
- Altmetric
SARS-CoV-2 and the resulting COVID-19 pandemic represents one of the greatest recent threats to human health, wellbeing and economic growth. Wastewater-based epidemiology (WBE) of human viruses can be a useful tool for population-scale monitoring of SARS-CoV-2 prevalence and epidemiology to help prevent further spread of the disease, particularly within urban centres. Here, we present a longitudinal analysis (March–July 2020) of SARS-CoV-2 RNA prevalence in sewage across six major urban centres in the UK (total population equivalent 3 million) by q(RT-)PCR and viral genome sequencing. Our results demonstrate that levels of SARS-CoV-2 RNA generally correlated with the abundance of clinical cases recorded within the community in large urban centres, with a marked decline in SARS-CoV-2 RNA abundance following the implementation of lockdown measures. The strength of this association was weaker in areas with lower confirmed COVID-19 case numbers. Further, sequence analysis of SARS-CoV-2 from wastewater suggested that multiple genetically distinct clusters were co-circulating in the local populations covered by our sample sites, and that the genetic variants observed in wastewater reflected similar SNPs observed in contemporaneous samples from cases tested in clinical diagnostic laboratories. We demonstrate how WBE can be used for both community-level detection and tracking of SARS-CoV-2 and other virus’ prevalence, and can inform public health policy decisions. Although, greater understanding of the factors that affect SARS-CoV-2 RNA concentration in wastewater are needed for the full integration of WBE data into outbreak surveillance. In conclusion, our results lend support to the use of routine WBE for monitoring of SARS-CoV-2 and other human pathogenic viruses circulating in the population and assessment of the effectiveness of disease control measures.

Introduction
The emergence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), and the resulting global Coronavirus disease 2019 (COVID-19) pandemic has had disastrous socio-economic and political consequences worldwide (Chakraborty and Maity, 2020). This led to the World Health Organisation (WHO) declaring the COVID-19 pandemic a global health emergency (WHO, 2020). In response to this, many countries implemented a range of mitigation strategies to reduce the spread of disease, including social distancing, restricted movement, use of personal protective equipment, contact tracing, shielding of vulnerable populations, local or national lockdowns, and community mass testing (Cirrincione et al., 2020; Iacobucci, 2020). These measures are of particular importance in urbanised areas where the spread of disease is most likely (Zhang and Schwartz, 2020). These measures proved to be largely effective at reducing the first wave of COVID-19, albeit not completely eliminating infections (Goscé et al., 2020; Jarvis et al., 2020). The occurrence of subsequent waves of COVID-19 is of significant concern, as countries seek to learn from the effectiveness of the mitigation measures used during the first wave of infection (Aleta et al., 2020).
A large proportion of SARS-CoV-2 infections are asymptomatic or result in only a mild infection (Nishiura et al., 2020). When symptoms do become apparent, this typically occurs 3–7 days after infection (Arons et al., 2020) and severity can vary widely across different sectors of society, disproportionately affecting the elderly (Wang et al., 2020). Evidence points towards the fact that individuals can transmit the virus unknowingly prior to developing symptoms. Furthermore, a- and pre-symptomatic individuals pose challenges to surveillance efforts to accurately estimate the presence and extent of infection in the community. In a more practical sense, both asymptomatic and pre-symptomatic individuals also pose a major threat to public health as they can unknowingly spread the virus to more vulnerable groups (He et al., 2020).
Although mass community testing has been instigated in many countries to estimate the prevalence of COVID-19 in the population, this is costly and the demand for tests frequently exceeds the capacity of testing facilities (Barasa et al., 2020). Focussing testing solely on symptomatic cases may also fail to capture asymptomatic and pre-symptomatic infections, and may focus on populations such as those who are hospitalised, meaning that surveillance is unavailable for the wider community. In some cases, it can also be difficult to obtain nasopharyngeal swabs from high-risk parts of the community due to a range of physical, logistical or cultural issues. Wastewater-based epidemiology (WBE) detects genome fragments of SARS-CoV-2 shed in faeces and urine, and represents an alternative strategy to monitor the levels of virus circulating at population-level scales (Farkas et al., 2020, Kitajima et al., 2020, Polo et al., 2020). WBE approaches have previously been successful in evaluating the prevalence of other viral diseases (e.g. polio-, norovirus) and also for tracking the use of illicit substances, pharmaceuticals and exposure to xenobiotics (Castiglioni et al., 2014; Ozawa et al., 2019; Zuccato et al., 2008). Monitoring viruses in wastewater also allows an evaluation of the potential risk posed by the discharge of treated and untreated wastewater into the wider environment. Overall, WBE may represent a cost-effective method for determining viral prevalence at the population-level, and has been used to monitor SARS-CoV-2 in a range of countries (Supplementary Table 1).
