
- Altmetric
RNA 2′-O-methylation is widely distributed and plays important roles in various cellular processes. Mycoplasma genitalium RNase R (MgR), a prokaryotic member of the RNase II/RNB family, is a 3′-5′ exoribonuclease and is particularly sensitive to RNA 2′-O-methylation. However, how RNase R interacts with various RNA species and exhibits remarkable sensitivity to substrate 2′-O-methyl modifications remains elusive. Here we report high-resolution crystal structures of MgR in apo form and in complex with various RNA substrates. The structural data together with extensive biochemical analysis quantitively illustrate MgR’s ribonuclease activity and significant sensitivity to RNA 2′-O-methylation. Comparison to its related homologs reveals an exquisite mechanism for the recognition and degradation of RNA substrates. Through structural and mutagenesis studies, we identified proline 277 to be responsible for the significant sensitivity of MgR to RNA 2′-O-methylation within the RNase II/RNB family. We also generated several MgR variants with modulated activities. Our work provides a mechanistic understanding of MgR activity that can be harnessed as a powerful RNA analytical tool that will open up a new venue for RNA 2′-O-methylations research in biological and clinical samples.
INTRODUCTION
RNA ribose methylation (2′-O-methylation) is one of the ubiquitous nucleotide modifications found in various RNA species from bacteria, archaea and eukarya (1–3). It plays important roles in the control of gene expression as well as various cellular processes (4–7). Eukaryotic RNA 2′-O-methylation regulates the translation (8–11) and is related to human diseases, such as cancers and autoimmune diseases (12,13). RNA 2′-O-methylation is also one of the most common modifications in various viruses including human immunodeficiency virus (HIV-1) (14) and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (15), which is crucial in modulating the viral replication and host antiviral immune responses (16–18). RNAs bearing 2′-O-methylations show enhanced stability (19–21) and increased resistance to degradation by some Ribonuclease Rs (RNase R) (22). RNase R from Escherichia coli (EcR) and a number of mycoplasma species such as Mycoplasma genitalium (MgR), Mycoplasma hyor (MhR), Mycoplasma canis (McR), are a major type of 3′-5′ exoribonucleases belonging to the RNase II/ RNB family that are involved in RNA degradation (23), maturation (24) and quality control (25,26) in all kingdoms of life. Prototypical of members of the family also include E. coli RNase II (EcII), yeast Rrp44 and human Dis3-like proteins. Eukaryotic Rrp44, Dis3L1 are critical components of exosome complexes involved in cytoplasmic RNA degradation (27,28). Dis3L2 is exosome-independent but functions in microRNA and non-coding RNA decay (28). Impairment of Dis3-like protein functions leads to human diseases (29,30). In prokaryotic cells, RNase II only acts on linear RNA (31), while RNase R can process structured RNA as it behaves as a helicase that independently unwinds and degrades duplex RNA with a 3′-overhang (32).
MgR is the only 3′-5′exoribonuclease identified in Mycoplasma genitalium that is essential for cell survival (33). MgR and EcR share 27% identity and 48% similarity in sequence and are functionally similar (22). However, distinct from EcR, MgR is significantly sensitive to RNA ribose methylation (22). Hydrolysis of RNA is hindered when MgR encounters a nucleotide containing a covalent adduct such as a methyl group at the ribose 2′-hydroxyl (34). The biological significance of MgR’s sensitivity to 2′-O-methylation has not been fully elucidated, but this unique feature of MgR makes it a potential analytical tool for studies of eukaryotic RNAs.
Methods for mapping 2′-O-methylation in RNA were developed to facilitate the study of RNA ribose modification, as loss of individual modifications generally was not adequate to address biological questions (35). Besides sequencing techniques such as RiboMeth-seq (36), quantitative measurement of MgR resistant RNA substrates may reveal the profile of 2′-O-methylation modification in all sorts of cellular RNA species with better specificity, efficiency and accuracy (35). Another technique, SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension), chemically modifies RNA 2′-hydroxyls to investigate secondary and tertiary structures (37). The RNA adducts are conventionally detected by Primer Extension, which has limitations in certain inaccessible regions of RNA for analysis, especially at the 5′ and 3′ ends (34). Utilization of MgR in RNase-detected SHAPE (selective 2′-hydroxyl acylation analyzed by protection from exoribonuclease) makes it possible for single nucleotide analysis of local nucleotide flexibility for most nucleotides in an RNA to overcomes these defects (38).
Multiple domains of RNase R work in collaboration to degrade different species of structural and linear RNAs (39). The members of RNase II/ RNB family bear similar domain organizations, consisting of N-terminal cold shock domains (CSD1 and CSD2) responsible for RNA binding, a central conserved RNB domain for RNA hydrolysis, and a S1 domain at the C terminus that also participates in RNA binding (40). MgR resembles EcR, as they contain a helix-turn-helix (HTH) domain at the N-terminus (22). Crystal structures of RNase II (41), Rrp44 (27) and Dis3L2 (42) in complex with single stranded (ss) RNA revealed a conserved RNA hydrolysis mechanism within the active site. The crystal structure of apo-EcR (32) revealed two potential RNA-binding sites, one ‘apical groove’ between CSD1 and S1 domains similar as in Dis3L2 and RNase II, and another ‘lateral groove’ between CSD1 and RNB domains as in Rrp44. However, it is still an open question how EcR or MgR interact with the various RNA species.
To investigate the molecular basis of the specific nuclease activity of RNase R, especially the remarkable sensitivity of MgR to RNA 2′-O-methylation, we performed extensive structural, enzymatic and biochemical studies on RNase R variants. We determined the crystal structures of apo-MgR and its RNA bound complexes, including single and double stranded RNAs, as well as substrates with 2′-O-methylation. Our three-dimensional models reveal a dynamic process where multiple domains of MgR work in concert to process structured RNAs differently compared with linear RNAs. The significant sensitivity of MgR to RNA 2′-O-methylation is likely due to a sophisticated arrangement of an array of amino acids in the catalytic pocket of RNB domain, and proline 277 is crucial for MgR to be outstanding in the family for the hindered ribonuclease activity towards 2′-O-methylation. Our work provides not only a mechanistic understanding for MgR activity and sensitivity to 2′-O-methylation, but also we generated MgR variants with higher sensitivity to ribose modification, which may be applicable for future routine treatment of biological and clinical samples that will open up a new venue for research of RNA 2′-O-methylations in biological and pathologic processes.
