescence. Per = ability to promote a model peroxidase reaction. Dim = ability to form dimers.
Importance of mutations in tetrads and changes in multimeric state
Our next goal was to explore mechanisms by which mutations in this library alter G-quadruplex biochemical specificity. A possible clue came from the observation that, for each of the five biochemical functions analyzed, the range of activity measurements for variants containing mutations in tetrads (i.e. in the tetrad library) was larger than for those containing mutations in loops (i.e. in the loop 1 library) (Figures 4A and B). This was also true with respect to specificity: for nine of the ten possible pairs of biochemical functions, the range of specificity scores was larger (and for seven pairs, this was at least an order of magnitude larger) for variants containing mutations in tetrads than for those containing mutations in loops (Figure 4C). Similar trends were observed in a smaller dataset of 41 library members that were confirmed to form G-quadruplexes using CD or NMR (Supplementary Figure S7). One way to understand this difference is to consider the effects of these two types of mutations on G-quadruplex structure. In the context of functionally active variants in our library, mutations in tetrads of the reference G-quadruplex induce multimer formation approximately five times more frequently than mutations in loops (37,43) (Figure 2). This suggested to us that multimerization might play a role in modulating the biochemical functions and specificities of these G-quadruplexes. To further explore this idea, we analyzed the multimeric states of six types of G-quadruplexes in our library: those specific for GTP-binding activity compared to fluorescence and vice versa (Figure 5A), those specific for GTP-binding activity compared to peroxidase activity and vice versa (Figure 5B), and those specific for fluorescence compared to peroxidase activity and vice versa (Figure 5C). For each pair of functions, patterns of specificity were different for monomers, dimers, and tetramers. This can be appreciated by noting the clustering of points corresponding to G-quadruplexes with different multimeric states in the full dataset (Figure 5 and Supplementary Figure S8) and in a smaller dataset made up of 41 library members that were confirmed to form G-quadruplexes using CD or NMR (Supplementary Figure S9). It is also reflected in differences in the average activities of monomers, dimers, and tetramers in both libraries (Supplementary Table S6). These differences are not present in scrambled control datasets in which multimeric states are randomly assigned to library members (Supplementary Figure S8 and Supplementary Table S6).

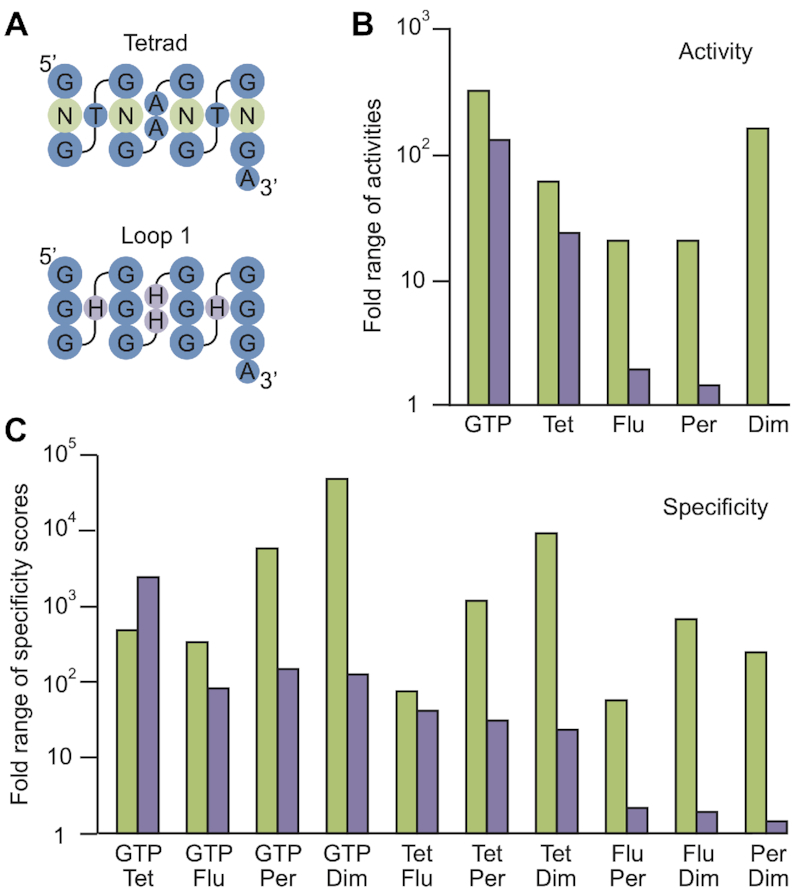
Mutations in tetrads have larger effects than mutations in loops on G-quadruplex biochemical function and specificity. (A) Positions of mutations in tetrads (in the tetrad library) and loops (in the loop 1 library) mapped onto the secondary structure of the reference construct. (B) For each of the five functions analyzed, the fold range of activities for variants containing mutations in tetrads (green bars) is compared to that for variants containing mutations in loops (purple bars). (C) For each of the ten possible pairs of functions, the range of specificity scores for variants containing mutations in tetrads (green bars) is compared to that for variants containing mutations in loops (purple bars). GTP = GTP-binding activity. Tet = ability to form tetramers. Flu = ability to generate fluorescence. Per = ability to promote a model peroxidase reaction. Dim = ability to form dimers.