Despite the simplicity of the approach, the quantitative recovery of viruses and viral nucleic acids from wastewater is notoriously difficult (Farkas et al., 2018a). For example, virus concentrations in wastewater can be heavily influenced by (i) dilution by rainfall and industrial inputs, (ii) the presence of compounds that may degrade the virus (e.g. detergents, pH, salt), (iii) the presence of substances that physically protect the virus (e.g. faecal matter), (iv) loss of viral RNA during long transit times through the wastewater network due to decay and sorption, (v) variable shedding rates in the community, and (vi) inhibitory substances in the wastewater that may interfere with quantitative (reverse transcription)-PCR (q(RT-)PCR) reactions (Polo et al., 2020). In addition to these factors, the protocols used to concentrate and purify viral nucleic acids from wastewater samples can have substantial impacts on recovery, leading to underestimation of the quantities of the virus present in the wastewater system. Consequently, there is a need to better understand the factors that influence observable levels of SARS-CoV-2 in wastewater to allow validation of the approach for surveillance purposes.
Large-scale efforts to monitor changes in the SARS-CoV-2 genome and track its circulation at national and global scales have largely relied on the analysis of high-throughput sequencing of the SARS-CoV-2 genome in symptomatic individuals (Islam et al., 2020; Meredith et al., 2020; Plessis et al., 2021). As retrospective screening of respiratory samples has detected asymptomatic cases of COVID-19 (Meredith et al., 2020), it suggests that lineages may appear in wastewater samples prior to observation in clinical cases. Because wastewater aggregates samples from across a community/area, sequencing of SARS-CoV-2 RNA recovered from wastewater is likely to contain multiple lineages and so analysis of this data also has the potential to assess the proportions of different lineages circulating in the wider population. This potentially enables the identification of lineages that are known to be present and early warning of new lineages not previously observed in a catchment.
Here, we present a 3.5-month longitudinal analysis of SARS-CoV-2 RNA prevalence and genetic diversity across six different urban centres during the imposition and gradual lifting of the first national lockdown period in the UK (March-July 2020). The aims of this study were to (i) investigate the use of WBE for tracking SARS-CoV-2 after the implementation of national lockdown measures at six urban centres of varying size within the UK, (ii) determine the influence of environmental factors (e.g. flow) on levels of SARS-CoV-2 RNA and a human faecal marker DNA virus (crAssphage) in wastewater, (iii) investigate the impact of wastewater treatment on the removal of SARS-CoV-2 RNA from wastewater, and (iv) assess the utility of WBE in understanding SARS-CoV-2 genetic variation through high-throughput sequencing.
Materials and methods
All laboratory procedures were carried out in line with Public Health England/ Public Health Wales advice on the handling of samples suspected of containing SARS-CoV-2.
Sampling sites and wastewater sampling
Untreated influent and treated effluent wastewater were collected from six wastewater treatment plants (WWTPs) located in Wales and Northwest England. The WWTPs served urban areas in the local authority areas of Gwynedd, Cardiff, Liverpool, Manchester, the Wirral and Wrexham, with a total combined population equivalent of ~3 million people (Suplementary Fig. 1). Untreated wastewater influent from the six WWTPs was sampled on a weekly basis between March and July 2020. Samples were collected in polypropylene bottles as single grab samples with the exception of the Wirral site, which was collected as a 24 h composite sample using an autosampler. Grab samples were collected on weekdays between 08.00 and 09.00 a.m. to ensure temporal comparability, and treated effluent was also collected periodically at the same time as influent. Samples were transported on either the same day, or overnight on ice, to the laboratory, stored at 4 °C and processed within 24 h of receipt. Aliquots of wastewater samples (1.5 ml) were also frozen in polypropylene vials at −80 °C for subsequent physico-chemical analyses and extraction of pre-concentration viral nucleic acids.
Wastewater physicochemical analyses
Wastewater samples were pasteurised before physicochemical analysis by heating to 60 °C for 90 min. Wastewater ammonium concentrations were determined colorimetrically using the salicylic acid procedure of Mulvaney (1996). Nitrate was determined colorimetrically using the vanadate procedure of Miranda et al. (2001) while molybdate-reactive phosphate (MRP) was determined according to Murphy and Riley (1962). All analysis was performed in a 96-well plate format using a PowerWave XS Microplate Spectrophotometer (BioTek Instruments Inc., Winooski, VT). Wastewater electrical conductivity (EC) was measured using a Jenway 4520 conductivity metre and pH with a Hanna 209 pH metre (Hanna Instruments Ltd., Leighton Buzzard, UK).
Wastewater concentration and nucleic acid extraction
Duplicate samples of 50–100 mL of unpastuerised wastewater influent underwent centrifugation (10,000 g, 30 min, 4 °C) and the supernatant and pellet retained. Supernatants were concentrated to 500 µL using Centriprep 50 kDa MWCO centrifugal concentrators (Merck KGaA, Germany). For wastewater effluent samples (see Supplementary Table 5), 1–2 L of each effluent was initially concentrated using tangential flow ultrafiltration with a 100 kDa PES membrane (Spectrumlabs, USA) as previously described (Farkas et al., 2018c), followed by secondary concentration using Centriprep concentrators as described above.
