
Competing Interests: The authors have declared that no competing interests exist.
- Altmetric
The CRISPR/Cas9 system is an efficient genome editing tool that possesses the outstanding advantages of simplicity and high efficiency. Genome-wide identification and specificity analysis of editing sites is an effective approach for mitigating the risk of off-target effects of CRISPR/Cas9 and has been applied in several plant species but has not yet been reported in pepper. In present study, we first identified genome-wide CRISPR/Cas9 editing sites based on the ‘Zunla-1’ reference genome and then evaluated the specificity of CRISPR/Cas9 editing sites through whole-genome alignment. Results showed that a total of 603,202,314 CRISPR/Cas9 editing sites, including 229,909,837 (~38.11%) NGG-PAM sites and 373,292,477 (~61.89%) NAG-PAM sites, were detectable in the pepper genome, and the systematic characterization of their composition and distribution was performed. Furthermore, 29,623,855 highly specific NGG-PAM sites were identified through whole-genome alignment analysis. There were 26,699,38 (~90.13%) highly specific NGG-PAM sites located in intergenic regions, which was 9.13 times of the number in genic regions, but the average density in genic regions was higher than that in intergenic regions. More importantly, 34,251 (~96.93%) out of 35,336 annotated genes exhibited at least one highly specific NGG-PAM site in their exons, and 90.50% of the annotated genes exhibited at least 4 highly specific NGG- PAM sites, indicating that the set of highly specific CRISPR/Cas9 editing sites identified in this study was widely applicable and conducive to the minimization of the off-target effects of CRISPR/Cas9 in pepper.
Introduction
In mutants, which are of great significance for both gene function analysis and crop genetic improvement, allelic variation mainly results from naturally or artificially induced mutation. Compared to natural variation, the most prominent advantage of artificially induced mutation is the high mutation frequency achieved. The main methods currently used for achieving artificially induced mutation include physical mutagenesis, chemical mutagenesis, random transposon insertion, and target gene editing technologies. Among these approaches, target gene editing, in which nucleotide variation is introduced at an appointed site and the target mutations are obtained accurately and efficiently, thereby speeding up the functional identification of target genes and genetic breeding improvement, is an ideal method for artificially inducing mutations [1].
A variety of target gene editing techniques, including the use of zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and the CRISPR/Cas system, have been developed to date [2]. The CRISPR/Cas system, which has entered the mainstream in recent years and been widely used in humans [3], animals [4], microorganisms [5] and plants [6], possesses the outstanding advantages of high simplicity and efficiency in contrast to the other two techniques. According to the number and functional characteristics of the Cas gene, CRISPR/Cas systems can be divided into 2 categories, including 6 different types (I to VI) [7–9]. The first category of CRISPR/Cas systems, including types I, III and IV, requires multiple Cas proteins to collaboratively interfere with the target gene, while the second category requires only one Cas protein. The type II CRISPR/Cas system, namely CRISPR/Cas9 system belongs to the second category and is now the most widely used gene editing system.
The CRISPR/Cas9 gene editing system is mainly composed of one Cas9 protein and one small guide RNA (sgRNA). The Cas9 protein from Streptococcus pyogenes (SpCas9) was first applied for use in the CRISPR/Cas9 system [10]; SpCas9 recognizes the protospacer adjacent motif (PAM) sequence 5′-NGG-3′ (where “N” can be any nucleotide base) in the target DNA, then cleaves the target DNA at 3 nt upstream of the PAM site, generating a blunt end, and gene editing is finally achieved by nucleotide insertion, deletion and substitution at the cleavage site mediated by the receptor cellular DNA repair machinery, including the nonhomologous end joining (NHEJ) and homologous recombination repair (HDR) mechanisms [11]. The sgRNA of the CRISPR/Cas9 system, artificially designed based on crRNA (CRISPR RNA) and the core sequence of trans-acting crRNA (tracrRNA), is a short single-stranded RNA that guides the Cas9/sgRNA complex to perform cleavage at 3 nt upstream of the PAM site through complementary base pairing between the 5’ end (~20 bp) of the sgRNA and the protospacer sequence of the target DNA, which determines the specificity of gene editing [12].