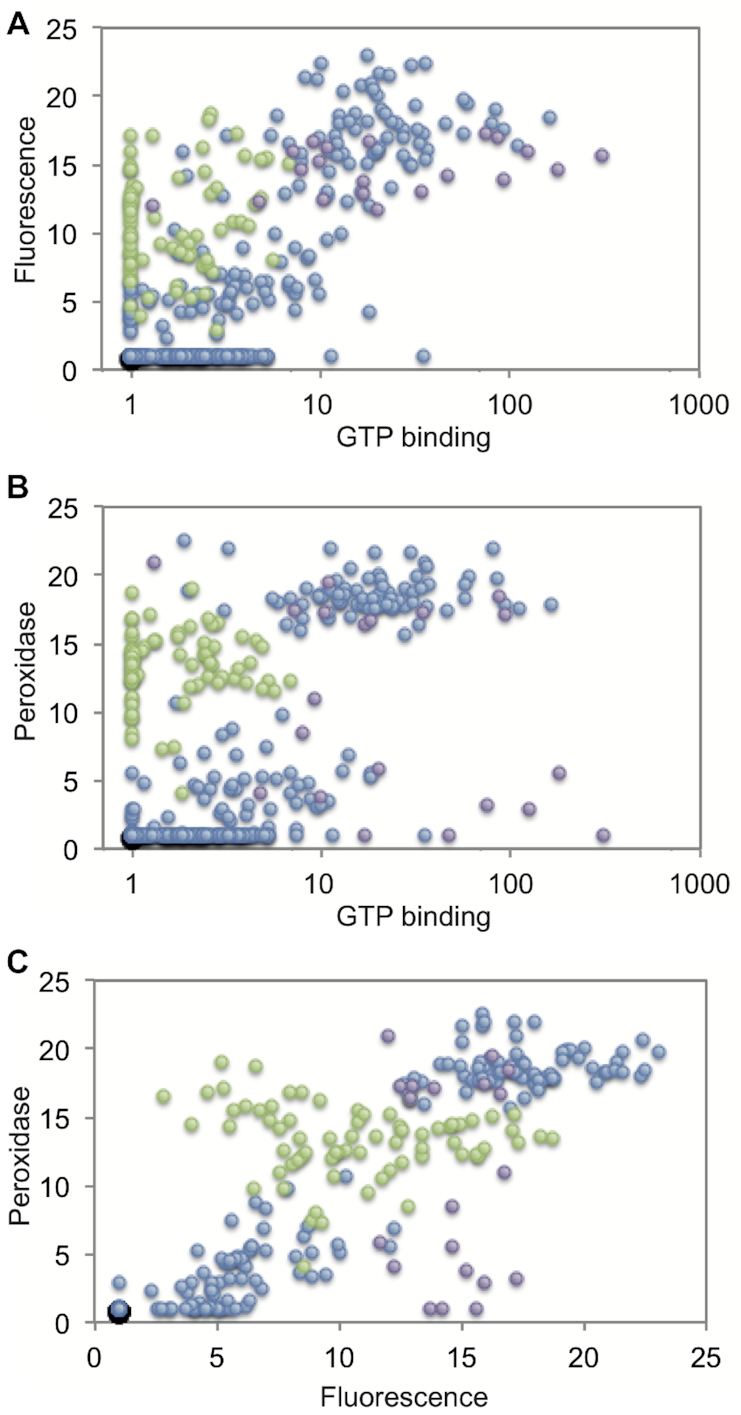
Correlation between G-quadruplex biochemical specificity and multimeric state. (A) Comparison of GTP-binding activity and fluorescence for sequences that form only monomers (blue), dimers but not tetramers (green), and tetramers but not dimers (purple). (B) Same, but for GTP-binding and peroxidase activity. (C) Same, but for fluorescence and peroxidase activity.
To further explore the relationship between multimeric state and biochemical function, we analyzed our dataset using principal component analysis (44–45 and Supplementary Methods). This method facilitates the identification of statistically significant patterns in complex datasets such as the one described here, and also makes it possible to reduce dimensionality while retaining much of the diversity of the original dataset. When applied to a dataset containing all 496 sequences, principal component analysis revealed a correlation between the ability of sequences in the library to form tetramers and to bind GTP, as well as between the ability to form dimers and to promote the peroxidase reaction (Supplementary Methods Figures S2–S5; see also references 35 and 37). These correlations were also observed in a restricted dataset made up of 41 sequences confirmed to form G-quadruplex structures using CD or NMR (Supplementary Methods Figures S21–S24). Principal component analysis also revealed mutations in the library that are correlated with different biochemical functions. For example, sequences in the tetrad library with the sequence GGHH (H = A, C or T, where H is preferably A) in the central tetrad of the reference G-quadruplex form tetramers, bind GTP efficiently, and are more fluorescent than average (Supplementary Methods Figures S6–S9). Conversely, sequences in the tetrad library with the sequence HNGG or NHGG (H = A, C or T; N = A, C, T or G) in the central tetrad of the reference G-quadruplex are all (except for A6) dimers and promote the peroxidase reaction (Supplementary Methods Figures S6–S9). Taken together, these experiments indicate that changes in the biochemical specificity of the G-quadruplexes in this library are correlated with changes in multimeric state, and suggest that this is one mechanism by which G-quadruplex biochemical specificity can be modulated.
