
These authors contributed equally
Lead Contact
- Altmetric
Drosophila provides a powerful model in which to study inflammation in vivo, and previous studies have revealed many of the key signaling events critical for recruitment of immune cells to tissue damage. In the fly, wounding stimulates the rapid production of hydrogen peroxide (H2O2).1,2 This then acts as an activation signal by triggering a signaling pathway within responding macrophages by directly activating the Src family kinase (SFK) Src42A,3 which in turn phosphorylates the damage receptor Draper. Activated Draper then guides macrophages to the wound through the detection of an as-yet unidentified chemoattractant.3, 4, 5 Similar H2O2-activated signaling pathways are also critical for leukocyte recruitment following wounding in larval zebrafish,6, 7, 8, 9 where H2O2 activates the SFK Lyn to drive neutrophil chemotaxis. In this study, we combine proteomics, live imaging, and genetics in the fly to identify a novel regulator of inflammation in vivo; the PTP-type phosphatase Pez. Pez is expressed in macrophages and is critical for their efficient migration to wounds. Pez functions within activated macrophages downstream of damage-induced H2O2 and operates, via its band 4.1 ezrin, radixin, and moesin (FERM) domain, together with Src42A and Draper to ensure effective inflammatory cell recruitment to wounds. We show that this key role is conserved in vertebrates, because “crispant” zebrafish larvae of the Draper ortholog (MEGF10) or the Pez ortholog (PTPN21) exhibit a failure in leukocyte recruitment to wounds. This study demonstrates evolutionary conservation of inflammatory signaling and identifies MEGF10 and PTPN21 as potential therapeutic targets for the treatment of inflammatory disorders.
•
Pez is a novel regulator of inflammation needed for macrophage recruitment to wounds
•During Drosophila inflammation, Pez acts in the H2O2/Src42a/Draper signaling axis
•Pez and Draper form dynamic clusters within macrophages in response to tissue damage
•Pez (PTPN21) and Draper (MEGF10) orthologs play conserved roles in fish leukocytes
Through the combination of proteomics, genetics, and live imaging, Campbell et al. identify Pez as a novel regulator of inflammation in vivo. Pez acts in the damage-sensing, H2O2/Src42a/Draper signaling axis, wherein it dynamically clusters with Draper to enable rapid leukocyte migration to epithelial wounds in both the fly and fish.
Results and Discussion
To identify further components of the H2O2-Src42A-Draper inflammatory signaling axis in Drosophila macrophages, we undertook a phosphoproteomics approach to identify phosphoproteins regulated downstream of H2O2 and Src42A. Control and src42A[E1] mutant stage 15 embryos were disaggregated by crushing to engage global inflammatory signaling (Figure S1A). Disaggregation was carried out both with or without catalase (to quench H2O2 signaling), and GFP-positive macrophages (srp-Gal4 driven upstream activating sequence [UAS]-GFP) were collected by fluorescence-activated cell sorting (FACS). The macrophage-specific peptides obtained were tandem mass tagged (TMT) labeled, phospho-enriched, and identified by liquid chromatography-mass spectrometry (Figure S1B). Finally, an organism-specific database search was conducted to identify the peptides isolated (Figures S1C–S1E). This revealed the protein tyrosine phosphatase (PTP)-type phosphatase Pez as differentially phosphorylated in the presence of both H2O2 and Src42A (Figures S1E and S1F). Because the ortholog of Pez (PTPN21) had previously been identified as an interactor and regulator of SFK signaling in other contexts,10, 11, 12 we chose to investigate Pez in inflammatory cell migration.
To determine whether Pez is expressed in embryonic macrophages, we used Pez-Gal4 (P{GawB}PezNP4748) to drive UAS-GFP and investigated GFP expression by immunofluorescence. Co-labeling with anti-singed (a macrophage marker in Drosophila)13 confirmed that Pez is expressed within macrophages at stage 15 of development (Figure S1G). We next sought to determine whether Pez plays a role in normal macrophage behavior using two independent Pez mutant lines (Figure 1A). Following their specification from the head mesoderm, macrophages follow a stereotypical migration pattern to become evenly distributed by the end of embryogenesis.14, 15, 16 This characteristic developmental dispersal of macrophages in Pez mutant embryos occurred normally, with macrophages following the expected dispersal routes at identical migratory speeds to controls (Figures S1H and S1I; Video S1).

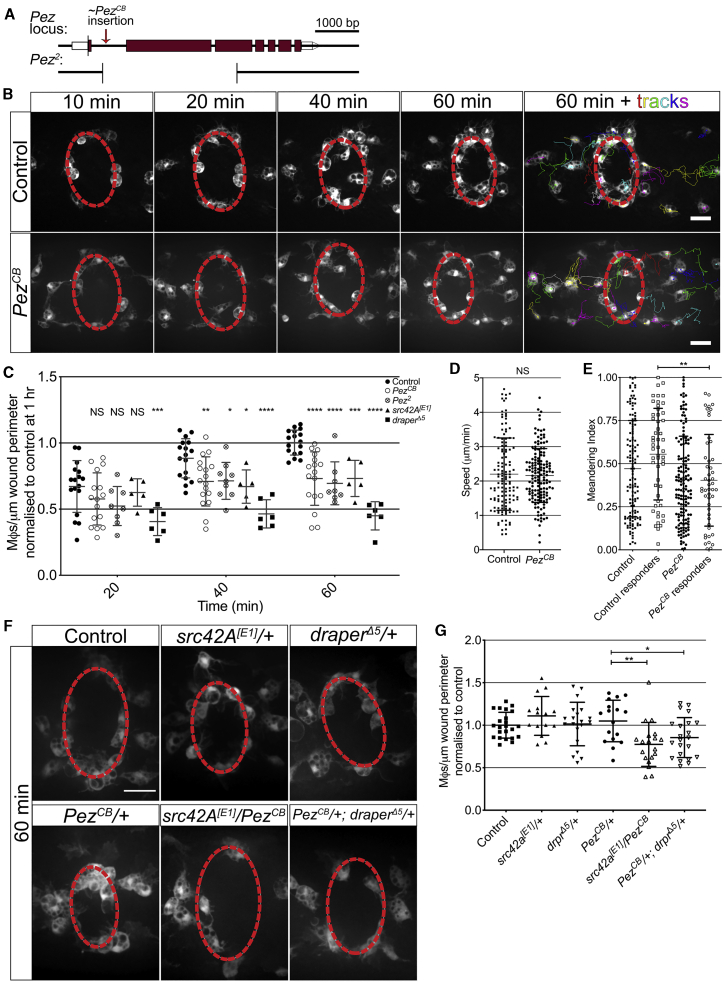
Pez Is Required for Macrophage Migration to Epithelial Wounds and Functions within the H2O2-Src42A-Draper Signaling Pathway
(A) Pez locus highlighting mutant alleles. Approximate CB insertion (6.056 kb) site is indicated. Pez2 deletion is marked below, adapted from Poernbacher et al.17
(B) Live imaging of inflammation following laser ablation reveals reduced macrophage recruitment in PezCB mutants. Wound margin is denoted by dashed red line. Cell tracks are shown at 1 h.
