
The authors wish it to be known that, in their opinion, the first three authors should be regarded as Joint First Authors.
- Altmetric
Pif1 is an SF1B helicase that is evolutionarily conserved from bacteria to humans and plays multiple roles in maintaining genome stability in both nucleus and mitochondria. Though highly conserved, Pif1 family harbors a large mechanistic diversity. Here, we report crystal structures of Thermus oshimai Pif1 (ToPif1) alone and complexed with partial duplex or single-stranded DNA. In the apo state and in complex with a partial duplex DNA, ToPif1 is monomeric with its domain 2B/loop3 adopting a closed and an open conformation, respectively. When complexed with a single-stranded DNA, ToPif1 forms a stable dimer with domain 2B/loop3 shifting to a more open conformation. Single-molecule and biochemical assays show that domain 2B/loop3 switches repetitively between the closed and open conformations when a ToPif1 monomer unwinds DNA and, in contrast with other typical dimeric SF1A helicases, dimerization has an inhibitory effect on its helicase activity. This mechanism is not general for all Pif1 helicases but illustrates the diversity of regulation mechanisms among different helicases. It also raises the possibility that although dimerization results in activation for SF1A helicases, it may lead to inhibition for some of the other uncharacterized SF1B helicases, an interesting subject warranting further studies.
INTRODUCTION
Helicases are vectorial enzymes that convert the chemical energy stored in ATP into mechanical force to destabilize the hydrogen bonds between complementary base pairs in duplex DNA and translocate along single-stranded nucleic acids. They are critical in many aspects of genome maintenance, including DNA repair, replication and recombination (1–3). Helicases are classified into six different superfamilies (SF) based on the conserved helicase motifs. SF1 helicases are further divided into two sub-classes according to their polarities: those that translocate along their nucleic acid substrates with 3′−5′ polarity (SF1A) and those with 5′−3′ polarity (SF1B) (2,3).
To perform diverse functions, helicases must be carefully regulated in cells through intra- and/or inter-molecular interactions, because unregulated helicases may be inactive, or highly active to unwind all nucleic acids encountered, consequently undermining genomic stability (4–6). Extensive structural and functional studies of some SF1 helicases have provided considerable evidences that their helicase activities may be regulated by spatial reorientations of subdomains through protein–protein interactions (3). Bacillus stearothermophilus PcrA, Escherichia coli Rep and UvrD are homologous SF1A helicases comprising two RecA-like domains, 1A and 2A, and two accessory domains, 1B and 2B (7–9). These helicases seem to contain all that is needed for helicase activity and their monomeric forms can translocate processively along ssDNA (10–12). Homo-dimerization or association with accessory proteins significantly activate helicase activity in vitro (13–15). In crystal structures, domain 2B of PcrA (7,16), Rep (8) and UvrD (9,17) either adopts an open conformation (corresponding to an open ssDNA binding cleft) or a closed conformation. The positions of domain 2B in the two conformations differ by a rotation of 130°−160° about a hinge region connecting it to domain 2A. Biochemical and biophysical studies revealed that domain 2B of these helicases is regulatory and its rotational conformational state can modulate helicase activity, and self-assembly or interaction with an accessory protein shifts it to a closed conformation and alleviates its autoinhibition effect (10,18–20). However, it is unknown whether dimerization has similar activation effect for SF1B helicases.
The Pif1 helicase family is a group of SF1B helicases that are evolutionarily conserved from bacteria to humans, playing multiple roles in maintaining genome stability in both nucleus and mitochondria (21–25). They variously affect telomeric (26,27), ribosomal (28) and mitochondrial DNA replication (29). Recent crystal structures of bacterial, yeast and human Pif1 helicases have revealed that Pif1 family helicases share close subdomain folding features with other SF1 helicases (30–34), reviewed in (25). However, Pif1 family also exhibits domain variability as, for instance, a large insertion in domain 2B exists in ScPif1 (32) and a WYL domain at the C-terminal part is found in Pif1 helicases from thermophilic bacteria Thermotoga elfii (33) and Deferribacter desulfuricans (Supplementary Figure S1, alignment of Pif1 proteins from bacteria, yeast and human). In addition, a structure of BaPif1 (Pif1 form Bacteroides sp, PDB code: 6L3G) dimer in complex with a forked dsDNA has been observed, with the two interacting molecules binding to the 5′ arm and 3′ ss/dsDNA junction, respectively, and regulating each other’s helicase activity (35). Extensive biochemical studies have revealed characteristics of Pif1 helicases in duplex DNA unwinding. ScPif1 (Pif1 from Saccharomyces cerevisiae) and hPif1 (human Pif1) require ssDNA for binding and robustly unwind forked dsDNA (36–39). ScPif1 unwinds duplex DNA in a single-base-pair step and prefers unwinding RNA/DNA hybrids over dsDNA (37,40,41). ScPif1 exists as a monomer in solution and may dimerize upon binding to ssDNA and forked DNA (36,42). Like other SF1 helicases, ScPif1 can translocate on ssDNA as monomers (43). Finally, ScPif1 can unwind dsDNA as a monomer (44), just as Pif1-like helicases T4 phage Dda and E. coli TraI (45,46).
In the present study, structural, biochemical and smFRET (single-molecule fluorescence resonance energy transfer) approaches are combined to determine the structural and mechanistic basis of Thermus oshimai Pif1 (ToPif1) helicase activity. We demonstrated that apo-ToPif1 is in a closed conformation and binding of partial duplex DNA results in an open conformation with a large movement of domain 2B/loop3. During DNA unwinding, repetitive switching of domain 2B between the open and closed conformations was observed. Furthermore, we determined dimeric structures of ToPif1 in complex with ssDNA and different ATP analogues. We revealed that, in contrast with SF1A helicases PcrA, Rep and UvrD, the ssDNA binding-induced dimerization of SF1B ToPif1 inhibits rather than enhances its helicase activity.
MATERIALS AND METHODS
Protein expression and purification
The gene encoding Thermus oshimai Pif1 (ToPif1, residues 64−507) was cloned into pET15b-SUMO and then transformed into the C2566H E. coli strain (New England Biolabs). When the culture reached early stationary phase (OD600 = 0.55−0.6) at 37°C, 0.3 mM IPTG (Isopropyl-β-D-thiogalactopyranoside) was added and the protein expression was induced at 18°C over 16 h. Cells were harvested by centrifugation (4500 g, 4°C, 15 min) and pellets were suspended in lysis buffer (20 mM Tris-HCl, pH 7.5, 500 mM NaCl, 10 mM imidazole and 5% glycerol (v/v)). Cells were broken with a French press and then further sonicated 2−3 times to shear DNA. After centrifugation at 12 000 rpm for 40 min, the supernatants were filtered through a 0.45-μm filter and loaded onto a Ni2+-charged IMAC column (GE Healthcare). After washing twice, the SUMO-ToPif1 was then eluted from the Ni2+ affinity column with elution buffer (20 mM Tris-HCl, pH 7.5, 500 mM NaCl, 300 mM imidazole and 5% glycerol (v/v)) at 4°C. The eluted protein was treated with SUMO protease (Invitrogen, Beijing) and simultaneously dialyzed against the lysis buffer at 4°C overnight. Then, the SUMO-digested protein was loaded on a Ni2+ affinity column (equilibrated in the lysis buffer) to remove the SUMO-tag and further purified by a HiTrap Heparin column (GE Healthcare). The eluted fraction containing ToPif1 was collected and concentrated. The final purified protein was dialyzed against the storage buffer (20 mM Tris-HCl, pH 7.5, 500 mM NaCl, 1 mM DTT) and concentrated to approximately 10 mg/ml for crystallization and was about 95% pure as determined by SDS-PAGE. Mutations and truncations were engineered by PCR overlapping-PCR protocol.
DNA substrate preparation
All the DNA substrates used in this study were chemically synthesized and HPLC-purified by Sangon Biotech (Shanghai) and are listed in Supplementary Table S1. The oligonucleotides used in dynamic light scattering (DLS) assay and small-angle X-ray scattering (SAXS) samples were prepared at 10 μM working concentration. The partial duplex DNAs used in the stopped-flow assay and SAXS analysis were prepared by heating the complementary single-stranded oligonucleotides to 95°C in stocking buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl) for 5 min and then annealed by slow cooling to room temperature.