Multimerization changes the properties of 5′ and 3′ ends of these G-quadruplexes
Our previously proposed models for monomers, dimers, and tetramers suggest that multimerization changes the properties of the 5′ and 3′ ends of the G-quadruplexes in this library: monomers contain stable tetrads at both the 5′ and 3′ ends of the structure, dimers contain stable tetrads only at the 3′ end of the structure, and tetramers contain stable tetrads only at the 5′ end of the structure (37,43) (Figure 6; note that the models for dimers and tetramers have not been confirmed using high-resolution structural methods). Effects of random sequence 5′ and 3′ overhangs on biochemical function were also sometimes different for G-quadruplexes with different multimeric states (Supplementary Figure S10), providing additional evidence that the 5′ and 3′ ends of these structures are distinct. Such differences could help to explain why changes in G-quadruplex biochemical specificity are correlated with changes in multimeric state. To further investigate this idea, we used a Cy5 fluorescence quenching assay (31) to probe binding of the porphyrin hemin to the 5′ and 3′ ends of three G-quadruplexes in the library with different multimeric states (sequences A1, A2 and A8 in Figure 2; sequences shown in the legend to Figure 6). Because porphyrins are known to stack on the terminal tetrads of G-quadruplexes (for example, reference 46), we reasoned that this approach would allow us to determine if a G-quadruplex with a certain multimeric state contained stable terminal tetrads. In a more general sense, this assay provides information about the functional properties of the 5′ and 3′ ends of these structures. Pilot experiments showed that Cy5 modifications at the 5′ terminus inhibited tetramer formation, but these modifications did not otherwise appear to affect multimeric state (Figure 6A and Supplementary Figure S11). For this reason, the 5′ end of tetramers could not be analyzed using this approach, although information could still be obtained about the 5′ end of the dimeric intermediate thought to form in the tetramer folding pathway (37) (Figure 6D). Titrations showed that the monomeric form of the G-quadruplex binds hemin the most efficiently, with a dissociation constant >1000-fold lower than that of a control oligonucleotide which cannot form a G-quadruplex (Figures 6B–D, Supplementary Figure S12, and Supplementary Table S7). In addition, they revealed that the 5′ and 3′ tetrads in the monomer are not equivalent: the affinity of hemin for the 5′ tetrad is ∼70-fold higher than for the 3′ tetrad (Figure 6B and Supplementary Table S7). We originally hypothesized that this difference was due to the 3′ adenosine overhang, which can potentially block the 3′ tetrad, but similar results were obtained for a variant lacking this adenosine (Supplementary Figure S13 and Supplementary Table S7). The dissociation constant of the highest affinity site in the dimer for hemin is ∼250-fold higher than that of the monomer (Figure 6C and Supplementary Table S7), and the dissociation constant of the highest affinity site in the dimeric intermediate of the tetramer is ∼60-fold higher than that of the monomer (Figure 6D and Supplementary Table S7). The preferred binding site in these two structures is also different: the dimer preferentially binds hemin at its 3′ end, while the dimeric intermediate of the tetramer preferentially binds hemin at its 5′ end. These experiments support our previous models for monomeric, dimeric, and tetrameric G-quadruplexes (37,43) and in particular suggest that monomers and the dimeric intermediate of tetramers contain accessible tetrads at their 5′ ends while monomers and dimers contain accessible tetrads at their 3′ ends (Figures 6B–D). They also show that the 5′ and 3′ ends of G-quadruplexes in our library with different multimeric states have distinct functional properties.

