
The authors wish it to be known that, in their opinion, the first five authors should be regarded as Joint First Authors.
- Altmetric
Maintenance of stem-cell identity requires proper regulation of enhancer activity. Both transcription factors OCT4/SOX2/NANOG and histone methyltransferase complexes MLL/SET1 were shown to regulate enhancer activity, but how they are regulated in embryonic stem cells (ESCs) remains further studies. Here, we report a transcription factor BACH1, which directly interacts with OCT4/SOX2/NANOG (OSN) and MLL/SET1 methyltransferase complexes and maintains pluripotency in mouse ESCs (mESCs). BTB domain and bZIP domain of BACH1 are required for these interactions and pluripotency maintenance. Loss of BACH1 reduced the interaction between NANOG and MLL1/SET1 complexes, and decreased their occupancy on chromatin, and further decreased H3 lysine 4 trimethylation (H3K4me3) level on gene promoters and (super-) enhancers, leading to decreased enhancer activity and transcription activity, especially on stemness-related genes. Moreover, BACH1 recruited NANOG through chromatin looping and regulated remote NANOG binding, fine-tuning enhancer–promoter activity and gene expression. Collectively, these observations suggest that BACH1 maintains pluripotency in ESCs by recruiting NANOG and MLL/SET1 complexes to chromatin and maintaining the trimethylated state of H3K4 and enhancer–promoter activity, especially on stemness-related genes.
INTRODUCTION
The pluripotency of embryonic stem cells (ESCs) is maintained by a network of transcription factors, including essential regulators such as organic cation/carnitine transporter 4 (OCT4), sex-determining region Y box 2 (SOX2) and NANOG homeobox (NANOG), as well as estrogen-related receptor beta (ESRRB), Kruppel-like factors 4 (KLF4), zinc finger protein 42 (ZFP42) and other ancillary factors (1–3). NANOG and other key transcription factors can form chromatin loops, bridging enhancers and promoters together, and activate target genes, especially on large tandem enhancer regions (called super-enhancers). Several enhancers and promoters are looped together and bound by transcription factors including NANOG, SOX2, OCT4 and mediator complexes. These super-enhancers have high transcriptional activity and regulate essential pluripotent genes (4,5). However, it is still under exploration how the super-enhancer activity is regulated and how it fine-tunes pluripotency.
Histone modifications at the promoters and enhancers of genes correlate with activation or repression of transcription. The promoters of actively transcribed genes typically contain high levels of histone H3 lysine 4 tri-methylation (H3K4me3) and histone H3 lysine 27 acetylation (H3K27ac), whereas the active enhancers contain histone H3 lysine 4 mono-methylation (H3K4me1) and H3K27ac (6). The establishment and conversion among H3K4 mono-, di- and tri-methylation are catalyzed by protein complexes of the SET domain containing 1 (SETD1A/B) and Mixed Lineage Leukemia (MLL1–4) families (7). These complexes are composed of a variety of subunits, including WD repeat domain 5 (WDR5), ASH2L, DPY-30 and RB binding protein 5 (RBBP5) (8). SETD1A is important for early mouse embryonic development, as well as the self-renewal and differentiation of ESCs (9,10), and WDR5 interacts with OCT4 and binds to the OCT4 promoter to regulate ESC pluripotency (11). A recent report showed that ASH2L recruited OCT4/SOX2/NANOG (OSN) to the super-enhancers and formed ASH2L/OSN complex to regulate pluripotency of mESCs (12). However, the molecules mediating the recruitment and bridging of MLL/SET1 complexes and OSN, and the mechanisms for the enrichment of OSN on super-enhancers remain to be identified.
The transcription factor BTB and CNC homology 1 (BACH1) is widely expressed in most mammalian tissues and has a crucial role in oxidative-stress response (13,14) and cell cycle, as well as mechanisms involved in heme homeostasis, reactive oxygen species production, angiogenesis, and cancer metastasis (15–18). We have shown that BACH1 interacts with NANOG, SOX2, and OCT4 in ESCs and recruits ubiquitin specific peptidase 7 (USP7) to stabilize NANOG (19); thus, the loss of BACH1 expression in ESCs reduces the cells’ pluripotency and initiates differentiation into the mesendodermal lineage. However, the role of BACH1 as a transcription factor in ESCs is still unclear. BACH1 also participates in the recruitment of chromatin-modifying enzymes such as histone deacetylase 1 (HDAC1) in endothelial cells (16) and the nonhistone chromatin modifier High-mobility Group AT-hook2 (HMGA2) in epithelial ovarian cancer cells (20), but whether BACH1 plays a role in the chromatin modifications that maintain ESC pluripotency is largely unknown. In addition, BACH1 was found to mediate chromatin looping in vitro by forming heterodimers with small Maf proteins, linking cis-regulatory elements together (21), so it is also intriguing whether BACH1 regulates the recruitment of transcription factors within chromatin loops, and further regulates enhancer–promoter activity in an ESC model.
Here, we explore the mechanisms underlying the BACH1-mediated regulation of pluripotency in mouse ESCs (mESCs). Our results suggest that BACH1 directly interacts and bridges NANOG and MLL/SET1 complexes, recruits them to chromatin, and maintains the level of histone H3 lysine 4 trimethylation (H3K4me3) on gene promoters and (super-) enhancers, especially on stemness-regulating genes. Moreover, BACH1 recruits NANOG through chromatin looping and regulates remote NANOG binding, fine-tuning enhancer–promoter activity and gene expression. Thus, BACH1 appears to be an enhancer–promoter regulator, which is important for the maintenance of stem-cell pluripotency.
MATERIALS AND METHODS
Cell culture
mESCs (E14Tg2A line) were kindly provided by Dr Fei Lan. mESCs were maintained in 90% Dulbecco's modified Eagle's medium (DMEM) (Life Technologies, Carlsbad), 10% fetal bovine serum (FBS) (Life Technologies, Carlsbad, CA, USA), 1% non-essential amino acid (NEAA), 1% penicillin and streptomycin (100× stock) (Gibco-Thermo Fisher, Waltham, MA, USA), 0.1 mM β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO, USA), and 200 Units/ml murine leukemia inhibitory factor (LIF) (ESGRO; Merck-Millipore, Germany). mESC expansion was performed in gelatin-coated flasks (BBI Life Sciences, China). hESCs of H7 (from the WiCell Research Institute) were cultured on hESC-qualified Matrigel (BD Biosciences, San Diego, CA, USA) supplemented with mTeSR1 (Stemcell Technologies, Vancouver). Human embryonic kidney (HEK) 293T cells were originally from the American Type Culture Collection and were cultured in DMEM with 10% FBS.
Construction of vectors
Mouse Bach1 gene was generated and cloned into pcDNA3.1. Briefly, the full-length cDNA of Mouse Bach1 or DNA binding-defective cDNA of mouse Bach1 [Bach1-ΔbZIP (1–553 aa)] was generated by PCR and verified by DNA sequencing. HA-epitope tags were introduced to the C termini of the Bach1 gene. Glutathione S-transferase (GST)-BACH1, GST-BACH1-ΔbZIP or GST-BACH1-ΔBTB fusion protein was constructed by inserting PCR-generated DNA fragments encoding regions of full-length BACH1 (1–739 aa) or BACH1-ΔbZIP (1–553 aa) or BACH1-ΔBTB (128–739 aa) into PGEX-6P-1 vector. HIS-tagged NANOG was constructed by inserting PCR-generated DNA fragments encoding Nanog into PET-28A vector. Primers for cloning mouse Bach1 or Nanog cDNA are listed in Supplemental Table S2. All constructs were verified by sequencing.