Crystallization of ToPif1-nucleic acid complexes
Crystallization trials of ToPif1 and its complexes with DNA and ATP analogs/ADP were performed at 20°C by the sitting-drop vapor diffusion method. Crystallization screening was carried out at 20°C using commercial screening kits (Hampton Research, Molecular Dimensions and Rigaku Reagents), where the ToPif1-DNA complex was mixed at a 1:1 ratio with the reservoir solution. The initial crystals were obtained by mixing 1 μl of the protein (0.2 mM ToPif1) with 1 μl of a reservoir solution comprising 0.1 M sodium Hepes-MOPS buffer system (pH 7.5), 1.25% ethylene glycol, 10% PEG4000 and 20% glycerol. Crystals of Q164C/E409C mutant were obtained by using 0.1 M sodium Hepes-MOPS buffer system (pH 7.5), 0.1 M NDSB-256, 2% 1,6-hexanediol, 2% 1-butanol, 2% 1,2-propanediol (racemic), 2% 2-propanol, 2% 1,4-butanediol, 2% 1,3-propanediol, 10% PEG8000 and 20% ethylene glycol. Crystals of ToPif1-dT15-no nucleotide complexes (ATPγS was not seen) were obtained by using 0.1 M sodium Hepes-MOPS buffer system (pH 7.5), 0.03 M NaF, 0.03 M NaI, 0.03 M NaBr, 10% PEG4000 and 20% glycerol as reservoir solution with a protein/DNA ratio of 1:1 and 1 mM ATPγS. Crystals of ToPif1-dT15-ADP·AlF4/ADP·MgF4/ADP·VO4 ternary complexes with protein/DNA ratios of 1:1 and 2:1 were obtained using 0.1 M MES monohydrate-imidazole buffer system (pH 6.5), 1.25% ethylene glycol, 8% PEG4000 and 16% glycerol as reservoir solution and 1 mM ADP·AlF4/ADP·MgF4/ADP·VO4 was added. Crystals of ToPif1-dT15-ADP with a protein/DNA ratio of 1:1 were obtained using 0.1 M Tris-HCl (pH 8.5), 0.03 M NaF, 0.03 M NaI, 0.03 M NaBr, 0.03 M MES, 10% PEG4000 and 20% glycerol as reservoir solution and 1 mM ADP was added. Crystals of ToPif1-S7D11-ADP·AlF4 with a protein/DNA ratio of 1:1.2 were obtained using 0.1 M sodium Hepes-MOPS buffer system (pH 7.5), 0.03 M CaCl2, 0.03 M MgCl2, 10% PEG8000 and 20% ethylene glycol as reservoir solution and 1 mM ADP·AlF4 was added. All these conditions were optimized by a grid search using 48-well Linbro plates at 20°C where 1 μl of protein sample and 1 μl of reservoir solution were mixed together and equilibrated with 60 μl of reservoir solution.
X-ray data collection, phasing and refinement
All X-ray diffraction data were collected on beamline BL19U-1 at Shanghai Synchrotron Radiation Facility using a Pilatus 6M detector (Dectris) and were processed using XDS (47). The initial apo-ToPif1 structure (crystals belonging to space group P21 with unit-cell parameters a = 74.27 Å, b = 59.41 Å, c = 117.15 Å, β = 91.82°, and two molecules per asymmetric unit) was solved by molecular replacement, performed in PHENIX (48) with Phaser (49) using the Bacteroides sp Pif1 (BsPif1) structure as template model (30). Manual reconstruction was done with COOT (50) and further refinement was performed in PHENIX. The other ToPif1 structures were then solved by molecular replacement using the apo structure as search model. Cell parameters and data collection statistics are reported in Supplementary Table S2.
Small-angle X-ray scattering
SAXS experiments were carried out at 20°C with SEC-HPLC coupled to SAXS data collection at beamline SWING (SOLEIL Synchrotron, Saint-Aubin, France). The samples of protein, DNA and protein–DNA complexes were injected at a concentration of 10 mg/ml on BioSEC3 HPLC column (Agilent) at a flow rate of 0.2 ml/min while using 25 mM Tris-HCl, pH 7.5, 100 mM NaCl and 5% glycerol as buffer. Scattering data were collected using a PCCD170 detector (Aviex) and data reduction and processing of images were done with Foxtrot (51). Analysis of the HPLC-SAXS profiles was performed using Foxtrot and US-SOMO HPLC-SAXS module of Ultrascan2 software (52). Briefly, I(q, t) stack of data frames I(q) collected over time t was transformed into I(t, q) stack allowing visualization of peaks at I(t, qmin) for the lowest value of q recorded (qmin = 0.007 Å−1). I(0) and radius of gyration (Rg) were calculated over the profile with Guinier approximation up to q limit satisfying qRg < 1.3. Peaks that were not baseline-resolved were deconvoluted and modeled with Gaussian approximation tools included in US-SOMO. SAXS profiles I (t, q) for the extracted peaks were generated, then frames with constant Rg were averaged to produce SAXS reference data I(q) for each peak. Pair distance distribution function (PPDF) and maximum particle dimension (Dmax) were calculated using the GNOM4 program from ATSAS 2.8 suite (53). Ab initio envelopes for isolated proteins and DNAs were determined using DAMMIF with experimental Rg and Dmax values as constraints. Atomic models derived from crystal structures were adjusted to SAXS data by flexible modeling with MODELLER. Profiles of atomic models were calculated and fitted to the experimental data using CRYSOL and aligned on ab initio bead models with SUPCOMB. All the SAXS parameters are summarized in Supplementary Table S3.
Stopped-flow unwinding assay
Fluorescence stopped-flow assay was performed as previously described (54). Briefly, unwinding kinetics was measured in a two-syringe mode, where ToPif1 and fluorescently labeled DNA substrate were pre-incubated at 37°C in one syringe for 5 min and the unwinding reaction was initiated by rapidly mixing ATP from another syringe. Each syringe contained unwinding reaction buffer A (25 mM Tris-HCl, pH 7.5, 70 mM NaCl, 2 mM MgCl2 and 2 mM DTT). All concentrations listed are after mixing unless noted otherwise. For converting the output data from volts to percentage of unwinding, a calibration experiment was performed in a four-syringe mode, where the helicase, the hexachlorofluorescein-labeled single-stranded oligonucleotides, the fluorescein-labeled single-stranded oligonucleotides and ATP were in four syringes, respectively. The fluorescent signal of the mixed solution from the four syringes corresponds to 100% unwinding. The standard reaction was performed with 4 nM DNA substrates, 1 mM ATP and 5−250 nM helicase in buffer A.
All stopped-flow kinetic traces were averages of over 10 individual traces. Some kinetic traces were analyzed using Bio-Kine (version 4.26, Bio-Logic, France) with the following Equation 1,
where A(t) represents the fraction of DNA unwound at time t; A1 and A2 are the unwinding amplitudes; k1 and k2 are the unwinding rate constants of the two phases; and t0 is the time when the fraction of DNA unwound starts to rise. From the four parameters obtained through fitting, we can get the total unwinding amplitude AT = A1 + A2 and the initial unwinding rate (i.e. the slope of the kinetic unwinding curve at early times) ku = k1A1 + k2A2.Single-molecule fluorescence data acquisition
The smFRET study was carried out with a home-built objective-type total-internal-reflection microscopy and performed as described previously (55). In brief, 50 pM fluorescently labeled DNA (or 100 nM S69C/S405C mutant) were added to the chamber containing imaging buffer composed of 20 mM Tris-HCl (pH 7.5), 2 mM MgCl2, 70 mM NaCl, 2 mM DTT and an oxygen scavenging system (0.8% d-glucose, 1 mg/ml glucose oxidase, 0.4 mg/ml catalase and 1 mM Trolox). After immobilization for 10 min, free DNA (or S69C/S405C) molecules were removed by washing with the imagining buffer. We used an exposure time of 100 ms for all recordings at a constant temperature of 22°C. The FRET efficiency was calculated using IA /(ID + IA), where ID and IA represent the intensities of donor and acceptor, respectively. Basic data analysis was carried out by scripts written in Matlab, and all data fittings were generated by Origin 9.0. Histograms were fitted by Gaussian distributions, with the peak positions unrestrained.