(C) Quantification reveals a significant decrease in macrophage numbers at wounds in the two Pez mutant lines at 40 and 60 min post-injury (n ≥ 10 wounded embryos/genotype; multiple t tests with Holm-Sidak multiple comparisons).
(D and E) Cell tracking reveals (D) macrophage speed post-wounding is unaffected in PezCB mutants (n ≥ 130 cells from ≥5 embryos/genotype; Mann-Whitney U test), and (E) meandering index is significantly reduced in responding (cells that reach the wound site at any point within 2 h) PezCB macrophages (n = 53 responders from ≥5 embryos/genotype; Mann-Whitney U test).
(F) Heterozygote (src42A[E1]/+, draperΔ5/+, and PezCB/+) and transheterozygote (src42A[E1]/PezCB and PezCB/+; draperΔ5/+) mutant embryos at 60 min post-wounding. Wound margin is denoted by dashed red line.
(G) Significantly reduced macrophage wound recruitment in transheterozygotes embryos versus PezCB/+ (n ≥ 15 wounded embryos/genotype; one-way ANOVA with multiple comparisons).
All error bars are mean ± SD. NS, not significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, and ∗∗∗∗p < 0.001. All scale bars represent 20 μm. See also Figure S2 and Videos S1, S2, and S3.
During this migration, macrophages actively clear developmentally generated apoptotic corpses, which are identifiable inside GFP-expressing macrophages as fluorescent-negative vacuoles.18 Live imaging of Pez mutant macrophages at stage 15 revealed normal cell morphology, with cells displaying lamellipodial protrusions and containing intracellular vacuoles (Figure S1J; Video S2). Quantification of vacuole numbers in Pez mutant macrophages revealed no significant defect in their phagocytic capability (Figure S1K). Finally, live imaging revealed that, following the completion of their dispersal, Pez mutant macrophages migrate at the same speed and in the same manner as control cells (Figures S2A–S2C; Video S2). Together, this demonstrates that Pez is dispensable for basal macrophage migration and function.
To investigate whether Pez plays a role in the inflammatory recruitment of macrophages to wounds, we carried out live imaging following laser ablation. In control animals, this leads to a rapid recruitment of macrophages to the wound site, with numbers peaking 1 h after insult (Figures 1B and 1C). Macrophage counts 1 h post-injury (1 hpi) revealed a significant reduction in macrophage recruitment in both PezCB and Pez2 mutant embryos when compared to controls (Figures 1B and 1C). This was despite there being significantly more macrophages within Pez mutant embryos (Figure S2D). Importantly, Pez mutant wounds closed at comparable rates to controls (Figure S2E). Interestingly, the Pez wound recruitment phenotype is comparable to that observed following loss of Src42a (Figure 1C).
To further investigate this inflammatory defect, PezCB mutant macrophages were tracked following live imaging (Video S3). This revealed that the reduction in the number of macrophages present at wounds in PezCB mutants was not due to a slower inflammatory migration speed (Figure 1D) but due to a lower meandering index in responding cells (Figure 1E). This corresponded to a later arrival time and lower wound residency of macrophages in PezCB mutants when compared to controls (Figures S2F and S2G).
Because we initially sought to identify novel interactors in the H2O2/Src42A/Draper inflammatory signaling pathway and the Pez phenotype is comparable to that of Src42a mutants and consistent with a macrophage navigational defect, genetic interaction studies were employed to determine whether Pez lies within the same signaling axis. We found no defect in macrophage recruitment to wounds made in heterozygous src42A[E1]/+, draperΔ5/+, or PezCB/+ embryos. However, wounds made to src42A[E1]/PezCB or PezCB/+; draperΔ5/+ embryos showed a significant reduction in the number of macrophages recruited at 1 hpi when compared to PezCB/+ heterozygotes (Figures 1F and 1G). Taken together, these data demonstrate that Pez is a novel component of the H2O2/Src42A/Draper signaling pathway and drives macrophage recruitment to wounds.
Because Pez is widely expressed in stage 15 embryos (Figure S1G), we next confirmed that the role of Pez in macrophage wound recruitment was cell autonomous. To achieve this, we used two macrophage-specific drivers (srp-gal4 and crq-gal4) to express one of two Pez-specific RNAi constructs and quantified macrophage recruitment to wounds. Macrophage-specific Pez RNAi led to a significant reduction in the number of macrophages at epithelial wounds at 1 hpi, demonstrating that Pez is required within macrophages for effective chemotaxis (Figures 2A and 2B). These RNAi constructs were validated and were sufficient to significantly reduce Pez protein levels in vivo (Figures 2C and 2D).

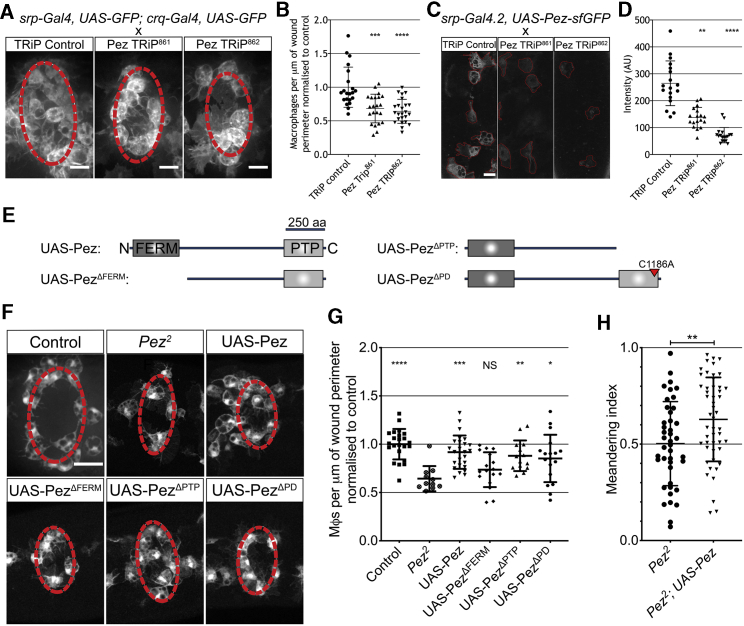
The Role of Pez in Macrophage Wound Recruitment Is Cell Autonomous and Dependent upon the FERM Domain
(A) Macrophage-specific expression of Pez-RNAi (TRiP constructs) impairs inflammatory recruitment to wounds (images 1 h post-wounding). Scale bars represent 10 μm. Wound margin is denoted by dashed red line.