Generation of Bach1-KO and DoxBach1 mESCs
Bach1-KO mESCs were generated by using CRISPR/Cas9 genome editing technology. sgRNAs (Supplemental Table S2) targeting the genomic regions of interest were designed with the CRISPR Design Tool (http://crispr.mit.edu/) and synthesized by Shanghai Sunny Biotechnology Co. Annealed sgRNA oligos were cloned into LentiCRISPR V2 (Addgene, Watertown, MA, USA), transiently transfected into mESCs with Lipofectamine 2000 (Thermo Fisher, Waltham, MA), and then selected with puromycin (Selleck Chemicals, Houston, TX, USA); single-cell clones were sorted 7 days later. Bach1-FLAG cDNA was cloned into the vector (pPB-TRE-EGFP-PGK-Neo). DoxBach1 mESCs were generated by using the piggyBac (PB) transposon system with the pCAG-T7-mPB, pCAG-rtTA-Puro and pPB-TRE-BACH1-PGK-Neo vectors (1:2:2 ratio) and puromycin selection (Selleck Chemicals, Houston, TX, USA). BACH1 expression was induced by treating DoxBach1 mESCs with 0.5 μg/μl doxycycline (Selleck Chemicals, Houston, TX, USA).
Viral transduction of mESCs
Lentiviral particles of Bach1 were generated by transfecting HEK293T cells with LvEP05- Bach1-FLAG vector and packing vectors psPAX2 and pMD2.G. mESCs were transduced with the concentrated lentivirus for 48 h.
RNA interference
mESCs were transfected with Bach1 siRNAs or control siRNAs by Lipofectamine 2000. The transfection was performed according to the instructions. siRNA sequences are listed in Supplementary Table S2.
Mouse embryonic fibroblast (MEF) reprogramming
Mouse embryonic fibroblast (MEF) reprogramming was performed as described previously (22). MEFs were isolated from 13.5 d.p.c. mouse embryos and cultured in DMEM containing 10% FBS. Lentiviral plasmids for the reverse tetracycline transactivator (FUW-M2rtTA, Addgene, Watertown, MA, USA) and the tetracycline-inducible expression of mouse OCT4, SOX2, Klf4 and Myc (TetO-FUW-OSKM, Addgene, Watertown, MA, USA) were packaged in HEK293T cells with psPAX2 and PMD2.G, respectively. MEFs (1 × 104 cells/well) were plated into six-well plates and cultured for 24 h; then, concentrated lentivirus solutions containing mouse Yamanaka factors and Bach1 were added, and the cells were cultured for another 24 h before addition of the reprogramming medium (Knockout DMEM, 10% Knockout Serum Replacement [Thermo Fisher, Waltham, MA, USA], 1% NEAA, 1% PS, 0.01% CHIR98014, 0.01% PD0325901 and 1 × 103 units/ml mouse LIF).
Alkaline phosphatase staining and colony assay
The alkaline phosphatase assay was conducted with an Alkaline Phosphatase Detection Kit (Sigma-Aldrich, St. Louis, MO, USA) as directed by the manufacturer's instructions. Cells (1 × 104 cells/well) were cultured in 12-well plates for 2–3 days, fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 1–2 min, and then rinsed with TBST solution (20 mM Tris–HCl, pH 7.4, 0.15 M NaCl, 0.05% Tween 20). FRV-Alkaline solution (30 μl) and sodium nitrite solution (30 μl) were mixed for 2 min at room temperature and then added with naphthol AS-BI Alkaline solution (30 μl) into water (1350 μl); then, the mixture (1 ml/well) was added to the cells, and the cells were incubated in the dark at room temperature for 15 min, rinsed with TBST and covered with PBS. Colonies were viewed under a microscope. Colonies containing undifferentiated (i.e. alkaline phosphatase-positive, red) cells, differentiated (colorless) cells and mixed populations of cells were counted.
Cell proliferation assay
Cells (1.3 × 104 cells/well) were seeded into gelatin-coated 12-well plates, cultured for the indicated periods, dissociated with 0.05% trypsin (Gibco-Thermo Fisher, Waltham, MA, USA), and then counted with a cell-counting chamber.
Embryoid body (EB) formation assay
mESCs were trypsinized and suspended in LIF-free EB medium at 1 × 105 cells per well and plated onto six-well low-attachment plates. The EB culture medium contains DMEM supplemented with 10% FBS, 1 mM l-glutamine, 1% NEAA, 0.1 mM β-mercaptoethanol. The culture medium was changed every 2 days. EBs were cultured with EB medium until the indicated time points.
Quantitative real-time PCR and RNA- sequencing
Quantitative real-time PCR and RNA- sequencing were performed as described previously (19). Total RNA was isolated using Trizol reagent, and complementary DNA (cDNA) was synthesized with the ReverTra Ace qPCR RT Kit (TOYOBO, Osaka, Japan, #FSQ-101). Real-time quantitative PCR was performed with the qPCR SYBR® Green Master Mix (Yeason, Shanghai, China), and the primer sequences are listed in Supplementary Table S2. The expression of each gene was analyzed by the Ct method and was normalized to the expression of Actb. Genome-wide gene expression analyses were performed using RNA-sequencing (RNA-seq). Non-ribosomal RNA was isolated from 1 μg total RNA by using a TrueLib Poly (A) mRNA Magnetic Isolation Module (Excell Bio, Shanghai, China), and sequencing libraries were prepared with the TrueLib mRNA Library Prep Kit for Illumina (ExCell Bio, Shanghai, China). Sequencing was performed on the NovaSeq 6000 and Hiseq XTen sequencer (Illumina Inc, San Diego, CA, USA) by Annoroad Company (Beijing, China) and Basepair Company (Suzhou, China).
Analysis of RNA-sequencing data
The FASTQ data were trimmed adaptor by trim_galore (bioinformatics.babraham.ac.uk), then mapped to mouse genome mm9 using HISAT2 (23), and assembled by HOMER(24) to get Transcripts per million (TPM) for each gene. The TPM value of genes was used to eliminate the statistical biases inherent in the RPKM measure (25). Differentially expressed genes were generated by limma (26) with fold change >2.0 and P <0.05. Gene Ontology analysis (27), KEGG Pathway analysis (28), and volcano plot were performed on www.omicsbean.cn.
Immunoprecipitation (IP) and western blotting
Cells were lysed in lysis buffer. For immunoprecipitation, cell lysates were incubated with the indicated antibodies and protein A/G plus agarose at 4°C overnight. After washing four times with PBS, protein SDS loading buffer was added and boiled for 5 min. Proteins were resolved on an SDS-PAGE gel, transferred onto a polyvinylidene fluoride (PVDF) membrane (Merck-Millipore, Germany), labeled with their respective antibodies. Blots were incubated with secondary antibodies and developed with ECL (Thermo Fisher Scientific, Waltham, MA, USA), and signals were detected via chemiluminescence on a Tanon-5500 Imaging System (Tanon Science & Technology Ltd, Shanghai, China). The intensity of the bands was measured via densitometric analysis with ImageJ software.
Immunofluorescence staining
Cells were fixed, permeabilized, incubated, stained and photographed as described previously (16). Briefly, cells were incubated with primary antibodies in blocking solution overnight and then with Cy3-conjugated donkey anti-rabbit/mouse IgG or FITC-conjugated donkey anti-goat secondary antibodies for 1 h (Jackson ImmunoResearch Laboratories, West Grove, PA, USA). Nuclei were counter-stained with DAPI, and then the cells were viewed under a fluorescence microscope (Olympus, Tokyo, Japan).
In vitro pull-down assay
The GST-tagged BACH1 or BACH1-ΔbZIP or BACH1-ΔBTB and HIS-tagged NANOG recombinant proteins or SETD1A complex recombinant proteins were expressed and purified with glutathione-Sepharose 4B beads. Purified GST-tagged recombinant proteins BACH1 or BACH1-ΔbZIP or BACH1-ΔBTB and purified HIS-tagged NANOG or SETD1A complex recombinant proteins were then incubated together for 2 h at 4°C, followed by three times of wash. Conjugated proteins were then eluted by SDS loading buffer, and examined by western blot analysis.