Dynamic light scattering (DLS) assay
DLS measurements were performed at 37°C using a DynaPro NanoStar instrument (Wyatt Technology Europe GmbH, Germany) equipped with a thermostated cell holder and disposable cuvettes (UVette, Eppendorf). All solutions were filtered using 0.22 μm filters. Purified ToPif1 (10 μM) was mixed with ssDNA in buffer B (20 mM Tris-HCl, pH 7.5, 10 mM NaCl, 1 mM MgCl2, 1 mM DTT, 1 mM ADP·AlF4) and then dialyzed in buffer A (final volume, 30 μl). The scattered light was collected at an angle of 90°. Recording times were typically between 3 and 5 min (20−30 cycles in average, 10 s in each cycle). The analysis was performed with the Dynamics 7.0 software using regularization method (Wyatt Technology). The molecular weight was calculated from the hydrodynamic radius using the following empirical Equation 2,
where Mw and RH represent the molecular weight (in kDa) and the hydrodynamic radius (in nm), respectively.Analytical size-exclusion chromatography (SEC)
Gel filtration experiments were performed using a 10/30 Superdex 200 GL gel filtration column (GE Healthcare) as described previously (32,54). Briefly, the column was equilibrated at a flow rate of 0.4 ml/min with 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM MgCl2, 1 mM DTT. About 100 μg of ToPif1 was loaded with a final concentration of 65 μM and the absorbance at 280 and 260 nm was recorded. The experiments in the presence of ssDNA were performed by preincubating ToPif1 and DNAs in buffer B for 30 min at 25°C. The calibration graph of logRS versus Kav was constructed using a high and low molecular weight calibration kit from Sigma: cytochrome c (12.4 kDa), carbonic anhydrase (29 kDa), albumin (67 kDa), phosphorylase b (97.4 kDa) and thyroglobulin (669 kDa). Assuming similar shape factors, the calibration plot of logMw versus Kav allowed the determination, in a first approximation, of the molecular weights of ToPif1 and its complexes with ssDNA.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)
One dimensional SDS-PAGE was carried out according to Laemmli (56). Ten percent of resolving slab gels were used (10 × 8 × 0.1 cm3). Samples were prepared for electrophoresis by mixing 10 μl of extracted protein, 2.5 μl of 2-mercaptoethanol, and 7.5 μl of 0.002% bromophenol blue in 0.0625 M Tris-HC1 (pH 6.8), containing 10% glycerol and 2% SDS. The PageRuler Prestained Protein Ladder for electrophoresis analysis was purchased from Thermo Scientific. All protein stainings were performed using Comassie Blue according to Hames and Rickwood (57).
Circular dichroism (CD) spectropolarimetry
CD experiments were performed according to the method described by Greenfield (58) with a Bio-Logic MOS-450/AF-CD optical system (BioLogic Science Instruments, Seyssinet-Pariset, France) equipped with a temperature-controlled cell holder, using a quartz cell with 1 mm path length; 20 μM solution of protein were prepared in 200 mM sodium phosphate buffer (NaH2PO4/Na2HPO4, pH 7.5). CD spectra were recorded in the UV region (180–260 nm) at 1-nm increment with an averaging time of 2 s at 25°C. Nitrogen was used to flush the sample compartment throughout the whole experiment.
Dimer crosslinking
Double mutant Q327C/W482C and dT15 were mixed at a protein/DNA ratio of 2.5:1 in a buffer containing 20 mM Tris-HCl (pH 7.5), 20 mM NaCl and 1 mM MgCl2, and equilibrated on ice for 2 days. After treatment with oxygen for 3 h, the complex was purified using a 10/30 Superdex 200 GL gel filtration column (GE Healthcare) and then a HiTrap Heparin column (GE Healthcare). The solution was heated in a 72°C water bath for 10 min, followed by 60% (w/v) ammonium sulfate precipitation and reconstitution in buffer C (20 mM Tris-HCl pH 7.5, 70 mM NaCl, 2 mM MgCl2). The crosslinked dimer was detected by SDS-PAGE.
RESULTS
ToPif1 in pre-catalytic closed conformation is inactive
Unliganded ToPif1 (64–507) was crystallized in space group P21 with two molecules in the asymmetric unit. The structure was solved by molecular replacement using Bacteroides sp Pif1 (BsPif1) structure (30) (PDB code: 5FTD) as a search model and refined to 3.2 Å resolution. All the residues 64−507 were clearly attributed. The N-terminal part (1–63) that is predicted to be unstructured was deleted in order to favor crystallization. Apo-ToPif1 is monomeric and contains four structurally defined domains, RecA-like domains 1A and 2A and accessory domains 1B and 2B, and the Pif1-specific signature motif (PFSS) (Figure 1A and B, crystal form I). Its domain organization resembles that of other SF1 family helicases. Domain 1B folds as an unstructured loop. Domain 2B folds as a SH3-like domain comprising five β-strands arranged in two perpendicular β-sheets as observed in typical SF1B helicases DrRecD2 (PDB 3E1S) and BsPif1 (30,59). Furthermore, an unusual long antiparallel β-hairpin (named as loop3) within domain 2B extends toward domain 1B and interacts with several residues of domain 1A (Y401-Q128/R126/T114, E409-Q164 and K411-K109), stabilizing the protein in a closed conformation (Figure 2A).

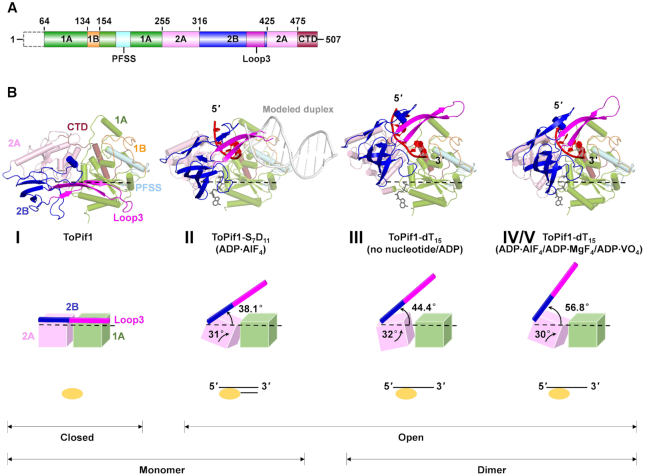
Overall structures of ToPif1 and its complexes with DNA/ATP analogues. (A) Schematic representation of ToPif1 domain structures, with the conserved domains 1A (green), 1B (orange), 2A (pink) and 2B (blue) with loop3 in magenta, Pif1-specific signature motif (PFSS, cyan) and C-terminal (CTD, brown). (B) Upper panel, overall structures of apo-ToPif1 (crystal form I) and ToPif1 in complex with DNA/ATP analogues (crystal forms II−V); lower panel, schematic representation of the spatial orientations of 2B/loop3 domain relative to the helicase core and the oligomerization states of ToPif1 in the different crystallized structures. The structures are shown in the same orientation, superimposed on domain 1A.

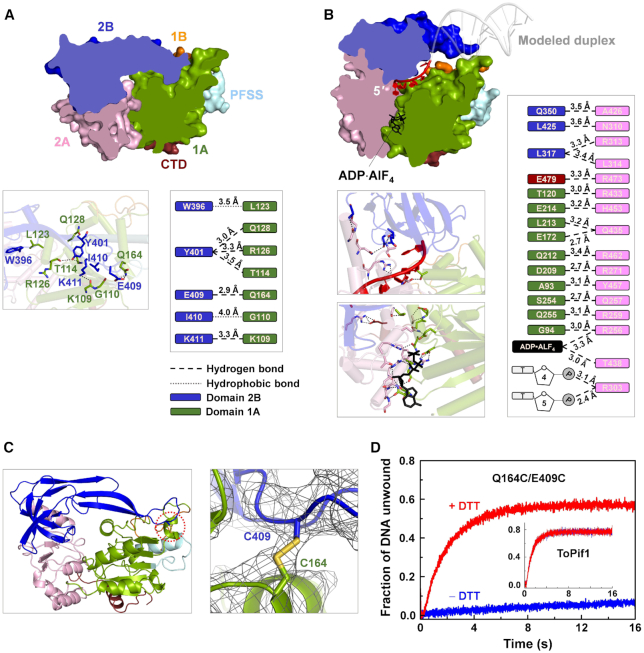
Interactions of domain 2B/loop3 and domain 1A stabilizing ToPif1 in a closed and inactive conformation. (A) Side (cutaway) view of the apo-ToPif1 structure (crystal form I) showing the closed state (upper panel) and the interactions between domains 2B/loop3 and 1A (lower panel). (B) Side view of the structure of ToPif1 in complex with a partial duplex DNA (crystal form II). Binding of the partial duplex DNA disrupts the interactions between domains 2B and 1A and stabilizes the protein in an open conformation. (C) Overview of crystal structure of double mutant Q164C/E409C with the disulfide bond between C409 and C164 marked by a dotted circle (left panel), and close-up view of the disulfide bond (right panel). (D) Stopped-flow unwinding kinetic curves of ToPif1 and Q164C/E409C. The experiments were performed as described in ‘Materials and Methods’ section with 100 nM protein, 4 nM fluorescently labeled partial duplex DNA (S8D16) and 1 mM ATP in the absence or in the presence of 3 mM DTT.
