
- Altmetric
Most mutations in cancer genomes occur in the non-coding regions with unknown impact on tumor development. Although the increase in the number of cancer whole-genome sequences has revealed numerous putative non-coding cancer drivers, their information is dispersed across multiple studies making it difficult to understand their roles in tumorigenesis of different cancer types. We have developed CNCDatabase, Cornell Non-coding Cancer driver Database (https://cncdatabase.med.cornell.edu/) that contains detailed information about predicted non-coding drivers at gene promoters, 5′ and 3′ UTRs (untranslated regions), enhancers, CTCF insulators and non-coding RNAs. CNCDatabase documents 1111 protein-coding genes and 90 non-coding RNAs with reported drivers in their non-coding regions from 32 cancer types by computational predictions of positive selection using whole-genome sequences; differential gene expression in samples with and without mutations; or another set of experimental validations including luciferase reporter assays and genome editing. The database can be easily modified and scaled as lists of non-coding drivers are revised in the community with larger whole-genome sequencing studies, CRISPR screens and further experimental validations. Overall, CNCDatabase provides a helpful resource for researchers to explore the pathological role of non-coding alterations in human cancers.
INTRODUCTION
Mutations in the cancer genome can be divided into drivers and passengers. Driver mutations are the ones that confer a selective advantage for the cancer cells to grow. Multiple databases of protein-coding drivers in cancer, such as COSMIC, Intogen, OncoKB and CIViC, have helped further follow-up investigations and enabled the utility of the driver catalog in numerous basic and translational research studies (1–4). Recent studies have shown that besides mutations in protein-coding regions, mutations in non-coding regions, such as promoters, enhancers, insulators and non-coding RNAs, can also act as cancer drivers (5–12). Although mutations at the TERT promoter are the most prominent example of non-coding drivers, evidence supporting the functional role of other non-coding mutations as cancer drivers is dispersed in several independent publications. Different computational approaches and experimental methods have used different signals to identify non-coding cancer drivers and it is hard to assess their consensus in the absence of a unified database. The lack of a database dedicated to non-coding cancer drivers hinders their further downstream computational analysis, functional characterization, and their utility for translational research. Here, we built CNCDatabase (Cornell Non-coding Cancer driver database), a manually curated database that contains detailed information about non-coding cancer drivers from published studies. The CNCDatabase contains 1201 genes with significant alterations in their non-coding regions in 31 cancer types from 25 published articles.
MATERIALS AND METHODS
Data model
The CNCDatabase has been designed as a relational database to store the collected non-coding cancer drivers from multiple sources. The detailed Entity-Relationship (ER) diagram and description of all tables are provided on the ‘Download’ page of the CNCDatabase website. The database schema follows a snowflake structure where the multidimensional data is connected to the centralized fact table. The design follows the database normalization rules for keeping the data integrity of multiple related entities, such as, non-coding driver evidence, functional element and gene associations, cancer type and reported study (Figure 1A and Supplementary Figure S1). The data structure employed in the CNCDatabase allows it to be extended to accommodate new data types without significant changes in the existing model. As a result, the database is highly scalable, an essential feature of a data integration project.