MATERIALS AND METHODS
Protein expression and purification
Full length of MgR, McR, MhR, EcR and RNase II were cloned into the expression vector pET19b containing an N-terminal 10× His-tag, respectively. MgR and EcR mutants were generated by quick-change mutagenesis kits (Agilent). Constructs were verified by DNA sequencing (Tsing Ke Biotech). The recombinant plasmids were transformed into BL21 (DE3) E. coli cells and induced with 0.2 mM IPTG in Terrific Broth media at 16°C overnight. The resulting His-tagged protein was purified by a HisTrap HP affinity column (GE Healthcare), eluted in 50 mM Tris (pH 7.5), 500 mM NaCl, 5 mM MgCl2, 5% (vol/vol) glycerol and 300 mM imidazole. The protein was further purified using a HiLoad 16/60 Superdex 200 prep grade column (GE Healthcare) equilibrated with 50 mM Tris (pH 7.5), 300 mM NaCl, 5 mM MgCl2 and 10% (vol/vol) glycerol. The protein purify was examined by SDS-PAGE. Proteins were pooled and concentrated to 15 mg/ml, flash frozen in liquid nitrogen and stored at −80°C.
Enzymatic activity and intermediate productivity determination
The enzymatic activity assays of MgR and its homologs were conducted under multiple turnover conditions, where we measured the linear initial rate of RNA degradation and calculated the enzymatic activity (nt degraded min−1 molecule of enzyme−1). The indicated amount of enzyme was incubated at room temperature with 25 μM 5′-carboxy fluorescein (FAM) labelled 30-nt oligo A substrate with a 2′-O-methylation at nucleotide 15 in reaction buffer (20 mM Tris pH 8.5, 100 mM NaCl, 0.01 mM ZnCl2 and DEPC-treated water) as suggested in the previous report (22). A 36 μl reaction was initiated and 6 μl was removed and quenched at 0.5, 1, 2, 4, 6 and 8 min time points in 2X formamide loading buffer (0.025% SDS, 95% formamide, 0.025% bromophenol blue, 0.025% xylene cyanol and 18 mM EDTA). Quenched samples were heated to 70°C for 5 min, divided into three parallel samples and resolved by denaturing urea PAGE. Reactions were run in triplicate. Products were analyzed by fluorescence imaging (Tanon 3500) and quantified with ImageJ software. We quantified the amount of substrate degraded (nt pmol) that equals to the amount of product formation including intermediate products and final products multiplying by n-15 or n-3 (where n is the number of nucleotides of the substrate, 3 is the length of the end product, and 15 is the length of the intermediate product). Then the amount of substrate reduction (nt pmol) at 0.5, 1, 2, 4, 6 and 8 min was plotted against time and linear regression was used to determine the initial rate (slope) of the linear portion of the curve with Graph Pad Prism 6. The initial rate was converted to enzymatic activity (nt/min/molecule of enzyme) by dividing the initial rate (nt pmol substrate hydrolyzed/min) by the pmol of enzyme used in each assay. Synthetic 30-nt RNA oligo rA, and 2′-O-methylated 30-nt RNA oligo rA substrates were purchased from Genscript BioTechnologies Company. For the intermediate productivity of each enzyme, we quantified the initial amount of intermediate product (pmol) at 0.5, 1, 2, 4, 6 and 8 min, respectively, then the intermediate product molar mass (pmol) was divided by the molar mass of total amount of substrate multiplying 100%. The intermediate productivity standard deviation (SD) and the significant difference were calculated by Statistical Product and Service Solutions (SPSS) 17.0 software. The dsRNA was prepared by annealing a 5′-FAM-labeled 40-nt RNA (5′-CCCGAGACCGAGACCGAGACGAC CGACCUUUUUUUUUUUU-3′) with a 28-nt RNA (5′-GGUCGGUCGUCUCGGUCU CGGUCUCGGG-3′) to generate a 28-basepair dsRNA with a 12-nt single stranded 3′-overhang.
Crystallization
Apo-MgR crystals were grown at 22°C using the micro-batch-under-oil method by mixing 1.6 μl wildtype MgR (in 20 mM HEPEs (pH 6.8), 25 mM NaCl, 1 mM MgCl2 and DEPC-treated water) with 1.2 μL crystallization buffer containing 100 mM KCl, 100 mM HEPES (pH 6.8), 15% PEG 5000MME. The best crystals were obtained by seeding with protein concentration at 17 mg/ml. The crystals of MgR-ssRNA and MgR-Me were obtained using inactive mutant MgR-D284A mixed with rA9 RNA and 2′-O-methylated oligo rA (5′-AAAmeA-3′), respectively, in a 1:1.2 molar ratio and incubated for 10 min at 37°C before subjected to crystallization described above. The dsRNA substrate was prepared by annealing a 13-nt oligo (5′-CCGUGUAAAAAAA-3′) with a 6-nt oligo (5′-ACACGG-3′) to generate a dsRNA with 6 bp with a 7-nt single stranded 3′-overhang. The MgR-D284A was incubated with a 1.2 molar excess of the dsRNA for 10 min at 37°C. Crystallization was carried out at 22°C by sitting-drop method by mixing the protein/RNA sample at 6 mg/ml with an equal volume of 100 mM CHES (pH 9.5), 20% PEG 8000. All crystals appeared in 1–2 days and were placed directly in a freshly prepared solution composed of the crystallization buffer supplemented with 20% glycerol and flash frozen in liquid nitrogen. Diffraction data were collected at the Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U1. Data were processed using HKL2000 (43). The data statistics are summarized in Table 1.