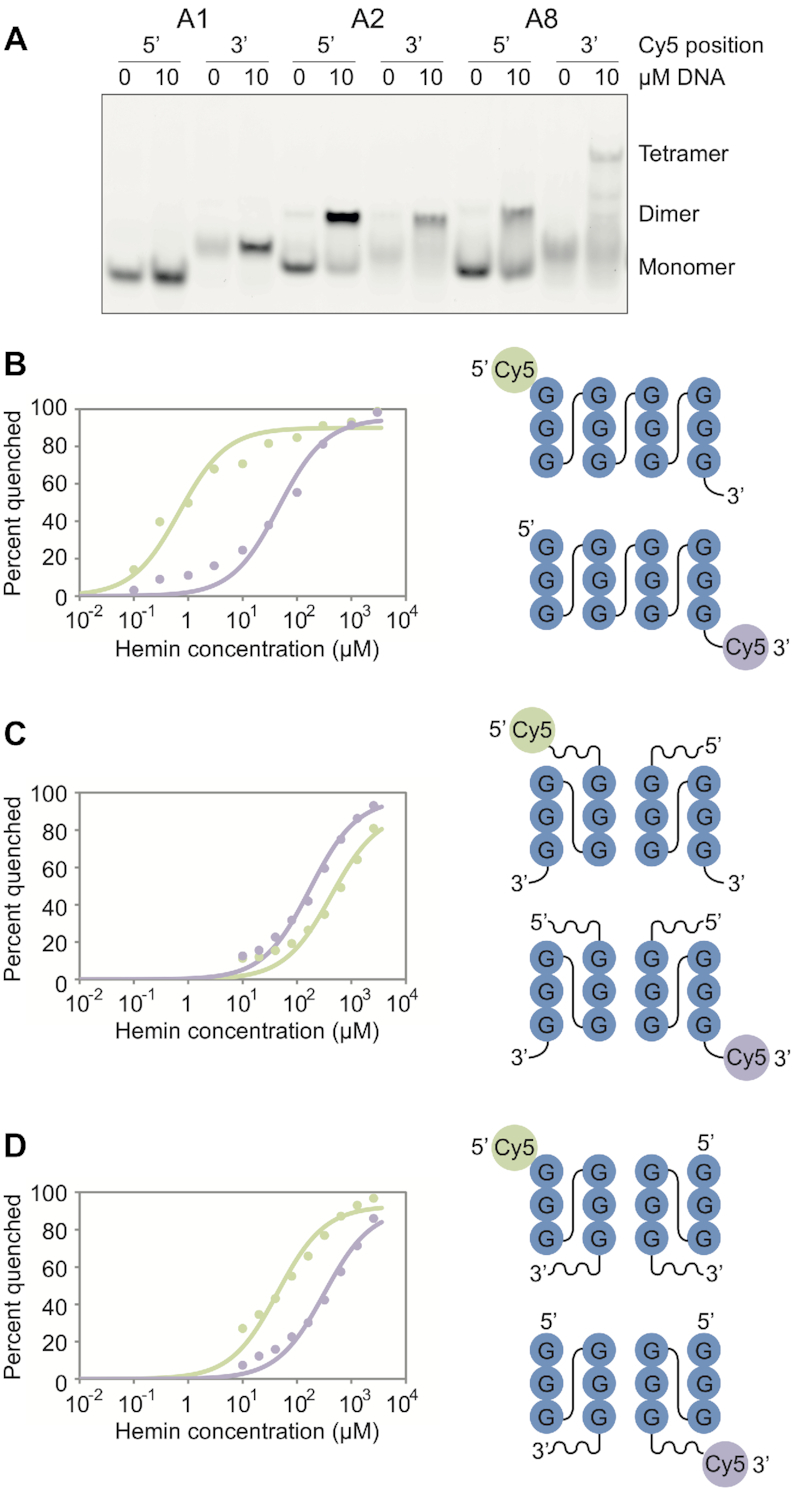
Multimerization changes the functional properties of G-quadruplex 5′ and 3′ ends. (A) Effect of 5′ and 3′ Cy5 modifications on formation of G-quadruplexes with different multimeric states. The gel was visualized using a Cy5 filter so that only Cy5 labeled DNA is visible. Experiments were performed using 0.1 μM Cy5 labeled DNA and 0 or 10 μM unlabeled DNA. (B) Left: fluorescence of a G-quadruplex that forms monomers (A1) modified at its 5′ (green) or 3′ end (purple) with Cy5 as a function of hemin concentration. Kd for 5′ tetrad is 0.69 ± 0.17 μM; Kd for 3′ tetrad is 46 ± 11 μM. Right: model of this G-quadruplex modified at the 5′ (above; Cy5 shown in green) or 3′ (below; Cy5 shown in purple) end with Cy5. This model is based on the solution structure shown in Figure 7. (C) Same, but with a G-quadruplex that forms dimers (sequence A2). Kd for 5′ tetrad is 428 ± 57 μM; Kd for 3′ tetrad is 174 ± 25 μM. This model can rationalize the differential binding of hemin to the 5′ and 3′ tetrad of the dimer, and is also supported by experiments described in reference 37, but its high-resolution structure has not yet been determined. (D) Same, but with a sequence that forms dimers and tetramers (sequence A8). Kd for 5′ tetrad is 44 ± 8 μM; Kd for 3′ tetrad is 321 ± 65 μM. The dimeric intermediate of this G-quadruplex is shown because the 5′ Cy5 modification prevents 5′-5′ stacking of the dimer to form a tetramer (37). This model can rationalize the differential binding of hemin to the 5′ and 3′ tetrad of the dimer and is also supported by experiments described in reference 37, but its high-resolution structure has not yet been determined. Experiments in panel B were performed using 0.1 μM Cy5 labeled DNA and 0 μM unlabeled DNA, while those in panels C and D were performed using 0.1 μM Cy5 labeled DNA and 10 μM unlabeled DNA. See also Supplementary Table S7. A1 = GGGTGGGAAGGGTGGGA. A2 = GAGTGGGAAGGGTGGGA. A8 = GGGTGGGAAGAGTGGGA.