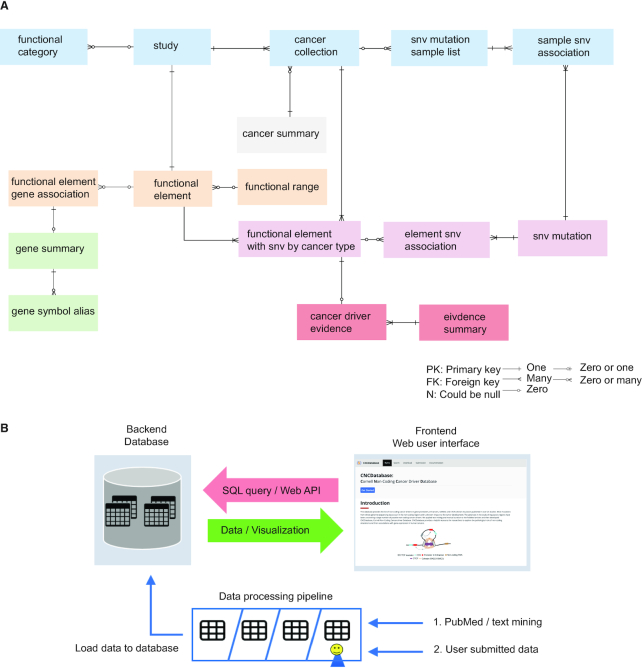
Data model and architecture of CNCDatabase. (A) Simplified database entity-relationship diagram (ERD). (B) The schematic data flow in the CNCDatabase between web interface in the frontend and PostgreSQL database in the backend. Manually curated cancer driver lists from PubMed or from users can be loaded into the database.
Data collection and processing
We collected the data from studies related to non-coding cancer drivers in PubMed by text mining within the title or abstract of the articles for the existence of combinations of keywords such as noncoding, driver, cancer, and their alternative terms, for example: noncoding[Title/Abstract]) OR non-coding[Title/Abstract]) AND driver[Title/Abstract]). After a manual review of the returned abstracts from the PubMed search, we extracted the non-coding driver evidence from the text and supplementary files of the 25 selected articles. We focused on the publications reporting non-coding alterations at the promoters, 5′ UTRs, 3′ UTRs, enhancers, splice sites, non-coding RNAs, and CTCF-cohesin insulators.
Because the CNCDatabase aims to catalog the comprehensive list of non-coding cancer drivers, we include the ones with at least one type of evidence: computational prediction, differential gene expression association from RNA-seq, and other experimental validation. The evidence term ‘computational prediction’ means the non-coding regions exhibit statistically significant signals of positive selection from whole-genome sequencing data. The term ‘differential gene expression association from RNA-seq’ means the mutations in the non-coding region are associated with differential gene expression between wild type and mutated samples from RNA-seq data. Finally, ‘other experimental validation’ means the mutations in the non-coding region have been validated for a molecular or cancer-related phenotype by either luciferase assay, CRISPR–Cas9 or some other experimental assays.
Architecture of CNCDatabase
CNCDatabase consists of a relational database server using PostgreSQL (version 9.6.6). It provides an application program interface (API) to access all stored data. The backend server is complemented with a frontend web-based user interface (UI) (Figure 1B). We use Node.js (version 10.15.3) and Express.js framework (version 4.16.4) to build the backend server. We use React.js (version 16.8.5) and Bootstrap4 (version 4.0.0) as the frontend web development framework for a responsive UI, which means the website is suitable for both desktop and mobile data viewing. The chart visualizations use the plotly.js (version 1.46.1) package.
The CNCDatabase is freely available (https://cncdatabase.med.cornell.edu/) and the content of the database is available for download. We provide the code at GitHub (https://github.com/khuranalab/CNCDatabase) for users to make use of all services locally.
DATABASE FEATURES AND USE
Summary of database content
A total of 1673 entries from 32 cancer types in CNCDatabase correspond to 90 non-coding RNAs and non-coding regions of 1111 protein-coding genes that have been identified as cancer drivers from computational predictions, 18 genes for which non-coding mutations are associated with differential gene expression from RNA-seq, and 21 genes with non-coding cancer driver evidence from other experimental validation (Figure 2). Out of the 1201 genes with non-coding driver evidence from computational predictions, 355 were identified from individual cancer type analysis and 684 from pan-cancer analysis only where samples from multiple cancer types are pooled together for statistical power. The number of genes for individual cancer types varies from 1 in rhabdoid tumor to 270 in melanoma (Figure 2A). The publication from Weinhold et al. contributes the largest number of non-coding cancer driver candidates (453 genes) from computational predictions (Supplementary Figure S2) (13).