Previous studies have found that even if the sgRNA imperfectly matches the protospacer, the Cas9 protein can still perform cleavage at 3 nt upstream of the PAM site, making gene editing possible in nontarget regions; thus, off-target effects can occur [13–15]. To reduce or eliminate the risk of off-target effect, the identification of candidate editing sites with high specificity is a prerequisite for the application of the CRISPR/Cas9 system. To date, a variety of tools based on whole-genome sequence similarity analysis have been developed for target site design and off-target risk assessment, such as CrisprGE [16], Cas-OFFinder [17], Cas-Designer [18], CRISPRdirect [19] and CRISPOR [20]. However, the majority of those tools have been mainly applied in humans and animals. Based on whole-genome reference sequences, the distribution and specificity of genome-wide CRISPR/Cas9 editing sites in Arabidopsis thaliana, Medicago truncatula, soybean (Glycine max), tomato (Solanum lycopersicum), Brachypodium distachyon, rice (Oryza sativa), Sorghum bicolor, maize (Zea mays) and grape (Vitis vinifera) have been systematically analysed and compared [12, 21], providing an important reference for choosing highly specific editing sites of related species.
Pepper (Capsicum spp.) belongs to the family Solanaceae and has a cosmopolitan distribution and considerable economic importance [22]. The reference genome sequences of pepper were first released in 2014 [23, 24], marking the transition of pepper research from structural genomics to functional genomics. The identification and functional analysis of important genes controlling agronomic traits have become a significant direction in molecular genetics research in pepper. With the development and continuous improvement of technologies for pepper regeneration in vitro and its genetic transformation [25, 26], the CRISPR/Cas9 gene editing system will become a powerful tool and will be widely used for the functional analysis of pepper genes. In this study, we first identified CRISPR/Cas9 editing sites at the genome-wide level in pepper and then evaluated the obtained specificity through whole-genome sequence alignment. The purpose of this study was to provide a reference for the selection of highly specific CRISPR/Cas9 editing sites and facilitate the application of CRISPR/Cas9-mediated gene editing in pepper.
Materials and methods
Genomic data and CRISPR/Cas9 editing site identification
The ‘Zunla-1’ (v2.0) pepper reference genome sequence and related genome annotations [23] were used for CRISPR/Cas9 editing site identification. There were two PAM sites recognized by the CRISPR/Cas9 system: 5'-NGG-3' and 5'-NAG-3', which were identified by using EMBOSS software [27] in both the positive and reverse strands of the Zunla-1 reference genome sequence. The 20-nt sequences before all 5'-NGG-3' and 5'-NAG-3' sites were extracted to form two protospacer sets, referred to as the GG_spacer set and AG-spacer set, respectively.