(B) Pez-RNAi significantly reduces macrophage recruitment to wounds compared to control (n ≥ 21 wounded embryos/genotype; Kruskal-Wallis with Dunn’s multiple comparisons).
(C) Pez-sfGFP expression in macrophages (outlined in red) co-expressing either control RNAi or either Pez-RNAi. Scale bars represent 10 μm.
(D) Both RNAi lines significantly reduce macrophage Pez-sfGFP intensity levels (n = 18 cells from 6 embryos/genotype; Kruskal-Wallis with Dunn’s multiple comparisons).
(E) UAS-Pez expression constructs. FERM domain and PTP domains noted and deletions depicted. For phosphatase dead construct (UAS-PezΔPD), the mutated cysteine is noted. Adapted from Poernbacher et al.17
(F) Images of wounded Pez2 embryos with macrophage-specific expression of indicated Pez constructs, 1 h post-ablation. Scale bar represents 20 μm. Wound margin is marked by dashed red line.
(G) Macrophage-specific expression of UAS-Pez, UAS-PezΔPD, and UAS-PezΔPTP (but not PezΔFERM) is sufficient to rescue Pez2 wound recruitment defect (n ≥ 13 wounded embryos/genotype; one-way ANOVA with Dunnett’s multiple comparisons to Pez2).
(H) Quantification of meandering index reveals specific expression of Pez rescues the inflammatory chemotaxis of Pez2 macrophages (n ≥ 42 cells from n ≥ 5 wounded embryos/genotype; unpaired t test).
All error bars are mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, and ∗∗∗∗p < 0.001. See also Figure S2 andVideo S4.
We next sought to investigate the mechanism by which Pez is acting within chemotaxing macrophages. As well as a PTP domain, Pez harbors an N-terminal FERM domain (Figure 2E). To determine which domain of Pez is functional during macrophage recruitment, we expressed truncated Pez constructs17 in macrophages alongside GFP in a Pez2 mutant background (Figures 2E and 2F). We re-expressed four Pez constructs in Pez2 mutant macrophages—full-length Pez (UAS-Pez), Pez lacking the FERM domain (UAS-PezΔFERM), Pez lacking the PTP domain (UAS-PezΔPTP), and a phosphatase-dead Pez construct (UAS-PezΔPD). As expected, macrophage-specific expression of the full-length construct rescued both the wound recruitment and chemotaxis defect seen at 1 hpi in Pez2 mutants (Figures 2F–2H). Interestingly, expression of either of the phosphatase mutant constructs also rescued the mutant phenotype (Figures 2F and 2G). However, the ability of Pez mutant macrophages to migrate to wounds was not restored following the expression of UAS-PezΔFERM—demonstrating a specific requirement for the FERM domain of Pez in driving macrophage wound recruitment (Figures 2F and 2G; Video S4). Intriguingly, it is the FERM domain of the human Pez ortholog PTPN21 that has been demonstrated to directly bind to Src family kinases.12
As FERM domains are involved in protein localization,19 we generated tagged UAS-Pez constructs to investigate Pez dynamics in macrophages in vivo (Figure 3A). Macrophage-specific expression of Pez-sfGFP was sufficient to rescue recruitment to wounds in a Pez2 mutant (Figure S2H), and live imaging of Pez-sfGFP-expressing macrophages revealed dynamic puncta that formed within the lamellipod of macrophages before rapidly shuttling back toward the cell body at a rate of 0.12 ± 0.01 μm/s (Figure 3B). Upon wounding, this process was dramatically stimulated in the lamellipods of macrophages undergoing inflammatory chemotaxis (Video S5), resulting in a transient pulse of lamellipodial Pez puncta in macrophages within the vicinity of the wound, which then collectively flowed into the cell body.

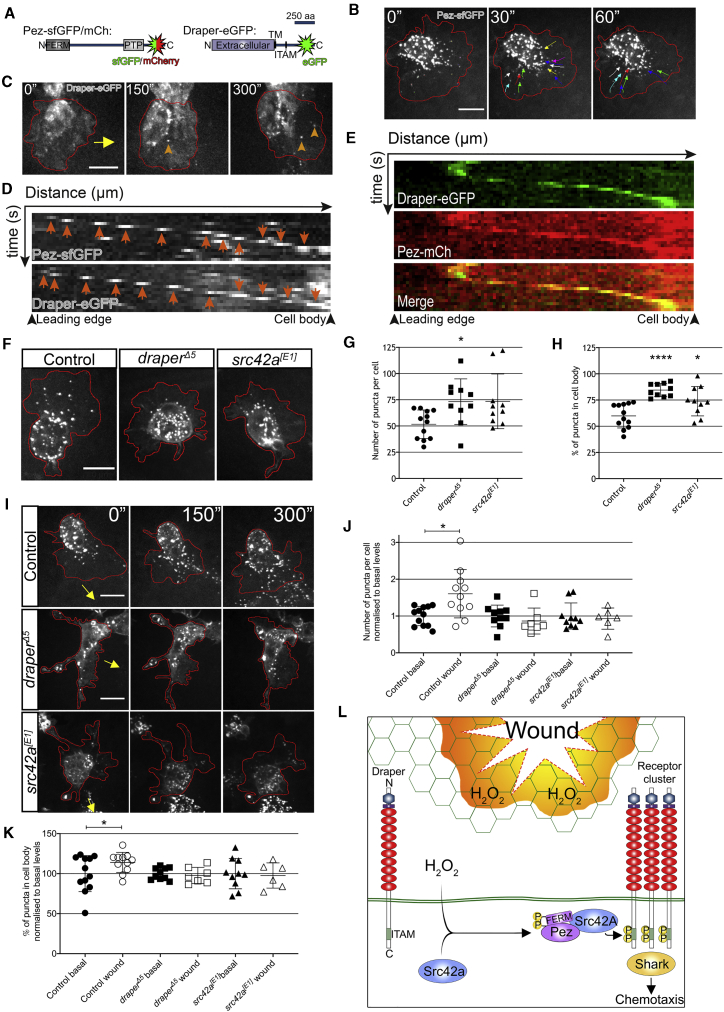
Dynamic Pez Puncta Are Stimulated upon Wounding in a Draper-Dependent Manner
(A) Diagrams of fluorescently tagged Pez and Draper constructs. For Pez, the FERM and PTP domains are shown. For Draper, the N-terminal extracellular domain is noted, along with the transmembrane domain (TM) and immunoreceptor tyrosine activation motif (ITAM).