| Data collection | apo-MgR | MgR-ssRNA | MgR-dsRNA | MgR-Me |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P212121 | P43212 | P212121 | P212121 |
| Cell dimensions | ||||
| a, b, c (Å) | 69.65, 80.77, 176.48 | 68.26, 68.26, 354.69 | 75.41, 96.09, 117.16 | 69.29, 80.78, 175.74 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution range* (Å) | 50.0–1.98 (2.01–1.98) | 50.0–2.24 (2.28–2.24) | 50.0–2.00 (2.03–2.00) | 30.0–1.90 (1.93–1.90) |
| Unique reflections | 70 690 (3444) | 41 557 (2028) | 57 987 (2865) | 78 483 (3891) |
| Completeness (%) | 99.9 (98.8) | 99.4 (98.2) | 99.9 (100) | 99.9 (99.9) |
| Redundancy | 13.1 (11.9) | 8.3 (7.0) | 10.6 (9.0) | 13.2 (13.4) |
| I/σI | 18.6 (1.1) | 18.0 (1.3) | 17.9 (1.5) | 18.0 (1.0) |
| R merge | 0.175 (2.931) | 0.098 (2.473) | 0.119 (1.087) | 0.148 (2.936) |
| Refinement | ||||
| Resolution range (Å) | 45.3–1.97 (2.04–1.97) | 48.3–2.24 (2.32–2.24) | 38.3–2.00 (2.07–2.00) | 28.0–1.90 (1.97–1.90) |
| No. of reflections (working/test) | 70 614/3689 | 41 433/1998 | 57 917/2000 | 78 365/1995 |
| R work/Rfree | 0.173/0.199 | 0.215/0.261 | 0.220/0.242 | 0.188/0.222 |
| Number of atoms | ||||
| Macromolecules | 5235 | 5388 | 4678 | 5265 |
| Ligand/ion | 1 | 1 | 1 | 1 |
| Water | 475 | 129 | 236 | 517 |
| B-factors | ||||
| Macromolecules | 43.4 | 72.1 | 64.32 | 45.6 |
| Ligand/ion | 49.3 | 49.5 | 25.8 | 82.2 |
| Water | 45.6 | 59.2 | 59.4 | 49.0 |
| r.m.s. deviations | ||||
| Bond lengths (Å) | 0.011 | 0.012 | 0.008 | 0.011 |
| Bond angles (°) | 0.98 | 1.53 | 1.16 | 1.00 |
Single crystal was used for each data collection and structure determination.
*Numbers in the brackets are for the highest resolution shell.
Structure determination and refinement
The structure of apo-MgR was solved by molecular replacement (44) using PHASER (45) with E. coli RNase R (32) (PDB ID 5XGU) as the search model. One molecule was identified in an asymmetric unit of the crystal. The initial model was auto-built by Buccaneer (46) and refined by iterative rounds of TLS refinement using Phenix (47), followed by rebuilding manually using Coot (48). The apo-MgR structure was then used as search model for other RNA complex structures determination and reference model during the refinement. Refinement statistics are summarized in Table 1. The final models were validated by MolProbity (49,50). Large area of extremely poor electron densities in the structure of MgR-dsRNA precluded accurate modelling of the positions of CSD1, S1 and part of the substrate, so we carefully avoided over interpretation of these regions, although we are confident of their presence and general location. All structural figures were generated with PyMOL (http://www.pymol.org). The coordinates and structure factors have been deposited in PDB.
MgR and its RNA complex preparation assay
To study complex formation, MgR and RNA were mixed and incubated at 37°C for at least 10 min prior to loading. Analytical size exclusion chromatography was performed at 4°C using a Superdex Increase 10/300 global column (GE Healthcare) pre-equilibrated in 20 mM HEPEs pH 6.8, 25 mM NaCl, 1 mM MgCl2, DEPC-treated water. Aliquots of 200 μl samples were injected and eluted at a flow rate of 0.75 ml/min.
Equilibrium binding assays
The binding of MgR and RNA substrates were determined by measuring the changes in fluorescence polarization using EnVision Multilabel Reader (Perkin Elmer). The 24-nt 5′-FAM labelled ssRNA (5′-CAAACAAAACCGmeUGmeUAAAAACAAmeA-3′) and the oligos with the same sequence but without 2′-O-methylation were used in the assays. RNA at a 10 nM final concentration was titrated with the indicated concentrations of MgR-D284A in binding buffer (20 mM HEPEs pH 6.8, 25 mM NaCl, 1 mM MgCl2, DEPC-treated water). Reactions were prepared at 37°C and incubated for 15 min, then read at 518 nm at an excitation of 492 nm. MST measurements were performed with 20 nM RNA with or without 2′-O-methylation as described in FP measurement and the indicated concentration of MgR-D284A in binding buffer containing 0.05% Tween. Thermophoresis was performed on a Monolith NT.115 (Nano Temper Technologies GmbH) set at 40% LED and 80% MST power at 25 °C and with 5s and 30s laser off and on times, respectively. We found that wild-type MgR is inactive in the presence of EDTA and no magnesium, but binds to RNA with the same affinity as the MgR inactive D284A mutant. Binding assays were conducted in triplicate, were quantified and fraction bound plotted versus free protein concentration. Kd values were determined by using nonlinear regression analysis with Graph Pad Prism 6 software.
Intermediate product analysis
MgR intermediate products were determined by liquid chromatography−mass spectrometry methods. Briefly, ssRNA substrates of 30-nt (5′-FAM-CAAAACAAAAC AAAAmeCAAAACAAAACAAAA-3′) with 2′-O-methylation at nucleotide rA15 digested by 1.5 μM final concentration of MgR at room temperature over time course of 16 min and products were analyzed by liquid chromatography and mass spectrometry (LC−MS). The LC−MS analysis was performed on a TripleTOF™ 5600 LC/MS/MS instrument (AB SCIEX, USA). The raw mass spectrometry data were analyzed with Thermo Scientific Proteome Discoverer. Intermediate product was confirmed by sequencing method using miRNA First Strand cDNA Synthesis kit (Sangon Biotech). Briefly, ssRNA substrates of 40-nt (5′-ACGCGCCTGGATACCGCAGCTAGAATG AmeGCTGAGATGAAA-3′) with 2′-O-methylation at nucleotide rA28 digested by 1.5 μM final concentration of MgR at room temperature over time course of 16 min and conducted poly A tailing reaction and cDNA synthesis reaction. Then cDNA cloned into the TA vector and verified by DNA sequencing (Tsing Ke Biotech). Different intermediates sequencing ratio determined by the sequencing repetitions of different intermediates divided by the total sequencing repetitions of intermediates and multiplied 100%.