Structural basis of G-quadruplex biochemical specificity
To complement these biochemical experiments, we used NMR to determine the high-resolution structure of the reference G-quadruplex (sequence A1 in Figure 2) (Figure 7, Supplementary Figures S14–S15, and Supplementary Table S1). We focused on this construct because it is one of the most active sequences in the library with respect to the ability to bind GTP, promote the model peroxidase reaction, and generate fluorescence. As expected based on previous studies (35,37), this G-quadruplex is monomeric and contains three tetrads (Figure 7C–F). Its general topology is similar to previously described G-quadruplexes with short loops for which structures have been determined (24,26,47–48). The twist of the helix is right-handed and each of the four strands is parallel. The glycosidic angles of the twelve guanines are in the anti conformation. The guanines in the 5′ tetrad are more flexible than those in other tetrads but are still predominantly anti, which is often (24) but not always the case (49–50) for the 5′ tetrad in parallel-strand structures of DNA G-quadruplexes with unmodified bases. The four loop nucleotides are orientated in a propeller conformation facing away from the main axis of the structure (Figure 7C–F). As has been previously observed for some G-quadruplexes with short loops (e.g. (47)), the loops in this G-quadruplex are flexible and sample a range of conformations (Supplementary Figure S16). On the other hand, the 3′ adenosine overhang is stably stacked on the 3′ tetrad, which means that the structural contexts of the 5′ and 3′ tetrads are not equivalent (Figure 7C–F). This does not appear to affect binding of hemin (Supplementary Figure S13 and Supplementary Table S7) but could facilitate preferential binding of other ligands to the 5′ tetrad. The proton NMR spectrum of this G-quadruplex is similar to that of other monomers in the library, suggesting that most or all adopt similar three-dimensional structures (Supplementary Figure S17). From this perspective, it can be thought of as a representative monomer.

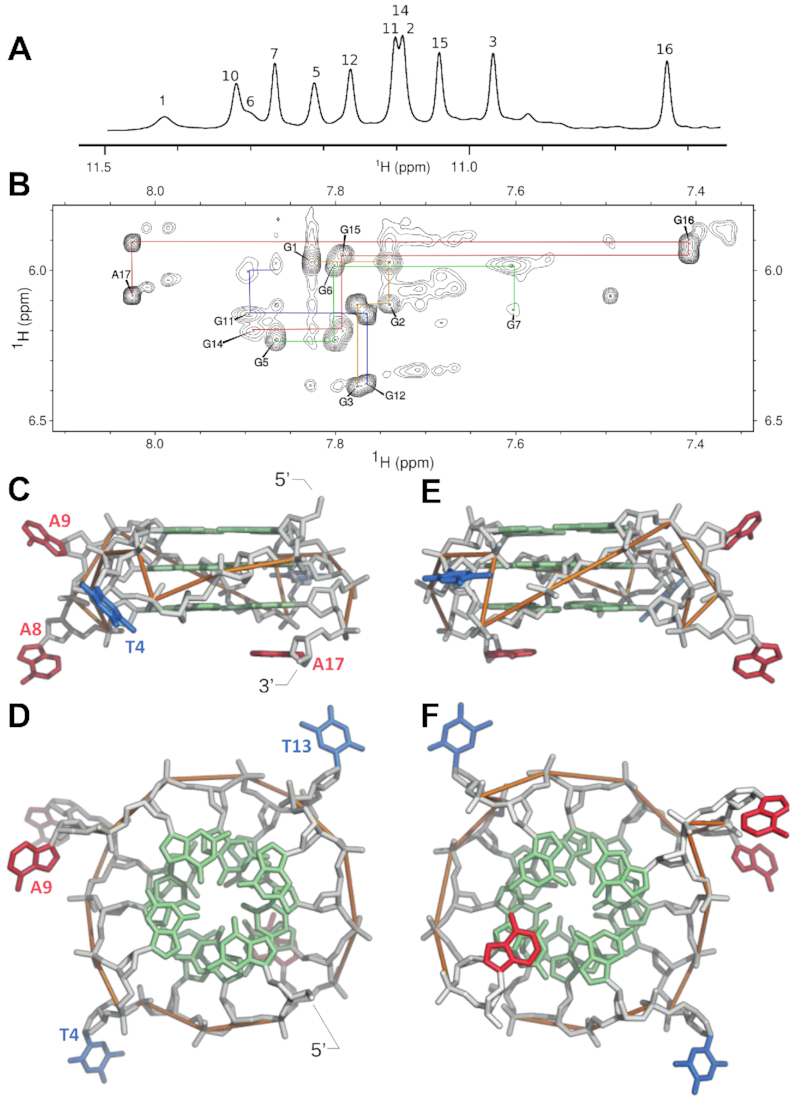
Solution structure of the reference G-quadruplex. (A) Imino proton spectrum of the G-quadruplex. (B) NOESY spectrum (mixing time 200 ms). H1′-H8 interactions are labeled and sequential correlations are indicated with lines (A17–G16–G15–G14, G12–G11–G10, G7–G6–G5, G3–G2–G1). NMR spectra were measured at 298K at a DNA concentration of 1.7 mM in a buffer containing 20 mM Tris, 200 mM KCl and 1 mM MgCl2. (C, E) View from the side of the G-quadruplex. The structure on the right (panel E) is rotated 180 degrees relative to the structure on the left (panel C). Adenines are shown in red, guanines in green, and thymines in blue. The structure contains three tetrads, and the 3′ adenine is stacked on the 3′ tetrad. (D, F) View from the top and bottom of the G-quadruplex. Left (panel D): view from the top of the G-quadruplex looking down on the 5′ tetrad. Right (panel F): view from the bottom of the G-quadruplex looking down on the 3′ tetrad. The right-handed twist of the helix, propeller loops, and guanines in the anti conformation are visible from this perspective. The sequence of this G-quadruplex is GGGTGGGAAGGGTGGGA, and it corresponds to A1 in Figure 2.
This structure provides a number of insights into the biochemical properties of the monomeric G-quadruplexes in the library. The three tetrads in the structure form extended aromatic systems likely to be responsible for its intrinsic fluorescence (13–15,51). They also provide potential binding sites for hemin (the cofactor in the peroxidase reaction) at both the 5′ and 3′ ends of the structure (46). Under the conditions used in our screen for peroxidase activity (0.5 μM hemin and 10 μM DNA), hemin probably stacks on the 5′ tetrad of the monomer (Figure 6B). The structure also helps to understand why mutations in loops of monomeric G-quadruplexes in the library have only small effects on fluorescence and peroxidase activity (sequences A1 and I9-K26 in Figure 2): these positions do not make contacts with tetrads, which are likely responsible for both functions. An important question that this structure does not answer is the location (or locations) of the GTP binding site in the G-quadruplex. Previous studies have shown that G-quadruplexes bind GTP by incorporating it into a tetrad (9,36,52–55). However, each of the tetrads in this structure contain four guanines. This could mean that at least one of the guanines is not stably incorporated and can be displaced by the GTP ligand, although it is also possible that GTP induces a more significant structural rearrangement. Taken together, these experiments indicate that the reference construct forms a monomeric, parallel-strand G-quadruplex containing tetrads which are accessible at both the 5′ and 3′ end of the structure. These features help to explain why monomers in this library tend to promote the model peroxidase reaction and generate fluorescence efficiently.