(B) Pez forms puncta within the cell body and lamellipod. Dynamic lamellipodial puncta flow inward from the cell periphery (denoted by red line). Colored arrows show puncta tracking over 1 min.
(C) Dynamic Draper-EGFP puncta (orange arrowheads) induced post-wounding. Red line donates cell periphery; yellow arrow indicates direction of wound.
(D) Kymographs of individual Pez-sfGFP and Draper-EGFP puncta (orange arrows) following wounding demonstrate similar dynamics over time.
(E) Kymograph of Draper-EGFP punctum reveals colocalization with Pez-mCh following wounding. For all kymographs, the x axes represent distance starting at lamellipod leading edge (174 nm/pixel; 17.4 μm total). The y axes represent time (10 s/pixel; 2.5 min total).
(F) Lamellipodial Pez-sfGFP puncta are suppressed in draperΔ5 and src42A[E1] mutant macrophages.
(G and H) Puncta number (G) and distribution (cell body versus lamellipod; H) significantly altered in mutants (n ≥ 10 cells from ≥5 embryos/genotype; Kruskal-Wallis with Dunn’s multiple comparisons and one-way ANOVA with Tukey’s comparisons, respectively).
(I) Images of control, draperΔ5, and src42A[E1] mutant macrophages (red outlines) 5 min post-wounding. Direction of wound marked by yellow arrow.
(J and K) Analysis of Pez puncta 5 min post-wounding reveals (J) a wound-induced significant increase in puncta number that is dependent on both Draper and Src42A (n ≥ 6 cells from ≥5 embryos/genotype for each condition; Kruskal-Wallis with Dunn’s multiple comparisons) and (K) a wound-induced significant increase in the proportion of puncta residing within the cell body of control cells that is absent in draper and src42A mutants (n ≥ 6 cells from ≥5 embryos/genotype for each condition; one-way ANOVA with Sidak’s multiple comparisons). All error bars are mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗∗p < 0.001. All scale bars represent 10 μm.
(L) Proposed role of Pez in wound-induced Draper clustering. Under basal conditions, Draper’s ITAM domain remains in an inactive state. Following H2O2-mediated Src42A activation, phosphorylated Pez is recruited to Draper clusters via its FERM-domain-mediated interaction with Src42a. Acting as an adaptor, Pez coordinates inflammatory Draper signaling via effectors such as Shark, leading to efficient macrophage chemotaxis.
Draper has also been shown to cluster into mobile puncta in Drosophila macrophage cell lines—a process that is proposed to drive its activation cycle akin to the mammalian T cell receptor.20 In order to investigate whether this occurs in vivo, we expressed Draper-EGFP in macrophages and visualized its localization through live imaging (Figure 3C). Limited Draper puncta were observed under basal conditions within the cell body of migrating macrophages. However, upon wounding, Draper puncta were observed forming at the leading edge of the lamellipod and flowing back toward the cell body (Figure 3D; Video S6)—which was highly reminiscent of that observed with Pez-sfGFP (Figures 3B and 3D). Co-expression of Draper-EGFP and Pez-mCherry revealed a clear colocalization of these two proteins at wound-induced puncta (Figures 3E and S2I).
We next investigated the localization of fluorescently tagged Draper or Pez in Pez, draper, and src42A mutants. Pez was not necessary for Draper puncta, consistent with the multimerization of Draper driving receptor clustering and implying that Pez instead plays a role in downstream signaling (Figure S2J). In the absence of either Draper or Src42A, macrophages under basal (unwounded) conditions retained Pez-sfGFP puncta, albeit with a slight increase in the absolute number of puncta per cell in draper mutant macrophages (Figures 3F and 3G). However, when compared to controls, the dynamic subcellular localization of Pez was strongly perturbed in both these mutants, wherein the Pez puncta were predominantly sequestered in the cell body (Figures 3F and 3H). Furthermore, in response to wounding, there was no stimulation of Pez clustering in either draper or src42A mutant macrophages as observed in control cells (Figures 3I–3K). These data imply that Pez dynamically relocalizes to the lamellipod in response to wound-induced Draper clustering and Src42a activity in order to potentiate inflammatory signaling.
Importantly, the few remaining lamellipodial Pez puncta within draper and src42A mutant macrophages appeared to behave normally and flowed toward the cell body with similar dynamics to those in controls (Figure S2K). This, together with the high basal number of Pez puncta present in either mutant relative to control, and the basal clustering of Pez in the control in the absence of detectable Draper puncta, is consistent with Pez having targets other than Draper. However, in response to the wound-induced surge in Draper clustering, Pez is co-opted into these puncta via its FERM-domain-mediated interaction with Src42a. The absence of any role for Pez’s catalytic activity in the Draper-mediated inflammation suggests that Pez is acting as an adaptor protein at Draper clusters. As such, Pez organizes these clusters into effective signaling hubs, allowing the critical threshold of activity to be met in order to drive inflammatory recruitment (Figure 3L).
Having identified a novel regulator of damage-induced inflammation in Drosophila, we sought to determine whether the activity of Pez in regulating chemotaxis is conserved in the vertebrate. We therefore investigated both the ortholog of Pez—PTPN21—and the ortholog of Draper—MEGF10—in a zebrafish leukocyte wound recruitment model. First, to confirm whether PTPN21 and MEGF10 are expressed in larval zebrafish leukocytes, we mined existing RNA sequencing (RNA-seq) datasets for transcript expression.21,22 This revealed that both ptpn21 and megf10 transcripts were enriched within neutrophils by 3 days post-fertilization (dpf) and macrophages by 2 dpf (Figures S2L and S2M).
To investigate what effects the loss of PTPN21 and MEGF10 have on the development of zebrafish leukocytes, we independently utilized the transgenic neutrophil line Tg(lysC:NLS-mScarlet)23 and macrophage reporter lines Tg(mpeg1.1:NLS-mScarlet) and Tg(mpeg1.1:EGFP) to generate CRISPR-Cas9-mediated mutant larvae (“crispants”; Figures S2N and S2P). Using restriction fragment length polymorphism analysis (RFLP),24 we were able to validate the successful generation of F0 crispant larvae (Figures S2O and S2Q). Imaging the entirety of the crispant fish revealed leukocyte distribution was unaltered when compared with wild type (Figure 4A). However, we found an increase in neutrophil number in PTPN21 crispants—akin to the macrophage phenotype we identify in Drosophila—and a 20% reduction in macrophage numbers in MEGF10 crispant fish (Figure 4B).