Chromatin immunoprecipitation (ChIP) and ChIP-seq
Chromatin immunoprecipitation (ChIP) and ChIP-seq were performed as described previously (29). Cells were cross-linked with 1% formaldehyde for 10 min, quenched with 0.125 M glycine for 10 min; then, the cell pellets were resuspended in high salt lysis buffer (50 mM HEPES/KOH, pH 7.5, 500 mM NaCl, 1% Trixon-X 100, 0.05% SDS, 10 mM EDTA and proteinase inhibitors), and sonicated with a Bioruptor (Diagenode) for 20 min in 30-s intervals to shear the chromatin into 200- to 400-bp lengths. Chromatin was immunoprecipitated with the indicated antibodies and Dynabeads protein A and protein G (Thermo Fisher, Waltham, MA, USA). For, BACH1-FLAG ChIP, DoxBach1 mESCs were treated with 0.5 μg/μl doxycycline (Selleck Chemicals, Houston, TX, USA) for 6 h. Then cells were cross-linked, quenched, and washed as described above. Cell nuclei were extracted before sonication. Furthermore, the sonicated chromatin was incubated with anti-FLAG affinity M2 agarose gel (Sigma-Aldrich, St. Louis, MO, USA) overnight at 4°C. Then beads were washed with high salt lysis buffer and low salt buffer (10 mM Tris–HCl pH 8.0, 250 mM LiCl, 1 mM EDTA, 0.5% NP40, 0.5% Na-Deoxycholate), and DNA was eluted in elution buffer (50 mM Tris–HCl pH 8.0, 10 mM EDTA, 1% SDS) with Proteinase K (Invitrogen-Thermo Fisher, Waltham, MA, USA) and RNase A (Thermo Fisher, Waltham, MA, USA) in 65°C for 4 h. Chromatin occupancy was quantified using qPCR and normalized to input DNA. The primers used in ChIP-qPCR assays are listed in Supplemental Table S2. For ChIP-seq experiments, ChIP DNA libraries and input DNA samples were prepared as directed by the protocol of KAPA Hyper Prep Kit for Illumina (Kapa Biosystems, Wilmington, MA, USA) and VAHTS Universal DNA Library Prep Kit for Illumina (Vazyme Biotech, Nanjing, China). Libraries were sequenced on Illumina Hiseq X Ten by Basepair Company (Suzhou, China).
Analysis of ChIP-sequencing data
The FASTQ data were trimmed adaptor by trim_galore (bioinformatics.babraham.ac.uk), and then they were mapped to mouse genome mm9 with Bowtie2 (30) in pair-end mode. Peaks were called by MACS1.4 (31) and MACS2 (32). Differential peaks were called by MACS2 -bdgdiff. ChIP-seq motif and ChIP-seq visualization were performed by HOMER tools (24) and visualized on UCSC genome browser. Heatmaps and signal plots were generated by deepTools (33) (3.3.0).
Chromatin loops and super-enhancers
Hi-C sequencing in mESCs was performed by Bonev et al. (34). Chromatin loops were generated from the 3D epigenome browser (35), and the loops were predicted by Peakachu (36). Chromatin loops combined with ChIP-seq data were visualized in WashU epigenome browser. Super-enhancers were defined by Benjamin Sabari, Alessandra Dall’Agnese et al. (37), in which ChIP-seq peaks of MED1 were used as input for ROSE to call super-enhancers.
Statistical analysis
Data are expressed as mean±SEM. Differences between two groups were evaluated for significance via two-tailed Student's t-test, and differences among three or more groups were evaluated for significance via one-way analysis of variance (ANOVA) followed by Tukey's multiple comparison test. Analyses were conducted with Prism software, and P <0.05 was considered significant.
RESULTS
BACH1 is required for the maintenance of mESC pluripotency
BACH1 colocalized with OCT4 in the inner cell mass of mouse blastocysts (Supplemental Figure S1A), and both BACH1 and OCT4 expression declined at day 6 and day 8 during embryoid body (EB) differentiation (Supplemental Figure S1B). Thus, we began to investigate whether BACH1 contributed to the maintenance of mESC pluripotency by generating Bach1-knockout (Bach1-KO) mESCs and doxycycline-inducible BACH1 overexpressing mESCs (DoxBach1 mESCs) (Supplemental Figure S2). Bach1 knockout induced dramatic morphological changes, as well as declines in alkaline phosphatase (AP) activity (Supplemental Figure S1C) and proliferation (Supplemental Figure S1D) in mESCs, and a greater proportion of Bach1-KO than WT mESC colonies were composed of primarily differentiated or mixed populations of cells (Supplemental Figure S1E). The expression level of pluripotency genes (SOX2, OCT4, NANOG, KLF4, stage-specific embryonic antigen 1 [SSEA1]) was also lower in Bach1-KO mESCs than in WT mESCs (Supplemental Figure S1F–G, Supplemental Figure S3A). When spontaneous differentiation was induced in mESCs via the withdrawal of leukemia inhibitory factor (LIF) for three days, Bach1-KO cells showed significantly higher levels of many differentiation genes, such as mesodermal (Brachyury [T], mesoderm posterior 1 [MESP1], MIXL1) and endodermal (GATA4, GATA6, SOX17) markers, but modestly lower expression levels of neuroectodermal-marker (SOX1 and NESTIN) (Supplemental Figure S3B). Furthermore, the effect of the Bach1-KO on pluripotency gene expression in mESCs could also be observed by transfecting the cells with Bach1 siRNA (Supplemental Figure S3C); whereas dox-induced BACH1 overexpression increased measures of AP activity (Supplemental Figure S3D) and the pluripotency gene expression (Supplemental Figure S3E); lentiviral-induced BACH1 expression largely reversed the effect of Bach1 knockout on mESC morphology (Supplemental Figure S1C) and pluripotency gene expression (Supplemental Figure S1G), and mesendodermal/neuroectodermal gene expression during spontaneous mESC differentiation (Supplemental Figure S3B).
RNA sequencing (RNA-seq) analysis indicated that 1883 genes were upregulated, and 2523 genes were downregulated in Bach1-KO mESCs than in WT mESCs (fold change > 2, P < 0.05) (Supplemental Figure S4A, Supplemental Table S1). Many of the downregulated genes were associated with signaling pathways regulating pluripotency (Supplemental Figure S4B). RT-qPCR assays further confirmed the decreased level of pluripotency genes (Supplemental Figure S1G). The upregulated genes were enriched for PI3K-AKT signaling and mitogen-activated protein kinase (MAPK) signaling pathways (Supplemental Figure S4C). Moreover, mouse embryonic fibroblasts (MEFs) transduced with lentivirus containing the BACH1 coding sequence together with four Yamanaka reprogramming factors had a significantly increased ability to form AP-positive mESC-like colonies (Supplemental Figure S1H–I), further demonstrating BACH1’s function in promoting pluripotency.
