
- Altmetric
The orchestration of ever larger conformational changes is made possible by the development of increasingly complex foldamers. Aromatic sheets, a rare motif in synthetic foldamer structures, have been designed so as to form discrete stacks of intercalated aromatic strands through the self‐assembly of two identical subunits. Ion‐mobility ESI‐MS confirms the formation of compact dimers. X‐ray crystallography reveals the existence of two distinct conformational dimeric states that require large changes to interconvert. Molecular dynamics simulation validates the stability of the two conformations and the possibility of their interconversion.
Unexpected conformational changes involving a global concerted rearrangement of the structure have been evidenced in large multistranded aromatic sheets. Increasing foldamer size thus gives access to new and defined conformational trajectories.
In biopolymers, conformational changes of large amplitude or with long‐range effects are associated with important molecular functions. They mediate cooperative binding between distant sites, for example the dioxygen binding sites of hemoglobin tetramers; [1] they allow for the remote transfer of information, as in G protein‐coupled receptors; [2] and they give rise to distinct protein–protein associations induced by, for example, domain swapping. [3] There is obvious interest in mastering and implementing such processes in synthetic chemical systems and important progress is being made in this direction using foldamers’ inherent conformational equilibria, including conformational changes induced by ligands, light, or electrons. [4] These efforts mark a transition from the mere control of molecular structures to the orchestration of dynamics. [5] As improvements in foldamer design and synthesis give access to increasingly large and complex objects,[ 6 , 7 ] one may expect novel types of conformational dynamics to emerge. Herein, we report the unexpected discovery of a complex conformational change in a multi‐stranded aromatic‐sheet foldamer. While aromatic foldamers have been known as notoriously rigid folded molecules, [8] we find that increasing their size gives access to new types of deformation.
We have reported on the folding of aromatic sheets stabilized by interactions between π systems and by rigid turns such as TS (Figure 1 b) that hold aromatic groups at a distance suitable for face‐to‐face stacking. [9] Interest for these systems stemmed from the low occurrence of sheets among foldamer structures when compared to helices, and the apparent greater difficulty to design discrete sheets that do not further aggregate and precipitate. [10] Compound 1 (Figure 1 a) was prepared as a representative of earlier designs. Its structure was fully characterized in solution through a complete assignment of its NMR spectra (Figure 2 b, Figures S16–S20) and in the solid state (Figure 2 a) to establish that the strand curvature associated with the A units (Figure 1 c) gives rise to a head‐to‐head arrangement of stacked aromatics within a curved two‐stranded sheet. [9c] A marker of the stability of the sheet conformation is the slow rotation on the NMR timecale of the dimethyl‐para‐phenylenediamine rings of TS (Figure S22).

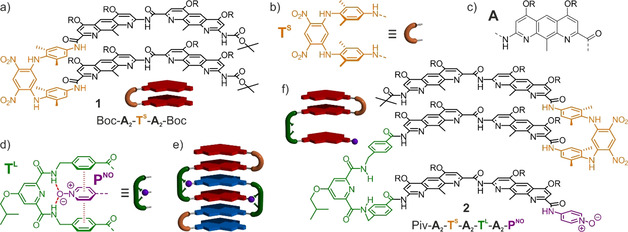
a) Chemical structure and schematic representation of two‐stranded aromatic sheet 1. b–d) Structures defining the letter code of TS, A, TL, and PNO, and association mode of the latter two (dashed lines depict hydrogen bonds while hashes show aromatic stacking). e) Schematic representation of a self‐assembled aromatic sheet. f) Structure and schematic representation of 2 (R=‐CH2CH(CH2CH3)2).