RESULTS
MgR is significantly sensitive to RNA ribose methylation in contrast to its homologs
To better understand the effect of methylated RNAs on the catalytic properties of MgR, we examined the exoribonuclease activity of it and related homologs. We expressed and purified full-length, wild-type MgR, MhR, McR, EcR and RNase II. The recombinant proteins all eluted as monodispersed, monomeric peaks from a size exclusion column with high purity (Supplementary Figure S1). To determine the catalytic activities of MgR to various RNA substrates, we then performed RNA degradation assays. All the samples tested effectively digested a 30-mer ssRNA in a sequence and length independent manner (Figure 1A and E), but with diverse hydrolysis rates (Figure 1A and Supplementary Figure S2). However, these enzymes exhibited different catalytic properties with RNA substrates containing a 2′-O-methyl modification (Figure 1B and C; Supplementary Figure S3A and B). Remarkably, MgR digestion of a 30-mer RNA with a 2′-O methylation at nucleotide 15 resulted in an additional intermediate band ∼15 nucleotides (nt) in length (Figure 1B). Only after prolonged incubation or an increase in enzyme concentration, MgR digested all RNA to the final product of ∼3 nt as previously reported (22). In contrast, other enzymes tested in the assay efficiently degraded the methylated RNAs, as the intermediate product was present at dramatically lower levels (Figure 1B and D; Supplementary Figure S3). RNase II showed about eight-times higher total hydrolysis rate towards the methylated RNAs compared to MgR (Figure 1C). Even the two enzymes from the other Mycoplasma species, MhR and McR, also showed about six- and five-times higher activity compared with MgR, respectively (Figure 1C). To quantitatively analyze the efficiency of enzymes generating intermediate product under our experimental conditions, here in the work we defined a parameter – intermediate productivity, which is the molar mass ratio of the intermediate product to the total RNA substrate ×100%. MgR’s intermediate productivity is about five times higher than all other homologs, which is statistically significant (95% confidence interval, P < 0.05) (Figure 1D and Supplementary Table S1).

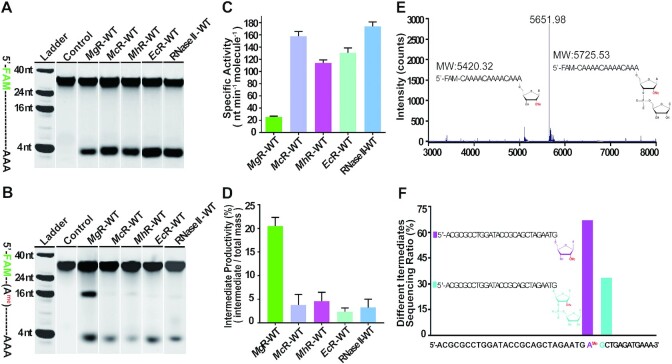
Enzymatic activity of MgR and its homologs. (A,B) RNA degradation assays of MgR and its homologs with substrates of 30-nt oligo rA without (A) or with (B) 2′-O-methylation at nucleotide rA15. The assays were performed at room temperature over time course of 0, 0.5, 1, 2, 4, 6, 8 or 16 min. Products were resolved by denaturing urea PAGE. The results at 4-min time point for each enzyme were collectively displayed. The uncropped gels with results of all the time points were shown in Supplementary Figures S2 and S3A. (C) Enzymatic activity (nucleotides (nt) min−1 molecule−1) of MgR compared with other homologs degrading the methylated substrate used in panel B and Supplementary Figure S3. Enzymatic activity was determined from the initial rate (pmol substrate degraded per minute) divided by the amount of enzyme (pmol) multiplied by (n-3 or n-15) (where n is the number of nucleotides of the substrate, 3 is the length of the end product, and 15 is the length of the intermediate product). (D) Intermediate productivity at 4 min time point of MgR compared with other homologs degrading methylated substrates used in panel B and Supplementary Figure S3. Intermediate productivity was determined using the initial intermediate product molar mass (pmol) divided by the total amount of RNA substrate molar mass (pmol) and multiplied by 100%. Mean ± S.D. are shown. (E) Mass spectrometry for the identification of MgR intermediate product. A 30-nt 5′-FAM labeled RNA substrate ((CAAAA)n, n = 5) with a 2′-O-methylation at nucleotide rA15 was used. The schematic figures of the 15-nt and 16-nt intermediate products are illustrated and molecular weights (MW) marked. (F) TA clone sequencing for the identification of MgR intermediate product. A 40-nt ssRNA substrate with 2′-O-methylation at nucleotide rA28.
To identify the intermediate product and the stall position of MgR hydrolysis on 2′-O-methylated RNA, we subjected it to mass spectrometry and sequencing analysis. These revealed a mixture of 16-mer and 15-mer RNAs, products either 1 nt downstream from the methyl-modification or exactly at the methylated position, respectively (Figure 1E and F). As reported previously (22), RNase R showed sensitivity to RNA ribose methylation and MgR is significantly more sensitive among other ribonucleases within the RNase II/RNB family.
The structures of MgR and RNA complexes illustrate the substrate binding mode
To better understand the molecular basis of RNase R processing of RNA substrates, we first determined the crystal structure of full-length MgR to a resolution of 1.98 Å by molecular replacement (Figure 2A and Table 1). One protein molecule bound with a Mg2+ ion was observed in the asymmetric unit of the crystal. Residues 82–725 are clearly resolved, including domains CSD1, CSD2, RNB, and S1 (Figure 2A). Residues 1–82, harboring the N-terminally His-tagged HTH domain, are not defined, suggesting a highly flexible bridging region connecting the HTH and CSD1 domains. We then generated an N-terminally truncated construct MgR△HTH (82–725) with unaffected catalytical activity (Supplementary Figure S4A and B) and determined its crystal structure, which showed no difference to the full-length. We also produced full-length MgR without the N-terminal His-tag which showed similar enzymatic activity (Supplementary Figure S5).