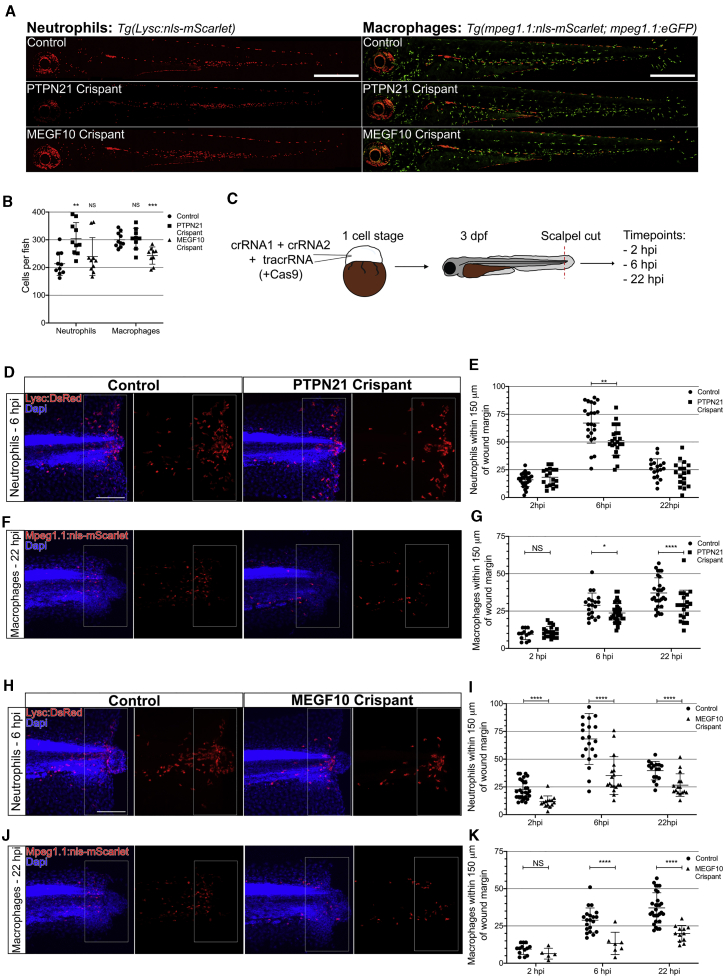
The Orthologs of Pez (PTPN21) and Draper (MEGF10) Are Required for Leukocyte Recruitment to Wounds in Zebrafish Larvae
(A) Representative images of entire control, PTPN21 crispant, and MEGF10 crispant zebrafish 3 dpf larvae expressing either lysc:nls-mScarlet (neutrophil marker) or mpeg1.1:nls-mScarlet and mpeg1.1eGFP (macrophage marker). Scale bars represent 500 μm.
(B) Quantification of leukocyte numbers revealed an increase in neutrophils in PTPN21 crispants (n = 10 larvae/genotype; Kruskal-Wallis with Dunn’s multiple comparisons) and a decrease in macrophage in MEGF10 crispants (n ≥ 7 larvae/genotype; one-way ANOVA with Dunnett’s multiple comparisons).
(C) For wound studies, zebrafish embryos (one cell stage) were injected with 2 CRISPR guide RNAs (crRNAs) alongside tracrRNA and raised to 3 dpf. Following tailfin transection, fish were stained at 2, 6, and 22 h post-injury (hpi).
(D) Images of wounded control larvae and PTPN21 crispants at 2, 6, and 22 hpi time points. Tg(lysC:DsRed) (red) zebrafish co-stained with DAPI (blue) are shown. Quantification zone of 150 μm proximal to the wound margin is marked by the white box across all images.
(E) Significantly reduced neutrophils recruited to the wound at 6 hpi in PTPN21 crispant larvae compared to control (n ≥ 18 wounded larvae/genotype for each time point; multiple t test).
(F) Images of wounded control larvae and PTPN21 crispants at 2, 6, and 22 hpi time points. Tg(mpeg1.1:nls-mScarlet) (red) zebrafish co-stained with DAPI (blue) are shown. Quantification zone of 150 μm proximal to the wound margin is marked by the white box across all images.
(G) Significantly reduced macrophages recruited to the wound at 6 hpi and 22 hpi in PTPN21 crispants (n ≥ 17 wounded larvae/genotype for each time point; multiple t test).
(H) Images of wounded control larvae and MEGF10 crispants at 2, 6, and 22 hpi. Tg(lysc:DsRed) (red) zebrafish stained with DAPI (blue) are shown. Quantification zone of 150 μm proximal to the wound margin is marked by the white box across all images.
(I) Significantly reduced neutrophils recruited to the wound at all time points in MEGF10 crispant compared to the control (n ≥ 15 wounded larvae/genotype for each time point; multiple t test).
(J) Images of wounded control larvae and MEGF10 crispants at 2, 6, and 22 hpi time points. Tg(mpeg1.1:nls-mScarlet) (red) zebrafish co-stained with DAPI (blue) are shown. Quantification zone of 150 μm proximal to the wound margin is marked by the white box across all images.
(K) Significantly reduced macrophages recruited to the wound at 6 hpi and 22 hpi in PTPN21 crispant (n ≥ 13 wounded larvae/genotype for each time point; multiple t test).
All error bars are mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗∗p < 0.001. All scale bars represent 100 μm. See also Figure S2.
We next investigated leukocyte recruitment to tailfin transection wounds made in 3 dpf control and crispant embryos (Figure 4C). In control animals, these large wounds trigger a robust inflammatory response—with neutrophil recruitment peaking at 6 hpi and remaining at the wound until 24 hpi and macrophage numbers continuing to increase over a 24 h period.25 Consistent with our findings in the fly, wounds made to PTPN21 crispant fish revealed a significant reduction in the peak number of neutrophils recruited to tail fin wounds at 6 hpi (Figures 4D and 4E) and a reduction in macrophage numbers at both 6 hpi and 22 hpi (Figures 4F and 4G).
