
The authors have declared that no competing interests exist.
- Altmetric
Salmonella is a major cause of foodborne disease globally. Pigs can carry and shed non-typhoidal Salmonella (NTS) asymptomatically, representing a significant reservoir for these pathogens. To investigate Salmonella carriage by African domestic pigs, faecal and mesenteric lymph node samples were taken at slaughter in Nairobi, Busia (Kenya) and Chikwawa (Malawi) between October 2016 and May 2017. Selective culture, antisera testing and whole genome sequencing were performed on samples from 647 pigs; the prevalence of NTS carriage was 12.7% in Busia, 9.1% in Nairobi and 24.6% in Chikwawa. Two isolates of S. Typhimurium ST313 were isolated, but were more closely related to ST313 isolates associated with gastroenteritis in the UK than bloodstream infection in Africa. The discovery of porcine NTS carriage in Kenya and Malawi reveals potential for zoonotic transmission of diarrhoeal strains to humans in these countries, but not for transmission of clades specifically associated with invasive NTS disease in Africa.
Healthy humans infected with non-typhoidal Salmonella (NTS) typically suffer from diarrhoeal disease which resolves without treatment. However, NTS infection of patients with an impaired immune system can lead to either bloodstream infection or infection in another part of the body; so-called ‘invasive’ NTS infection. Over the last twenty years, NTS have been the most prevalent bacteria to be isolated from human blood in sub-Saharan Africa.
It is well known that pigs are able to carry a wide range of different NTS serovars without showing signs of disease themselves. Carrier pigs may be able to intermittently shed NTS in their faeces which may have the potential to cause disease in humans. If good hygiene protocols are not followed during the porcine slaughter process, pork intended for human consumption may become contaminated with NTS. In the next few years, pork consumption is projected to increase dramatically in sub-Saharan Africa. The degree of asymptomatic carriage of NTS by pigs in this region has never been investigated in detail.
In this study, we report that pigs in sub-Saharan Africa are able to carry a wide variety of non-typhoidal Salmonella serovars, such as S. Typhimurium, that have the potential to cause diarrhoeal disease in humans. No clades of NTS which have previously been associated with invasive NTS disease in sub-Saharan Africa were detected.
Introduction
Infection with non-typhoidal Salmonella (NTS) in healthy humans is typically associated with self-limiting enterocolitis, but in immunocompromised patients can lead to bloodstream or focal metastatic infections [1]. However over the past two decades, NTS have been the most prevalent bacteria to be isolated from human blood in sub-Saharan Africa (sSA) [2–5]. The main risk factors for invasive NTS (iNTS) disease are HIV [6], malaria [7], and malnutrition [8]. The emergence of iNTS disease has been associated with specific, multidrug resistant clades of Salmonella [9,10]. Despite an increasing amount of evidence suggesting human adaptation [11,12], the reservoir for these novel lineages has not been established.
Pigs act as a reservoir for NTS as they can carry a diverse range of Salmonella serovars asymptomatically in the tonsils, intestine and mesenteric lymph node (MLN) tissue [13]. Carrier pigs intermittently shed potentially pathogenic Salmonella bacteria via faeces, and pork may become contaminated during slaughter processes if proper procedures and hygiene are not observed, for example, incorrect hanging of carcasses for evisceration or contact between the meat and a soiled floor can transmit Salmonella [14]. Extensive work has been undertaken to investigate Salmonella carriage and excretion by pigs in Europe and the USA, where the most common Salmonella serovars isolated are S. Derby, S. Enteritidis and S. Typhimurium [15–19]. These serovars frequently cause human infection, accounting for 43.6% of cases of gastroenteritis-associated salmonellosis in southern Europe [20]. Consequently, porcine carriers of Salmonella are considered to pose a threat to public health.
Currently the published data from sSA countries is limited [21]. Pork consumption and supply in Kenya is estimated to rise by 268% between 2010–2050 [22] a trend that is predicted to be replicated across sSA [23]. Pigs in many rural areas of Kenya and Malawi are free-roaming with access to human faeces in areas where open-defecation occurs, and often defecate in close proximity to human domestic environments, which may facilitate zoonotic transmission [24]. To investigate the prevalence and diversity of NTS in pigs in sSA, we isolated Salmonella from the faeces and MLN of pigs at slaughter in Kenya and Malawi.
Materials and methods
Location and sampling
Pigs included in this study were those brought for slaughter on the day of sample collection at designated slaughterhouses in three study sites: Busia (Kenya), Nairobi (Kenya) and Chikwawa (Malawi). Samples were collected between October 2016 and May 2017. See S1 Table for more details.