BACH1 interacts with NANOG, OCT4, SOX2 and MLL/SET1 complexes
Co-immunoprecipitation (co-IP) in mESCs (Figure 1A) and hESCs (Supplemental Figure S5A) indicated that BACH1 interacted with several components of MLL/SET1 complexes, including MLL1, WDR5, RBBP5, ASH2L and SETD1A. Consistent with our previous report in hESCs, BACH1 also interacted with OCT4, SOX2, and NANOG (OSN) in mESCs (Figure 1A and Supplemental Figure S5B–D). BACH1 colocalized with MLL/SET1 complexes subunits, RBBP5 and SETD1A, as well as OSN in the nucleus of mESCs (Supplemental Figure S5E). To elucidate whether the interaction of BACH1 and MLL/SET1 complexes depends on OSN, we conducted a co-IP assay in HEK293T cells that expressed FLAG-tagged BACH1. In the absence of OSN, exogenous BACH1 still interacted with MLL/SET1 complexes (Figure 1B). To further verify the direct protein-protein interaction, recombinant BACH1 protein was purified from bacteria and incubated with SET1 complexes or recombinant HIS-tagged NANOG protein. In vitro pulldown assays confirmed that GST-tagged BACH1 interacted directly with RBBP5, ASH2L, SETD1A or NANOG (Figure 1C–E). NANOG interacted directly with GST-tagged full-length BACH1 but not with BACH1 lacking BTB domain (BACH1-ΔBTB) (Figure 1D). SETD1A coimmunoprecipitated with the full-length BACH1 but not with BACH1-ΔBTB or bZIP domain (BACH1-ΔbZIP) (Figure 1E). To elucidate whether BACH1 affects the interaction between OSN and MLL/SET1 complexes, we compared the interaction between NANOG and RBBP5/SETD1A in WT and Bach1-KO mESCs. Notably, loss of BACH1 reduced the interaction between NANOG and RBBP5/SETD1A when a similar amount of NANOG protein was pulled down (Figure 1F). These findings indicate that BACH1 interacts with MLL/SET1 complexes and NANOG directly and individually, and that BACH1 bridges the interaction between NANOG and MLL/SET1 complexes. Moreover, we observed that only WT-BACH1 but not BACH1-ΔBTB or BACH1-ΔbZIP can rescue the effect of Bach1 knockout on mESC morphology and proportion of undifferentiated cells (Figures 1G and 6D).

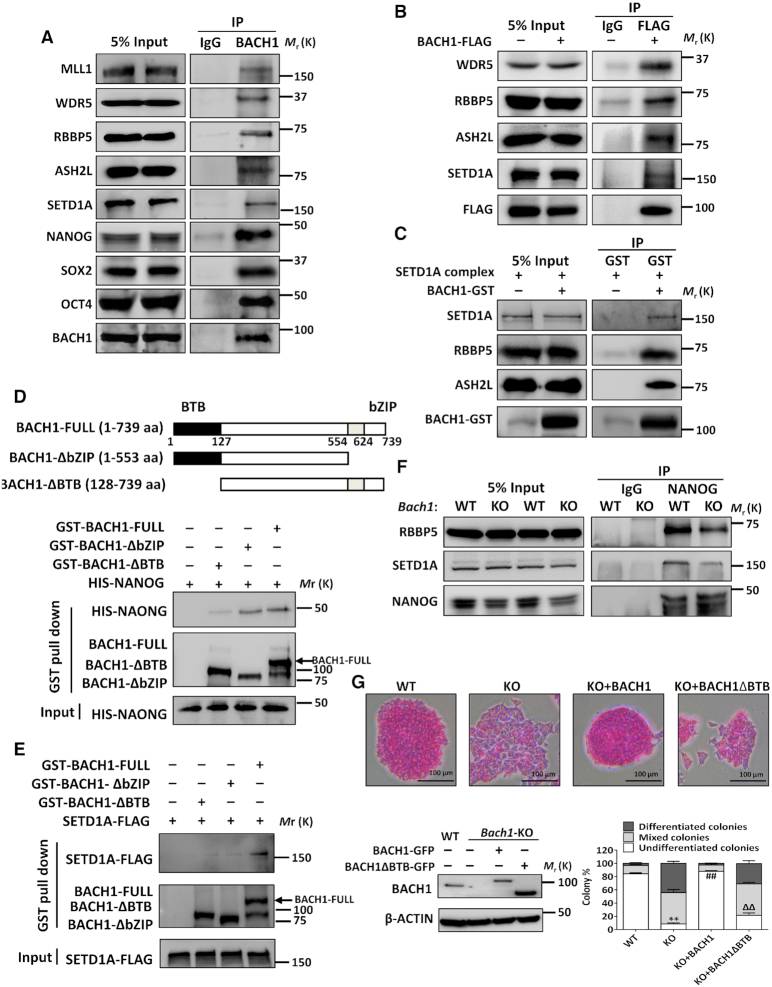
BACH1 interacts with NANOG, OCT4, SOX2 and MLL/SET1 complexes. (A) Co-immunoprecipitation of BACH1, MLL1, WDR5, RBBP5, ASH2L, SETD1A, NANOG, SOX2 and OCT4 in WT mESCs. (B) Co-immunoprecipitation of WDR5, RBBP5, ASH2L, SETD1A, BACH1-FLAG in BACH1-FLAG overexpression HEK293T cells. (C) Bacteria expressed GST-tagged BACH1 was incubated with SETD1A complex (SETD1A, ASH2L, RBBP5) recombinant proteins, and then GST-pulldown assay and immunoblot detected BACH1-GST and SETD1A complex (SETD1A, ASH2L, RBBP5). (D–E) Bacteria expressed NANOG-HIS (D) and/or HEK293T cells expressed SETD1A-FLAG (E) was incubated with GST-tagged full-length BACH1 protein (GST-BACH1-FULL) or GST-tagged BACH1 lacked bZIP domain (GST-BACH1-ΔbZIP) or BTB domain (GST-BACH1-ΔBTB); GST-pulldown assay and immunoblot detected the interaction between SETD1A-FLAG or NANOG-HIS and GST-BACH1-Full, GST-BACH1-ΔbZIP, or GST-BACH1-ΔBTB. (F) Co-immunoprecipitation of RBBP5, SETD1A and NANOG in WT and Bach1-KO mESCs. (G) Alkaline phosphatase (AP) staining of colonies (upper), the proportion of total cell clones in undifferentiated, partially differentiated, and fully differentiated cells; and BACH1 protein level in WT and Bach1-KO mESCs transfected with BACH1 FULL or BACH1-ΔBTB or empty plasmid (n = 3; ** P < 0.01 versus WT, ##P < 0.01 versus KO, ΔΔP < 0.01 versus KO + BACH1) (lower).
Colocalization of BACH1 and NANOG at gene promoters and enhancers in mESCs.
Chromatin immunoprecipitation and sequencing (ChIP-seq) analyses of mESCs expressing a dox-inducible, FLAG-tagged version of BACH1 confirmed that BACH1 preferentially bound to active gene promoters and active enhancers (Figure 2A), and KEGG pathway analysis indicated that BACH1-bound genes are enriched in the pathways of proteasome and pluripotency (Supplemental Figure S6A). Motif analysis showed that BACH1 binding sites were enriched not only in the binding motif of itself, but also in the motif of pluripotency factors such as KLF4, NANOG, OCT4, SOX2 and ESRRB (Figure 2B), suggesting their colocalization. Notably, the ChIP-seq binding heatmaps showed that NANOG, ASH2L, H3K4me1, H3K4me3 and H3K27ac, but not H3K27me3, were highly co-occupied on BACH1 binding active enhancers and promoters (Figure 2C). Venn diagram showed that 1,909 genes were co-occupied by BACH1, NANOG, H3K4me1 and H3K4me3 (Figure 2D). Representative snapshots of ChIP-seq tracks show the colocalization of BACH1 and NANOG on the H3K4me1+ H3K27ac+ active enhancers neighboring Tns3 and Lefty2, and on the H3K4me3+ H3K27ac+ active promoter of Jarid2 (Figure 2E).