Selected wastewater concentrates, centrifugation pellets and unconcentrated wastewater samples were spiked with approximately 4 × 105 genome copies (gc) of murine norovirus (MNV) as a viral RNA extraction control. Positive and negative nucleic acid control extractions of nuclease-free water with or without the same quantity of MNV spike-in were used to quantify MNV recovery by q(RT-)PCR and to check for cross-contamination during the nucleic acid extraction process or q(RT-)PCR assay setup (described in Section 2.4). The MNV was cultured in BV2 cells in Dulbecco's modified Eagle's minimum essential medium supplemented with 2% foetal bovine serum (FBS) at 37 °C in 5% CO2 for two days. Viruses were harvested by three cycles of freeze-thawing (−20 °C/+37 °C) followed by centrifugation and 100 × dilution of the supernatant in phosphate-buffered saline pH 7.4. Aliquots of MNV stock were stored at –80 °C until use. The MNV and BV2 tissue stocks were kindly provided by Prof Ian Goodfellow (University of Cambridge, UK).
Nucleic acids were extracted using the NucliSENS MiniMag Nucleic Acid Purification System (BioMérieux SA, Marcy-l'Étoile, France) according to the manufacturer's protocol as described elsewhere (Farkas et al., 2021) in a final volume of 50 (last week of March 2020) or 100 µL (April-July 2020) of elution buffer. Extracted nucleic acids were stored at –80 °C prior to q(RT-)PCR quantification. The nucleic acid extractions and q(RT-)PCR assay preparation were carried out in separate laboratories inside class II microbiological safety cabinets to minimise the risk of contamination.
q(RT-)PCR and qPCR assays
The q(RT-)PCR assays were carried out in a QuantStudio® Flex 6 Real-Time PCR System (Applied Biosystems, USA) using primers, probes and reaction conditions described in Supplementary Table 2. SARS-CoV-2 N1 and MNV RNA were quantified using a duplex q(RT-)PCR assay or in triplex with SARS-CoV-2 E gene, as described in Farkas et al. (2021). The 25 μL reaction mix contained 1 × RNA Ultrasense Reaction Mix with 1 µL RNA Ultrasense Enzyme Mix (Invitrogen, USA), 12.5 pmol of the forward and the reverse primers, 6.25 pmol of the probe/probes, 0.1 × ROX reference dye, 1.25 µg bovine serum albumin (BSA) and 2–5 μL of the extracted wastewater RNA, molecular grade water as a negative control or virus standards. Initially, 5 µL of extracted RNA was tested for wastewater samples. If the MNV recovery was lower than 1%, samples were retested with 2 µL sample/reaction to assess inhibition of the q(RT-)PCR assay, however this was found to be detrimental to assay sensitivity. All data-points used in the analysis came from assays of 5 μL of extracted nucleic acids.
CrAssphage was used as a marker of human faecal abundance/loading in the wastewater (Farkas et al., 2019; Stachler et al., 2018). CrAssphage DNA was quantified using a singleplex qPCR as described previously (Farkas et al., 2019). The 20 µL reaction mix contained 1 × KAPA Probe Force qPCR mix (KAPA Biosystems, USA) with 10 pmol of the forward, 10 pmol of the reverse primers, 5 pmol of the probe, 1 µg bovine serum albumin, and 2 µL and 4 µL of the concentrated and original wastewater nucleic acid extracts or controls.
A serial dilution of DNA standards within the range of 105–100 gc µL−1 was used for quantification. For SARS-CoV-2, commercially available circular plasmids carrying the N gene or E gene were used (Integrated DNA Technologies Inc., Coralville, IA). Plasmid DNA concentrations were halved when setting up serial dilutions to account for ssRNA producing half the fluorescence signal of dsDNA at the same concentration. For MNV and crAssphage, custom-made, single-stranded oligo DNA sequences carrying the target region were used (Life Technologies, USA). Negative controls (molecular grade water) were included in each run. All samples, standards and controls were run in duplicate and the mean value for each extraction replicate used for further analysis.
The limit of detection (LoD) and limit of quantification (LoQ) of the triplex q(RT-)PCR assays were determined previously (Farkas et al., 2021) by running wastewater samples spiked with low concentrations of SARS-CoV-2 (1–150 gc µL−1N1 CDC and 1–200 gc µL−1E Sarbeco) and MNV RNA (1–80 gc µL−1) in ten replicates. The q(RT-PCR) assay LoD (the lowest concentration where all replicates were positive) were 1.7, 3.8 and 3.1 gc µL−1 for the N gene, E gene and MNV, respectively. The LoQ (the lowest concentration where the coefficient of variance was below 0.25) were 11.8, 25.1 and 32.1 gc µL−1 for the N gene, E gene and MNV, respectively.
q(RT-)PCR data analysis and visualisation
Data were analysed using QuantStudio™ Real-Time PCR Software, version 1.3 (Applied Biosystems, USA). The baseline (cycle threshold; Ct) was manually adjusted after each run, when necessary. Viral concentrations were expressed as mean gc 100 ml−1 wastewater calculated from two q(RT-)PCR duplicates of two extraction duplicates (n = 4) per sampling timepoint. Statistical analyses and data visualisation was performed in R v4.0.2 (R Core Team, 2020; Wickham, 2016). Supplementary Table 3 contains a full list of packages used in the data analysis.