DISCUSSION
In this study, we used biochemical and structural approaches to characterize the activity and specificity of each sequence in a 496-member library of variants of a monomeric reference G-quadruplex with respect to five biochemical functions. This library contains both canonical G-quadruplexes (those that match the G3–5N1–7G3–5N1–7G3–5N1–7G3–5 consensus sequence) and noncanonical ones (those that do not match this consensus sequence). It also contains sequences that are inactive with respect to these five functions. To rule out the possibility that our conclusions are due in part to sequences in the library that do not form G-quadruplexes but are nonetheless active in these assays, we also analyzed a smaller dataset made up of 41 library members that were shown to form G-quadruplexes by CD (Supplementary Table S2) or NMR (Supplementary Figure S17). These two datasets gave similar results (compare Figure 3 with Supplementary Figure S6, Figure 4 with Supplementary Figure S7, Figure 5 with Supplementary Figure S9, Figure 8 with Supplementary Figure S18, and Supplementary Materials Figures S2–S5 with Supplementary Materials Figures S21–S24). One important conclusion of our study is that the sequence requirements of G-quadruplexes with different functions are overlapping but distinct (Figures 2, 3, and 8). Another is that multimerization appears to be one mechanism by which changes in G-quadruplex function and specificity can be modulated (Figures 4 and 5). This can perhaps be seen most clearly by comparing the functional properties of G-quadruplexes in the library with different multimeric states. Functional monomers (i.e. those with at least one function among the ones we investigated) tend to bind GTP, promote the model peroxidase reaction, and generate fluorescence (for example, mutant A1 in the tetrad library and mutants I9-K26 in the loop 1 library). Dimers typically promote the peroxidase reaction and generate fluorescence, but do not bind GTP efficiently (for example, mutants A14-A22 in the tetrad library and mutants K27-N13 in the loop 2 library). Conversely, tetramers generally bind GTP and generate fluorescence, but do not promote the peroxidase reaction as efficiently as monomers or dimers (for example, mutants B28-C5 in the tetrad library and mutants N14-P31 in the loop 3 library). These groupings are consistent with principal component analysis, which supports a correlation between the ability to form tetramers and to bind GTP as well as between the ability to form dimers and to promote a model peroxidase reaction. They are also observed in a smaller dataset made up of 41 library members shown to form G-quadruplexes using CD or NMR (Supplementary Methods Figures S21–S24). Although our experiments do not fully explain the basis for these differences, they suggest that changes in the functional properties of the 5′ and 3′ ends of G-quadruplexes with different multimeric states could play a role (Figure 6). The pattern of these changes recapitulates one of the major trends in the data: dimers in our library tend to be specific for peroxidase activity compared to GTP binding and contain a stable 3′ tetrad, while tetramers tend to be specific for GTP binding compared to peroxidase activity and contain a stable 5′ tetrad instead. In the case of monomers, we can further understand these differences from the perspective of a high-resolution structure (Figure 7). This shows that monomers contain three tetrads, including accessible tetrads at both ends of the structure. These features likely explain why monomers in the library tend to promote the model peroxidase reaction and generate fluorescence efficiently. Ongoing experiments in our group seek to further characterize the mechanistic basis of these changes in function and specificity from the perspective of representative dimeric and tetrameric structures.