Faecal and MLN samples were taken from pigs post mortem. Between 1 and 25g faeces were taken manually directly from each pig rectum. Once the entire gastrointestinal tract had been removed during meat processing, between 1-4g of MLN tissue was excised using a sterile scalpel. At least five individual MLN were sampled per pig. Approximately three of the lymph node samples were taken from the mesentery of the ileum and jejunum, and two samples were taken from the colonic mesentery, and samples from each animal were pooled (total 1 to 4g). Slaughtermen in Chikwawa were extensively trained in the sampling methodology prior to commencing the study. Samples were processed in the laboratory within four hours of collection.
Additional metadata were collected on paper (Malawi) and electronically (Kenya) using a ‘Field Information Support Tool’ developed from a Case Report Form by the Kestrel Technology Group (Kestrel Technology Group, LLC) on a Nexus 5 Android device. This questionnaire included name of the village where the pig was reared, previous antibiotic treatment, age, sex, breed of pig and method of transport of the pig to slaughterhouse. The GPS location of each of the slaughterhouses in Kenya and butcheries in Malawi was recorded.
Microbiological methods (Fig 1)
NTS were isolated by culture using standard procedures (International Organisation for Standardisation (ISO 6579:2002). The exterior of the surface of each of the MLN samples was placed briefly into a flame to remove any residual exterior contamination. 1g of MLN and 1g of faeces from each pig were placed into separate stomacher bags containing 9ml 2% buffered peptone water. Samples were homogenized and each sample was incubated for 18hours (h) at 37°C in air. Following pre-enrichment, 0.1ml of each sample was placed into a sterile bijou containing 9.9ml Rappaport-Vassiliadis broth, and were incubated for 24h at 42°C. Each enriched sample was inoculated onto both Brilliant Green agar and Harlequin ABC Salmonella plates. Following 24h incubation, positive colonies were inoculated onto nutrient agar plates and incubated for a further 24h at 37°C prior to antisera agglutination and antimicrobial susceptibility testing. Salmonella agglutination was carried out using Salmonella antisera (Polyvalent O-antigen and H Phase 1 and 2 or H Phase 2 antigens). Isolates which showed positive agglutination with Poly O Antigen and either H Phase 1 and 2, or H Phase 2 antigens were submitted for Whole Genome Sequencing (WGS) as presumptive salmonellae.

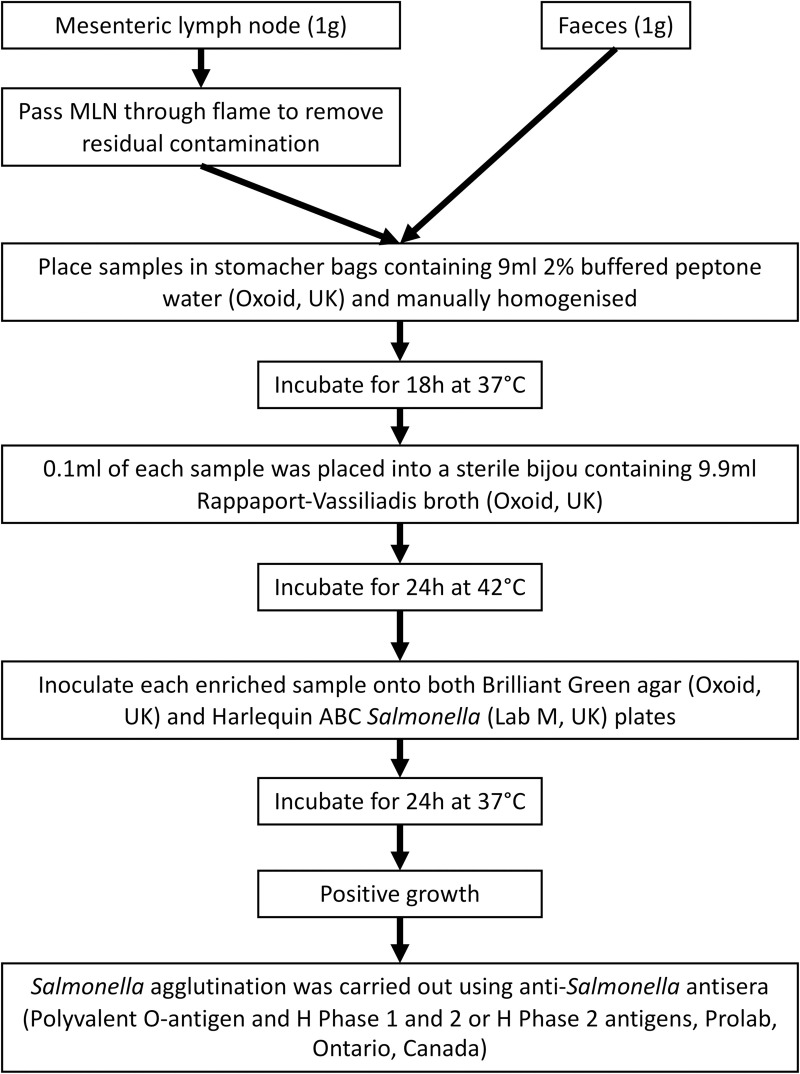
Microbiological Methods.