SARS-CoV-2 RNA amplicon sequencing and data processing
RNA from 84 extraction duplicates from 42 time-points, plus no-template negative controls, were treated with DNase, and used to generate cDNA (NEB Luna Script). Subsequently, SARS-CoV-2 cDNA underwent PCR amplification using V3 nCov-2019 primers (ARTIC) generating 400 bp amplicons tiling the viral genome (Quick and Loman, 2020). Amplicon generation was followed by sequencing library construction (NEB Ultra II DNA), with equimolar pooling of samples and quantification. Final library size was assessed on a Bioanalyser high sensitivity DNA chip, and DNA concentration determined by Qubit double-stranded DNA high sensitivity assay, and then by qPCR using the Illumina Library Quantification Kit from Kapa (KK4854) on a Roche Light Cycler LC480II according to the manufacturer's instructions. Libraries were sequenced on an Illumina MiSeq generating 2 × 250 bp paired end reads. An average of ca 291,000 reads (ca 146 Mbp) per sample were mapped using bwa-mem against the SARS-CoV-2 genome reference (MN908947.3) within the ncov2019-artic-nf v3 pipeline (https://github.com/connor-lab/ncov2019-artic-nf). SNPs and indels were identified using Varscan v2.4.4 with default settings and summary statistics for coverage and diversity were generated in R v4.0.2 (R Core Team, 2020; Wickham, 2016). Sites were filtered to remove SNPs and indels with a coverage of less than 50 × and a variant frequency of less than 10% per sample. The number of SNP and indel sites were calculated per sample.
The relationship between SNP and indel site frequency and the proportion of the genome with coverage at greater than 50 × coverage and the log10 gc µL−1 were examined with Spearman's correlations. An index of SNP plus indel frequency per sample was calculated by taking the number of SNP and indel sites and dividing by the proportion of the genome with coverage at greater than 50 reads. A mean SNP and indel frequency index were then calculated per pair of wastewater samples to examine the effect of the number of positive tests in the previous 7 days in the local authority area, sample date and WWTP site on the number of SNPs and indels discovered, using a general linear model using the ‘glm’ function and type II ANOVA using the R package ‘car’. A Spearman's correlation was used to examine the relationship between the index of SNP and indel frequency and the log population equivalent served by each wastewater treatment plant. Variants at SNP and indel sites were compared to those recorded in clinical samples using the ‘cov_glue_snp_lineage’ function from R package ‘sars2pack’.
Results and discussion
Study description and q(RT-)PCR assay development
We monitored the SARS-CoV-2 RNA concentration in influent wastewater at six wastewater treatment plants (WWTPs) using q(RT-)PCR over a period of 3.5 months during the imposition and gradual lifting of the first UK-wide lockdown, and compared these data to the numbers of positive clinical tests and deaths reported by the Office for National Statistics (ONS), UK Government and Public Health Wales for lower tier local authority areas within which the WWTPs were located (HM Government, 2020; Office for National Statistics, 2020; Public Health Wales, 2020). WWTPs represent a range in size (population equivalents from 40 thousand to 1.1 million) and spatial distribution (see Supplementary Fig. 1) and all implemented combined stormwater, domestic and trade wastewater collection. Influent wastewater grab samples were collected at the same time each week with the exception of The Wirral WWTP which was sampled from a 24 h composite autosampler. Limits of detection (LoD) and quantification (LoQ) were determined as described in Farkas et al. (2021).
Results for SARS-CoV-2 RNA concentrations from q(RT-)PCR quantification are displayed as unadjusted mean genome copies (gc) 100 ml−1 of wastewater rather than normalised by crAssphage concentrations as factors such as extraction efficiency can vary depending on the virus used (Medema et al., 2020). Although studies suggest that 24 h composite sampling is more representative than grab sampling, it has been shown that grab samples are accurate to within an order of magnitude (Ahmed et al., 2021a, Curtis et al., 2020). Further, our previous work has shown limited diurnal variability, particularly in large wastewater catchments where transit times can be up to 24 h and where large amounts of mixing occurs within the network (Farkas et al., 2018b). Transit times may also influence observable virus quantities due to degradation of viral nucleic acids as they pass through the sewage system; however, SARS-CoV-2 RNA has been shown to be relatively stable in wastewater under environmental conditions, with a T90 of 24 or 28 days at 15 or 4 °C (Ahmed et al., 2020b).
We compared mean SARS-CoV-2 RNA concentrations to daily flow and influent wastewater chemistry but found no statistically significant correlations (see Supplementary Table 4). The highly abundant bacteriophage crAssphage was used as a human faecal marker. No correlation was found between crAssphage and SARS-CoV-2 nucleic acid concentrations (Spearman, p = 0.8341). No effect on crAssphage concentration was observable from sampling week (Kruskal-Wallis, p = 0.9042), but a significant effect was found between crAssphage concentration and WWTP site (Kruskal-Wallis, p = 0.01751). These data indicate that faecal loading was constant throughout the study period and that different WWTPs have different balances of human waste and industrial/ other domestic wastewater sources.