Identification of highly specific CRISPR/Cas9 editing sites
Based on the method reported previously, the specificity of CRISPR/Cas9 editing sites in pepper was evaluated. Class 0.0 and Class 1.0 spacers were expected to provide high specificity in CRISPR/Cas9 gene editing [12] and were thus classified as highly specific sites in this study. Since the sgRNA/Cas9 complex showed much less affinity and tolerance toward mismatches at the NAG-PAM site [5], in this study, we only assessed the specificity of the GG_spacers, for which the possibility of off-target effects was evaluated by using the AG_spacer set. The method is outlined as follows:
The hard-masking function of USEARCH [28] was used to mask and remove GG_spacers containing low-complexity sequences;
GG_spacers with the same sequences at the 6~20-nt region were removed;
GASSST [29] and UBLAST [28] were used to generate a pairwise alignment for the remaining GG_spacers. According to the GG_spacer position and the minimum number of mismatches (minMM_GG, including InDel and SNP) between each GG_spacer and other GG_spacers, the GG_spacers were graded into three classes: Class 0 spacers shared no significant matching sequence with other GG_spacers; Class 1 spacers showed no fewer than four mismatches (minMM_GG≥4) or three mismatches adjacent to PAM sites; Class 2 included the other GG_spacers;
For Class 0 and Class 1 GG_spacers, pairwise alignments were performed with AG_spacers, which were further graded into four classes as follows according to their position and the minimum number of mismatches (minMM_GG, including InDel and SNP) between each GG_spacer and other AG-spacers: Class 0.0 spacers exhibited no fewer than three mismatches with AG_spacers (minMM_AG≥3) or shared no significant matching sequence with AG_spacers; Class 0.1 spacers exhibited fewer than three mismatches with AG_spacers; Class 1.0 spacers exhibited no fewer than three mismatches with AG_spacers (minMM_AG≥3) or shared no significant matching sequence with AG_spacers; Class 1.1 spacers exhibited fewer than three mismatches with AG_spacers.
PCR verification and sequence analysis
Primer pairs flanking the selected target sites were designed by using the Primer3web (version 4.1.0; http://primer3.ut.ee/) tool. PCR reaction was performed in a 20 μL mixture including 2.0 μL DNA template (50 ng/μL), 2.0 μL PCR buffer (10×), 2.0 μL Mg2+ (25 mM), 1.5 μL forward and reverse primer (1 μM), 0.2 μL dNTPs (10 mM), and 1U Taq DNA polymerase. PCR procedure was as follow: 94°C for 3 min, 32 cycles of 94°C for 30 s, 55°C for 30 s, and 1 min at 72°C; and a final extension at 72°C for 10 min. PCR amplication of each sites were repeated three times and then the products were directly sequenced and assembled. Alignment of each sequence to the reference genome was conducted by using the local blastn:2.9.0+.
Results and discussion
Content and composition of CRISPR/Cas9 editing sites in pepper genome
A total of 603,202,314 CRISPR/Cas9 editing sites, containing 229,909,837 (~38.11%) NGG-PAM sites and 373,292,477 (~61.89%) NAG-PAM sites, were detected in the pepper genome. This was approximately 4.63 times greater than the number identified in another Solanaceae species, tomato (130,302,150), conforming to the law that the larger the size of a genome, the greater the number of CRISPR/Cas9 editing sites it contains [12]. The average density of NGG-PAM and NAG-PAM in pepper was 69.75/Kb and 112.56/Kb (Table 1), respectively, which were similar to those in tomato (63.30/Kb and 103.43/Kb, respectively), but the density of NGG-PAM in pepper was much less than that in monocot species such as rice (101.69/Kb) and maize (119.22/Kb) [12].

| Chr. | NGG | NAG | Subtotal | ||
|---|---|---|---|---|---|
| No. | Density | No. | Density | ||
| P1 | 22,489,579 | 74.71 | 33,673,230 | 111.86 | 56,162,809 |
| P2 | 11,839,695 | 72.21 | 18,417,689 | 112.33 | 30,257,384 |
| P3 | 17,618,433 | 67.37 | 29,560,783 | 113.04 | 47,179,216 |
| P4 | 15,393,261 | 71.36 | 24,658,517 | 114.32 | 40,051,778 |
| P5 | 15,303,392 | 70.43 | 24,853,200 | 114.39 | 40,156,592 |
| P6 | 15,305,364 | 69.72 | 25,109,431 | 114.38 | 40,414,795 |
| P7 | 15,309,150 | 68.93 | 24,810,273 | 111.70 | 40,119,423 |
| P8 | 11,278,024 | 73.57 | 17,545,055 | 114.45 | 28,823,079 |
| P9 | 16,539,878 | 69.26 | 27,445,080 | 114.93 | 43,984,958 |
| P10 | 14,506,128 | 70.51 | 23,477,296 | 114.11 | 37,983,424 |
| P11 | 15,159,289 | 68.80 | 24,592,215 | 111.61 | 39,751,504 |
| P12 | 15,974,353 | 69.47 | 26,241,216 | 114.12 | 42,215,569 |
| P0 | 43,193,291 | 60.43 | 72,908,492 | 102.00 | 116,101,783 |
| Total | 229,909,837 | 69.75 | 373,292,477 | 112.56 | 603,202,314 |
With respect to the composition of the PAM sites, the TGG and CGG types accounted for the highest (~38.88%) and lowest proportions (~7.44%) of total NGG-PAM sites, respectively (Fig 1A), similar to the composition pattern found in the grape genome [21]. For NAG-PAM sites, the AAG type was the most abundant, with a proportion of ~36.07%, followed by TAG, GAG and CAG, accounting for 29.55%, 19.54% and 14.84% of the total NAG-PAM sites, respectively (Fig 1B).