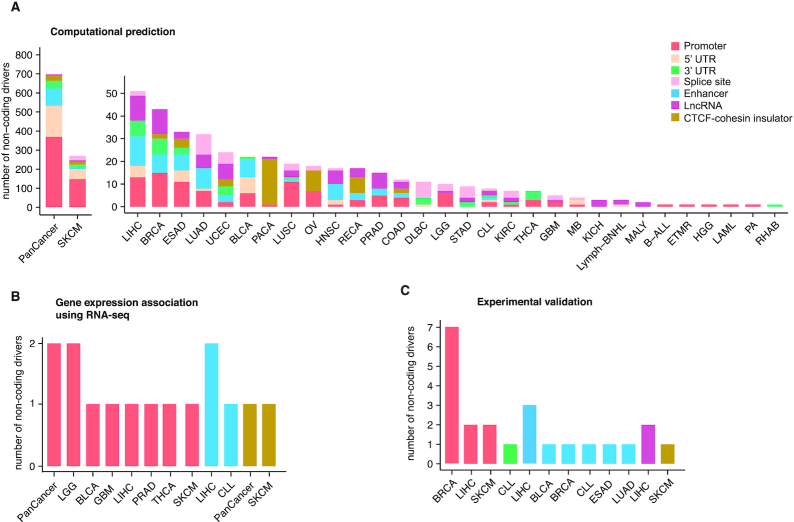
Summary of number of non-coding drivers. (A) Number of non-coding drivers in each cancer type by computational prediction. Cancer types include pan-cancer (PanCancer), skin cutaneous melanoma (SKCM), liver hepatocellular carcinoma (LIHC), breast invasive carcinoma (BRCA), esophageal carcinoma (ESAD), lung adenocarcinoma (LUAD), uterine corpus endometrial carcinoma (UCEC), bladder urothelial carcinoma (BLCA), pancreatic ductal adenocarcinoma (PACA), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), head and neck squamous cell carcinoma (HNSC), kidney renal papillary cell carcinoma (RECA), prostate adenocarcinoma (PRAD), colon adenocarcinoma (COAD), lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), low grade glioma (LGG), stomach adenocarcinoma (STAD), chronic lymphoctytic leukemia (CLL), kidney renal clear cell carcinoma (KIRC), papillary thyroid carcinoma (THCA), glioblastoma multiforme (GBM), medulloblastoma (MB), kidney chromophobe (KICH), b-cell non-hodgkin lymphoma (Lymph-BNHL), malignant lymphoma (MALY), B-cell acute lymphoblastic leukemia (B-ALL), embryonal tumor with multilayered rosettes (ETMR), high-grade glioma (HGG), acute myeloid leukemia (LAML), pilocytic astrocytoma (PA) and rhabdoid tumor (RHAB). (B) Number of non-coding drivers in each cancer type that show differential gene expression in samples with mutations vs. without from RNA-seq data. (C) Number of non-coding drivers in each cancer type with support from other functional validation, such as CRISPR/Cas9 or luciferase reporter assay.
Web user interface
CNCDatabase provides an intuitive web interface that facilitates browsing and searching through four main sections including ‘Home’, ‘Search’, ‘Download’, ‘Submission’ and ‘Documentation’ (Figure 3). The landing page (‘Home’) provides abstract graphics of the available information. From there, a simple button (‘Get started’) immediately allows the launch of the user's custom query. All data in the CNCDatabase can be downloaded from the ‘Download’ section as text format files or database contents for further downstream analysis.