Temporal trends in SARS-CoV-2 RNA concentration in wastewater and comparison to COVID-19 epidemiology
For each WWTP, 64% ± 6.8 q(RT-)PCR tests (mean ± standard error (SEM), sites = 6, n = 90) detected SARS-CoV-2 in influent wastewater above the LoD, with SARS-CoV-2 RNA concentrations in wastewater influent having quantities above the LoQ in 28.9% ± 2.2 of samples (see Supplementary Fig. 2). No sites showed SARS-CoV-2 concentrations in WWTP effluent above the LoQ and only one above the LoD (Wrexham, 19/05/20, n = 22, see Supplementary Table 5). Fig. 1 a shows a drop in wastewater SARS-CoV-2 RNA concentration, new positive clinical tests and COVID-19 related deaths following the imposition of the UK-wide lockdown beginning in late March 2020. A number of spikes in clinical cases can be observed without corresponding spikes in wastewater, e.g. Wrexham in late June. These can occur due to surge testing following local workplace-related outbreaks and changes in testing eligibility during the study, highlighting the inherent difficulties in comparing wastewater loads to positive tests when testing is both limited and non-random.

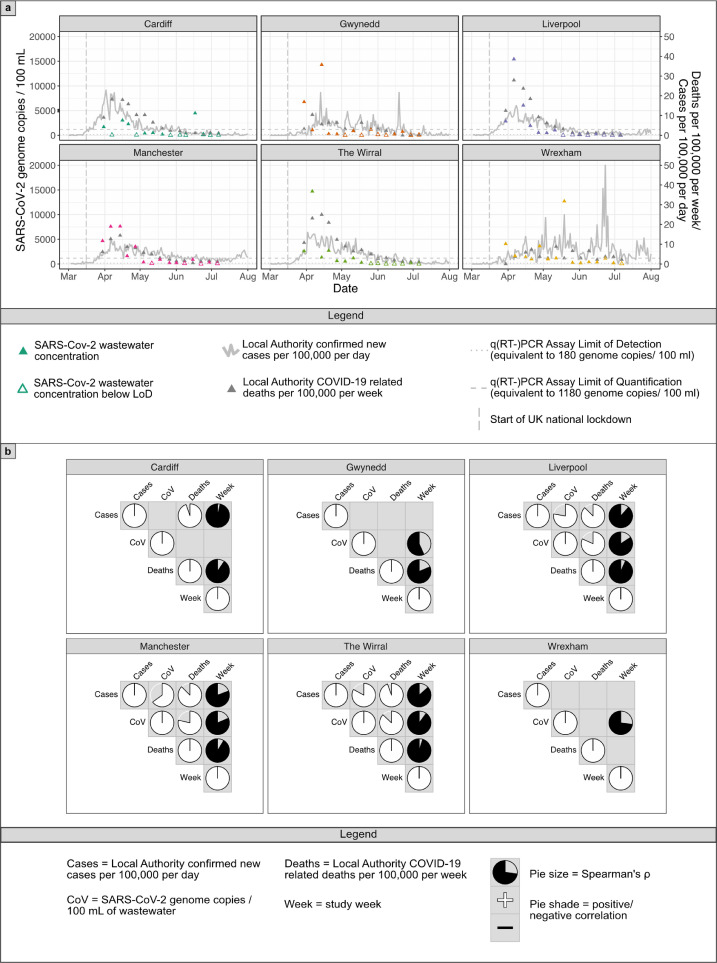
(a) Temporal trend of the recorded number of COVID-19 infections and deaths at six urban centres in the UK and the corresponding levels of SARS-CoV-2 in wastewater. The coloured triangles represent levels of SARS-CoV-2 in influent wastewater, with open triangles being below LoD. Grey triangles represent the number of COVID-19 reported deaths and the solid line represents the number of COVID-19 cases reported in each study region. The dashed and dotted horizontal lines represent the assay LoQ (scaled to 1180 genome copies/ 100 ml) and LoD (180 genome copies/ 100 ml) respectively, scaled for a sample volume of 100 mL. The dashed vertical line represents the imposition of UK-wide lockdown measures. (b) Correlation of SARS-CoV-2 RNA concentration (CoV) in influent wastewater with COVID-19 related cases and deaths at six urban centres in the UK. Pie charts represent Spearman correlation ρ where p < 0.05 with fullness indicating degree of correlation and colour representing positive (white) or negative (black) correlations.
WWTPs in Manchester, Liverpool and the Wirral showed strong correlations between SARS-CoV-2 RNA concentration and daily positive tests (Fig. 1b and Supplementary Fig. 3). Negative correlations were also observed between viral concentrations in all sites and time following the implementation of national lockdown, except Cardiff, indicating these measures lowered the prevalence of the virus in local populations. The Cardiff, Gwynedd and Wrexham WWTPs did not show the same trends between viral RNA concentrations and tests/ deaths, potentially due to several different factors such as water chemistry or lower, broader peaks in SARS-CoV-2 prevalence. Gwynedd is also a popular holiday destination and sees regular weekend influxes of holiday makers from other parts of the UK, which could affect WWTP SARS-CoV-2 concentrations either positively (through visits from asymptomatic/ pre-symptomatic individuals) or negatively (through people commuting from rural areas outside of the WWTP catchment area). Additional factors such as transit time within the sewage network, catchment flow dynamics, and differences between local authority reporting areas for positive tests and WWTP sewershed coverage could affect viral RNA recovery. In contrast to the Gwynedd site, the Wirral site showed the strongest correlation between SARS-CoV-2 RNA concentrations and the number of positive clinical tests/ COVID-19 related deaths, and is of a size inbetween that of the Wrexham and Gwynedd WWTPs (see Supplemental Fig. 1), suggesting that the use of 24-hour composite sampling may improve the correlation between SARS-CoV-2 wastewater quantification and local clinical cases.