Because Pez and Draper work together to drive inflammation in Drosophila macrophages, we investigated whether MEGF10 is also required for leukocyte recruitment to wounds. Indeed, neutrophils in MEGF10 crispants showed a significantly reduced wound recruitment as early as 2 h post-wounding, and in macrophages, MEGF10 crispant showed nearly 50% reduction at 6 and 22 h post-wounding (Figures 4H–4K). This provides compelling evidence that both PTPN21 and MEGF10 regulate inflammation in zebrafish and that the H2O2-Src42A-Pez-Draper signaling axis is an evolutionarily conserved signaling pathway that directs the earliest innate immune inflammatory response to damage in vivo. Further studies are required to identify more components of this inflammatory signaling axis, but from this study, PTPN21 and MEGF10 emerge as key regulators of inflammation and should now be explored as potential therapeutic targets for the treatment of inflammatory disorders.
STAR★Methods
Key Resources Table

| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| α-GFP | Abcam | Abcam Cat# ab13970; RRID: AB_300798 |
| α-singed | DSHB | DSHB Cat# sn 7C; RRID: AB_528239 |
| α-armadillo | DSHB | DSHB Cat# N2 7A1 ARMADILLO; RRID: AB_528089 |
| α-mCherry | Abcam | Abcam Cat# ab125096; RRID: AB_11133266 |
| α-chicken AF488 | Invitrogen | Molecular Probes Cat# A-11039; RRID: AB_142924 |
| α-mouse AF568 | Invitrogen | Molecular Probes Cat# A-21124; RRID: AB_141611 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Multisite Gateway Three Fragment vector construction kit | Invitrogen | 12537023 |
| Catalase | Sigma | C1345 |
| Trypsin | Sigma | T1426 |
| 16% Methanol Free Paraformaldehyde | Alfa Aesar | 11490570 |
| Heptane | Sigma | 34873 |
| Triton X-100 | Sigma | T8787 |
| BSA | Sigma | A4503 |
| Vectashield Mounting Media | Vector Labs | H-1000 |
| Voltalef oil | VWR | 24627.188 |
| NLS-Cas9 | NE Biolabs | M0646 |
| RNase free water | Sigma | W4502 |
| DNeasy Blood and Tissue Kit | QIAGEN | 69504 |
| MyTaq Red Mix | Meridian Bioscience | BIO-25043 |
| BslI | NEBiolabs | R0555 |
| MWoI | NEBiolabs | R0573 |
| Tricaine/MS-22 | Sigma | E10521 |
| Horse serum | Sigma | H0146 |
| Critical commercial assays | ||
| Multisite Gateway Three Fragment vector construction kit | Invitrogen | 12537023 |
| Experimental Models: Organisms/Strains | ||
| DROSOPHILA | N/A | |
| P{GawB}PezNP4748 | Kyoto Stock Center | RRID: DGGR_104771 |
| serpentHemoGal4 | 23 | N/A |
| serpentHemoGal4.2 | This work | N/A |
| croquemort-Gal4 | 24 | N/A |
| UAS-GFP | BDSC | RRID: BDSC_6874 |
| UAS-2xeGFP | BDSC | RRID: BDSC_6658 |
| UAS-PezTRiP861 | BDSC | RRID: BDSC_33918 |
| UAS-PezTRiP862 | BDSC | RRID: BDSC_33919 |
| UAS-TRiPLuciferase | BDSC | RRID: BDSC_31603 |
| UAS-Pez | 15 | N/A |
| UAS-PezΔFERM | 15 | N/A |
| UAS-PezΔPTP | 15 | N/A |
| UAS-PezΔPD | 15 | N/A |
| UAS-Pez-sfGFP | This work | N/A |
| UAS-Pez-mCherry | This work | N/A |
| UAS-Draper-eGFP | This work | N/A |
| w1118 | BDSC | RRID: BDSC_3605 |
| PezCB | Kyoto Stock Center | RRID: DGGR_123596 |
| Pez2 | 15 | N/A |
| src42A[E1] | [25] BDSC | RRID: BDSC_6408 |
| draperΔ5 | 26 | N/A |
| ZEBRAFISH | N/A | |
| Tg(lysC:DsRed2) | 23 | N/A |
| Tg(mpeg:NLS-Scarlet)ed207 | This work | N/A |
| Tg(lysC:NLS-mScarlet)ed229 | This work | N/A |
| Tg(mpeg1,1:EGFP)gl22 | 27 | N/A |
| Oligonucleotides | ||
| crRNA MEGF10 1: GCTACAGAACGGCCTATCGC | Sigma | Custom |
| crRNA MEGF10 2: TGTCAGTGTGAGCCGGGCTG | Sigma | Custom |
| crRNA PTPN21 1: GGTGGCATCATGTAGGGCTG | Sigma | Custom |
| crRNA PTPN21 2: GAATCAGGGCGCTGTGCCGG | Sigma | Custom |
| crRNA MEGF10 locus 1 genotyping fwd: aaccgaaaacaaatcaaaggagggc | Eurofins | Custom |
| crRNA MEGF10 locus 1 genotyping rev: acattgtaaaagcgctacagaaacaaa | Eurofins | Custom |
| crRNA MEGF10 locus 2 genotyping fwd: tgcttgtgtttgtttgcttg | Eurofins | Custom |
| crRNA MEGF10 locus 2 genotyping rev: tgaatggcttttgtcactcg | Eurofins | Custom |
| crRNA PTPN21 locus 1 genotyping fwd: gcagttcactataaaggcagc | Eurofins | Custom |
| crRNA PTPN21 locus 1 genotyping rev: gtggccgttaaagtgcatc | Eurofins | Custom |
| crRNA PTPN21 locus 2 genotyping fwd: gatgtcctccaacccaagca | Eurofins | Custom |
| crRNA PTPN21 locus 2 genotyping rev: aaaggatactgtcctgcgcc | Eurofins | Custom |
| tracrRNA | Sigma | TRACRRNA05N |
| Software and Algorithms | ||
| GraphPad Prism V8.4.1 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ/FIJI | National Institute of Health | https://imagej.nih.gov/ij/ |
| Volocity | PerkinElmer | https://www.perkinelmer.com/lab-products-and-services/resources/cellular-imaging-software-downloads.html |
| Zen Black | Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
| Photoshop | Adobe | https://www.adobe.com/uk/products/photoshop.html |
| Illustrator | Adobe | https://www.adobe.com/uk/products/illustrator.html |
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Will Wood w.wood@ed.ac.uk
Materials availability
Plasmids and transgenic lines generated in this study are available by request.
Data and code availability
This study did not generate datasets/code.
Experimental Model and Subject Details
Drosophila stocks and genetics
Drosophila stocks were maintained according to standard protocols26. Embryos for live imaging and fixation were collected from apple juice agar plates from overnight laying cages (all incubated at 22°C, with the exception of RNAi experiments, which were kept at 29°C overnight to boost expression).