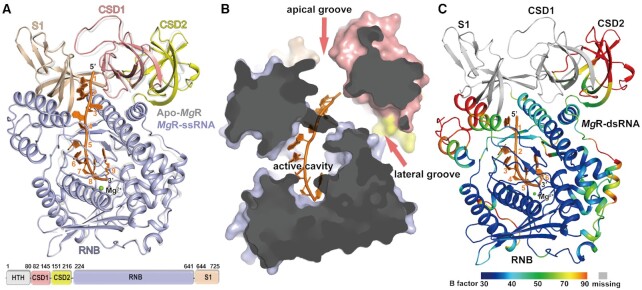
Crystal structures of MgR and its RNA complex. (A) Overall structure of apo-MgR compared with MgR-ssRNA and the domain schematic. Apo-MgR (grey) is superimposed with MgR–ssRNA (domains colored). The 9-nt linear RNA substrate is colored in orange and each nucleotide from 5′ to 3′ is labeled. The Mg2+ ion is shown as green sphere. (B) Surface representation of MgR-ssRNA displaying the apical groove, lateral groove, and the active cavity in the RNB domain. The image has been clipped to facilitate observation of the cavity. The surface is colored according to domains as in panel A. The clipping plane is blanked in gray. (C) Overall structure of MgR–dsRNA. The ribbon is colored according to B-factors from blue with low values to red with high values. The missing part of the structure was drawn according to MgR–ssRNA and colored in grey.
This apo structure (apo-MgR) revealed high conservation of individual domains to the rest of the RNB family, but with variation in domain arrangements. The RNB domain of MgR aligned well with that of EcR with an average RMSD of 1.5 Å for 385 Cαs. When RNB domains were superimposed, CSD1 and CSD2 domains of MgR positioned relatively in place as in EcR with mass center of each domain shifted around 3 Å, but varied in relative orientation to the RNB domain with an average RMSD of 6.7 Å for CSD1 and 8.9 Å for CSD2 (Supplementary Figure S6A). As observed in the EcR structure (32), two potential RNA binding grooves are also present in MgR, including an apical one between the S1 and CSD1 domains and a lateral one between the RNB and CSD1 domains (Figure 2B).
We then co-crystallized MgR with a series of different RNA substrates. To prevent substrate hydrolysis, we used the catalytically inactive D284A mutant (Supplementary Figure S4). Interestingly, ssRNA fragments longer than 9 nt failed to co-crystalize with MgR as it disrupted the pre-formed apo-form crystals. Analysis of the crystal packing of apo-MgR revealed extensive molecular contacts between symmetry related molecules involving residues around the apical groove, but not the lateral groove (Supplementary Figure S7A). This suggests that ssRNA substrates bind the apical groove and prohibit crystal packing of the RNA-bound complex (Supplementary Figure S7B). Eventually, we determined the crystal structures of MgR in complex with 9 nt ssRNA (MgR–ssRNA, 2.2 Å), dsRNA with a 3′-ss overhang (MgR-dsRNA, 2.0 Å), and 4 nt ssRNA oligos with 2′-O-methylation (MgR-Me, 1.9 Å) (Table 1). Each protein molecule bound to one RNA substrate, which was confirmed by size exclusion chromatography (Supplementary Figure S8).
Binding of ssRNA to MgR does not induce conformational changes to the overall protein structure. MgR-ssRNA and apo-MgR are nearly identical (RMSD 0.95 Å). We observe density for all nine nucleotides of the polyA (rA9), threading from the opening of the apical groove all the way into the active site in the RNB domain (Figure 2A). rA1 just touches the neighboring molecule that is involved in crystal packing interactions at the apical groove (Supplementary Figure S7B). rA2–rA4 form a bend in the RNA chain with almost no contacts to surrounding protein residues. The final five nucleotides (rA5–rA9) are well-ordered in the cavity within the catalytic RNB domain (Figure 2A and Supplementary Figure S7B).
MgR undergoes dramatic domain rearrangements degrading dsRNA
RNase R can carry out structured dsRNA degradation independently, which was confirmed by our activity assay on MgR, EcR and McR, using a synthetic substrate of 28-bp dsRNA with a 12 nt 3′-ss overhangs (Supplementary Figure S9). To understand how RNase R recognition of dsRNA differs from ssRNA, we solved the crystal structure of MgR bound to a 6 bp dsRNA with a 7 nt 3′- ss overhang to 2.0 Å (Figure 2C and Table 1). Each protein molecule bound to one dsRNA substrate, confirmed by size exclusion chromatography during complex preparation (Supplementary Figure S8C). We observed that the 3′-ss overhang bound to the RNB domain is nearly identical to the MgR-ssRNA structure with an RMSD of 0.5 Å. However, no density was observed for the duplex RNA (Figure 2C). Astonishingly, all the RNA binding related domains—CSD1, CDS2 and S1—were very disordered despise our high-resolution data. For example, the electron densities for the CSD1 and S1 domains were extremely poorly defined such that only a few residues could be modeled while the CSD2 domain could only be modeled but with exceptionally high B factors. We noticed that crystals of MgR-dsRNA, MgR-ssRNA and apo-MgR were with distinct unit cells and/or space groups (Table 1). Similar to the crystal packing of apo-MgR, crystal of MgR-ssRNA was held largely by extensive molecular contacts between symmetry related molecules involving residues around the apical groove. However, such crystal packing was not existing in MgR-dsRNA, presumably due to the bulkier substrate bound to the apical groove which prohibited the crystal packing around regions including CSD1 and S1 domains. It is a likely explanation for the poorly-defined electron densities. It might also suggest that the dramatic rearrangement of the RNA binding domains caused by dsRNA binding is likely due to a highly dynamic status of accessory domains when dsRNA anchors in the apical groove, with CSD1 and S1 playing a more direct and vivid part in the whole process. It implies distinct mechanisms of MgR nuclease activity for single stranded and structured dsRNA.