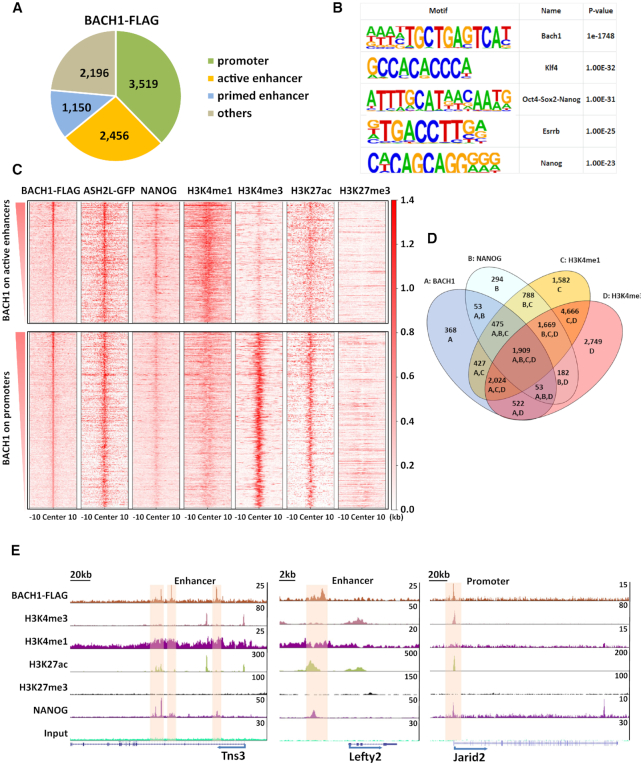
Colocalization of BACH1 and NANOG on gene promoters and enhancers in mESCs. (A) BACH1-FLAG ChIP-seq data in mESCs were analyzed to identify the genomic distribution of BACH1 binding sites in genomic DNA. Promoter: ±2 kb of transcription start sites (TSS). Active enhancer: H3K4me1+; H3K27ac+ promoter negative distal region. Primed enhancer: H3K4me1+; H3K27ac− promoter negative distal region. (B) The known transcription factors’ motifs at BACH1-FLAG enriched regions. (C) Heatmaps showing the genome-wide ChIP-seq binding profiles of BACH1-FLAG, ASH2L-GFP, NANOG, H3K4me1, H3K4me3, H3K27ac and H3K27me3 at BACH1-FLAG enriched regions on active enhancers (BACH1-FLAG+ H3K4me1+ H3K27ac+ distal regions) and BACH1-FLAG enriched regions on promoters (BACH1-FLAG+ regions within TSS±2 kb) (region center ±10 kb). (D) Venn diagram of genes enriched by BACH1-FLAG, NANOG, H3K4me1 and H3K4me3. (E) Representative snapshots of ChIP-seq tracks for BACH1-FLAG, H3K4me1, H3K4me3, H3K27ac, H3K27me3 and NANOG on the enhancers neighboring Tns3 and Lefty2 and on the promoter of Jarid2.
Next, we performed ChIP-seq analysis to evaluate the binding of NANOG in WT and Bach1-KO mESCs. Loss of BACH1 reduced the binding of NANOG at 2,365 regions (Supplemental Figure S6B–C, cluster 1), while 1,616 regions de novo built NANOG occupancy (Supplemental Figure S6B-C, cluster 3). The reason could be that knockout of Bach1 released NANOG from BACH1 recruited locus and rearranged it into novel locations. Furthermore, we found that NANOG enrichment was decreased in 1,361 genes in Bach1-KO mESCs, and these genes were involved in the development process and signaling pathways regulating pluripotency of stemness (Supplemental Figure S6D and E), consistent with our RNA-seq analyses.
Loss of BACH1 reduces the recruitment of NANOG and SETD1A to the targeting promoters and enhancers
We next overlapped ChIP-seq data of BACH1-binding genes with RNA-seq data from Bach1-KO versus WT mESCs. We identified 926 BACH1-targeting genes, which were directly bound by BACH1, and Bach1-KO led to their expression changes (Figure 3A). Among these 926 genes, we found that 538 genes were down-regulated and 388 genes were up-regulated upon Bach1 knockout (Figure 3A and B). Since BACH1 interacted with NANOG, MLL/SET1 complexes and Bach1-KO reduced their interaction (Figure 1A–F), we then asked whether loss of BACH1 affected their recruitment to chromatin. Indeed, we found that the chromatin occupancies of NANOG, RBBP5, and SETD1A near the BACH1-binding sites were lower in Bach1-KO than in WT mESCs (Figure 3C and D), especially on the enhancer and promoter of pluripotency-regulating genes.

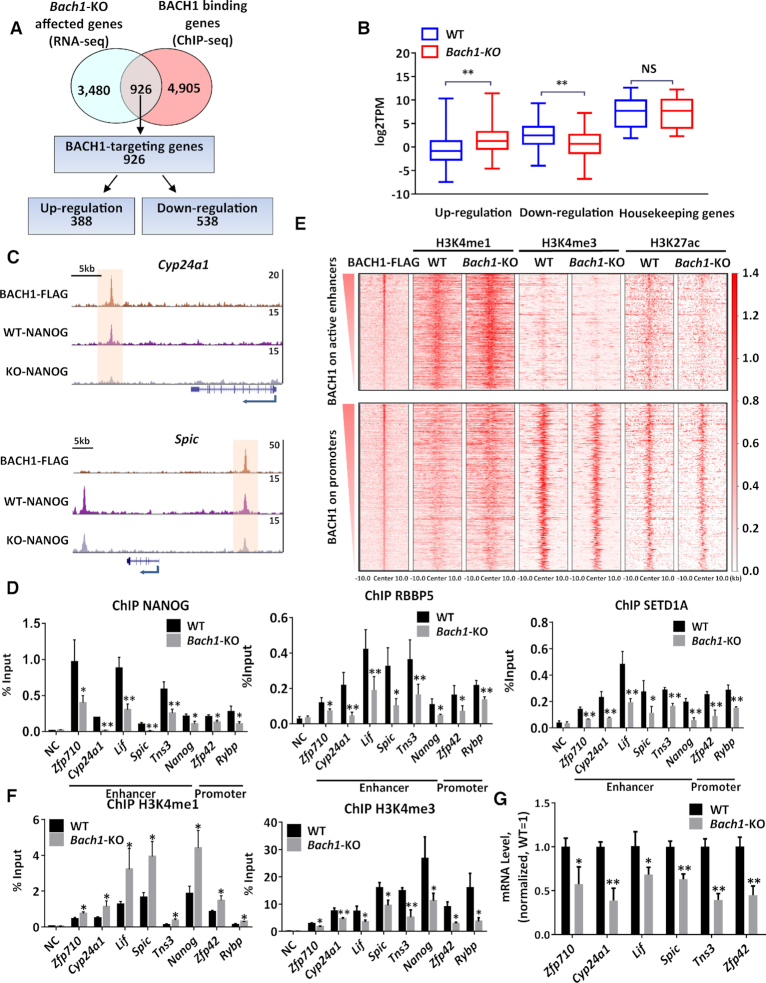
BACH1 recruits NANOG, RBBP5 and SETD1A to the BACH1-targeting enhancers and gene promoters, and maintains the tri-methylated state of H3K4. (A) Venn diagram showing the corresponding numbers of BACH1-bound genes analyzed from ChIP-seq, genes whose expression was affected by Bach1 knockout analyzed from RNA-seq. A total of 926 genes in the intersection were identified as BACH1-targeting genes, including 388 upregulated genes and 538 downregulated genes. (B) Box plot showing the expression of 926 BACH1-targeting genes and housekeeping genes in WT and Bach1-KO mESCs. (C) Representative snapshots of ChIP-seq tracks for BACH1-FLAG and NANOG in WT and Bach1-KO mESCs at the Cyp24a1 and Spic locus. (D) ChIP-qPCR analysis to validate the reduced recruitment of NANOG, RBBP5 and SETD1A in Bach1-KO mESCs vs WT mESCs on BACH1 target enhancers and gene promoters (n = 3; *P < 0.05, ** P < 0.01 versus WT). (E) Heatmaps of BACH1-FLAG, and H3K4me1, H3K4me3 and H3K27ac at BACH1 enriched regions on active enhancers or promoters in WT and Bach1-KO mESCs. Each horizontal line shows a separate BACH1-FLAG-bound peak (peak center ±10 kb). (F) ChIP-qPCR analysis to validate the increase of H3K4me1 and decrease of H3K4me3 in Bach1-KO mESCs vs WT mESCs on BACH1-targeting enhancers and gene promoters (n = 3; *P <0.05, **P < 0.01 versus WT). (G) RT-qPCR analysis of the genes expression in WT and Bach1-KO mESCs. Results were normalized to measurements in WT mESCs (n = 3; *P < 0.05, ** P < 0.01 versus WT).