Further exploration of site-specific factors and improved access to higher resolution spatial distributions of positive test locations is required to improve the accuracy of WBE in predicting COVID-19 prevalence amongst local populations as part of national monitoring programmes. Previous studies have corrected SARS-CoV-2 RNA concentration for WWTP flow (Gonzalez et al., 2020), and adjusted cases or positive tests for differences between local authority populations and WWTP catchment areas (Medema et al., 2020). Statistically, we found no benefit of correcting for these factors on Spearman correlation coefficients between WWTP SARS-CoV-2 RNA concentration and positive tests/ COVID-19 related deaths (see Supplemental Fig. 3), however due to differences between WWTP sites and sewersheds, we would caution against making extensive quantitative comparisons between sites.
Our data confirm that SARS-CoV-2 RNA is readily detectable in wastewater influent across a range of concentrations from <1.2 × 103 (<LoQ) to the highest recorded concentration of 1.5 × 104 gc 100 mL−1. This highlights how site-specific factors, concentration and quantification protocols, and sampling strategies can complicate quantitative comparisons between WWTPs within the same study, and when making comparisons to other international studies. There is a need to standardise SARS-CoV-2 wastewater quantification and take WWTP site identity into account when expanding WWTP monitoring programmes to national and international scales (Chik et al., 2021; Pecson et al., 2021). Nonetheless, this study demonstrates the longitudinal benefit of using WBE to monitor viral prevalence and the impact of public health interventions, particularly in the early stages of a novel disease outbreak.
Effect of window size/ offset on correlations
Due to shedding of SARS-CoV-2 from asymptomatic and pre-symptomatic individuals, a key driver of WBE research is the potential to detect upcoming spikes in infection in wastewater before increase in positive clinical tests. Consequently, several studies have used modelling approaches to assess if the wastewater concentration of SARS-CoV-2 preceded new spikes in clinical cases of COVID-19 (Ahmed et al., 2021b; D'Aoust et al., 2021). However, this is challenging due to variabilities in the point of an infection cycle at which a person gets tested, the severity and duration of symptoms, and the variability in viral shedding. The effect of varying the difference between the number of days between wastewater sampling and testing date and the number of days over which to sum the number of positive tests on the correlation between wastewater SARS-CoV-2 concentrations and cases was examined (Fig. 2 ). If only considering daily clinical testing data, the SARS-CoV-2 wastewater RNA concentration leads testing data by 2–4 days but this can be extended by approximately 1 day by using a rolling sum of positive clinical test cases over a series of days leading up to the clinical testing date being considered. It should be noted that the overall effect of varying these parameters is not large in that the correlation coefficients stay between 0.8 and 0.9 over a range of permutations.

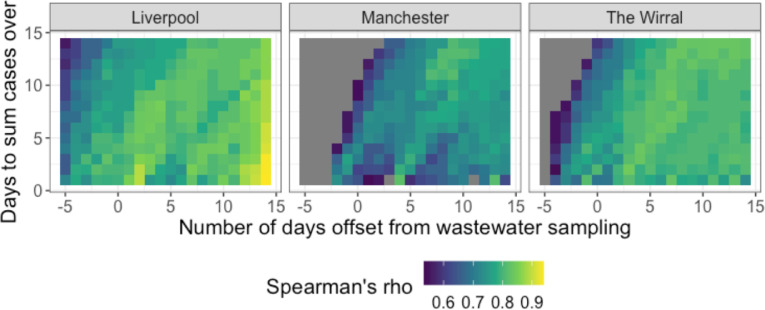
Effects of varying the number of days between wastewater sampling date and clinical testing date (x axis) and the number of days over which to sum cases over (y axis) on the strength of correlation between wastewater SARS-CoV-2 concentration and local authority positive tests. Quantities are shown where a false discovery rate corrected p-value was below 0.05.
Sequencing detects mutations in the SARS-CoV-2 genome comparable to those observable in clinical cases
WBE can also be used to monitor the genetic diversity SARS-CoV-2 circulating in the wider population. To this end, SARS-CoV-2 RNA was amplified using the ARTIC protocol primers in both extraction duplicates, where at least one of which showed q(RT-)PCR amplification were sequenced. In these samples, between 25 and 75% of the SARS-CoV-2 genome was recovered (Fig. 3 a), with coverage randomly distributed across the genome (Fig. 3b). This included samples that showed no amplification (8.3%) or amplification below the LoD (3.6%) of the N1 q(RT-)PCR assay (n = 84), suggesting that multi-locus amplicon sequencing based monitoring of wastewater for WBE may be of significant use in the early stages of future viral outbreaks. The proportion of the genome sequenced positively correlated with the amount of template (Spearman's ρ = 0.376, p = 0.0004, Fig. 3c).