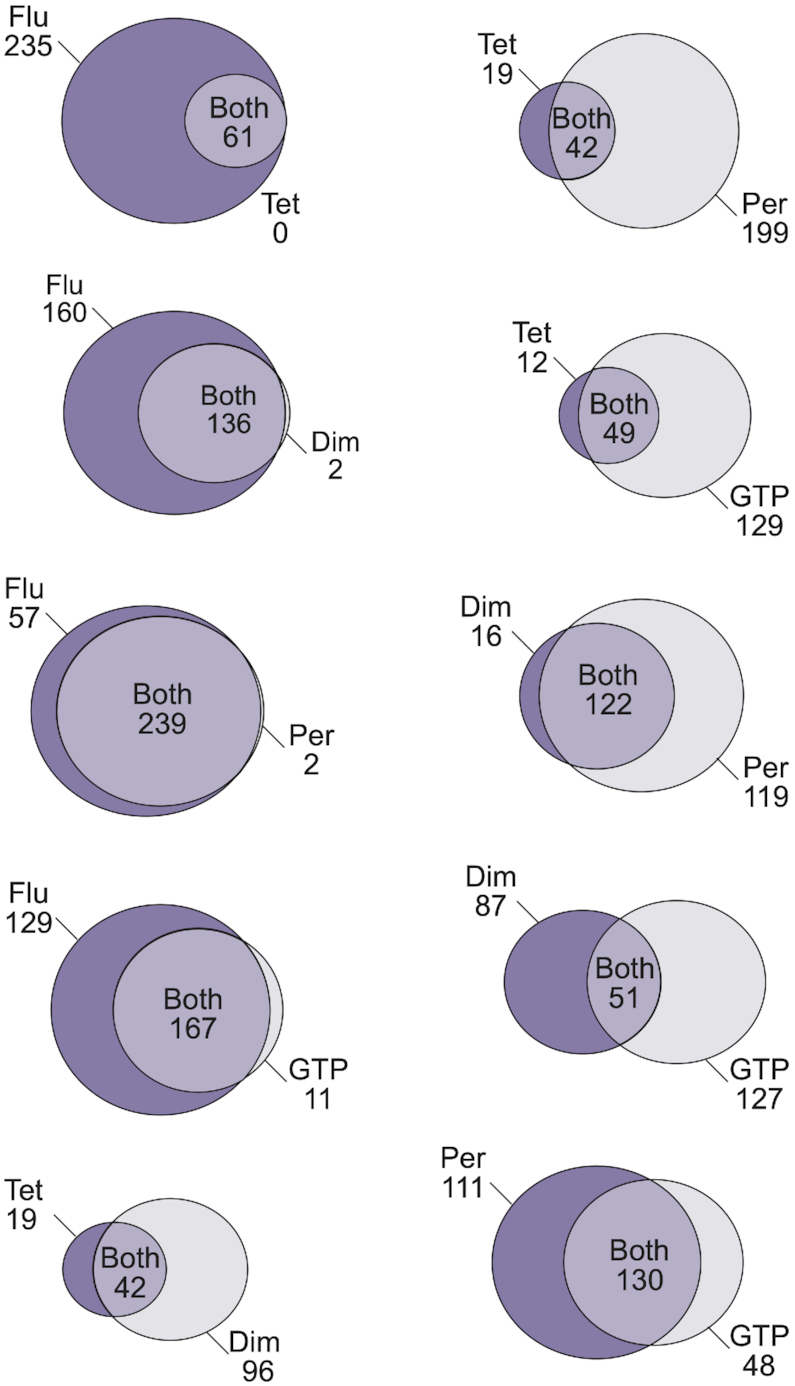
Overlapping model of G-quadruplex biochemical specificity. Comparison of the sequence requirements of G-quadruplexes in the library with different biochemical functions. The area of each circle is proportional to the number of sequences in the class it represents. Cutoffs for activity were the same as those used in Figure 2 (blue squares indicate active sequences, and white squares indicate inactive sequences). GTP = GTP-binding activity. Tet = ability to form tetramers. Flu = ability to generate fluorescence. Per = ability to promote a model peroxidase reaction. Dim = ability to form dimers.