An outline of the microbiological methods followed in the laboratory to undertake sample processing.
Whole genome sequencing
Presumptive Salmonella samples were submitted for WGS to the Earlham Institute, Norwich as part of the 10,000 Salmonella Genomes Project (https://10k-salmonella-genomes.com/) [25]. An individual Salmonella colony was inoculated into each 0.7ml FluidX 2D tri-coded jacket tube (FluidX Ltd, UK) containing 100μl 2% buffered peptone water solution (Oxoid), and incubated at 37°C for 24h. The FluidX 2D Tubes were then placed in a 95°C oven for 20 minutes to inactivate the isolates. DNA extraction was carried out by the Earlham Institute and library preparation was performed using a modified Illumina Nextera XT DNA Library Prep Kit (Illumina, FC-131-1096). Illumina short-read sequencing was carried out on these samples to achieve 150bp paired-end reads using the HiSeq 4000. Sequencing was multiplexed using 768 unique barcode combinations per sequencing lane. The insert size was approximately 180bp, and the median depth of coverage was 30x.
Quality control and read trimming
Paired-end reads were subjected to stringent quality checks using FastQC v0.11.5 (www.bioinformatics.babraham.ac.uk/projects/fastqc/) and MultiQC v1.0 (http://multiqc.info). Potentially contaminated sequences were detected using Kraken v0.10.5-beta [26] against the MiniKraken 8gb database, using a Salmonella abundance cut-off of 70%. The paired-end reads were adapter-trimmed using palindromic Trimmomatic v0.36 [27], and quality trimmed using SEQTK v1.3-r106 (https://github.com/lh3/seqtk).
Assembly and annotation
Unicycler v0.3.0b [28] was used to produce high quality genome assemblies which were assessed using QUAST. Genomes that exceeded the quality control metrics defined by Enterobase [29] were designated as high quality, and used for subsequent analysis. Annotation was performed using Prokka v1.12 [30] against a custom-made database of Salmonella-specific genes.
In silico typing
In vitro Salmonella serotyping was confirmed using the Salmonella in Silico Typing Resource (SISTR)[31]. The strains were also assigned a Multi Locus Sequence Type (MLST) using the software tool MLST v2.10 [32] based on the conservation of seven housekeeping genes.
Core gene-based phylogenetics
To investigate the relationship between the diverse set of 121 high-quality pig-derived Salmonella genomes a maximum likelihood phylogeny was inferred from a core gene SNP alignment. Core genes were defined as present in at least 99% of genomes. Roary v3.11 [33] and SNP sites v2.3.3 software were used to generate the alignment which comprised of 3,010 core genes and 208,657 sites. The maximum likelihood tree was built using RAxML-NG v0.4.1 BETA [34], with the general time reversible (GTR) model and gamma distribution for site-specific variation and 100 bootstrap replicates to assess support. To infer the position of the root, the phylogeny was rebuilt using S. bongori as an outgroup. The finished phylogeny (Fig 2) was then rooted according to the inferred position using the Interactive Tree Of Life (iTOL) v4.2 [35].

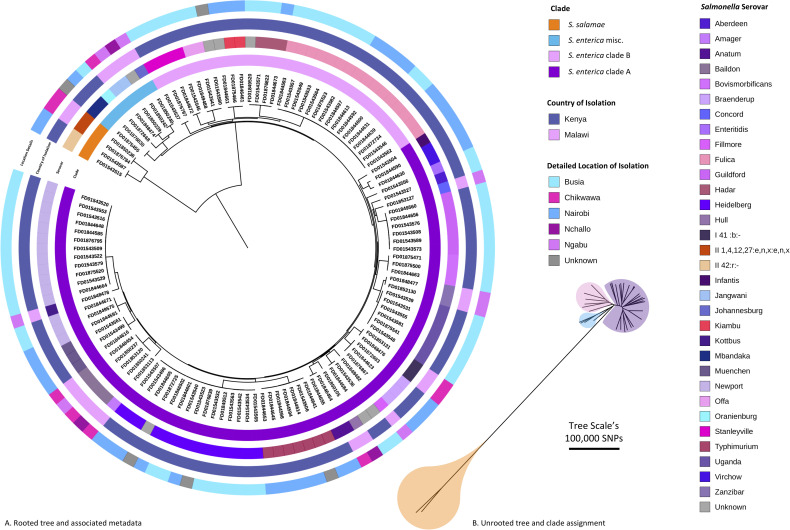
The diversity of pig-derived Salmonella identified in Kenya and Malawi.