Loss of BACH1 leads to a switch of H3K4me3 and H3K4me1 at both typical and super-enhancers
Since MLL/SET1 complexes catalyze the trimethylation of histone H3 lysine 4 (H3K4me3), the decrease of their chromatin localization may lead to changes on histone modifications. Although the global levels of histone modifications H3K4me1, H3K4me3, H3K27me3 and H3K27ac did not significantly change between WT and Bach1-KO mESCs (Supplemental Figure S7A), upon loss of Bach1 H3K4me1 enrichment increased, while H3K4me3 enrichment declined at BACH1 binding active enhancers and promoters, and H3K27ac enrichment had modest decrease on BACH1 binding active enhancers but largely unchanged (Figure 3E, Supplemental Figure S7B and C). These observations were validated by ChIP-qPCR at the promoters and enhancers of BACH1-targeting genes (Zfp710, Cyp24a1, Rybp, Spic and Tns3), especially stemness-related genes such as Nanog, Zfp42 and Lif (Figure 3F). Importantly, the expression level of these genes decreased in Bach1-KO mESCs (Figures 3G and 5D).
Notably, we found BACH1 was strongly enriched at super-enhancers in mESCs (called by Benjamin Sabari, Alessandra Dall’Agnese, et al. (37), through MED1 ChIP-seq in mESCs) (Figure 4A and B), and loss of BACH1 led to H3K4me3 partially switched to H3K4me1 at the super-enhancers (Figure 4C–E), consistent with the decreased chromatin occupancy of RBBP5 and SETD1A (Figure 3D). H3K4me3 partially switching to H3K4me1 suggested a reduction of super-enhancer activity. These BACH1 bound super-enhancers were neighboring the genes regulating embryo development and gene expression (Figure 4F), indicating their prominent role in regulating pluripotency. Collectively, these observations suggest that BACH1 is required for full level chromatin binding of NANOG and MLL/SET1 complexes in mESCs, and is important for the maintenance of H3K4me3 and activity of target promoters and (super-) enhancers to promote downstream gene expression, especially on pluripotency-regulating genes.

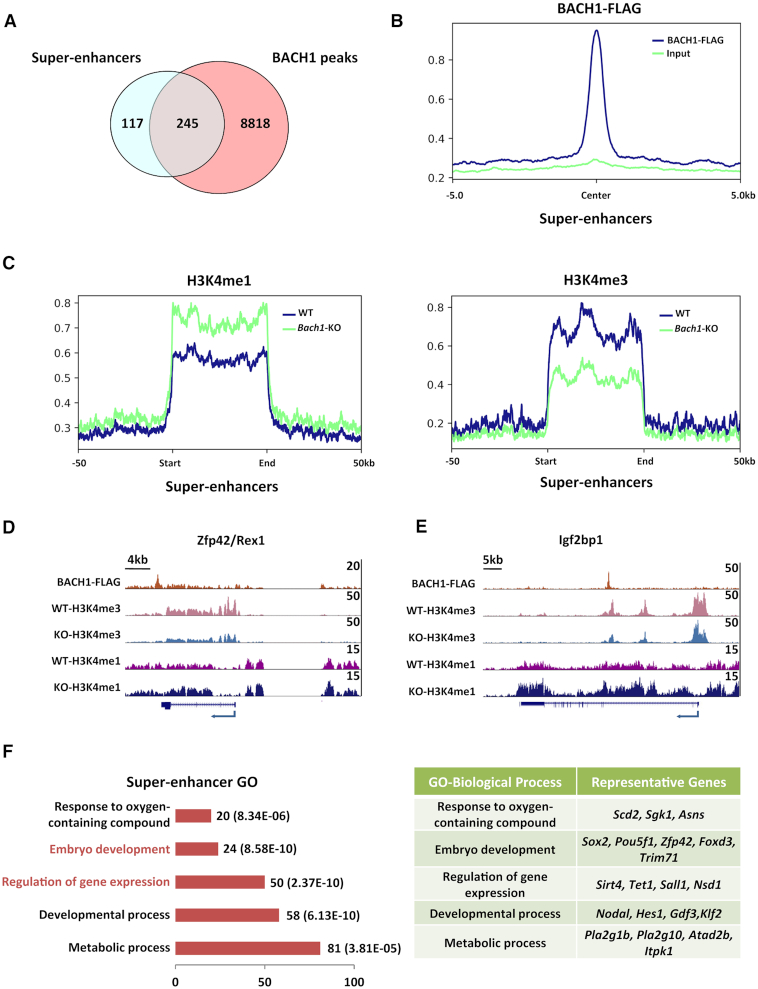
BACH1 localizes at super-enhancers, and loss of BACH1 leads to H3K4me3 partially switched to H3K4me1 at super-enhancers. (A) Venn diagram of BACH1-FLAG enriched peaks and super-enhancers in mESCs. (B) Signal profiles of BACH1 and input at super-enhancers in mESCs. (C) Signal profiles of H3K4me1 (left) and H3K4me3 (right) in WT and Bach1-KO mESCs at super-enhancers. (D, E) Representative snapshots of H3K4me3 and H3K4me1 in WT and Bach1-KO mESCs at BACH1 bound super-enhancers. (F) The Gene Ontology analysis of genes neighboring BACH1 bound super-enhancers in mESCs (left) and the representative genes (right).
BACH1 and NANOG strongly co-bind at chromatin loop anchors, and BACH1 regulates the remote NANOG binding
In ChIP-seq tracks, we found that Bach1 knockout led to the decrease of NANOG binding not only at peaks colocalized with BACH1 (direct recruitment), but also at peaks close to BACH1 binding regions, possibly through the effect mediated by chromatin looping (Figure 5A and B, Supplemental Figure S7D). In order to test this hypothesis, we used published HiC data in mESCs (34) and found that these regions indeed formed chromatin loops with the BACH1 bound sites (Figure 5A and B, Supplemental Figure S7D). Loss of BACH1 led to the decrease of enhancer RNAs (eRNAs) at these chromatin loop anchors (Figure 5C), and reduced the expression of neighboring genes including stemness-regulating genes Lif and Rybp (Figure 5D), indicating the reduction of enhancer activity. Interestingly, we also found that NANOG occupancy was also higher in chromatin loop anchors which contained BACH1 (Figure 5E). These data suggested that BACH1 can also regulate the recruitment of NANOG on chromatin interaction sites, and further fine-tune the enhancer activity and gene expression, including stemness-regulating genes.