As we have mentioned, domain 2B is regulatory in SF1A helicases PcrA, Rep, and UvrD, which was highlighted especially by the interesting work showing that a Rep monomer with undetectable unwinding activity can be converted into a superhelicase by blocking its domain 2B in the closed conformation through crosslinking (18). To probe whether domain 2B of ToPif1 behaves in the same manner, we also made a similar crosslinking with ToPif1 by taking advantage of its unique cysteine-free sequence feature. We introduced two cysteine residues at positions of Q164 on domain 1A and E409 on domain 2B/loop3. The spatial distance between Q164 and E409 is 2.9 Å, which is suitable to establish a disulfide bond between the two residues and lock domain 2B in the closed conformation (Figure 2A). The crystal structure of the double mutant Q164C/E409C shows that a disulfide bond between Q164C and E409C has been correctly formed (Figure 2C). To confirm that the expected intramolecular but not intermolecular disulfide bond was also formed in solution, both SDS-page and DLS analyses were performed with the single (Q164C and E409C) and double (Q164C/E409C) mutants, demonstrating that these mutants exclusively remain as monomers in solution (Supplementary Figure S2A and C). Then, fluorescent stopped-flow helicase activity assay was performed with the wild-type ToPif1 and the three mutants. While the wild-type helicase and the two single mutants displayed normal helicase activities in the absence or presence of DTT (insert of Figure 2D and Supplementary Figure S2B), the unwinding activity of the double mutant Q164C/E409C was negligible (Figure 2D) in the absence of DTT, indicating that ToPif1 is inactive in the closed conformation. Interestingly, the unwinding activity of Q164C/E409C could be rejuvenated with 3 mM DTT and achieved a level comparable to the wild-type helicase (Figure 2D). These results, altogether, illustrate that in sharp contrast to SF1A helicases, SF1B ToPif1 is inactive when its domain 2B is blocked in the closed conformation.
Binding of partial duplex DNA induces an open conformation of domain 2B/loop3
ToPif1 displays a directional polarity from 5′ to 3′. To capture the structural conformation of ToPif1 during initial strand separation, a ternary complex of ToPif1 was then crystallized, with a partial duplex DNA bearing a 7-nt 5′ ssDNA overhang and a 11-bp dsDNA (S7D11) in the presence of Mg2+ and ADP·AlF4. As in the above apo structure, the crystal also belongs to P21 space group, and diffracted up to 1.9 Å (crystal form II, Figure 1B). The crystal structure has two molecules in the asymmetric unit, but the buried area (263 Å2) is notably too small to determine any significant intermolecular interactions. Furthermore, the electron densities corresponding to the dsDNA and the extremity of loop3 (residues 402−410) are not strong enough for determining their structures, indicating that these structures are disordered either statically or dynamically.
Comparison of the apo structure to the ternary complex bound to partial duplex DNA and ADP·AlF4 shows an important conformational change of domains 2A and 2B of ToPif1. Binding of ssDNA with 5′ end on 2A and 3′ end on 1A domains induces a rotation of the 2A domain that allow stacking of residues H432 and F451 on ribose and interaction of R302 and R303 with the phosphate backbone (Supplementary Figure S4A). Nucleotide binding induces also a structural rearrangement of the linker between 1A and 2A domains (Q255, R256). Residues E172 and Q212 from domain 1A and R256 and R462 from domain 2A participate to the coordination of AlF4 mimicking phosphate position of ATP (Supplementary Figure S3A−F). When the complete ATP-binding site is formed, domain 2A undergoes a 31° rotation relative to domain 1A (Figure 1B). This movement upon ATP analog binding is the same as previously described for BsPif1, BaPif1 and hPif1 (30,31,34) and the nucleotide binding site is remarkably conserved (Supplementary Figure S3G and H). Following the rotation of domain 2A, domain 2B/loop3 undergoes a 38.1° opening movement (Figure 1B). As detailed in Figure 2A and B, these conformational changes disrupt all interactions between domains 2B/loop3 and 1A observed in the apo structure and consequently establish a new subset of interactions between domains 2A and 1A/CTD, and possibly between the extremity of loop3 and the dsDNA. Therefore, the fact that both structures of the dsDNA and the extremity of loop3 appear to be mobile and disordered may imply loop3 interacts with the dsDNA and contributes actively to dsDNA unwinding. The conformation and configuration of ssDNA in the complex are in general agreement with models for helicase action, where ssDNA recognition by the helicase involves mostly the sugar-phosphate backbone but very few base-specific interactions (Supplementary Figure S4A).
Dimeric structures are induced by ssDNA binding
To get structural snapshots of ToPif1 in different transition states with its substrates, ToPif1 was crystallized with a polyT (dT15) in combination with ADP and different ATP analogs, including, ADP·AlF4, ADP·MgF4 and ADP·VO4. Though ATPγS was added during crystallization, it is not visible in the electron density and this structure is considered as containing no nucleotide. New crystal forms III, IV and V were obtained (Figure 1B) and the best diffracting structures were refined to 1.9 Å resolution. In each structure, the adenine base is recognized and stabilized by Q72 in the Q-motif (Supplementary Figure S4B, left panel). Especially, as Q-motif’s presence in a helicase generally increases its ATP specificity (60), we observed that both of the DNA unwinding rate (corresponding to hydrolysis rate) and efficiency of ToPif1 are higher with ATP than with other nucleotides (Supplementary Figure S4C). In contrast, BsPif1 lacks a functional Q-motif, because the amino acid equivalent to Q72 in ToPif1 is replaced with M10 in BsPif1 (Supplementary Figure S4B, right panel) (30), and thus it displays general NTPase activities (54).
Unexpectedly, in the three distinct crystal forms with different crystal packings, a dimeric form of ToPif1 was always crystallized, either through 2-fold crystallographic symmetry or with two molecules in the asymmetric unit related by a 2-fold non-crystallographic symmetry (NCS) (Figures 1B, 3A and Supplementary Figure S5), indicating that dimerization is an intrinsic structural property of ToPif1 upon ssDNA binding. This finding is consistent with our results of DLS analysis in conjunction with SEC, which show that the protein also forms a dimer with these ssDNA and long-tailed ss/dsDNA substrates in solution (Supplementary Figure S6 and Table S4). The dimeric configurations in the three types of crystals (forms III, IV and V) are virtually the same and can be superimposed with rmsd's of all Cα atoms varying between 1.97 and 2.29 Å.

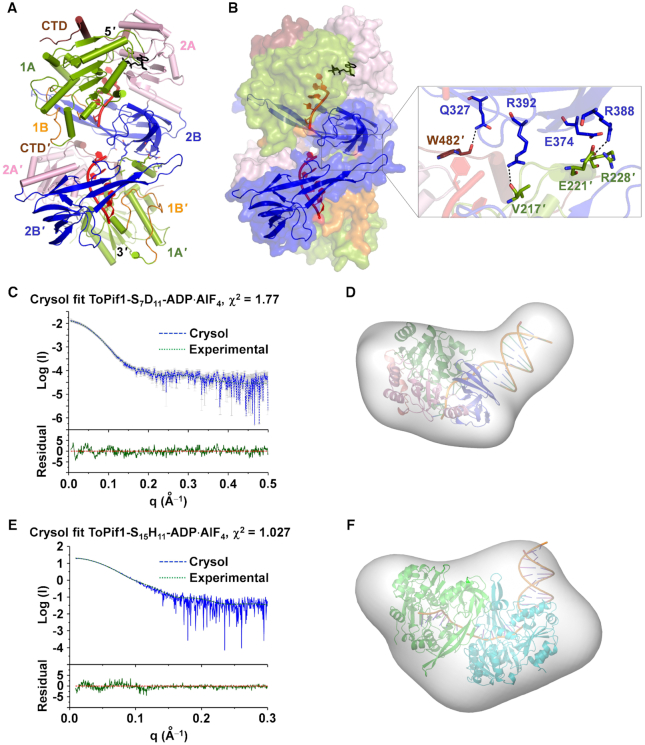
Dimerization of ToPif1. (A) Overall structure of the dimeric ToPif1-dT15-ADP·AlF4 ternary complex (crystal form V). The bound ADP·AlF4 is shown as black sticks. (B) Surface representation of the structure with domains 2B, 2B′ and DNA shown in ribbon diagram (left panel), and details of the interactions between the two ToPif1 molecules (right panel). (C andD) Fit curve of the SAXS data of ToPif1-S7D11-ADP·AlF4 with a modelled monomer of ToPif1 bound to the 5′ overhang of S7D11 calculated with Crysol, and the model superimposed on the ab initio envelope calculated with DAMMIF. (E and F) The fit curve and the model of a ToPif1 dimer bound to the 5′ overhang of S15H11 (ToPif1-S15H11-ADP·AlF4) superimposed on the ab initio envelope calculated with DAMMIF.