A maximum likelihood phylogenetic tree based on core gene SNPs. The tree was rooted at the inferred position of the outgroup S. bongori. B. Maximum likelihood phylogenetic tree (unrooted). Note the colours refer to clade designation. Both visualised using ITOL (https://itol.embl.de).
Contextualising pig-derived S. Typhimurium ST313 isolates
We describe below the finding of S. Typhimurium sequence type 313 (ST313) in some of our samples. Given the specific public health importance of this ST, and to place the pig-derived S. Typhimurium ST313 isolates into a global context, a phylogeny was constructed that included 207 published ST313 genomes (S2 Table)[36–40]. A single nucleotide polymorphism (SNP) alignment was inferred from 2,004 core genes using Roary v3.11 [33] and SNP sites v2.3.3 [41]. The alignment comprised of 4,999 SNP sites. The final maximum likelihood tree was built using RAxML-NG v0.4.1 BETA [34] with 100 bootstrap replicates to assess support. The relatedness of the pig-derived ST313 was visualised with iTOL v4.2 [35].
The Short Read Sequence Typing for Bacterial Pathogens (SRST2) v0.2.0 [42] software tool was used to detect the presence of plasmid and prophage sequences associated with ST313, using a custom-made database based on plasmid and prophage sequences present in the ST313 reference strain D23580 and known variants. Reporting of gene presence is based on 90% coverage against the reference sequences. For pairwise comparison, pig-derived ST313 contigs were ordered against the ST313 reference genome D23580 using ABACAS v1.3.1 [43]. A pairwise comparison file was then generated between the ordered assemblies using BLASTn with default parameters, and visualised with the Artemis Comparison Tool v10.2 [44] (S1 Fig).
Antimicrobial resistance (AMR) testing
Genetic determinants for antimicrobial resistance were identified using Staramr v0.5.1 (https://github.com/phac-nml/staramr) against the ResFinder [45] and PointFinder [46] databases. An acquired AMR gene was considered to be present in a genome if percentage nucleotide homology was >90%. Confirmatory phenotypic antimicrobial susceptibility testing was carried out by disk diffusion on any isolates that contained antimicrobial-resistance determinants according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines [47]. Isolates were tested in duplicate for susceptibility to seven antibiotics (pefloxacin 5μg, trimethoprim/sulfamethoxazole 25μg, tetracycline 30μg, fosfomycin/glucose6phosphate 200μg, ceftriaxone 30μg, ampicillin 10μg and gentamicin 10μg) (all disks from Mast Group). Plates were incubated for 18-24h at 37°C, and the zones of inhibition were read for each disk to the nearest millimetre. According to EUCAST breakpoint tables for Enterobacteriaceae, isolates were classified as either susceptible or resistant to each antibiotic [48]. Phenotypic results were correlated with the genome-derived identification of antimicrobial resistance genes for each isolate.
Statistical analysis
Descriptive statistics with 95% confidence intervals were used to describe the prevalence and diversity of NTS detected using Microsoft Excel version 15.31. The frequency and diversity of the antimicrobial susceptibility phenotypes and genotypes of the NTS detected were also analysed using descriptive statistics with a 95% confidence interval.
Ethics
Ethical approval for this study was obtained from the University of Liverpool Veterinary Research Ethics Committee (Reference number VREC465), the Kenya National Commission for Science Technology and Innovation accredited International Livestock Research Institute Institutional Animal Care and Use Committee, Nairobi, Kenya (IACUC reference number 2016.19) and the College of Medicine Research Ethics Committee, Malawi (reference number P.02/17/2124).
Results
Descriptive epidemiology
Faeces and MLN were sampled from 647 pigs across the three study areas (Busia = 276, Nairobi = 306 and Chikwawa = 65). Isolates that showed positive agglutination using the Salmonella O antigen test were obtained from 259 pigs. All 259 isolates were submitted for whole genome sequencing, of which 149 were genotyped initially as being NTS. 28/149 isolates failed quality control checks, therefore 121 isolates were genotyped as being NTS. This gives an overall prevalence of NTS of 12.7% (95% confidence interval (CI); 8.75–16.6%) in Busia, 9.2% (95%CI; 5.9–12.4%) in Nairobi and 24.6% (95%CI; 14.1%-35.1%) in Chikwawa (See Table 1, S2 Fig). Several pigs were found to be carrying more than one serovar of NTS; 5.7% of pigs from Busia, 7.15% from Nairobi and 6.3% from Chikwawa (Table 1, S2 Fig).