The following driver lines were combined with UAS constructs: Pez-Gal4 (P{GawB}PezNP4748, Kyoto), serpentHemoGal428, serpentHemoGal4.2 (srp-Gal4.2, an enhanced expression construct generated in the lab by Dr Kate Comber and Dr Fred Rodrigues) and croquemort-Gal4 (crq-Gal4;29). The following UAS constructs were used in this study: UAS-GFP, UAS-PezTRiP861 (VDRC), UAS-PezTRiP862 (VDRC), UAS-Pez, UAS-PezΔFERM, UAS-PezΔPTP, UAS-PezΔPD (all a kind gift of Dr. Hugo Stocker17), UAS-Pez-sfGFP and UAS-Draper-eGFP (both generated in this study – synthesized and cloned into pUASt by GeneArt and commercially injected by Best Gene Inc.). The mutant alleles used in this study were: w1118 (as a control background), PezCB (P{RS3} insert of 6.046 Kb - Kyoto), Pez2 (3083 bp deletion – line a gift from Dr. Hugo Stocker17), src42A[E1] (EMS point mutant30) and draperΔ5 (1379 bp deletion31).
Zebrafish lines and rearing
All zebrafish lines were kept and raised under standard conditions32 and all experiments were approved by the British Home Office (project license No PEE579666). Tg{lysC:DsRed2}23 and Tg(lysC:NLS-mScarlet)ed229 lines were used to label neutrophils, whereas macrophages were visualized through Tg(mpeg1,1:EGFP)gl2232 and Tg(mpeg1.1:NLS-mScarlet)ed207. The Tg(mpeg:NLS-Scarlet)ed207 and Tg(lysC:NLS-mScarlet)ed229 line was generated using the Multisite Gateway Three Fragment vector construction kit (Invitrogen 12537023). In brief a 5′ Entry vector containing 1.85K mpeg1.1 promoter fragment or 8K lysC promoter (gift from Prof. Steve Renshaw), pME-nlsScarlet, p3E-(SV40)PolyA and a pDest-Tol2-polyA vector (Tol2 kit33) were added into a LR reaction according to manufacturer’s instruction. The recombinational cloning resulted in the final pDest-Tol2-mpeg1.1::nls-Scarlet-polyA and pDest-Tol2-lysC::nls-Scarlet-polyA vector. The final transgenic DNA plasmids were used to generate F0 founder fish. F1 adult fish was out crossed with wild-type fish, brightly labeled larvae were selected as F2. All experiments described were using F3 larvae from the F2 in cross.
Method Details
Proteomics screen
Following overnight laying, stage 15 w∗;srp-GAL4,UAS-GFP and w∗;src42A[E1],srp-Gal4,UAS-GFP dechorionated embryos were collected in both the presence and absence of catalase. For the catalase treatment a 100x solution of 0.1 g catalase (Sigma C1345) in 1.9 mL of PBS was added to all solutions cells came into contact with. 250-280 embryos per sample were placed into the tip of a cold loose-fitting Dounce homogenizer. Embryos were then pestled gently in 250 μL of Seecof buffer.34 The pestle was washed with 750 μL dissociation media34 and transferred to an Eppendorf tube. The embryonic suspension was then sieved through at 40 μm nylon mesh and collected into a cold Eppendorf tube. The mixture was then centrifuged for 5 min at 350 rcf. at 4°C, the supernatant removed, and cells resuspended in 250 μL cold Seecof. Samples were kept on ice at all times.
Macrophages were then sorted by gating single/live/GFP+ cells into lysis buffer and kept at −80°C until further analysis. A total of 6 samples per treatment containing between 376,000-466,000 total cells were then pooled. Pooled samples were then trypsin (Sigma T1426) digested, and TMT labeled at the peptide level. All samples where then combined and phospho-enriched using a TiO2 column. Finally, phospho-enriched and TiO2 flow through (containing the non-phosphorylated peptides) were sent to LC-MS analysis. Returned peptide spectra were then compared to Drosophila melanogaster databases to obtain protein information. Ratios of peptide abundances were compared across sample type. Due to low overall protein abundance, the dataset was adjusted by normalizing to the median protein ratios of total protein levels between samples. See Figure S1.
Drosophila Fixation and immunostaining
Dechorionated embryos were collected in a 2 mL glass vial containing a 1:1 4% PFA:heptane mixture. Embryos were left tumbling in fixative for 30 minutes at room temperature, washed with PBS-Tx-BSA and incubated in primary antibodies at 4°C overnight. After washing with PBS-Tx-BSA and blocking with horse serum (2% v/v, Sigma-Aldrich) for 30 minutes, embryos were incubated with secondary antibodies for 1 hour at room temperature. Washed embryos were then mounted in Vectashield mounting medium. Primary antibodies: α-GFP (1:500, Abcam Ab13970), α-singed (Fascin, 1:100 DSHB Sn 7C) and α-armadillo (β catenin, 1:25, DSHB N2 7A1). Secondary antibodies: α-chicken AF488 (1:250 Invitrogen A11039) and α-mouse AF568 (1:250 Invitrogen A21124).
Drosophila Live imaging
Dechorionated embryos were staged and genotyped (by selecting against fluorescent balancer chromosomes) before being mounted in a droplet of VOLTALEF oil (VWR) on a glass slide, flanked by supporting coverslips with a bridging coverslip sealed on top as previously described.35 Images at z-slice intervals of 0.5 μm were acquired with a spinning disc confocal microscope (Perkin Elmer Ultraview) with either a 63x (NA 1.4) or a 40x (NA 1.3) objective. Epithelial wounds were generated by laser ablation as previously described36 using a nitrogen-pumped Micropoint ablation laser (Andor Technologies).
RNaseq data mining
Existing RNaseq datasets21,22 were accessed through Gene Expression Omnibus to mine for expression of PTPN21 and MEGF10 in 3 dpf larval neutrophils and 2 dpf larval macrophages. For both datasets, the raw counts matrix was used to calculate Transcripts Per Million (TPM) to account for sequencing depth and gene transcript length.