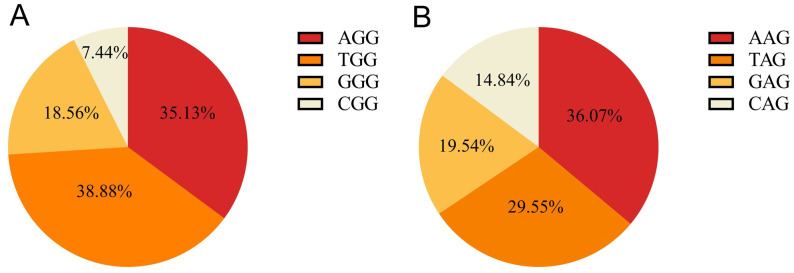
Composition of pepper PAM sites.
A, NGG-PAM; B, NAG-PAM.
Distribution characteristics of CRISPR/Cas9 editing sites in pepper genome
The CRISPR/Cas9 editing sites (NGG-PAM and NAG-PAM) were uniformly distributed on all 12 chromosomes (P1~P12) of pepper (Fig 2). With the exception of chromosome P0, P1 and P8 exhibited the most and least CRISPR/Cas9 editing sites, respectively (Table 1). The number of NGG-PAM and NAG-PAM sites on the pepper chromosomes was significantly positively correlated (R2 = 0.997) with chromosome length (Fig 3). The density of NGG-PAM sites on different chromosomes (not including P0) ranged from 67.37/Kb (chromosome P3) to 74.71/Kb (chromosome P1). The densities of NAG-PAM sites on different chromosomes (excluding P0) were relatively similar to each other, with the minimum and maximum densities of 111.61/Kb (P11) and 114.93/Kb (P9), respectively (Table 1).


Distribution of different kinds of CRISPR/Cas9 editing sites in the pepper genome.
A, NGG-PAM+NAG-PAM site; B, NGG-PAM site; C, NAG-PAM site; D, Class 0.0; E, Class 0.1; F, Class 1.0; G, Class 1.1; H, Class 2.

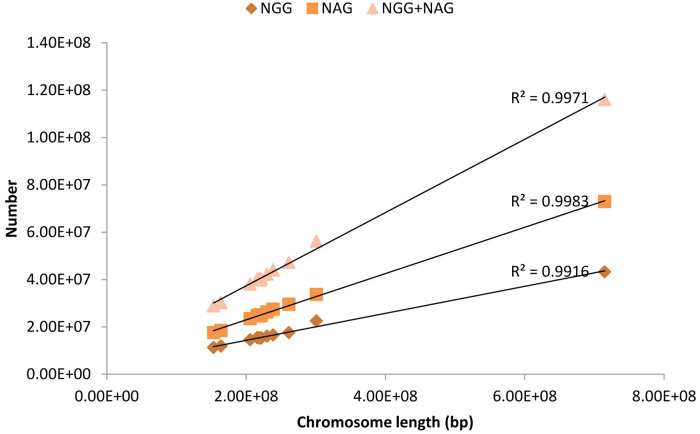
Correlation between the number of CRISPR/Cas9 editing sites and chromosome length in pepper.