| Sampling location | Total number of NTS isolates | Number of pigs in which NTS was detected | Percentage pigs carrying NTS (%) | Number of NTS isolates detected from mesenteric lymph node samples | Number of NTS isolates detected from faecal samples | Number of pigs in which more than 1 NTS isolate was detected* | Number of pigs carrying more than 1 serovar of NTS |
|---|---|---|---|---|---|---|---|
| Busia (n = 276) | 61 | 35 | 12.7 (8.7–16.6) | 44 | 16 | 22 | 2 |
| Nairobi (n = 306) | 40 | 28 | 9.1 (5.9–12.4) | 21 | 19 | 12 | 2 |
| Chikwawa (n = 65) | 20 | 16 | 24.6 (14.1–35.1) | 18 | 2 | 9 | 1 |
* This means that more than one NTS isolate was isolated during the culture method or from the faecal and mesenteric lymph node samples from one pig.
Serotypes and phylogenetic-relatedness of pig-derived Salmonella
To visualise the diversity of Salmonella identified, the genomic data were used to derive serotype information from 121 isolates, and a core gene SNP-based phylogeny was constructed (Fig 2). Thirty-two different Salmonella serovars were identified from two sub-species, S. enterica and S. salamae. Most serovars were unique to a single sampling site, seven serovars were found in two sampling sites; Nairobi and Busia (n = 5), Nairobi and Chikwawa (n = 1) and Busia and Chikwawa (n = 1). No serovars were detected in all three study sites (Fig 3). In total 8 isolates of S. Typhimurium were identified; 6 isolates of S. Typhimurium ST19 isolated from 4 pigs, and 2 isolates of S. Typhimurium ST313 isolated from 2 pigs.

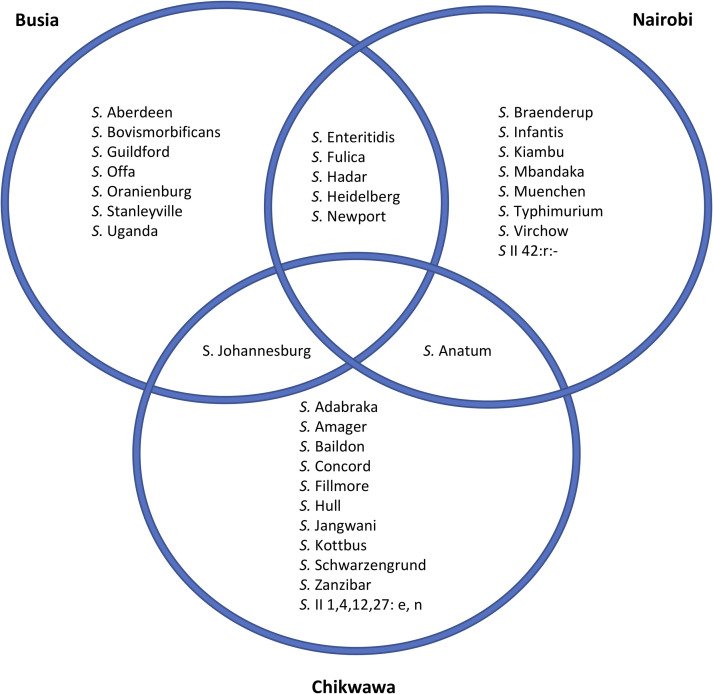
The Salmonella serovars detected in each of the study locations.
Phylogenetically, the Salmonella isolates belonged to four broadly defined groups, which corresponded to previously characterised Salmonella subspecies and clades (Fig 2, S3 Table). The largest clade (n = 80) included 66% of the isolates and 20 serovars, and is known as Clade A of S. enterica subspecies enterica [49], a grouping that contains serovars responsible for the majority of human disease. A second group of 30 S. enterica subspecies enterica isolates belonged to clade B [49], and originated from pigs in the two Kenyan study sites, 50% of which were the S. Fulica serovar. A third, smaller cluster of 7 S. enterica isolates did not belong to clade A or clade B. Four of these isolates were typed as S. salamae.