CRISPR-Cas9 gene editing of zebrafish embryos
CRISPR/Cas9-mediated mutant larvae “Crispants” were generated as described previously24. Briefly, CRISPR guide RNA (crRNA) sequences in which restriction enzyme recognition sequences overlapped the Cas9 cut site were identified in PTPN21 and MEGF10 exons and commercially synthesized (Sigma-Aldrich). 1 μL of each crRNAs were injected together into the embryo at the single-cell stage alongside 1 μL tracrRNA (Sigma-Aldrich), 0.3 μL NLS-Cas9 (NE Biolabs) and 1.7 μL RNase free water (Sigma-Aldrich). For neutrophil controls, Cas9 was omitted and replaced with a further 0.3 μL RNase free water. For macrophage experiments wound recruitment was compared to uninjected clutch-mates. Genotyping to confirm successful gene editing was performed following DNA extraction from individual larvae (95°C in 50mM NaOH for 1 hr, followed by addition of 0.5 M Tris-HCl pH 8.0) as previously described24). PCR of the edited region was performed using MyTaq Red Mix (Meridian Bioscience) and fragments were subsequently digested over night by the addition of 1 μL BslI or MwoI (NEBiolabs) directly to the reaction. Fragments were then resolved on a 2% agarose gel.
To quantify leukocyte numbers throughout the entire zebrafish larvae, control and Crispant fish were raised to 3 dpf and imaged using the VAST BioImager microscope platform as previously described37. Briefly, anesthetised live fish were mounted in glass capillaries and imaged laterally using a 1.6x post-magnification adaptor combined with a C-Plan-Apochromat 10x (NA 0.5) dipping lens (Carl Zeiss) and dual AxioCam 506 m CCD cameras (Carl Zeiss). Stitched maximum intensity projections of the entire larvae were imported into FIJI (NIH) and cell counter used to manually count fluorescent leukocyte nuclei.
Tailfin transection, fixation and staining
3 dpf larvae were anaesthetised by the addition of 0.02% buffered 3-aminobenzoic acid ethyl ester (Tricaine/MS-222) into the embryo medium and were left until paralysed. Using a scalpel, the entire tail fin and a small portion of the trunk distal to the end of the vasculature was removed. The embryos were then placed in fresh medium and allowed to recover. At 2 hours post injury (hpi), 6 hpi and 22 hpi larvae were culled using excess Tricaine. Culled larvae were then placed in an Eppendorf containing 4% PFA, 0.4% Triton-X diluted in PBS and fixed overnight at 4°C or at room temperature for 2 hours.
Wholemount immunostaining of zebrafish larvae was performed as described previously38. Wash buffer comprising PBS containing 0.1% Triton-X (PBST Sigma-Aldrich) and 5% horse serum (Sigma Aldrich) was used for blocking. Both primary and secondary antibodies were diluted in PBST containing 2%–5% horse serum and were left to incubate over night at 4°C. DAPI was added to secondary antibodies to visualize the entire tissue. Primary antibody: α-mCherry (1:500, Abcam Ab125096). Secondary antibody: α-mouse AF 568 (1:250 Invitrogen A21124). Stained samples were mounted laterally in Vectashield on glass slides and imaged on a Zeiss LSM880 confocal microscope using a 25x objective (NA 0.8).
Quantification and Statistical Analysis
All images were imported into FIJI (NIH). Vacuoles were counted in raw images before z-projection as fluorescent negative areas within the cell body. For cell tracking, the manual tracking plugin was used, and data was exported to Microsoft Excel to obtain mean cell speed and distance traveled. Meandering index was calculated as Euclidean distance/Total distance traveled and responding cells were defined as those that reached the wound site at any point within 2 hr. To quantify macrophages recruited to wounds in Drosophila embryos, the outline of the wound was defined using bright field images and was then drawn across all Z slices. Inflammatory recruitment was defined as any macrophage that contacted (specifically via its cell body) the wound perimeter over the time course of imaging following wounding. For wound recruitment analysis, macrophage numbers recruited to wounds in Drosophila embryos were divided by the wound perimeter to account for differing wound sizes due to variation in laser ablation. To quantify wound closure, wound perimeter was recorded over time and analyzed as a function of wound size at 10 minutes – the earliest time point at which the wound outline can be accurately measured by brightfield imaging.
Following live imaging of fluorescently tagged constructs within macrophages, lamellipods were outlined manually for visualization using the Freehand selection tool. Puncta were tracked using manual tracking and counted using the cell counter plugin within FIJI (NIH). Kymographs were generated using the reslice tool along a line (10 pixels wide) following the path of an individual punctum (line drawn using segmented line tool to accommodate non-linear path of the puncta). In each kymograph, the x axis represents distance starting at the lamellipod leading edge on the left, toward the cell body on the right (174 nm/pixel, 17.4 μm total). The y axis represent time (10 s/pixel, 2.5 min total).
For quantification of zebrafish larvae tailfin transection, a 150 μm area was outlined extending from the wound margin. All DsRed2/mScarlet positive leukocytes within this area were counted manually.
Raw data was collated using Microsoft Excel and imported into Prism 8 (GraphPad) for statistical analysis and graphing. All datasets underwent Normality tests to ensure the appropriate statistical tests were performed. For normally distributed data Unpaired t tests were performed, with Welch’s correction where variances were significantly different (determined by F-test). For data not normally distributed, Mann-Whitney U tests were performed to confer significance. ANOVA tests were performed for datasets with more than two groups for comparison. For data with comparable variances (F-tested) Tukey’s or Sidak’s multiple comparisons were performed as recommended by the software. Brown-Forsythe and Welch ANOVA tests were used were variances significantly differed.
References
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
Acknowledgments
We would like to thank Kate Comber for help with fly stock maintenance and technical support and Adam Williamson for sharing information prior to publication and for stimulating discussions. We also thank Kate Heesom and the Proteomics Facility at the University of Bristol. For Drosophila stocks, we thank Hugo Stocker, the Bloomington Stock Centre (Indiana University, USA), Kyoto Stock Center (DGRC), and the Vienna Drosophila Resource Center. For zebrafish work, thanks to Nikolay Ogryzko for generating the Tg(mpeg:NLS-scarlet) macrophage reporter and to Isabel Bravo for assistance in husbandry and techniques. This work was funded by a
Author Contributions
Conceptualization, J.S.C., F.S.L.M.R., and W.W.; Methodology, J.S.C., A.J.D., Y.F., D.A.L., and F.S.L.M.R.; Validation, J.S.C.; Investigation, J.S.C., A.J.D., H.T., A.M.E., J.J.E., and F.S.L.M.R.; Writing, J.S.C., A.J.D., H.T., and W.W.; Supervision, Project Administration, and Funding Acquisition, W.W.
Declaration of Interests
The authors declare no competing interests.
 PTPN21/Pez Is a Novel and Evolutionarily Conserved Key Regulator of Inflammation In Vivo
PTPN21/Pez Is a Novel and Evolutionarily Conserved Key Regulator of Inflammation In Vivo