The two ToPif1 molecules (referred to as molecules M and M′) within a dimer in the asymmetric unit are essentially identical. They are arranged in a head-to-tail manner and adopt a highly compact overall shape with a total buried area of 745 Å2 in the interface (Figure 3A and B). The DNA-binding site size of ToPif1 is about 5−7 nt according to the ternary crystal structures (Supplementary Figure S4D). Since crystal forms III and IV (Supplementary Table S2) contain only one molecule of ToPif1 bound to ssDNA in the asymmetric unit the resulted head-to-tail dimer generated by 21 crystallographic symmetry is not bound by one continuous single dT15 molecule, but rather, each monomer is bound independently by one dT15 molecule (Figure 3A and B). In the head to tail configuration, the 3′ end of one ssDNA is close to the 5′ end of the other one, mimicking a single continuous ssDNA. In crystal form V, the asymmetric unit contains a dimer of ToPif1 and there is an ambiguity if there are two dT15 molecules related by NCS (non-crystallographic symmetry) or a single dT15 spanning across the dimer. In order to know if a single ssDNA could be bound by a ToPif1 dimer in crystal form V, we used an ssDNA oligo (dGR17) containing T and G with a non-symmetric sequence. As ToPif1 exhibits comparable binding activities with G-rich and polyT oligos (Supplementary Figure S8A), we got a high-resolution (2.21 Å) structure of ToPif1 in complex with G-rich ssDNA (dGR17) in the presence of ADP·AlF4. The structure provides important additional information: (i) the structural superposition of the two complexes of ToPif1/dGR17 and ToPif1/dT15 demonstrated that the two structures are essentially the same in terms of global conformation, DNA and ATP bindings, and dimer interactions, except that an additional pair of residues (hydrogen-bonded K329/L480′) are implicated in the dimer interaction in the presence of G-rich oligos (Supplementary Figure S8B and C); (ii) the G-rich ssDNA-induced dimer is arranged in a head-to-tail manner. Though the density for ssDNA between the two ToPif1 molecules is weak, most of the bases T and G can be fitted unambiguously in the electron density map, according to the sequence of the oligonucleotide (Supplementary Figure S8E). We can thus conclude that the two ToPif1 molecules are bound by one continuous single G-rich ssDNA molecule (Supplementary Figure S8C and D).
To confirm the dimeric nature of ToPif1 when binding to DNA substrates in solution, SAXS coupled with SEC was used to determine the structures of ToPif1 in complex with different DNAs and ATP analogs mentioned above. From the SEC profiles of the different complexes, the radius of gyration and I(0) can be calculated by Guinier analysis, allowing an accurate determination of the molecular weight. The ternary complex with partial duplex DNA (ToPif1-S7D11-ADP·AlF4) has a molecular weight corresponding to a monomer of ToPif1 bound to ss/dsDNA. The monomeric model derived from X-ray structure fits the SAXS data with χ2 = 1.77 and is well fitted by the ab initio molecular envelope (Figure 3C and D). With the same analysis, the ToPif1 complexed with a DNA hairpin bearing a 15-nt 5′-ssDNA overhang (ToPif1-S15H11-ADP·AlF4) fits well with a dimeric model (χ2 = 1.027), in which the 15-nt ssDNA is bound with a head-to-tail dimer while the dsDNA is located at the putative catalytic site (Figure 3E and F and Supplementary Figure S7A and B). In addition, the comparison with BaPif1 bound to a DNA fork (35) shows that the two dimers are very different and different surfaces are involved (Supplementary Figure S7C−E).
Domain 2B/loop3 is blocked in a more open conformation with dimerization
The dimerization interface between the two ToPif1 molecules involves domain 2B of molecule M and domain 1A′ and the C-terminal of molecule M′, with a network of interactions connecting these domains in which loop3 plays a central role. Superposition of the structures of apo-ToPif1 (crystal form I) and the ternary complexes (crystal form II, III, IV or V) on domain 1A reveals that domain 2B/loop3 gradually undergoes an opening from 38.1° to 56.8° with loop3 rotating by 25° due to the dimerization (Figure 1B and Supplementary Figure S9). Importantly, the largest conformational change of domain 2B/loop3 is observed when ToPif1 is in complex with ssDNA and ATP analogues. In these complexes, the loop3 is in the open conformation with 56.8° (Figure 1B, crystal form IV/V), which otherwise would clash with domain 2A′ of molecule M′ (Supplementary Figure S9). Two new salt bridges (R388−E221′ and E374−R228′) and two new hydrogen bonds (R392−V217′ and Q327−W482′) block loop3 in the new spatial configuration (Figure 3B). The above structural features indicate that while loop3 in the monomeric ternary complex (ToPif1 bound with ss/dsDNA and ADP·AlF4, Figures 1B and 2B) has no interaction with the rest of protein and may be free to move, it is completely blocked in a more open conformation in the dimeric ternary complexes (Figure 3A and Supplementary Figure S5).
Dimerization inhibits helicase activity
Previous studies of SF1A helicases Rep, UvrD and PcrA have demonstrated that dimerization may induce a significant movement of the 2B domain, relieving its autoinhibitory effect and consequently stimulating helicase activity (10,18,19). To clarify the functional significance of the structurally determined monomer and dimers of ToPif1, we performed a series of DNA unwinding kinetics experiments to probe whether ToPif1 helicase activity can be similarly stimulated by dimerization as SF1A helicases. We first used fluorescently labeled DNA substrates possessing a 16-bp duplex linked with a 5′ ssDNA tail of varying length (S8D16, S14D16 and S26D16), with donor (fluorescein) and acceptor (hexachlorofluorescein) fluorophores covalently attached separately to the two strands at the blunt end of the duplex (Supplementary Table S1). This DNA substrate allows the DNA unwinding kinetics to be followed in real time in stopped-flow experiments by monitoring the change in FRET between the two fluorophores.
For the 8-nt tailed substrate (S8D16) that can only accommodate one ToPif1 molecule, the unwinding amplitude increases from 0.12 to 0.95 as ToPif1 concentration is increased from 5 to 250 nM. However, using the same duplex DNA bearing 14- or 26-nt 5′ ssDNA tail (substrate S14D16 or S26D16) that can accommodate more than one ToPif1 molecule, the unwinding amplitude abruptly reaches the maximum level (0.85−0.95) with increasing protein concentration (from 5 to 10 nM) and then starts to decrease as the protein concentration further increases (Figure 4A). At 250 nM, the unwinding amplitudes for 8-, 14- and 26-nt 5′-tailed substrates are 0.83 ± 0.05, 0.38 ± 0.03 and 0.14 ± 0.03, respectively. According to the DNA binding site size of ToPif1 (5−7 nt), while the 8-nt 5′ tail allows binding of only one monomer, the 14- and 26-nt 5′ tails are long enough to accommodate two or more ToPif1 molecules. Thus, the above results demonstrate that dimerization or oligomerization of ToPif1 inhibits, rather than stimulates, its helicase activity. This is in sharp contrast with the other previously reported SF1A helicases (13–15).