Antimicrobial resistance
We identified that 28/121 (23.1%) NTS isolates carried antimicrobial resistance (AMR) genes. These include 15/40 (37.5%) isolates from Nairobi, 12/61 (19.7%) isolates from Busia, and 1/20 (5.0%) isolate from Malawi. To determine the concordance between phenotypic and genotypic characterisation in our study, we analysed the antibiotic susceptibility phenotype of 26 genotypically resistant isolates (two of the isolates were unavailable for testing) (Table 2). Phenotypically, 16/26 isolates were susceptible to all antibiotics tested, 6/26 isolates were resistant to one antibiotic, 4/26 NTS isolates were resistant to two classes of antibiotics and none of the isolates were classified as multi-drug resistant (resistant to three or more classes of antibiotics).

| Pefloxacin | Ceftriaxone | Fosfomycin | Tetracycline | Trimethoprim-Sulfamethoxazole | Ampicillin | Gentamicin | |
|---|---|---|---|---|---|---|---|
| FD01853127 | dfrA14 sul2 | aph(3'')-1b aph(6)-1d | |||||
| FD01543571 | tet(A) | dfrA14 sul2 | aph(3'')-1b aph(6)-1d | ||||
| FD01849520 | tet(A) | dfrA14 sul2 | aph(3'')-1b aph(6)-1d | ||||
| FD01844591 | sul2 | aph(3'')-1b aph(6)-1d | |||||
| FD01844594 | tet(A) | sul2 | aph(3'')-1b aph(6)-1d | ||||
| FD01844598 | tet(A) | sul2 | aph(3'')-1b aph(6)-1d | ||||
| FD01844614 | tet(A) | sul2 | aph(3'')-1b aph(6)-1d | ||||
| FD01844645 | tet(A) | sul2 | aph(3'')-1b aph(6)-1d | ||||
| FD01844653 | tet(A) | sul2 | aph(3'')-1b aph(6)-1d | ||||
| FD01876797 | sul2 | blaTEM-1B | |||||
| FD01543496 | fosA7 | ||||||
| FD01543507 | fosA7 | ||||||
| FD01543523 | fosA7 | ||||||
| FD01543532 | fosA7 | ||||||
| FD01543534 | fosA7 | ||||||
| FD01543540 | fosA7 | ||||||
| FD01543542 | fosA7 | ||||||
| FD01543563 | fosA7 | ||||||
| FD01543565 | fosA7 | ||||||
| FD01844601 | fosA7 | ||||||
| FD01844605 | fosA7 | ||||||
| FD01846502 | fosA7 | ||||||
| FD01849512 | fosA7 | ||||||
| FD01872668 | fosA7 | ||||||
| FD01872725 | fosA7 | ||||||
| FD01876839 | fosA7 | ||||||
| FD01543506 | gyrA (D87Y) | fosA7 | |||||
| FD01844630 | tet(J) |
We found that 16/121 (13.2%) isolates carried fosA, the genetic determinant for resistance to fosfomycin. The presence of this gene was strongly associated with S. Heidelberg (15/16), but all 16 isolates were phenotypically susceptible to fosfomycin (Table 2). There were 8/121 (6.6%) isolates that harboured tetracycline resistance genes tetA (5.8%) or tetJ (0.8%), however 6/7 isolates carrying tetA (85.7%) and the single isolate carrying tetJ were phenotypically susceptible to tetracycline. One isolate showed phenotypic resistance to tetracycline, despite the absence of genomic predictions of known tetracycline resistance genes. 10/121 (8.3%) isolates contained sulphonamide resistance gene sul2, of which three carried dfrA14, and all three were phenotypically resistant to cotrimoxazole. One isolate that carried sul2 but not dfrA14 was also phenotypically resistant to co-trimoxazole. One isolate from a pig in Nairobi carried blaTEM-1B. The isolate was resistant to ampicillin and sensitive to ceftriaxone on phenotypic testing. One additional isolate showed phenotypic resistance to ampicillin but did not carry any known ampicillin resistance genes. We identified 9/121 (7.4%) isolates that carried the genes aph(3”)-1b and aph(6)-1d, both of which are associated with resistance to aminoglycosides; none of these isolates demonstrated phenotypic resistance against gentamicin. One isolate from a pig in Busia had a point mutation (D87Y) in gyrA, associated with resistance to fluoroquinolones and this was confirmed by testing the isolate against pefloxacin (Table 2).
S. Typhimurium ST313 analysis
Two S. Typhimurium ST313 were isolated from the MLN of individual pigs slaughtered in Nairobi (FD01844610 and FD01844641), and differed by 110 core gene SNPs. Neither isolate contained AMR genes, and both were shown to be antibiotic-susceptible phenotypically (Table 2). To determine the similarity between the two ST313 isolated here and S. Typhimurium ST313 currently causing an epidemic of iNTS in Africa, a core gene SNP-based phylogeny of the two genomes alongside published ST313 genomes was constructed (Fig 4). The resulting phylogeny showed that both isolates were related to a diverse group of isolates associated with human gastrointestinal disease in the United Kingdom, and most closely-related to the ST313 isolates U7 and U9, from the UK [36].