RNA binding to the RNB domain reveals residues critical for MgR catalytic efficiency
To understand how the RNB domain of MgR recognizes RNA, we then compared the active sites across all our RNA-bound structures. In the active site of the RNB domain in RNA containing structures, the five nucleotides upstream of the 3′-end (termed as rA5—rA9 below) are extensively engaged in hydrogen bonds with surrounding residues mainly through the phosphate groups, which explains the sequence-independent characteristic cleavage manner of MgR (Figure 3A). The structure also illustrates that rA5, rA6, rA8, rA9 form hydrogen bonds between ribose hydroxyl and side chains of S562, E463, D276, Y387, S280 and H331. The ribose hydroxyl of rA7 interacts indirectly with surrounding residues Y387 and E459 via a bridging structured water (Figure 3B and C). Packing of the 5 bases and extensive hydrogen bonding to the surrounding conserved residues facilitate the proper orientation of RNA and subsequently the cleavage of rA9 by an active enzyme. When RNA substrate is 4 nt or shorter, the interactions are not as strong as required, the processivity starts to lose gradually. MgR appears to make more extensive interactions with the RNA via ribose 2′-hydroxyls than EcII (Supplementary Figure S6B), which might explain the major end product of 3 nt for MgR versus 4 nt for EcII (41). As seen in homologous structures, the Mg2+ ion is coordinated by the conserved residues D276, D285, O3′ of rA8, O1P of rA9 and two water molecules in the octahedral geometry (Figure 3A and Supplementary Figure S4C). This suggests a conserved catalysis mechanism across the RNase II family, as proposed previously (41).

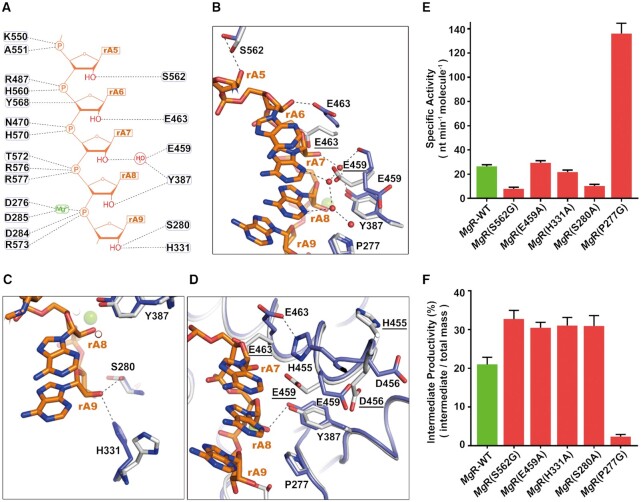
Critical RNA-interacting residues in the active cavity of MgR RNB domain. (A) A schematic view of the interactions between RNAs and the RNB domain of MgR. The dashed lines show the hydrogen bond interactions between rA5–rA9 (phosphate and ribose backbone in orange and 2′-hydroxyl groups in red) and side chains of residues (labeled ovals) in RNB domain. Mg2+ (colored in green) and structured water (colored in red) are labelled. (B–D) Structural conformational changes upon RNA binding by superimposing apo-MgR (in grey and residues marked with underlines) to MgR-ssRNA (in slate). Interactions between RNA ribose hydroxyls and surrounding residues are marked with dashed lines. RNA is shown as sticks in orange. Protein is shown as ribbon with side chains of related residues shown as sticks. Water molecules are shown as red spheres. Mg2+ is shown as green sphere. (E) Enzymatic activity (nt min−1 molecule−1) of various MgR mutants degrading a 30-nt rA with a 2′-O-methylation at nucleotide rA15. (F) Intermediate productivity (%) at 4-min time point of various MgR mutants degrading a 30-nt rA with a 2′-O-methylation at nucleotide rA15. The uncropped gels with results of all the time points were shown in Supplementary Figure S14.
We then compared active sites between the apo-MgR to the RNA-bound structures. We found three dramatic conformational changes, all related to the ribose hydroxyl interactions. (i) In the apo conformation, the side chain of the conserved E463 occupies the space of rA7. Upon RNA binding, it swings away from the cavity to avoid steric clashing and subsequently forms a hydrogen bond with H455, which moved ∼6 Å from its apo conformation (Figure 3B). (ii) In the absence of RNA, E459 sequesters an ordered water molecule that, in the RNA-bound state, forms a hydrogen bond with the ribose hydroxyl of rA7. Upon RNA binding, E459 swings away from the cavity and in turn pushes the side chain of D456 away from its apo conformation, to allow formation of a sophisticated water network between MgR and RNA (Figure 3D). (iii) The side chain of H331 moves towards the RNA 3′-end, which enables the imidazole ring to engage in hydrogen bonding with the hydroxyl group of rA9 (the nucleotide at the 3′-end) (Figure 3C). H331 likely participates in the optimal orientation of the substrate to facilitate release of the cleaved nucleotide. This is unique to MgR, as H331 and other nearby structural conformations are not conserved compared to EcR or EcII, suggesting a distinct and key role of this residue (Figure 4A and Supplementary Figure S10).

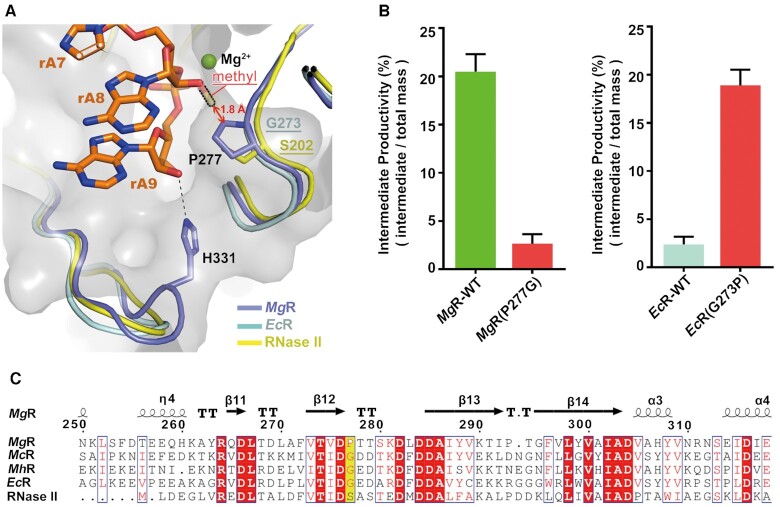
Molecular basis of MgR for recognition and significant sensitivity to RNA 2′-O-methylation. (A) The active cavity region of MgR is shown as grey surface and cartoon with side chains of residues P277 and H331 as sticks. The structure of MgR (slate) is superposed to those of EcR (cyan) and RNase II (yellow). The aligned EcR residue G273 and RNase II residue S202 are marked accordingly. A methyl group is modeled to the 2′-hydroxyl of rA8 to show the steric hindrance to MgR P277. (B) Mutagenesis studies on MgR P277 and EcR G273. Enzymatic activity assays were conducted on both the wild-types and mutants degrading RNA substrates with 2′-O-methylation. The intermediate productivity was determined as described in Figure 1C. (C) The structure-based sequence alignment including MgR, McR, MhR, EcR and E. coli RNaseII. The secondary structure elements (arrows for β strands and cylinders for α helices) and residue numbers of MgR are as shown. Those residues aligned to MgR P277 are highlighted in yellow. The conserved residues around the active site are highlighted in red. The sequences were aligned with Clustal Omega (64) and visualized using ESPript (65).