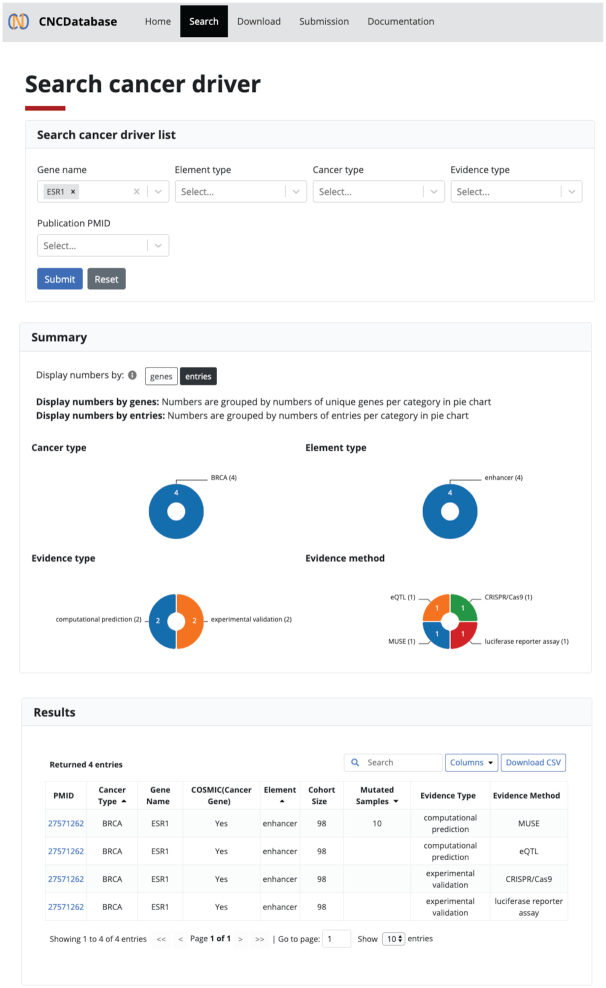
Web user interface and supported functionality in the CNCDatabase. User can use the combination of gene name, element type, cancer type, evidence type or publication id (PMID) to query the non-coding cancer driver list from the backend database. The returned result shows graphical summary and list in tabular format.
Searching for non-coding cancer drivers
In the ‘Search’ section, users can apply the fuzzy query to retrieve the non-coding driver entries. The query fields also support the auto-complete function so that users can quickly pick a valid gene name or cancer type. The database can be searched using multiple query types, including gene name, element type (e.g., promoter or enhancer), cancer type, evidence type, and publication PMID. If users do not select a specific cancer type, the system will return results from all cancer types by default. After clicking the ‘Submit’ button, the query results are displayed in a report organized into several components. The ‘Summary’ section provides pie chart representations to display the numbers in each category, including cancer type, element type, evidence type and evidence method. The ‘Results’ section shows the retrieved entries in a tabular format including publication id (PMID), cancer type, gene name, whether the gene is annotated as a cancer gene based on Cancer Gene Census (CGC) from COSMIC (catalog of somatic mutations in cancer), non-coding element type, element and element-to-gene association description, cohort size, fraction of mutated samples, evidence type, evidence method and evidence description. Users can also further refine the search results by entering the targeted value in the search field of the returned result table (Figure 3). The search results can also be downloaded in the CSV format.
Data submission and curation
Through the ‘Submission’ page of the web interface, users can submit new non-coding cancer drivers to the CNCDatabase. A valid data submission contains a text file with columns for the publication id (PMID), cancer type, gene name, non-coding element type, element-to-gene association description, cohort size, fraction of mutated samples, evidence type, evidence method and evidence description. Following file upload, users will receive email notifications to track data submission progress. The curators of CNCDatabase will manually check the submitted files to ensure the data is consistent with the database annotation format. When submitted data passes the curator check, the CNCDatabase data pipeline (https://github.com/khuranalab/CNCDatabase/tree/master/data_pipeline) will be used to split and store submitted data into the PostgreSQL database and the new data will be included in the next release of CNCDatabase. Thus, the CNCDatabase can serve as a central hub of information about non-coding cancer drivers for the cancer research community regardless of users’ bioinformatics expertise level.