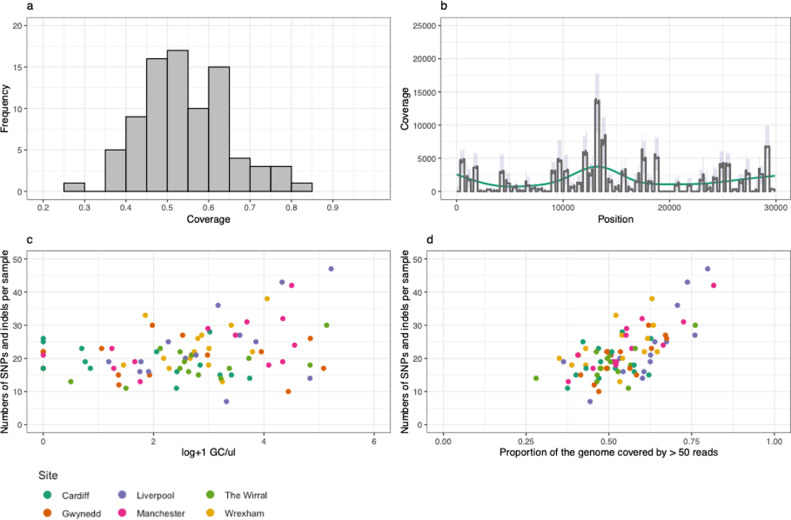
Coverage of the SARS-CoV-genome from reads recovered from wastewater samples. a) Frequency of the proportion of the genome sequenced at 50 × depth or greater. b) Coverage across the genome, median plotted in dark grey, interquartile ranges in purple and a smoothed GAM spline in green. c) Proportion of the genome sequenced relative to the estimated number of genome copies estimated from (RT)-qPCR. Note that sequence was obtained in several samples where the (RT)-qPCR for this locus was negative, reflecting the ability of the protocol to sequence genomes of low copy number. d) The number of SNP and indel sites detected relative to the proportion of the genome that was sequenced at 50 × or higher.
In total, 702 unique SNP sites and 267 indels were detectable across the 84 samples after filtering to remove sites with less than 50 reads and a variant frequency within a sample of less than 10%. The number of SNPs found correlated positively with the proportion of the genome that was sequenced (Spearman's ρ = 0.581, p < 0.0001; Fig. 3d).
Preliminary modelling suggests that the rate of positive tests in the source population and sampling week did not affect the mean number of SNPs and indels controlled for genome coverage (p > 0.05; Fig. 4 a and b), but a reduced model suggested that there was heterogeneity amongst sites (X 2 = 11.57, df = 5, p = 0.041; Fig. 4c). The index of SNP plus indel frequency was not related to log population equivalent served by each wastewater treatment plant (Spearman's ρ = 0.251, p = 0.251; Fig. 4d). This is explained by the presence of multiple viral lineages present within the sample, corresponding to the diverse infections in the population represented in the wastewater sample. A substantial fraction of the detected SNPs has previously been identified in clinical samples across the UK, and has the potential to be informative for distinguishing viral lineages (Supplementary Table 6).

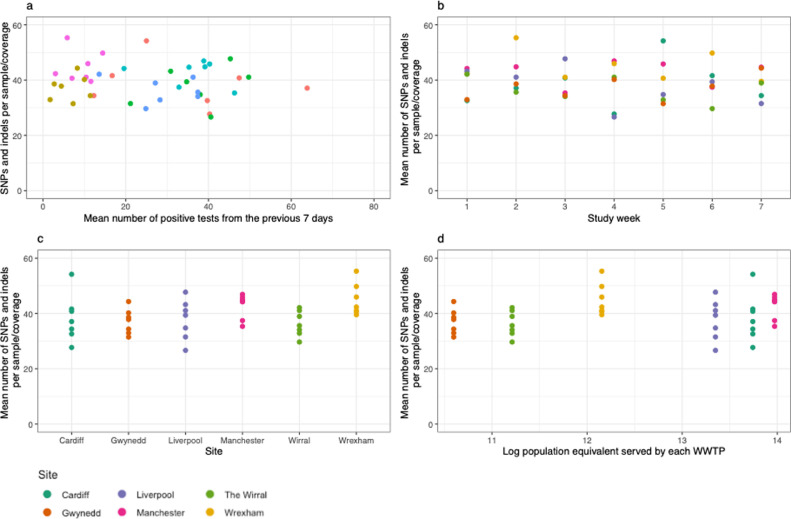
Comparison of the mean number of SNP/ INDELs sites divided by genome coverage to (a) positive tests in the previous 7 days in the local authority, (b) sample date, (c) WWTP site and (d) log10 population equivalent.