Little systematic information is available about the relationship between G-quadruplex sequence and function. The results described here support a model in which the sequence requirements of G-quadruplexes with different functions are overlapping but distinct (Figure 8). For most pairs of functions, sequences specific for either one function or the other were present in the library, while in several cases the sequence requirements of one function was a subset of that of the other (Figure 8). Similar patterns were observed in a smaller dataset consisting of 41 library members shown to form G-quadruplexes using CD or NMR, and the degree of overlap in the two datasets with respect to each of the ten pairs of functions was strongly correlated (R = 0.95) (Supplementary Figure S18). This probably indicates that a range of structurally distinct G-quadruplexes are present in our library, each with a different activity profile with respect to these five functions. From the perspective of biological regulation, our model is consistent with the idea that functionally distinct G-quadruplexes have different sequence requirements, although it does not rule out contributions from other factors. This model also has implications for the bioinformatic analysis of G-quadruplexes in genomes. Current algorithms to identify G-quadruplexes use models in which structurally distinct classes of G-quadruplexes, such as those with different strand polarities, are grouped together (20, 21). Our findings suggest that such models are too general in some cases because different subsets of sequences that match the G-quadruplex consensus motif can have distinct functional properties. In the context of our library, sequences A1 and I9-K26 illustrate this point the most clearly: each of these variants is classified as a G-quadruplex by standard models, and although the ability of these sequences to promote the model peroxidase reaction and generate fluorescence is similar, they differ by more than 100-fold in their ability to bind GTP (Figure 2 and Supplementary Table S3). Even larger differences are observed when noncanonical G-quadruplexes in the library (i.e those that differ from the G3–5N1–7G3–5N1–7G3–5N1–7G3–5 consensus sequence) are also considered. A relatively easy way to improve such algorithms would be to incorporate information about parameters already known to affect G-quadruplex structure and function. For example, because G-quadruplexes with longer loops are more likely to contain antiparallel strands than those with shorter ones (56), the specificity of some searches could likely be improved by sorting the results according to loop length. A more sophisticated approach would be to perform a series of functional screens (e.g. for the ability of different cellular proteins to bind G-quadruplexes) using libraries in which parameters such as tetrad number, loop length, and loop sequence are systematically varied. Bioinformatic studies using models derived from such studies would likely reveal associations with genomic features that are undetectable due to noise from the more general models currently used. A better understanding of functional classes of G-quadruplexes could also lead to the development of novel classes of G-quadruplex ligands. Such ligands are typically generated by targeting a specific G-quadruplex of interest, such as the Pu22 sequence in the c-MYC promoter (23). A disadvantage of such an approach is that it is difficult to obtain ligands which are specific for the desired G-quadruplex, which can potentially lead to off-target effects. By instead focusing on classes of functionally related G-quadruplexes, and in particular sequences that do not occur in multiple classes, it might be possible to identify ligands that target groups of G-quadruplexes with similar biological functions. Such ligands would be useful for understanding the biological roles of G-quadruplexes, and their effects might be more specific than those obtained by conventional approaches.
DATA AVAILABILITY
The structures and NMR data were deposited in the PDB (accession code: 6YY4) and BMRB (accession code: 34516) database (see also Supplementary Table S1).
ACKNOWLEDGEMENTS
We thank Vanda Lux for help with ion-exchange chromatography, Stanislava Matějková for ICP-OES analysis, Fernaldo Winnerdy and Anh Tuan Phan for advice regarding structure calculations, Jaroslav Kurfürst for assistance with Figure 8, and colleagues at the IOCB for useful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
IOCB Interdisciplinary Grant (to E.A.C.); ‘Chemical biology for drugging undruggable targets (ChemBioDrug)’ [CZ.02.1.01/0.0/0.0/16_019/0000729] from the European Regional Development Fund (OP RDE). Funding for open access charge: IOCB Interdisciplinary Grant (to E.A.C.).
Conflict of interest statement. None declared.
REFERENCES
2.
3.
5.
6.
7.
8.
9.
10.
11.
12.
13.
14.
15.
16.
17.
18.
19.
20.
21.
22.
23.
24.
27.
28.
29.
30.
31.
32.
33.
34.
35.
36.
37.
38.
39.
40.
41.
42.
43.
44.
45.
46.
47.
48.
49.
50.
51.
52.
53.
54.
55.
 Overlapping but distinct: a new model for G-quadruplex biochemical specificity
Overlapping but distinct: a new model for G-quadruplex biochemical specificity