Overview of data in CNCDatabase
One of the many uses of CNCDatabase is that it will help researchers prioritize the non-coding candidates for functional validation follow-up and look up which non-coding mutations have already had functional validation evidence (Supplementary Tables S1 and S2). Analysis of data in CNCDatabase reveals that the promoters of TERT, WDR74, PLEKHS1 and CCDC107 have support as non-coding drivers by computational predictions from more than four publications (Figure 4A). TERT, WDR74 and PLEKHS1 promoter mutations also have evidence supporting their role as non-coding drivers from RNA-seq data or other experimental assays. It will be interesting to interrogate the function of CCDC107 promoter mutations in breast, lung and rectal cancers in future studies (Figure 4B). In the 3′UTR regions, only NOTCH1 in CLL has functional assay evidence (Figure 5A). API5, DRD5, FAM230A and PCMTD1 could be good candidates for follow-up functional validations at 3′UTR regions. While many studies have identified candidate drivers at enhancers, TP53TG1 is the only gene that has support from multiple publications (Figure 5B). For lncRNAs, MALAT1 and NEAT1 are the genes with support both from computational predictions and from functional assays (Figure 5C). Although mutations at the 5′UTRs and splice sites do not have any support as cancer drivers from functional assays in any published study yet, there are multiple genes (WDR74, C16orf59, MED31, MTG2, PTDSS1, TBC1D12 and UMPS) with support from computational predictions from multiple publications (Figure 5D). The splice site mutations of TP53 and STK11 are the most promising candidates to conduct follow-up validations (Figure 5E).

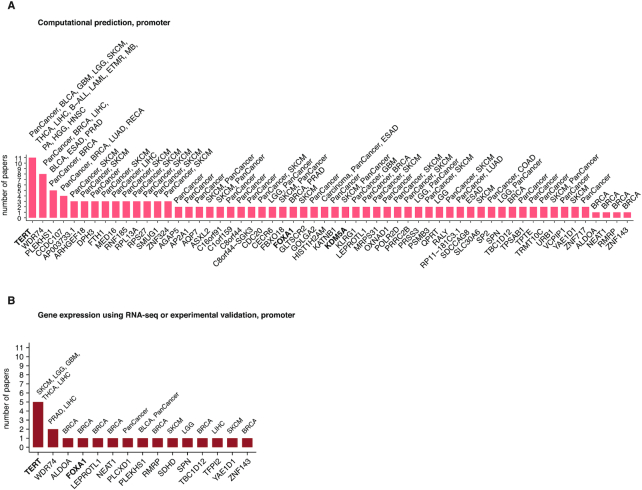
Non-coding cancer driver candidates at promoter regions. (A) based on computational predictions (B) based on association of mutations with differential gene expression or other experimental validation. In the results from computational predictions, for cancer driver candidates reported in only one publication, we only show candidates with support from experimental validation or those associated with cancer genes in COSMIC census list. The cancer genes are highlighted in bold.