Key residue identified for MgR recognition of RNA 2′-O-methylation
To test whether the methyl group on the RNA ribose would influence the substrate binding affinity to mediate sensitivity, we measured the affinity of MgR D284A (which is not involved in direct cation or RNA binding) to the FAM-labeled ssRNA 24-mer by fluorescence polarization. The binding affinity for ribose methylated ssRNA was comparable to the non-modified substrate with a Kd of 6.63 ± 0.69 and 6.62 ± 0.70 nM, respectively, and confirmed with microscale thermophoresis (Supplementary Figure S11A and C). MgR△HTH displayed similar binding affinities for the various RNA substrates compared to the full-length enzyme (Supplementary Figure S11B and D). Together, this shows that neither ribose methylation nor lack of the HTH domain affects binding of RNA to MgR.
We then tested whether methylation discrimination was conferred by the RNA binding domains. We generated a chimera containing the MgR RNB domain and the CSD1, CSD2, and S1 domains of EcR (Supplementary Figure S12A), which is poorly sensitive to substrate ribose methylation. This chimeric enzyme exhibited higher nuclease activity compared to wild-type MgR, but still produced a comparable intermediate product as MgR when digesting ribose methylated RNA (Supplementary Figure S12B). Thus, we concluded that the residues responsible for the 2′-O-methylation sensitivity of MgR reside in its RNB domain.
In order to check how the methyl group on ribose would influence the active site, we solved a 1.9-Å-resolution crystal structure of MgR in complex with a 4-nt ssRNA methylated on the second rA from the 3′-end (Table 1). The 4-nt ssRNA overlaid roughly to the positions of rA5-rA8 in MgR-ssRNA (Supplementary Figure S13). The methylated nucleotide overlaid well with rA7 of MgR–ssRNA substrate, but disrupted the structured water network observed in the nonmethylated RNA-bound structures. The structure suggests that the methyl group is likely to disrupt a number of specific interactions with the substrate ribose hydroxyl when sliding through the active site, subsequently influent the processivity of the enzyme. Thus, we mutated ribose-hydroxyl-interacting residues to their EcR homolog and measured their nuclease activities (Figure 3E and F; Supplementary Figure S14A and B). All these mutants still generated intermediate products when incubated with methylated RNA substrates (Supplementary Figure S14A). None of them abolished the sensitivity to the ribose methylation, but rather affect the hydrolysis rate and increase extent of sensitivity. For example, the intermediate productivity of S562G almost doubled that of the wild-type (Figure 3F). Considering the stall position of methylated RNA substrate hydrolyzed by MgR identified in our biochemical assays (Figure 1E and F), we focused on residue P277, which happens to be very unique within the highly conserved active site compared to other homologs such as EcR and RNase II (Figure 4A and C; Supplementary Figure S10). To investigate the role of P277 involved in recognition of ribose methylation, we generated a mutant MgR-P277G, mimicking its counterpart (G273) in EcR. Remarkably, in the nuclease activity assays P277G showed a 5-fold increase in the nuclease activity, suggesting a higher processivity of the enzyme, and the amount of intermediate product produced was dramatically decreased to a similarly low level as EcR (Figure 4B and 3E; Supplementary Figure S14A). Analysis of MgR–ssRNA structure revealed that P277 is ∼3 Å away from the ribose hydroxyl of rA8 (Figure 4A). When the substrate sliding through the active site, the methyl group on ribose would cause a steric clash with P277, likely decreasing the processivity of the substrate and the overall reaction rate (Figure 4A). Interestingly, a single mutation of G273P on EcR dramatically increased its sensitivity to ribose methylation (Figure 4B and Supplementary Figure S14A). Thus, we concluded that all the residues involved in RNA ribose hydroxyl interactions contribute collectively to the sensitivity of MgR to ribose methylation, but P277 is the most crucial residue which is responsible for the significant sensitivity of MgR within RNase II/RNB family.
DISCUSSION
RNA degradation is central to RNA quality control and recycling of the nucleotide pool in the cell. 2′-O-methylation protects RNA from degradation and modifies RNA tertiary structure flexibility. Compared with EcII, RNase R in E. coli or M. genitalium exhibits much higher activity on structured RNA, which makes it critical in RNA quality control, in terms of degrading the defective rRNAs (51) and tRNAs (52,53). Mycoplasmas 23S rRNAs are relatively prominent in ribose methylations, but markedly deficient in base-methylated residues compared with E. coli patterns (54). Despite the limited genome size of M. genitalium, it encodes a methyltransferase that catalyzes the 2′-O-methylation of the ribose of cytidine 1402 in 16S rRNA (55). MgR is incapable of fully degrading ribose-methylated rRNAs efficiently, whereas it is the only putative exoribonuclease identified up to date in M. genitalium (22), there might be other enzymes or cofactors which are relevant to the rRNA degradation. It has been suggested that MgR is responsible for the maturation of tRNAs (24) and possibly rRNAs, as incorrectly ribose-methylated rRNA molecules may be susceptible to MgR digestion.
RNase R, particularly MgR, exhibits the ability to discriminate substrates with 2′-O-methylations, make it biologically and practically important to RNA metabolism. However, previous biochemical knowledge on RNase R was mainly derived from EcR, which showed poor sensitivity to 2′-O-methylation. Lack of structural information on RNA binding to EcR provided limited insight on the mechanism of substrate binding and recognition by RNase R. In this study, we present high resolution crystal structures of MgR bound to various RNAs and answered several important mechanistic questions about the ribonuclease activity of this valuable enzyme.