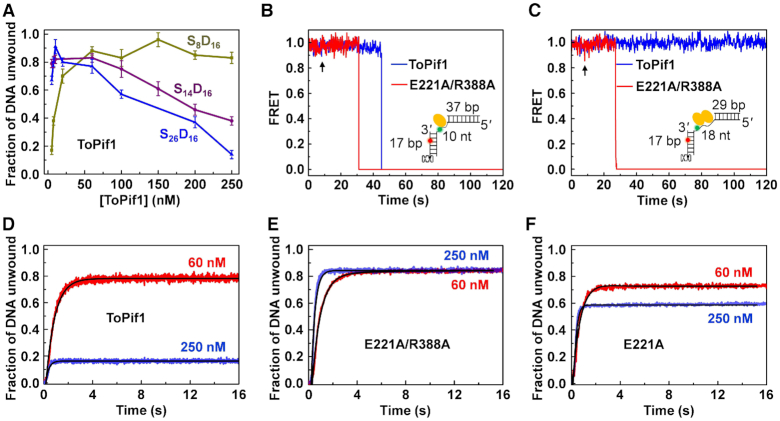
Functional analyses demonstrating dimerization inhibits the helicase activity. (A) Fraction of duplex DNA unwound versus ToPif1 concentration, determined by the stopped-flow assay with 4 nM partial duplex DNA and 1 mM ATP. (B and C) FRET traces for duplex unwinding by ToPif1 and E221A/R388A with substrate D17S10D37 or D17S18D29. About 1 μM protein and 10 μM ATP were used. Black arrows indicate the time of adding proteins. (D−F) Stopped-flow DNA unwinding kinetics of ToPif1 and its mutants (E221A/R388A and E221A). About 4 nM partial duplex DNA (S26D16), 60 (or 250) nM protein and 1 mM ATP were used. See ‘Materials and Methods’ section for more details.
Previously, ScPif1 was shown to catalyze annealing of complementary ssDNAs and this activity could counteract duplex unwinding, leading to an apparently inefficient helicase (44,61). Therefore, the decrease of unwinding efficiency at increasing ToPif1 concentration as observed above might arise from an annealing activity of ToPif1. To exclude this possibility, we then carried out annealing experiments by mixing complementary ssDNAs with ToPif1. The complementary ssDNAs are labeled with FAM and HEX, respectively, and their annealing would lead to a decrease of the fluorescence signal of FAM due to the FRET effect between the two fluorophores. Actually, however, we observed that the fluorescence signal does not decrease and it is essentially invariable in the absence of ATP; in the presence of ATP, it increases due to the ssDNA translocation activity of ToPif1 on the FAM-labeled ssDNA (Supplementary Figure S10). As no protein trap was used, the fluorescence signal first rises, then it remains at a steady-state level rather than decreases, just as observed with ScPif1 (43).
We then used smFRET to probe whether the unwinding activity of ToPif1 is gradually weakened with potential multiple intermediates during the state transition from monomeric to dimeric. To this end, we constructed two DNA substrates consisting of a fluorescently labeled dsDNA (17 bp) linked with another dsDNA through a 10- and 18-nt ssDNA (Figure 4B and C, inserts), which are referred to as D37S10D17 and D29S18D17, respectively. For these substrates, ToPif1 preferentially binds to the ssDNA segment that serves as a 5′ overhang for the fluorescently labeled dsDNA that is tethered to the PEG surface through biotin. Such DNA designs only allow one ToPif1 molecule to bind D37S10D17 and two ToPif1 molecules to bind D29S18D17. The two fluorophores are close to each other before DNA unwinding and become separated after unwinding, which correspond to high and low FRET signals, respectively. From the individual FRET trajectories, we found that ToPif1 rapidly unwinds D37S10D17, even with an ATP concentration as low as 10 μM (Figure 4B). However, ToPif1 may be completely inactivated with D29S18D17 that can accommodate a ToPif1 dimer (Figure 4C). Such an ‘all or none’ unwinding behavior indicates that dimerization quickly and totally inhibits ToPif1’s unwinding activity, possibly through blocking loop3 in the open conformation as shown above (Figure 3A and B). By analyzing >200 FRET trajectories with each substrate, we obtained the DNA unwinding kinetics data of ToPif1 (Supplementary Figure S11A and B). Obviously, ToPif1 is much more efficient in unwinding D37S10D17 than D29S18D17.
To further investigate the relationship between dimerization and regulation of helicase activity, six mutants E221A, R388A, E221A/R388A, R228A, Q327A and R392A were designed to disrupt the interactions that appear to stabilize the dimerization (Figure 3B). The mutants were purified and then characterized by CD, DNA binding and DNA unwinding measurements. All the six mutants are well folded as judged from the CD spectra (Supplementary Figure S12). They display similar DNA unwinding activities as the wild-type, except for R392A that exhibits a moderate reduction in helicase activity (red lines in Figure 4D−F and Supplementary Figure S11C−F). We then further investigated how protein concentration affects their unwinding activity. Under the same experimental conditions with substrate S26D16, while the unwinding amplitude of the wild-type ToPif1 decreases from 0.78 to 0.16 as the protein concentration is increased from 60 to 250 nM (Figure 4D), the five ‘good’ mutants, especially E221A/R388A, still display robust unwinding activity (blue lines in Figure 4E and F and Supplementary Figure S11C−F, and Supplementary Table S5). This clearly indicates that the physical interactions controlling the dimerization are crucial for the inhibition of helicase activity. In accordance with the above observations, smFRET assay also showed that the inhibition effect may be eliminated by mutations of these residues implicated in dimerization. As an example, the mutant E221A/R388A unwinds both D37S10D17 and D29S18D17 rapidly and efficiently (Figure 4B and C, Supplementary Figure S11A and B). In fact, E221A/R388A appears to be even more efficient than the wild-type ToPif1 in unwinding D37S10D17. We think the possible reason is that, for the wild-type helicase, the dimerization-induced inhibition phenomenon still has a probability to occur with the D37S10D17 substrate: a second monomer may bind instantaneously to the elongated ssDNA segment after the first monomer has started but not completed unwinding the 17-bp duplex.
As dimerization of ToPif1 inhibits its helicase activity, it is expected that a crosslinked ToPif1 dimer should be inactive in DNA unwinding. Previously it has been demonstrated that a crosslinked Rep dimer retains DNA helicase activity (62), in accordance with the fact that Rep is a dimeric helicase. For the ToPif1 dimer crosslinking, we introduced two cysteine residues at positions of Q327 on domain 2B and W482 on the CTD. The hydrogen bond between Q327 and W482′ in the dimerization interface (Figure 3B) thus will be replaced with a disulfide bond in the dimer formed by the double mutant Q327C/W482C. The dimer formation was induced by using dT15 (‘Materials and Methods’ section). SDS-PAGE showed the main part of the final reaction product was crosslinked ToPif1 dimer and which could be disrupted by DTT (Supplementary Figure S13A). As expected, the helicase activity of the crosslinked dimer is significantly compromised but it can be restored upon the addition of DTT (Supplementary Figure S13B).
Finally, to determine whether the inhibitory effect of ToPif1 still exists at the high temperatures of the host ecosystems, we performed DNA unwinding kinetics experiments under the same conditions as above but at 50°C that is the optimum growth temperature of T. oshimai. As the protein concentration was increased from 60 to 250 nM, we obtained a similar decrease of the unwinding amplitude of the wild-type ToPif1 (data not shown), just as that shown in Figure 4D at 37°C. This implies that dimerization also occurs at the high temperatures of the host ecosystems.
ToPif1 unwinding activity is correlated with the rotation of domain 2B/loop3
Our above structural and functional analyses demonstrate that ToPif1 is not active when its domain 2B/loop3 is restrained in the closed conformation by crosslinking (Figure 2D) or when it is in the dimeric form, in which domain 2B/loop3 of each molecule is blocked in the open conformation. This raises the possibility that domain 2B/loop3 may undergo dynamic movement between the two conformations for efficiently unwinding DNA substrates. Our structural analysis shows that the distances between residue S69 of domain 1A and S405 of loop3 corresponding to the closed and open conformation are 35.5 and 58.2 Å, respectively (Figure 5A and B). We therefore prepared a new double mutant S69C/S405C in which residues S69 and S405 were replaced with cysteine and then stochastically labeled with Cy3 and Cy5. By immobilizing S69C/S405C on PEG-passivated surface via a biotinylated His-tag antibody (Figure 5C), FRET signals of individual S69C/S405C molecules were monitored. Histograms constructed from smFRET trajectories show clearly that there exist two conformational states with FRET peaks at EFRET = 0.42 and 0.75, respectively (Figure 5D, upper panel), suggesting that the 2B domain of apo ToPif1 undergoes dynamic and reciprocating transition between the open (P1, EFRET = 0.42) and the closed (P2, EFRET = 0.75) conformational states in solution. Furthermore, ssDNA alone or with ADP·AlF4 biases ToPif1 toward the open conformation (Figure 5D, middle and lower panels).