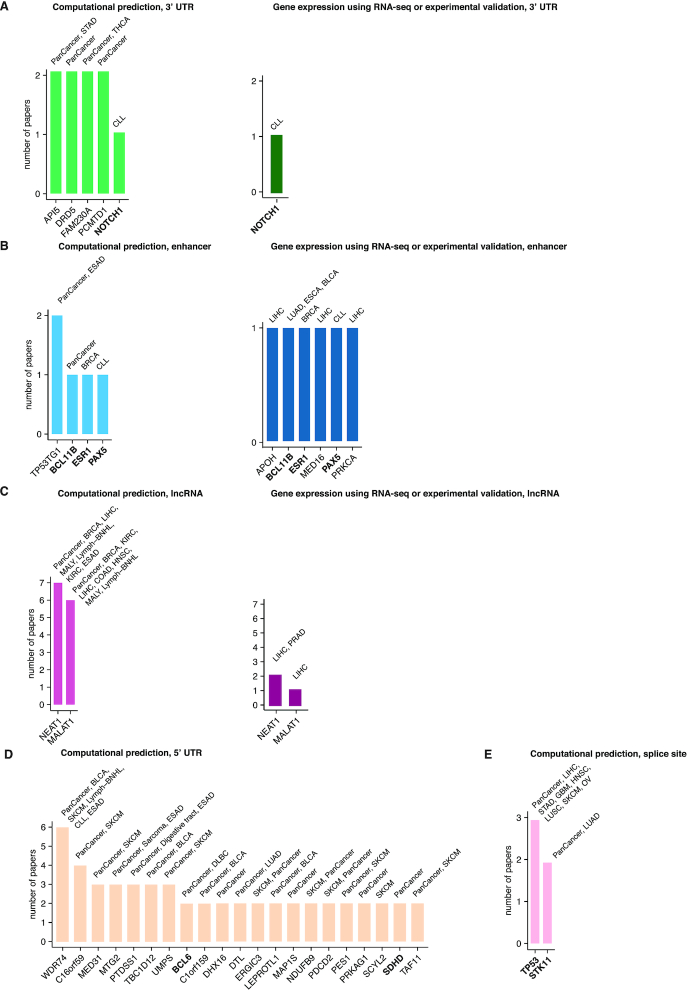
Non-coding cancer driver candidates from computational predictions and candidates with functional validations. (A) 3′ UTR, (B) enhancer, (C) lncRNA, (D) 5′UTR and (E) splice site. In the results from computational predictions, for cancer driver candidates reported in only one publication, we only show candidates with support from experimental validation or those associated with cancer genes in COSMIC census list. The cancer genes are highlighted in bold.
DISCUSSION AND FUTURE PERSPECTIVES
We report the CNCDatabase that integrates the functional evidence reported for non-coding cancer drivers in many independent publications. To create this comprehensive catalog, we used combinations of keywords to select relevant articles from PubMed and manually extracted the evidence from each article. At the time of writing this publication, most comprehensive studies of non-coding drivers have focused on single nucleotide variants and small insertions and deletions in cancer genomes. In the future, as more comprehensive studies focusing on large structural variants at non-coding regions that act as drivers are reported, we will incorporate them in the database (11). CNCDatabase contains 1300 non-coding cancer drivers with support from either computational prediction of positive selection, mutational association with gene expression or other experimental validation. It aims to advance the understanding of non-coding alterations in cancer for both basic and translational scientists and users with all levels of bioinformatics skills. Interactive queries can be used to browse the evidence supporting non-coding cancer drivers and search results can be exported for further custom analysis.
To regularly update CNCDatabase with the studies of non-coding cancer drivers, we will use a combination of automated text mining (Kindred relation classifier) and manual curation (14,15). The first version of the database has allowed us to annotate a set of sentences/words associated with the reports of noncoding drivers (Supplementary Table S3), which will be improved in every subsequent version. The automated text-mining tools can extract the non-coding cancer driver evidence from title, abstracts and full-text articles from PubMed and PubMed Central (PMC). With the advances in CRISPR screening technology, we expect more functionally validated non-coding cancer drivers will be reported in the future. In fact, CNCDatabase can help scientists pick the relevant lists of non-coding alterations for CRISPR validations whose results can be then added to the database to augment the functional evidence supporting or rejecting those drivers. In conclusion, CNCDatabase will serve as a valuable resource for the cancer community to complement the studies of oncogenic mechanisms that are currently mostly centered on protein-coding mutations.
ACKNOWLEDGEMENTS
We thank Douglas Duckworth from Weill Cornell Supercomputing Unit for help in the website deployment. We also thank Ann Palladino, Sandra Cohen, Tawny Cuykendall, Erica Duo Xu and Andre Forbes in the Khurana lab for valuable suggestions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01CA218668 to E.K.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
1.
2.
3.
4.
5.
6.
7.
8.
9.
10.
11.
12.
13.
14.
15.
 CNCDatabase: a database of non-coding cancer drivers
CNCDatabase: a database of non-coding cancer drivers