The vast majority of NGG-PAM (~94.41%) and NAG-PAM (~94.42%) sites were located in the intergenic regions of the pepper genome, while 8,661,656 (~3.77%) and 3,425,476 (~1.49%) NGG-PAM sites were located in intron and exon regions, respectively, and the rest (~0.32%) were located in UTRs and splicing regions (Table 2). Regarding the distribution pattern in different genomic regions, the pattern of NAG-PAM sites was similar to that of NGG-PAM sites (Table 2). The density of CRISPR/Cas9 editing sites in genic regions (including UTRs, exons, introns and splicing sites,) was lower than that in intergenic regions for NGG+NAG-PAM (159.03/Kb versus 180.68/Kb, Fig 4A), NGG-PAM (60.55/Kb versus 68.87/Kb, Fig 4B) and NAG-PAM (98.49/Kb versus 111.81/Kb, Fig 4C), which differs from the situation in grape [21].

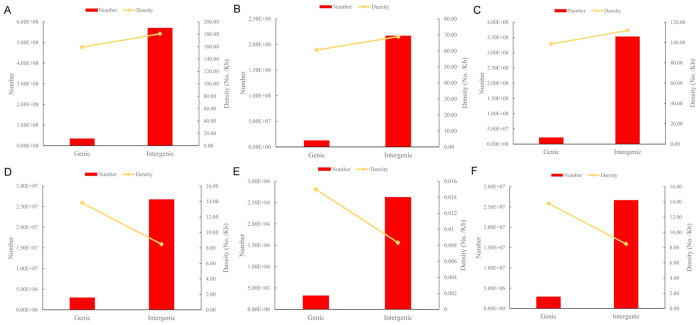
Comparison of the number and density of CRISPR/Cas9 editing sites between genic and intergenic regions.
A, NGG-PAM+NAG-PAM site; B, NGG-PAM site; C, NAG-PAM site; D, Class 0.0+Class 1.0; E, Class 0.0; F, Class 1.0.

| Genomic Region | NGG+NAG | NGG | NAG | |||
|---|---|---|---|---|---|---|
| No. | Percentage | No. | Percentage | No. | Percentage | |
| Intergenic | 569,505,881 | 94.41% | 217,081,038 | 94.42% | 352,424,843 | 94.41% |
| 5'UTR | 975,566 | 0.16% | 373,564 | 0.16% | 602,002 | 0.16% |
| 3'UTR | 937,645 | 0.16% | 340,106 | 0.15% | 597,539 | 0.16% |
| Exon | 8,487,423 | 1.41% | 3,425,476 | 1.49% | 5,061,947 | 1.36% |
| Intron | 23,217,393 | 3.85% | 8,661,656 | 3.77% | 14,555,737 | 3.90% |
| Splicing | 78,406 | 0.01% | 27,997 | 0.01% | 50,409 | 0.01% |
| Total | 603,202,314 | 100.00% | 229,909,837 | 100.00% | 373,292,477 | 100.00% |
Content of highly specific NGG-PAM sites in pepper genome
Through filtering and alignment analysis, 30,402,397 (~13.22%) NGG-PAM sites were successfully graded based on their specificity (Table 3). The total number of highly specific NGG-PAM sites in pepper, including those belonging to Class 0.0 and Class 1.0, was 29,623,855, which was 4.50 times higher than that in tomato, accounting for ~12.88% of the total NGG-PAM sites (Table 3), which was in line with the general rule that the number of specific gRNA spacers is positively correlated with genome size in eudicot species [12]. On average, there were 8.81/Kb highly specific sites in the pepper genome, which is comparable to that in the tomato genome (8.42/Kb, Table 3).