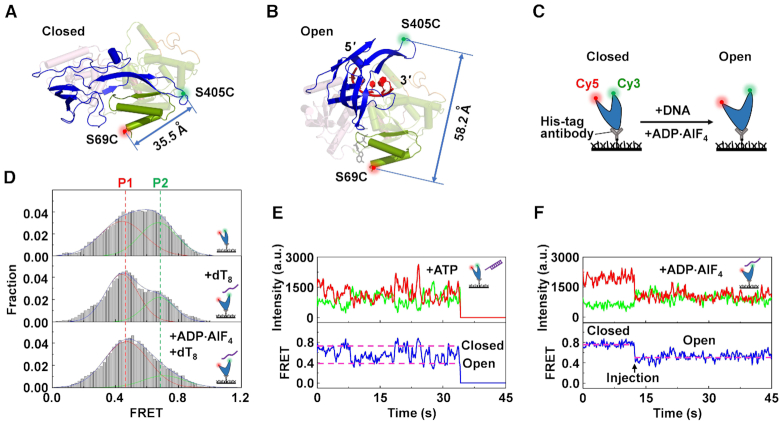
Conformational changes of domain 2B/loop3 determined by smFRET. (A andB) Cartoon representations of ToPif1 monomers, stochastically labelled with Cy3 and Cy5 at positions of S69C and S405C and referred to as S69C/S405C, in closed and open conformations. (C) Schematic of the expected conformational change of S69C/S405C due to ligand binding. The helicase was immobilized on PEG-passivated surface via a biotinylated His-tag antibody. (D) FRET histograms constructed from about 300 individual records of S69C/S405C alone (upper panel), S69C/S405C with dT8 (middle panel) or with dT8 and ADP·AlF4 (lower panel). Gaussian fittings yield two populations peaked at EFRET = 0.42 and 0.75. (E andF) Individual FRET traces recorded with 1 μM partial duplex DNA (S8D38) or ssDNA (dT8) in the presence of 1 mM ATP or ADP·AlF4. The detailed experimental conditions are described in ‘Materials and Methods’ section.
To compare whether the measured FRET values correspond to the crystallographically determined closed and open conformations, the expected FRET efficiencies were estimated using EFRET = [1 + (R/R0)6]−1, with R0 = 54 Å for the Cy3/Cy5 pair (63), and R = 35.5 and 58.2 Å for the closed and open conformations, respectively. The expected FRET efficiencies are ∼0.93 and ∼0.39, which are comparable to the measured results. The slightly lower measured value (∼0.75) for the closed state suggests that the 2B domain of ToPif1 in solution probably assumes a less closed conformation than that observed in the apo structure.
More interestingly, under unwinding conditions with both ATP and partial duplex DNA (S8D38) addition, ToPif1 induced FRET fluctuations between ∼0.42 and ∼0.75 (Figure 5E). In contrast, when ADP⋅AlF4 and ssDNA were added, the FRET signal decreased from ∼0.75 to ∼0.42 and then kept constant without significant fluctuations (Figure 5F), corroborating the structures in which ssDNA binding induces the opening of domain 2B/loop3 (Figure 1). Taken together, the above results show that the helicase activity of ToPif1 is correlated with repetitive opening and closing movements of domain 2B/loop3, which in turn confirms our conjecture that dimerization inhibits the helicase activity through blocking domain 2B in the open conformation.
DISCUSSION
In the present work, monomeric and dimeric crystal structures of ToPif1 alone and bound with partial duplex DNA or ssDNA are determined. The Pif1 helicase is evolutionarily conserved from bacteria to humans. Accordingly, the monomeric structures of ToPif1 are very similar to the structures of other Pif1 helicases, including bacterial BsPif1 (PDB code:5FTD) (30) and BaPif1 (PDB code:5FHG) (31), yeast ScPif1p (PDB code: 5O6E) (32) and human hPif1 (PDB code:6HPU) (31,34), in terms of overall fold and orientations of individual domains. In the monomeric apo structure of ToPif1, domain 2B/loop3 adopts a closed conformation by interacting with both domains 1A and 2A, occluding the ssDNA binding site (crystal form I, Figure 1B). Similarly, in the absence of DNA, BsPif1 (30), BaPif1 (31) and human hPif1 (31,34) also have such domain arrangements as apo-ToPif1, and when the movement of domain 2B is restricted by mutations of residues located in the hinge regions connecting domains 2A and 2B, BaPif1 becomes defective in ssDNA binding and duplex unwinding activities (31). By smFRET experiments, we demonstrated that domain 2B/loop3 of ToPif1 exhibits repetitive rotation/movement between the closed and open conformations during DNA unwinding (Figure 5E), which is likely to be also the case for other Pif1 helicases. Furthermore, when domain 2B/loop3 is locked in the closed conformation with intramolecular crosslinking, ToPif1 is inactivated; whereas addition of DTT converts the unwinding activity of the crosslinked mutant from undetectable to a level comparable to that of the wild-type enzyme (Figure 2C and D). Taken together, the above structural and mutational observations indicate that the conformational change of domain 2B/loop3 should be essential for the enzymatic activities of Pif1 helicases.
In the monomeric crystal structure of ToPif1 complexed with a partial duplex DNA and ADP·AlF4 (crystal form II, Figure 1B), domain 2B/loop3 undergoes a 38.1° opening movement from its orientation in the apo structure, which is consistent with the repetitive switching of domain 2B/loop3 between the open and closed conformations as observed in the smFRET experiments. In fact, ssDNA binding also leads to the same closed to open conformational change (38.1°) of domain 2B in BsPif1 (30). For BaPif1, its structures in complex with ssDNA and ss/dsDNA are essentially the same with the 2B domain in an open conformation and which differs from the closed conformation in the apo structure by 50° (31).
Within the above-mentioned structure of ToPif1 complexed with ss/dsDNA, both structures of the dsDNA and the extremity of loop3 could not be determined due to their poor electron densities. We think they are likely to be mobile and disordered, and interact with each other during the duplex unwinding. Note that both structures are clearly observed in our SAXS model (Figure 3C and D and Supplementary Figure S7A). Interestingly, these two parts are also missing in the crystal structure of BaPif1 complexed with ss/dsDNA (31), which further supports our speculation. It should be noted that those authors suggest the 12-nt complementary strand has been displaced from the tracking strand by the helicase, but we prefer to think that the DNA duplex is still intact although it is not structurally determined. The reason is that, if the short complementary strand is indeed displaced, then the two structures of BaPif1 with ssDNA and with ss/dsDNA would have no obvious difference. Actually, however, the extremity of loop3 is structurally captured in the ssDNA-bound structure and the tracking strand in the ss/dsDNA-bound structure displays an unusual ∼90° bend just at the position where the ss/dsDNA junction should locate, while such a bend is not observed in the ssDNA-bound structure (31).
We have observed dimeric structures (crystal forms III−V, Figure 3A and Supplementary Figure S5) of ToPif1 in complex with 15-nt ssDNA and different ATP analogues, and revealed in detail the structural and mechanistic basis of how homodimerization of a SF1 helicase directs the conformational change of domain 2B. In a head-to-tail manner, the two ToPif1 molecules dimerize just through specific residue interactions, two salt bridges and two hydrogen bonds, between domain 2B of molecule M and the C-terminal and 1A domains of molecule M′ (Figure 3B), which make ToPif1 dimer be stable enough to be structurally captured. Compared with the ss/dsDNA-bound monomeric structure, domains 2B/loop3 of both molecules are sterically blocked in a more open conformation in these dimeric structures (Figure 1B and Supplementary Figure S9).
Although these ssDNA binding-induced dimeric structures are not always observed on a single contiguous ssDNA (for crystal forms III and IV), an additional crystal structure with dGR17 oligo and DLS analysis in conjunction with SEC confirmed that ToPif1 forms a dimer on a 15-nt ssDNA substrate or on the 15-nt 5′-overhang of an ss/dsDNA substrate in solution (Supplementary Figure S6). A dimeric structure is also clearly observed in our SAXS model with an ss/dsDNA substrate bearing a 15-nt 5′-overhang (Figure 3E and F and Supplementary Figure S7B). Dimerization in solution is dependent on ssDNA length since dimerization was not observed without ssDNA (apo) or with ssDNA as short as 7nt (S7D11) (Supplementary Table S3). Since the oligos used are poly dT, we can rule out the possibility that dimerization is induced by duplex formation between different oligos. The most probable hypothesis is that an ssDNA, if long enough, can allow simultaneous binding of two ToPif1 molecules. More importantly, stopped-flow unwinding and smFRET assays showed that the helicase activity of ToPif1 is inhibited at high enzyme concentrations with ss/dsDNA substrates bearing a ssDNA long enough to load more than one ToPif1 monomer, whereas single or double mutations of the interacting residues observed in the dimerization interface may partially or even totally eliminate the inhibition effect (Figure 4 and Supplementary Figure S11). All together, these results demonstrate that the dimeric crystal structures indeed revealed an intrinsic structural property of ToPif1: binding to a single ssDNA induces its dimerization with the 2B domain blocked in an open conformation and the dimerization inhibits its helicase activity.
The biologic relevance of the dimerization effect for ToPif1 in DNA transaction is currently unclear. It is noteworthy that several key residues implicated in ToPif1 dimerization are not conserved across Pif1 family in prokaryote (Supplementary Figure S1). Modeling of a BsPif1 dimer based on ToPif1 dimer clearly shows that the interface in BsPif1 is not compatible with a dimer formation: no steric complementarity exists and the residues facing each other have the same charge (Supplementary Figure S14). However, because of the high sequence variability of these regions, dimer formation, though not a general shared property, may be possible for certain members of the family. Indeed, it was shown that yeast Pif1 also dimerizes upon DNA binding (32). In addition, we have expressed and purified additional Pif1 family helicases from Deferribacter desulfuricans and Anaerobaculum hydrogeniformans, DdPif1 and AhPif1. The DNA unwinding kinetics assays showed that, while the extent of inhibition by higher protein concentrations for DdPif1 is similar to that observed with ToPif1, that for AhPif1 is only moderate (data not shown). In contrast, the monomeric Pif1-like T4 Dda helicase displays a cooperative mode of unwinding and streptavidin displacement when multiple molecules are bound to the same ssDNA substrate (64,65). Our structural and mechanistic studies presented here can be used as a new tool to investigate how dimerization affects/regulates DNA replication, recombination and repair.
The previous dimeric structure of BaPif1 in complex with a forked dsDNA (PDB code: 6L3G) has revealed how two BaPif1 molecules coordinate with each other to unwind a forked dsDNA. In this dimeric structure, the BaPif1 molecule bound to the 5′ arm and the other one bound to the 3′ ss/dsDNA junction dimerize through residue interactions between domain 2B of the former and domain 2A of the latter (35). Unlike ToPif1, the dimerization does not alter the domain orientations of the two BaPif1 molecules and each of which is an active helicase. However, the dimerization of BaPif1 also has inhibiting effect on helicase activity: the protein interactions of the two molecules at the DNA fork regulate their individual helicase activity by limiting their processivities (35). When the residue interactions in the dimerization interface are reduced by mutations, the BaPif1 dimer may separate and each molecule may be free to unwind DNA independently. Note that the two BaPif1 molecules bound to the junction regions of the fork are related by a rotation of 68° rather than 180° and thus, as mentioned by the authors, the structure cannot be considered as a conventional dimer (35).
It is worthwhile to make a comparison of ToPif1 and SF1A helicases (Rep/UvrD/PcrA). ToPif1 is a monomeric helicase and its domain 2B/loop3 repetitively switches between the open and closed conformations during DNA unwinding. Accordingly, constraining domain 2B/loop3 in the closed conformation by crosslinking inhibits the helicase activity of ToPif1; homodimerization, on the other hand, also inhibits the helicase activity because domain 2B/loop3 is sterically blocked in the open conformation. Rep, UvrD and PcrA are dimeric helicases and their domain 2B is autoinhibitory and plays a regulatory rather than a catalytic role. As a result, constraining domain 2B in a closed conformation by crosslinking activates the helicase activity of a Rep monomer, even converting it to a highly processive helicase (18); binding of a second monomer shifts domain 2B of the leading monomer, which is bound at the ss/dsDNA junction, to a closed conformation and an active UvrD dimer is then formed (19). Obviously, ToPif1 and SF1A helicases exhibit opposite behaviors in certain aspects.
In a previous structural study of Rep, a dimer was crystallized with the two molecules sitting adjacent to each other on a 16-base ssDNA (PDB code: 1UAA) (8). The difference between the two molecules, one in an open and the other in a closed conformation, comprised a large rigid body rotation of domain 2B by approximately 130°. Unfortunately, this ssDNA-bound Rep dimer was excluded as a functional form for several reasons (8); particularly, Rep is a 3′ to 5′ helicase, but in the dimeric structure, the molecule close to the 5′ ssDNA end (i.e., the leading molecule) is in the inactive open conformation while the other one is in the active closed conformation. Even so, we noticed the Rep and ToPif1 dimers share some similar structural features. The two molecules are arranged in a head-to-tail manner in these dimers; while domain 2B of molecule M interacts with domain 1A′ and the C-terminal of molecule M′ in the dimeric ToPif1 (Figure 3A), domain 2B of the Rep molecule in the closed conformation interacts with domains 1A and 1B of the other one in the open conformation.
Then, what is the potential physiological significance that the unwinding activity of ToPif1 is limited by dimerization? This may be relevant to one of the functions of Pif1 helicases that are involved in the Okazaki fragment processing. From the study of eukaryote Okazaki fragment maturation, it is well established that primer extension by DNA polymerase δ displaces the downstream fragment into a flap. Most short flaps are cleaved by flap endonuclease I (FEN1). The homolog of FEN1 in a prokaryote is the 5′ exonuclease of DNA polymerase I (66). Although the precise role of Pif1 in Okazaki fragments processing remains unknown, genetic evidence demonstrated that Pif1 lengthens the displaced flaps and Dna2 nuclease cleaves an RPA-bound flap (67). However, a homolog of Dna2 is not yet identified and probably is absent in prokaryote and a longer flap cannot be processed. It is reasonable to postulate that in the prokaryote organisms, the dimerization of Pif1 provides an inhibitory regulation mechanism that allows flaps to be processed into an appropriate length, otherwise a longer flap cannot be processed due to the fact that no homolog of Dna2 exists in prokaryote organisms. Therefore, it appears that regulation of the unwinding activity by dimerization is the best way to provide an appropriate length of flaps for their cleaving by the 5′ exonuclease of DNA polymerase I.
In summary, we have reported monomeric and dimeric crystal structures of apo and differently complexed ToPif1. Combined with kinetics, smFRET and mutational studies, we have revealed structural features and helicase properties of ToPif1 that are distinct from other helicases. Especially, ssDNA binding induces ToPif1 dimerization and which has an auto-inhibitory effect on its helicase activity. This work should be helpful for further studies of the functioning of ToPif1 and other Pif1 helicases and may shed new light on the structural basis for dimerization and regulation of different helicases.
DATA AVAILABILITY
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 6S3E, 7ADA, 6S3H, 6S3I, 6S3M, 6S3N, 6S3O, 6S3P and 7BIL.
ACKNOWLEDGEMENTS
We are grateful for access to the SOLEIL (SWING) synchrotron radiation facility for SAXS data collections. We thank the beamline scientists at BL17U1 of the Shanghai Synchrotron Radiation Facility (China) and at BL18U1 and BL19U1 of the National Center for Protein Sciences, Shanghai, for assistance with data collections.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31870788, 11574252, 11774407]; CNRS LIA (‘Helicase-mediated G-quadruplex DNA unwinding and genome stability'). Funding for open access charge: National Natural Science Foundation of China [31870788, 11574252, 11774407].
Conflict of interest statement. None declared.
REFERENCES
1.
2.
3.
4.
5.
6.
7.
8.
9.
10.
11.
12.
13.
14.
15.
16.
17.
18.
19.
20.
21.
22.
23.
24.
25.
26.
27.
28.
29.
30.
31.
32.
33.
34.
35.
36.
37.
38.
39.
40.
41.
42.
43.
44.
45.
46.
48.
49.
50.
51.
52.
53.
54.
55.
56.
57.
58.
59.
60.
61.
62.
63.
64.
65.
66.
 Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition
Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition