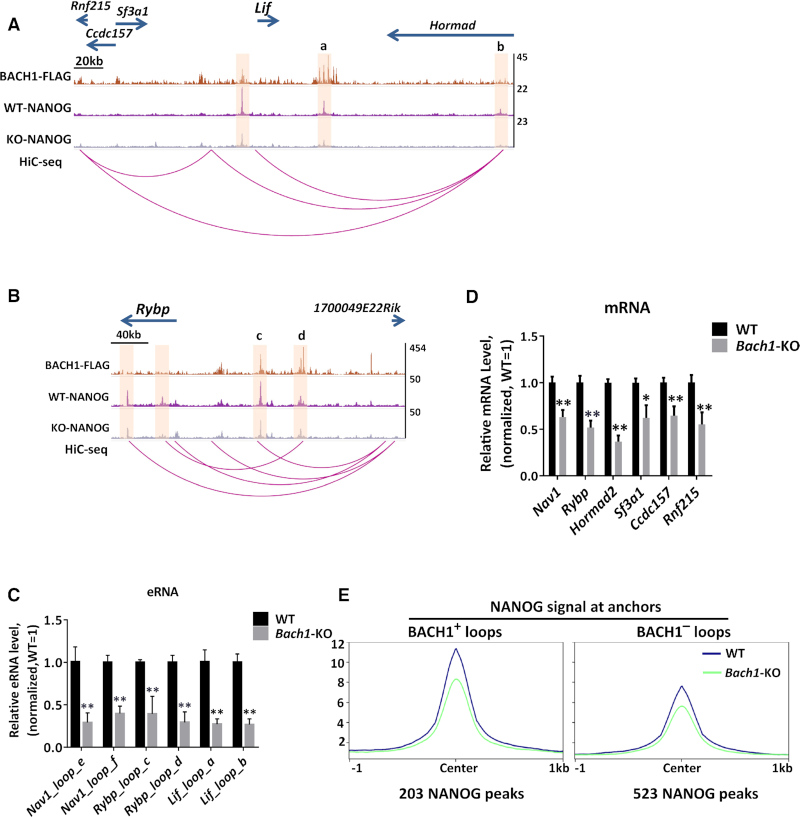
BACH1 and NANOG strongly co-bind at chromatin loop anchors, and BACH1 regulates the remote NANOG binding and enhancer activity. (A, B) Representative snapshots of chromatin loops and ChIP-seq tracks for BACH1-FLAG and NANOG in WT and Bach1-KO mESCs. Genome regions to be explained were highlighted. Chromatin loops were generated by Hi-C sequencing in mESCs and predicted by Peakachu. (C) RT-qPCR analysis of enhancer RNAs (eRNAs) transcribed from highlighted chromatin loop anchors in (A, B and Supplemental Figure S7D) in WT and Bach1-KO mESCs. Normalized by internal control gene Actb and normalized to WT mESCs (n = 3; *P < 0.05, **P < 0.01 versus WT). (D) RT-qPCR analysis of expression of genes regulated by BACH1 and chromatin looping in (A, B) in WT and Bach1-KO mESCs. Normalized by internal control gene Actb and normalized to WT mESCs (n = 3; *P < 0.05, ** P < 0.01 versus WT). (E) Signal profiles of NANOG ChIP-seq signal at NANOG peaks within BACH1-positive or BACH1-negative chromatin loop anchors. BACH1+ loops: either one anchor of a chromatin loop contained BACH1; BACH1– loops: none of the chromatin loop anchors contained BACH1.
BACH1 DNA binding domain is crucial for the recruitment of NANOG at chromatin and the enhancer activity
To determine if BACH1 directly affects NANOG binding to chromatin rather than merely reducing its expression, exogenously FLAG-tagged NANOG proteins were expressed at similar levels in WT and Bach1-KO mESCs. We found that the chromatin occupancy of exogenous FLAG-tagged NANOG was lower in Bach1-KO than in WT mESCs (Figure 6A). Moreover, only WT-BACH1 but not DNA binding-defective BACH1 (BACH1-ΔbZIP) rescued the decrease of the occupancy of NANOG, the level of enhancer RNAs (eRNAs) at the chromatin loop anchors, and the proportion of undifferentiated cells in Bach1-KO mESCs (Figure 6B–D), although BACH1-ΔbZIP could restore the NANOG expression in Bach1-KO mESCs as WT-BACH1 (Figure 6B, lower). Collectively, these data demonstrated that BACH1 directly affects the recruitment of NANOG at chromatin and maintains its enhancer activity, and the DNA binding domain of BACH1 is required.

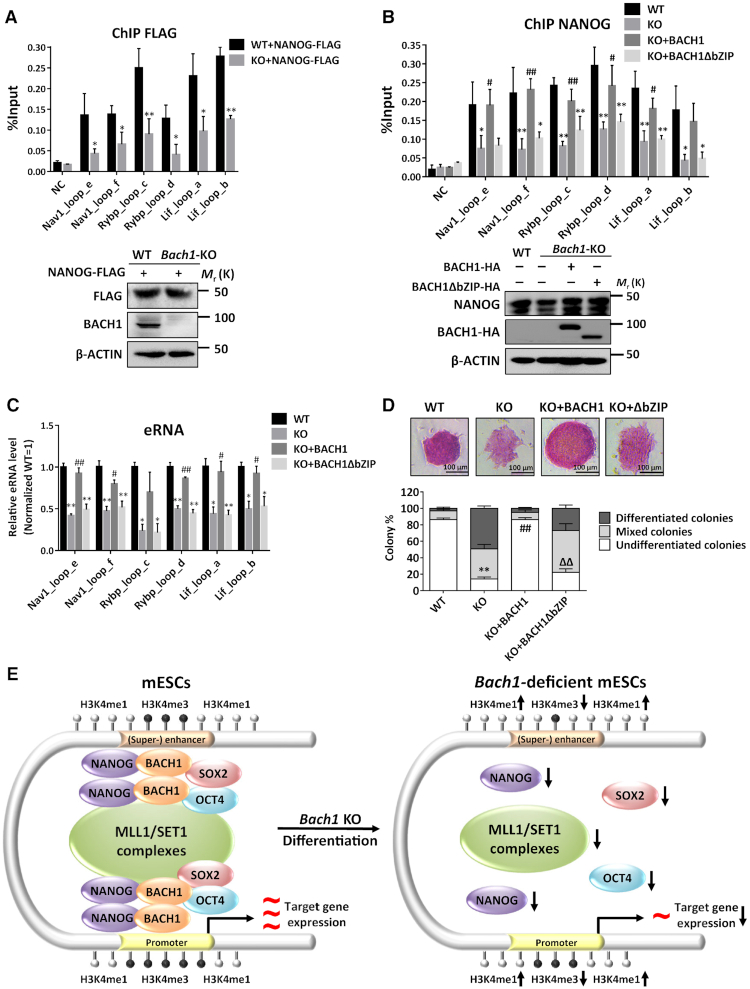
BACH1 DNA binding domain is crucial for the recruitment of NANOG at chromatin and the enhancer activity. (A) ChIP-qPCR analysis of exogenous FLAG-tagged NANOG in Bach1-KO mESCs overexpressing NANOG-FLAG vs WT mESCs overexpressing NANOG-FLAG on BACH1 target chromatin loop anchors (n = 3; *P < 0.05, ** P < 0.01 versus WT) (upper). Immunoblot verified the expression of exogenous FLAG-tagged NANOG and BACH1 (lower). (B) ChIP-qPCR analysis of endogenous NANOG in Bach1-KO mESCs rescued with full-length BACH1, BACH1-ΔbZIP or empty plasmid versus WT mESCs on BACH1 target chromatin loop anchors (n = 3; *P < 0.05, **P < 0.01 versus WT, #P <0.05, ##P < 0.01 versus Bach1-KO) (upper). Immunoblot verified the expression of exogenous NANOG and rescued exogenous HA-tagged BACH1 (lower). (C) RT-qPCR analysis of enhancer RNAs (eRNAs) transcribed from highlighted chromatin loop anchors in (Figure 5A, B and Supplemental Figure S7D) in WT and Bach1-KO mESCs transfected with full-length BACH1, BACH1-ΔbZIP or empty plasmid. Normalized by internal control gene Actb and normalized to WT mESCs (n = 3; *P < 0.05, **P < 0.01 versus WT, #P < 0.05, ##P < 0.01 versus Bach1-KO). (D) Alkaline phosphatase (AP) staining of colonies (upper), and the proportion of total cell clones of undifferentiated, partially differentiated and fully differentiated cells and BACH1 protein level in WT and Bach1-KO mESCs transfected with full-length BACH1, BACH1-ΔbZIP or empty plasmid (n = 3; ** P < 0.01 versus WT, ##P < 0.01 versus KO, ΔΔP < 0.01 versus KO + BACH1) (lower). (E) Schematic review showing BACH1 recruits NANOG and histone H3 lysine 4 methyltransferase MLL/SET1 complexes to activate enhancer–promoters and maintains pluripotency in embryonic stem cells.