| Chr. | Class 0.0 | Class 1.0 | Highly specific* | Class 1.1 | Class 2 | Subtotal | |
|---|---|---|---|---|---|---|---|
| No. | Density | ||||||
| P1 | 2,707 | 2,782,878 | 2,785,585 | 9.25 | 7,070 | 64,000 | 2,856,655 |
| P2 | 1,795 | 1,631,097 | 1,632,892 | 9.96 | 3,625 | 33,562 | 1,670,079 |
| P3 | 2,640 | 2,648,954 | 2,651,594 | 10.14 | 5,968 | 57,210 | 2,714,772 |
| P4 | 1,962 | 2,193,502 | 2,195,464 | 10.18 | 5,542 | 51,577 | 2,252,583 |
| P5 | 1,957 | 2,136,291 | 2,138,248 | 9.84 | 5,550 | 52,706 | 2,196,504 |
| P6 | 2,091 | 2,211,372 | 2,213,463 | 10.08 | 5,509 | 51,797 | 2,270,769 |
| P7 | 1,758 | 1,862,107 | 1,863,865 | 8.39 | 4,909 | 44,974 | 1,913,748 |
| P8 | 1,443 | 1,643,765 | 1,645,208 | 10.73 | 3,616 | 36,134 | 1,684,958 |
| P9 | 2,121 | 2,403,863 | 2,405,984 | 10.08 | 6,308 | 59,730 | 2,472,022 |
| P10 | 1,949 | 2,041,454 | 2,043,403 | 9.93 | 5,316 | 49,347 | 2,098,066 |
| P11 | 1,788 | 1,905,155 | 1,906,943 | 8.65 | 4,871 | 46,188 | 1,958,002 |
| P12 | 2,124 | 2,271,133 | 2,273,257 | 9.89 | 5,621 | 55,596 | 2,334,474 |
| P0 | 5,072 | 3,862,877 | 3,867,949 | 5.41 | 11,075 | 100,741 | 3,979,765 |
| Total | 29,407 | 29,594,448 | 29,623,855 | 8.81 | 74,980 | 703,562 | 30,402,397 |
*, equal to the sum of Class 0.0 and Class 1.0; the number of Class 0.1 spacers on all chromosomes is 0.
To validate the specificity of target sites belonging to the class 0.0 and class 1.0, a random set of 19 sites were chosen to be amplified by PCR, and then the PCR products were directly sequenced and assembled. After aligning them back to the Zunla-1 reference genome, all of the products were matched to one unique location in the genome (Fig 5, S1 Table and S1 Fig), indicating that the target sites of class 0.0 and class 1.0 had low risk of off-target.

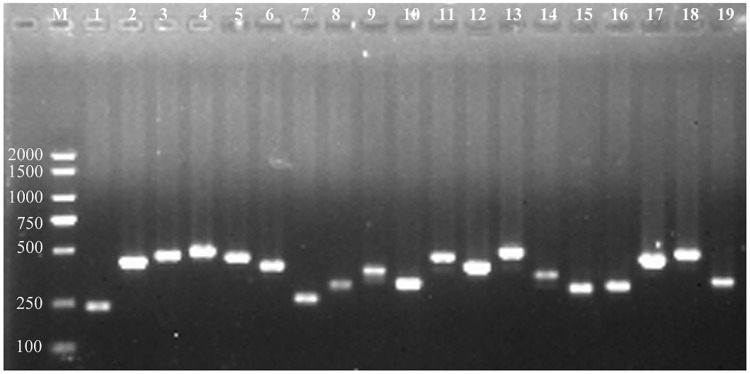
PCR amplification of 19 highly-specific target sites.
M, DL2000 plus, 1 to 10 represent A1 to A10 belonging to class0.0; 11 to 19 represent B1 to B9 belonging to class 1.0 (S1 Table).
Characterization of highly specific NGG-PAM sites’ distribution in pepper genome
The highly specific NGG-PAM sites were evenly distributed on all 12 chromosomes (P1~P12) of pepper (Fig 2). With the exception of P0, chromosomes P1 and P2 contained the maximum and minimum number of highly specific NGG-PAM sites, respectively (Table 3). The number of highly specific NGG-PAM sites in different genomic regions is shown in Table 4. Similar to the distribution of all NGG-PAM sites, there were a total of 26,699,387 (~90.13%) highly specific NGG-PAM sites located in intergenic regions, which was 9.13 times greater than the number in genic regions (Fig 4D). However, the average density of highly specific NGG-PAM sites in genic regions was higher than that in intergenic regions on the whole (13.80/Kb versus 8.47/Kb, Fig 4D) for Class 0.0 (0.015/Kb versus 0.008/Kb, Fig 4E) and Class 1.0 (13.79/Kb versus 8.46/Kb, Fig 4F). The same phenomenon occurs in the grape genome [21].

| Genomic Region | Class 0.0 | Class 1.0 | Total | |||
|---|---|---|---|---|---|---|
| No. | Percentage | No. | Percentage | No. | Percentage | |
| Intergenic | 26,234 | 89.21% | 26,673,153 | 90.13% | 26,699,387 | 90.13% |
| 5'UTR | 255 | 0.87% | 91,403 | 0.31% | 91,658 | 0.31% |
| 3'UTR | 107 | 0.36% | 99,099 | 0.33% | 99,206 | 0.33% |
| Exon | 450 | 1.53% | 939,750 | 3.18% | 940,200 | 3.17% |
| Intron | 2,347 | 7.98% | 1,783,696 | 6.03% | 1,786,043 | 6.03% |
| Splicing | 14 | 0.05% | 7,347 | 0.02% | 7,361 | 0.02% |
| Total | 29,407 | 100.00% | 29,594,448 | 100.00% | 29,623,855 | 100.00% |
We calculated the percentage of annotated genes that contained highly specific NGG-PAM sites identified in this study and found that 34,251 (~96.93%) out of 35,336 annotated genes exhibited at least one highly specific NGG-PAM site in their exons, and 90.50% of annotated genes exhibited at least 4 highly specific NGG- PAM sites (Fig 6 and S2 Table), indicating that the set of highly specific CRISPR/Cas9 editing sites identified in this study was widely applicable and will contribute to the minimization of off-target effects of CRISPR/Cas9 in pepper.

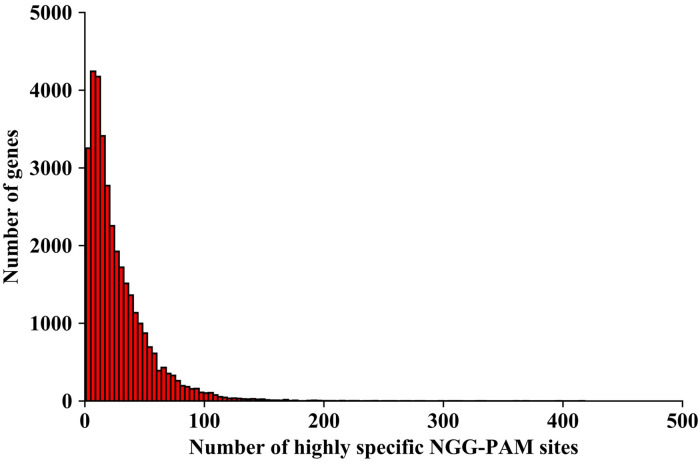
Histogram plots of gene numbers according to the number of exon-targeted highly specific NGG-PAM sites.
Acknowledgements
We thank Guojun Ouyang for his technical assistance in the analysis of highly-specific editing sites.
References
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
 Genome-wide identification and analysis of highly specific CRISPR/Cas9 editing sites in pepper (Capsicum annuum L.)
Genome-wide identification and analysis of highly specific CRISPR/Cas9 editing sites in pepper (Capsicum annuum L.)