Multiple SARS-CoV-2 lineages can be present within a single wastewater sample. Samples have the potential to contain viruses from both symptomatic and asymptomatic individuals within the community, as SARS-CoV-2 has been detected in the faeces of both asymptomatic and symptomatic individuals (Jones et al., 2020; Tang et al., 2020). Previous studies have sequenced SARS-CoV-2 genomes from wastewater (Ahmed et al., 2020a; Izquierdo-Lara et al., 2021; Martin et al., 2020; Nemudryi et al., 2020). We have shown not only that viral genome sequences can be recovered from wastewater samples, but that they exhibit substantial diversity across dozens of samples. Sequencing the genomes therefore has the potential to assess the diversity of viral infections in the wastewater catchment population and to identify emerging genetic variants before they are seen in clinical samples. In support of this, preliminary analysis suggests that the detected SNPs were consistent with those detected previously in clinical samples (see Supplementary Table 6). However, because the SNPs from wastewater samples are not phased across the genome, and because the genome coverage is imperfect, assigning viral lineages to samples will require a bespoke statistical framework to be developed.
Use of wastewater-based epidemiology in COVID-19 and future pathogen surveillance
Attempting to quantitatively link observed viral RNA concentrations to detectable cases is challenging (Medema et al., 2020). Many assumptions need to be made regarding the persistence of SARS-CoV-2 in wastewater, quantities of the virus shed in faeces and the influence of water chemistry (Ahmed et al., 2020a).
Sample processing methodology can also be a substantial source of variability. Concentration method, qPCR assay design and inter-lab variation can create variation in detectable SARS-CoV-2 RNA quantities (Pecson et al., 2021; Westhaus et al., 2021). Use of appropriate process controls is necessary to monitor the effects of these factors when making intra- and inter-laboratory comparisons. Choice of process control is complex as a closely related surrogate virus should be used where available and further global collaboration and co-ordination is required to widen access to WBE technologies (Polo et al., 2020). In addition to this, the effects of SARS-CoV-2 on global supply chains and the need to perform WBE at scale create additional pressures where sub-optimal protocols may become necessary in the future to achieve testing scale desired for national monitoring programs.
Despite the possible sources of variability mentioned above, we have demonstrated that WBE is suitable for quantitatively tracking the course of the early stages of the SARS-CoV-2 pandemic and the effects of public health interventions, even in the early stages of a novel outbreak, where lack of surge capacity prevents optimal sampling. We highlight how tiled primer array sequencing complements q(RT-)PCR based detection of SARS-CoV-2 and enhances the sensitivity and usefulness of WBE in detecting the presence of novel mutations in the SARS-CoV-2 genome. Early detection of viral pathogens by q(RT-)PCR requires a suitable assay and routine monitoring of WWTPs however alternative technologies such as viral metagenomics may be more suited to initial detection of emerging and unknown pathogens (Farkas et al., 2020). Our results suggest that viral amplicon sequencing could be more sensitive than q(RT-)PCR for detection of known pathogens. In future, monitoring could be targeted towards ports of entry and major metropolitan centres to maximise the likelihood of detection (Medema et al., 2020).
Conclusions
•Our results demonstrate that levels of SARS-CoV-2 RNA in wastewater generally correlated well with the abundance of clinical COVID-19 cases recorded within the community in large urban centres.
•At the population level, wastewater-based epidemiology was used to confirm the success of lockdown measures (i.e. restricted movement and human-to human contact) implemented at the national scale to control the transmission of SARS-CoV-2.
•The genetic diversity of SARS-CoV-2 from wastewater suggests that multiple genetically distinct clusters were co-circulating in the local populations, and that the genetic variants observed in wastewater reflect similar SNPs observed in samples from nasopharyngeal swabs taken contemporaneously at clinical testing centres.
•A greater understanding of the factors that affect SARS-CoV-2 RNA quantification in wastewater is still required to enable the full integration of wastewater-based epidemiology data into wider outbreak surveillance programmes.
•Our results lend support to the use of routine wastewater-based epidemiology to monitor SARS-CoV-2 and other human pathogenic viruses circulating in the population and to assess the effectiveness of disease control measures.
Data availability
q(RT-)PCR and chemical data recorded in this study is available as supplementary information and from the Environmental Information Data Centre (EIDC, www. eidc.ceh.uk). DOI:10.5285/ce40e62a-21ae-45b9-ba5b-031639a504f7.
Sequencing read files analysed in this study can be accessed from the European Nucleotide Archive (project PRJEB42191).
Author contributions
DLJ, LSH, KF, SKM and JEM conceived the project. LSH, JT, MAD and KF undertook the experimental work. LSH and KF undertook the processing and analysis of the q(RT-)PCR data. KHM, TB and SP undertook the processing and analysis of the sequencing data. LSH, KF and DLJ led the data interpretation and writing of the manuscript. All other authors contributed to the final draft of the article.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
Acknowledgements
This work was funded by UK Research and Innovation (UKRI) under the COVID-19 Rapid Response Programme (projects NE/V004883/1 and NE/V010441/1) and the Centre for Environmental Biotechnology Project funded though the European Regional Development Fund (ERDF) by Welsh Government. LSH was supported by a Soils Training and Research Studentship (STARS) grant from the Biotechnology and Biological Sciences Research Council (BBSRC) and
 Monitoring SARS-CoV-2 in municipal wastewater to evaluate the success of lockdown measures for controlling COVID-19 in the UK
Monitoring SARS-CoV-2 in municipal wastewater to evaluate the success of lockdown measures for controlling COVID-19 in the UK