DISCUSSION
The regulation of (super-) enhancer activity is essential for the maintenance of ESC pluripotency. However, the transcription factor network orchestrating (super-) enhancer activity in ESC pluripotency maintenance was not fully understood. In the present study, we demonstrated that BACH1 directly interacted with and bridged NANOG and MLL/SET1 complexes, and recruited them to (super-) enhancers and promoters, maintaining a high level of histone H3 lysine 4 trimethylation (H3K4me3) and enhancer–promoter activity. It further promoted downstream gene expression especially on stemness-related genes, such as Nanog, Zfp42 and Lif. Moreover, BACH1 directly affects NANOG binding to chromatin loop anchors and regulates remote NANOG binding, fine-tuning enhancer–promoter activity and gene expression. Thus, it suggests that BACH1 forms a novel complex, bridging master pluripotent transcription factors OSN and MLL/SET1 family proteins at (super-) enhancers and promoters to control the pluripotency network of stem cells (Figure 6E).
Our previous study suggested that BACH1 interacted with and stabilized ESC identity markers OSN(19), but the role of BACH1 as a transcription factor in ESCs is still unclear. Super-enhancer forms giant chromatin loop with several enhancers and promoters, which is enriched for OSN. It suggests us to explore whether BACH1 binds to and regulates super-enhancers and enhancer–promoter activity. BACH1 was found to mediate chromatin loops in vitro by linking multiple Maf recognition elements (MAREs) together (21). Recent studies showed that MLL4 and H3K4me1 could also regulate chromatin looping (38), but the recruitment mechanism is not clear. Here we found that BACH1 recruited NANOG to chromatin loop anchors and interacted with OSN and MLL/SET1 complexes. BACH1 depletion reduced the interaction between NANOG and MLL/SET1 complexes, and decreased the recruitment of NANOG and MLL/SET1 complexes on chromatin. Importantly, DNA binding-defective BACH1 (BACH1-ΔbZIP) decreased the occupancy of NANOG with NANOG total protein level unchanged, indicating that BACH1 directly affects NANOG recruitment at chromatin rather than merely reducing its expression. Thus, the results presented here indicate that BACH1 may act as a mediating protein in chromatin loops, recruiting key pluripotent factor NANOG (could also be SOX2 and OCT4) and histone-modifying enzymes MLL/SET1 complexes, fine-tuning the promoter-enhancer activity, especially the activity of super-enhancers, promoting downstream gene expression, and finally contribute to pluripotency maintenance.
The distribution and conversion of mono-, di-, and tri-methylation states of H3K4 appear to regulate gene transcription by establishing physical boundaries that recruit different transcription regulators and co-activators (39). For example, although the chromatin readers ING1 and ING2 interact with both di- and tri-methylated H3K4, they preferentially bind H3K4me3 (40), and H3K4me3 is located closest to the transcription start site of active promoters, with H3K4me2 and H3K4me1 located progressively farther away (41). H3K4me1 has been reported to demarcate the recruitment of these factors spatially and hence restrict the transcriptional activity (39). Loss of the H3K4me3-specific demethylase KDM5C leads to the increase of H3K4me3 at (super-) enhancers and their overactivation, which upregulates the transcription of nearby genes (29). These reports suggest that a switch from H3K4me1 to H3K4me3 may lead to higher transcriptional activity. Moreover, a previous report has shown that BACH1 degradation by hemin decreased the enrichment of dimethylation and trimethylation of H3K4 at the enhancer and promoter of HO-1 (42). The results from these earlier reports are consistent with our observations that Bach1-KO led to a switch from H3K4me3 to H3K4me1 in promoters and (super-) enhancers, which suppressed transcriptional activity, likely due to the reduced recruitment of H3K4me3 methyltransferase complexes MLL/SET1. Since the H3K4me1 and H3K4me3 methyltransferases compete for the H3K4 substrate, BACH1 appears to regulate the methylation state of H3K4 via recruiting MLL/SET1 complexes.
BACH1 can function as both a transcriptional activator and a repressor. It activates the expression of Hexokinase 2 and GAPDH in lung cancer cells (43) and the epithelial-mesenchymal transition (EMT) genes in ovarian cancer cells (20), but suppresses the expression of electron transport chain (ETC) genes in triple-negative breast cancer (44) and Wnt/β-catenin target genes in human endothelial cells (16). BACH1-induced transcriptional activation or repression may be associated with the coactivators or corepressors interacted with BACH1. HDAC1 seems to be a corepressor for BACH1-mediated repression of transcription (16,45). However, how BACH1 activates gene transcription is largely unknown. Our findings unravel the role of BACH1 in activating gene transcription by recruiting NANOG and MLL/SET1 complexes to enhancers/promoters and maintaining the trimethylated state of H3K4. BACH1 lacking the BTB domain or bZIP domain cannot bind to the OSN and MLL/SET1 complexes and failed to rescue the decreased pluripotency in Bach1-KO mESCs, indicating that both the BTB domain and bZIP domain of BACH1 are essential for the maintenance of ESC pluripotency by BACH1. We have shown that BACH1 repressed the transcription of mesendodermal genes via recruiting PRC2 to catalyze trimethylated H3K27 on these genes’ promoters in hESCs (19). However, in the present study, BACH1 is not colocalized with H3K27me3 in mESCs. This discrepancy between human and mouse ESC remains unknown. One possibility is that human and mouse ESCs have different pluripotent states (primed vs. naïve) and may rely on different transcription regulators and epigenetic pathways to maintain pluripotency.
In summary, the results presented here suggest that BACH1 maintains pluripotency in ESCs by bridging and recruiting the essential pluripotency regulator NANOG and the MLL/SET1 complexes to maintain the trimethylated state of H3K4 on promoters and (super-) enhancers, and upregulates the expression of downstream genes especially pluripotency-regulating genes. Collectively, these observations suggest that BACH1 has a key regulatory role in maintaining stem-cell pluripotency.
DATA AVAILABILITY
The accession numbers reported in this paper are: RNA-seq and ChIP-seq data of WT & Bach1-KO mESCs (GEO: GSE142519), ChIP-seq data of H3K27me3 in mESC (GEO: GSM1893474) and ChIP-seq data of ASHL-GFP in mESC (GEO: GSM1258240).
ACKNOWLEDGEMENTS
We thank Dr W. Kevin Meisner for providing editorial assistance and Dr. Ruitu Lv for the advise on RNA-seq analysis.
Author contributions: M.D. and L.F. conceived and designed the project. N.C., W.S.Q., G.J.Y., W.X.X., J.M.P., C.Z.X., G.W.X. and L.M. performed experiments and analyzed the data. M.D. and W.S.Q. wrote the manuscript. G.J.Y. and Q.Y. edited the manuscript. W.X.H., Z.X.L., C.S.F., G.M.X., Z.J.Y. and H.J.D. provided valuable comments and interpreted the results. M.D. supervised the whole study.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Great Program [92068202]; General Program [81873469] of the National Natural Science Foundation of China; Shanghai Science and Technology Commission of China [19JC1411300]; Program of Shanghai Academic/Technology Research Leader [20XD1400600]; National Key Research and Development Program of China [2018YFC2000202 to D.M., 2018YFA0108700 to F.L.]; Innovative research team of high-level local universities in Shanghai and a key laboratory program of the Education Commission of Shanghai Municipality [ZDSYS14005]. Funding for open access charge: Great Program [92068202] of the National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
1.
2.
3.
4.
5.
6.
7.
8.
9.
10.
11.
12.
13.
14.
15.
16.
17.
18.
19.
20.
21.
22.
23.
24.
25.
26.
27.
28.
29.
30.
31.
32.
33.
34.
35.
36.
37.
38.
39.
40.
41.
42.
43.
44.
 BACH1 recruits NANOG and histone H3 lysine 4 methyltransferase MLL/SET1 complexes to regulate enhancer–promoter activity and maintains pluripotency
BACH1 recruits NANOG and histone H3 lysine 4 methyltransferase MLL/SET1 complexes to regulate enhancer–promoter activity and maintains pluripotency