In our nuclease activity assays, RNase R from three Mycoplasma species and E. coli were tested on their sensitivity to 2′-O-methylated RNA substrates. As reported previously, we also observed that MgR is not the most active enzyme in the family (22) but produced the most distinct intermediate RNA products when incubated with 2′-O-methylated RNA substrates. The stall position of 2′-O-methyl RNA substrate digestion by MgR was reported to be 2 nt downstream of the modification (22). We showed that it is only one or no nucleotide before the methylated site. The discrepancy might due to systems with different methodologies and analyses. However, it also suggested that the stall is not a sudden effect of a specific position which blocked the passage of the RNA, but rather an accumulated effect where the methyl group alters the interactions between RNA and MgR during the processive procedure.
Exoribonucleases from the RNase II/RNB family are defined by containing a highly conserved RNB domain, which we demonstrated to harbor the hydrolysis activity and sensitivity to 2′-O-methylation. The accumulation of intermediate products during RNase R digestion could be due to either the dramatically decreased catalysis rate at the active site or/and the processivity of the enzyme via weakening the translocation rate of the substrate. The catalytic active site of RNB family, especially the residues around the Mg2+ ion and the last two nucleotides at the 3′-end, are fairly conserved. When enzymes approach the methylated position, the catalytic rate would likely be decreased as the aspartic acid coordinating Mg2+, which also interacts with the ribose hydroxyl, would lose its optimal orientation with the ribose. In this case, we explained that all enzymes we tested here exhibit sensitivity to the 2′-O-methylation to a certain extent. This was previously observed for the nuclease activity of EcR, as it generated a trace amount of the intermediate product (22,38).
Structural comparison between apo-MgR and the RNA complexes together with structures of other RNB family members revealed a series of critical residues related to the protein-RNA interactions (Supplementary Figures S6B and S10). Surprisingly, in the apo form of EcR structure, the corresponding residues adopt a more similar conformation as in the RNA complex of MgR. This suggests EcR has fewer conformational rearrangements upon RNA binding that allow for easier substrate translocation and thus higher nuclease activity. Indeed, P277 in MgR functions as a gate keeper which greatly influences the translocation rate of substrate. This is most apparent when the substrate is methylated. Mutation to glycine or serine at the corresponding position in EcR or RNase II, respectively (Figure 4A), opens up the passage to easier accommodate a bulkier methyl group, resulting in a higher catalytic activity at the expense of lower methylation sensitivity. These snapshots provided a framework to understand how the enzyme functions, and give a structural basis for the prediction of sensitivity to 2′-O-methylations of other RNB family members (Supplementary Figure S10).
The feature of MgR to discriminate 2′-O-methylations makes it intriguing as an analytical tool for RNA modifications. Ribose 2′-O-methylations are present in various RNA species such as transfer (t) RNA, ribosomal (r) RNA, small nuclear (sn) RNAs, message (m) RNAs and microRNAs (56–59). This modification has been shown to play more and more critical roles, including RNA stabilization under extreme temperatures and pH, regulation of gene expression, as well as various other cellular processes (4–7). However, the exact role of each ribose 2′-O-methylation remained elusive, because its labile chemical nature limits sensitivity and precludes mapping in different RNAs. The current advances in high-throughput detection of 2′-O-methylation include chromatography and mass-spectroscopy (60,61), RTL-P-2′-O-Me-Seq (62), and RibOxi-Seq (63), etc. All these traditional methods are not particularly specific for 2′-O-methyl modified residues and are extremely laborious and time-consuming. Therefore, the major challenges in exploring the function of 2′-O-methylation of RNAs are the careful mapping in different RNAs as well as precise quantification of the modification rate for every given site. MgR treatment might become the key to opening up the next RNA frontier. MgR has the potential to provide quantitative data at single‐nucleotide resolution because the method is based on the difference in the nucleophilicity of a 2′‐hydroxyl and a 2′‐O‐methyl group. Selective 2′-O-methylation analyzed by protection from MgR with a simple one-tube exoribonuclease degradation facilitates detecting sites of covalent modification in RNA. Furthermore, the MgR mutants generated in this work can be used as improved tools for the specific sample treatment. For example, the S562G mutant has higher sensitivity to ribose methylation and could help enrich for methylated RNAs intermediate product. Application of these MgR variants deserves further experimental trials with designated design. Our work provided detailed structural information of MgR binding to RNAs, which will be the premise for a rational design and alteration of the enzyme for other special needs.
DATA AVAILABILITY
The atomic coordinates have been deposited in the Protein Data Bank under the codes ID 7DCY (apo-MgR), 7DIC (MgR-ssRNA), 7DOL (MgR-dsRNA) and 7DID (MgR-Me).
ACKNOWLEDGEMENTS
We thank Dr Henry Nguyen, Dr Guohui Li for valuable discussion, and Hongjuan Chen, Yao Tang for technical assistance. We also thank the staff at beamline BL17U1 of the Shanghai Synchrotron Radiation Facility (SSRF). We thank Dr Qihan Chen for providing the initial plasmids with coding sequences of the ribonucleases.
Author contributions: A.A. and X.J. designed the experiments; A.A., X.L, X.Q., T.Y., Y.L., H.G. and T.L. performed the experiments; A.A. and X.J. analyzed the data; A.A. and X.J. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This research is supported by the National Key Research and Development Program of China [2018YFA0507100, 2016YFD0500300], National Natural Science Foundation of China [Grant No. 81871639] and the Fundamental Research Funds for the Central Universities.
Conflict of interest statement. None declared.
REFERENCES
1.
2.
4.
5.
6.
7.
8.
9.
10.
11.
12.
13.
14.
15.
16.
17.
18.
19.
20.
21.
22.
23.
24.
25.
26.
27.
28.
29.
30.
31.
32.
33.
34.
35.
36.
37.
38.
39.
40.
41.
42.
43.
44.
45.
46.
47.
48.
49.
50.
51.
52.
53.
54.
55.
56.
58.
59.
60.
61.
62.
63.
64.
 Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation
Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation