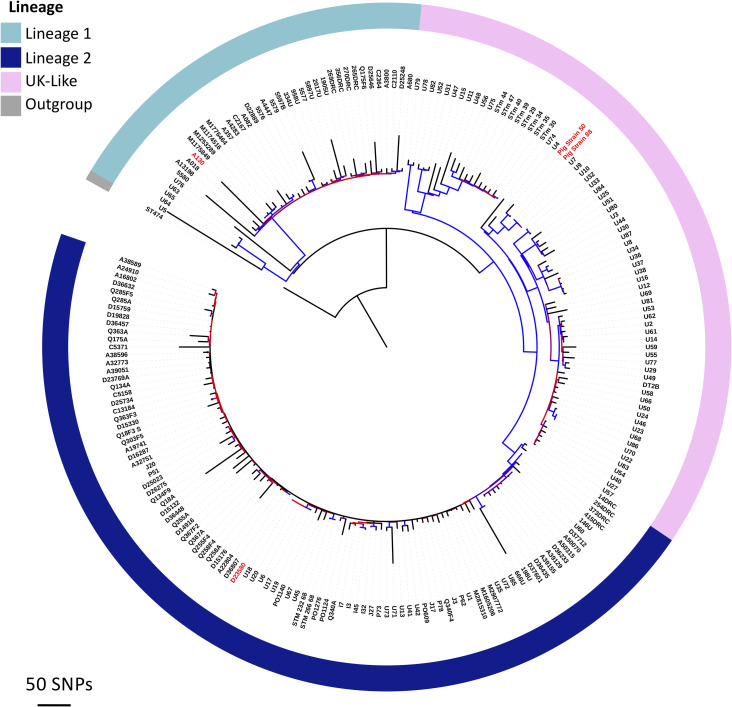
Two pig-derived S. Typhimurium ST313 isolates in the context of published ST313 genomes.
The A maximum likelihood phylogenetic tree based on core genome SNPs. Phylogeny is outgroup-rooted to S. Typhimurium ST19 strain 4/74 [57]. Visualised using ITOL (https://itol.embl.de).
Accessory genome analysis revealed that the two pig-derived S. Typhimurium ST313 shared a similar prophage and plasmid repertoire to other ST313 isolates responsible for gastrointestinal disease in England and Wales. Importantly, neither belonged to African S. Typhimurium ST313 Lineage 2, which is currently causing the epidemic of iNTS in sSA (Fig 5). In terms of the African ST313 lineage 2 prophages, the two pig-derived ST313 carry Gifsy-2, ST64-B, Gifsy-1 and BTP5, but lack BTP1. In relation to the African ST313 lineage 2-associated plasmids [50] the two pig-derived ST313 isolates carry pBT2 and pBT3, but pBT1 is absent. The two pig-derived ST313 isolates carried the large S. Typhimurium virulence plasmid (pSLT), which did not contain the 10kb MDR gene cassette characteristic of African Lineage 2 ST313.

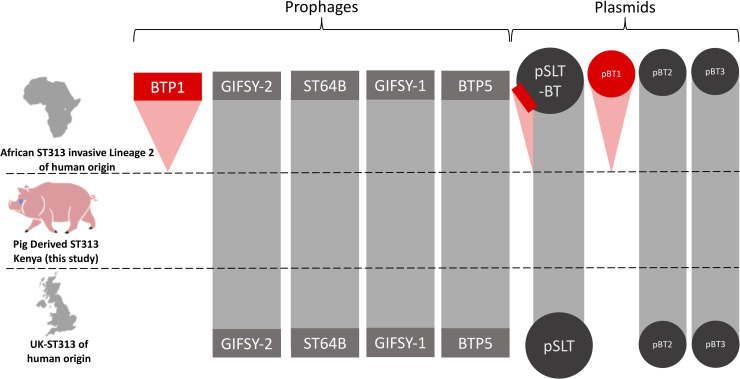
Comparison of the plasmid and prophage repertoires of S. Typhimurium ST313 variants.
The presence and absence of Salmonella prophages BTP1, Gifsy-2 ST64B, Gifsy-1 and BTP5 and Salmonella plasmids pSLT-BT, pBT1, pBT2 and pBT3 are shown on three variants of S. Typhimurium ST313. Grey indicates similarity above 95% to African ST313 lineage 2 reference genome D23580. Red indicates absence compared to African ST313 lineage 2 reference genome D23580. The red rectangle on plasmid pSLT-BT represents the multidrug resistance cassette which is present in African ST313 lineage 2, but absent from UK-ST313 and the pig-derived ST313 isolates FD01844610 and FD01844641.
Discussion
Our findings provide evidence of Salmonella carriage by pigs in Kenya and Malawi, and reveals the potential for pigs to act as a reservoir for human disease associated NTS serovars. We demonstrate that pigs in Kenya and Malawi carry and excrete a diverse range of Salmonella serovars to the environment, the majority of which (66%, 21/32) have previously caused gastroenteritis in Africa [39]. We found no evidence that pigs are a reservoir for the novel lineages of S. Typhimurium and S. Enteritidis particularly associated with invasive disease in sSA.
There is extensive knowledge of Salmonella in pigs in the USA and Europe [15–19], however, there is limited data from sSA [21]. As the African market for pork continues to grow, and because Salmonella is one of the most common causes of foodborne illness worldwide [37,51], increased knowledge of Salmonella excreted by pigs in sSA is required. In Europe and the USA, management strategies have been developed in pig farming to reduce the spread of this Salmonella, which have been most successful in Scandinavia [52]; locally relevant strategies are necessary for sSA.
Seven isolates of S. Typhimurium ST19 were detected from pigs slaughtered in Nairobi. Metadata indicates that three of the host pigs were reared within a five-kilometre radius of each other and differed by less than 5 core genome SNPs, raising the possibility that the ST19 strain had been transmitted between these pigs during rearing, transport or slaughter and could pose a threat to human health. The consequent opportunity for human exposure to zoonotic Salmonella at any of these stages raises serious public health concerns, and highlights the need for on-farm studies. There is clearly scope to better understand the transmission dynamics of such strains in situ.
Two isolates of particular relevance were S. Typhimurium ST313, a sequence type responsible for almost two thirds of iNTS cases in Malawi, and never previously found in food animals in Africa. Core gene phylogenetic analysis showed that the isolates from pigs in the Nairobi abattoir were related to a diverse group of ST313 isolates which currently cause gastrointestinal disease in humans in England and Wales [36] (Fig 4). Importantly from a public health perspective, the two pig-derived S. Typhimurium ST313 isolates were not closely related to lineages of ST313 associated with iNTS disease in Africa. We have not found that pigs are a reservoir for Salmonella strains which are strongly associated with Salmonella causing invasive disease in Africa, i.e. African ST313s.
S. Enteritids and S. Typhimurium are responsible for nearly 90% of all human NTS infections in sSA [4,53]. Within this study only a small number of S. Enteritidis and S. Typhimurium isolates have been detected. Therefore, only a limited number of the isolates detected are of critical importance to humans as a potential zoonoses.
The majority of pig-derived Salmonella isolates were susceptible to all antibiotics tested, and no isolates were classified as multidrug-resistant (resistant to three or more classes of antibiotics). One gyrA mutation was identified in a single pig-derived isolate indicating genotypic resistance to fluoroquinolones. Fluoroquinolone antibiotics are increasingly being used in African human clinics [54], but evidence from Nairobi suggests that fluoroquinolones are used less frequently to treat animals in the veterinary sector [55].
This study reports porcine reservoirs of zoonotic diarrhoeal-causing NTS serovars in sSA, and did not find evidence of pigs as a reservoir for lineages of ST313 associated with invasive disease. As Salmonella is one of the most common causes of foodborne illness worldwide [56] there is a need for coordinated national epidemiological surveillance programmes to monitor food borne pathogens in pork production in sSA, especially as this industry expands. Such information will facilitate the development of intervention strategies aimed at limiting the cases of human Salmonella disease linked to transmission of Salmonella spp. from pigs in sSA.
Acknowledgements
The support of Dr Laura Falzon, Erin Furmaga and Milicent Ojang, as well as the whole Zoonotic and Emerging Disease Group’s assistance with the field sample collection for this project was invaluable.
Microbiological analysis was carried out in the following laboratories; Directorate of Veterinary Services-International Livestock Research Institute (DVS-ILRI) Zoonoses Laboratory in Busia, the ILRI Laboratories in Nairobi and the Malawi-Liverpool Wellcome Trust Clinical Research Programme (MLW) in Blantyre, Malawi.
References
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
 Salmonella identified in pigs in Kenya and Malawi reveals the potential for zoonotic transmission in emerging pork markets
Salmonella identified in pigs in Kenya and Malawi reveals the potential for zoonotic transmission in emerging pork markets