![a) Crystal structure of 1 (CCDC 2040478). b) Part of the 1H 400 MHz NMR spectrum of 1 in CDCl3 at 298 K. c) Part of the 1H 700 MHz NMR spectrum of 2 in [D8]toluene at 298 K. d) Collision cross section (DTCCSHe) of 22 measured by drift tube ion mobility ESI‐MS (black circles) in He and calculated from snapshots sampled from a molecular dynamics simulation (green bars). A molecular model of 22 with some fraying at one end (red arrow) is shown at the right. The corresponding calculated CCS is indicated in the graph, illustrating that the experimental and calculated CCS distribution of 22 are both narrow and indicative of a very compact conformation.](/dataresources/secured/content-1765948956412-59035533-911f-430b-8f6c-6bdd144cb4a9/assets/ANIE-60-2574-g002.jpg)
a) Crystal structure of 1 (CCDC 2040478). b) Part of the 1H 400 MHz NMR spectrum of 1 in CDCl3 at 298 K. c) Part of the 1H 700 MHz NMR spectrum of 2 in [D8]toluene at 298 K. d) Collision cross section (DTCCSHe) of 22 measured by drift tube ion mobility ESI‐MS (black circles) in He and calculated from snapshots sampled from a molecular dynamics simulation (green bars). A molecular model of 22 with some fraying at one end (red arrow) is shown at the right. The corresponding calculated CCS is indicated in the graph, illustrating that the experimental and calculated CCS distribution of 22 are both narrow and indicative of a very compact conformation.
As an extension of this work, we devised that turn TL (Figure 1 d) might promote the intercalation of an aromatic group and thus give access to unprecedented discrete self‐assembled aromatic sheets (Figure 1 e). Derivatives of N,N′‐dibenzyl 2,6‐pyridine‐dicarboxamide have indeed been shown to bind to aryl compounds, including within macrocyclic structures, through both hydrogen bonding and aromatic stacking. [11] Using a pyridine N‐oxide PNO as a hydrogen bond acceptor, we thus designed sequence 2 (Figure 1 f), comprised of both a short turn TS and a long turn TL. Based on energy‐minimized molecular models, 2 was expected to dimerize through a reciprocal intercalation.
The synthesis of 2 was carried out using classical aromatic amide coupling steps and is presented in the supporting information. Its NMR spectrum in [D8]toluene showed sharp resonances indicating the presence of well‐defined species (Figure 2 c). However, the number of peaks was larger than expected for a C 2 symmetrical dimer. In other nonpolar solvents, the lines tended to broaden, indicating faster dynamics, but the spectrum remained complex regardless of temperature (Figures S23–S28). Dilution experiments down to 0.54 mm [12] did not produce any change, suggesting that if an aggregate was present, its dissociation constant was below this value (Figure S21). The ESI mass spectrum clearly showed the formation of a 22 dimer. Furthermore, an ion mobility analysis [13] generated a collision cross‐section profile that matched well with the calculated profile of a compact interdigitated sheet conformation (Figure 2 d, Figure S38). However, these data alone did not allow to ascertain the very structure of 2 and reasons for NMR signal multiplicity.
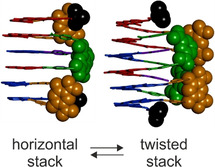
Essential evidence came from two crystal structures of 2, which validated the initial design but also uncovered unanticipated structural variations (Figure 3). Both structures showed the expected dimerization as depicted in Figure 1 e, in particular the reciprocal intercalation, the hydrogen bonding of PNO N‐oxide function of one strand to the TL amide NH of the other strand (d O–HN≈2.1 Å, Figure 3 e,j), and the formation of a face‐to‐face stack of six A2 segments. In addition, the structures also demonstrated two conformations, C1 (Figure 3 a–e, crystallized from CH2Cl2/n‐hexane) and C2 (Figure 3 f–j, crystallized from CHCl3/MeOH) that display considerable differences, as highlighted by dashed lines throughout Figure 3. Through a complex and large amplitude sliding of the various A2 segments with respect to one another (Figure 3 d,i), the TS, TL, and pivaloyl (piv) units undergo a large swing, while the overall architecture is preserved. Among notable differences between C1 and C2 is the fact that A2 segments are perfectly flat and stacked perpendicular to the structure main axis in C1 (Figure 3 c), whereas they are tilted and slightly M‐helically twisted in C2 (Figure 3 h). [14] The complex (Figure 2 c) or broad (Figure S23–S28) NMR spectra of 2 are consistent with the large conformational changes required to interconvert C1 and C2. Furthermore, one cannot exclude the existence of intermediate or alternate conformational states, for example a P‐helically twisted diastereomeric analogue of C2. [14]

![Top views (a,f), front views (b,g), side views (c,h), details of π–π stacking (d,i), and hydrogen bonding (e,j) of two crystal structures of 22 (CCDC 2040479 (C1) und 2040480 (C2)) demonstrating distinct conformers C1 (a–e) and C2 (f–j).
[14]
In (a–d) and (f–i), TS, TL, and the terminal tert‐butyl groups are shown as orange, green, and black space filling models, respectively. The rest of the molecules are shown in tube representation. The A units are shown in red or blue to distinguish the two molecules within each 22 dimer. Included solvent molecules, side chains, and hydrogen atoms (except amide NH in e and j) have been omitted.](/dataresources/secured/content-1765948956412-59035533-911f-430b-8f6c-6bdd144cb4a9/assets/ANIE-60-2574-g003.jpg)
Top views (a,f), front views (b,g), side views (c,h), details of π–π stacking (d,i), and hydrogen bonding (e,j) of two crystal structures of 22 (CCDC 2040479 (C1) und 2040480 (C2)) demonstrating distinct conformers C1 (a–e) and C2 (f–j). [14] In (a–d) and (f–i), TS, TL, and the terminal tert‐butyl groups are shown as orange, green, and black space filling models, respectively. The rest of the molecules are shown in tube representation. The A units are shown in red or blue to distinguish the two molecules within each 22 dimer. Included solvent molecules, side chains, and hydrogen atoms (except amide NH in e and j) have been omitted.
We endeavored to investigate C1 and C2 using molecular dynamics simulations (MD) to assess their stability (Figures S29–S37). Energy minimization of the C1 and C2 structures from the crystal data did not give rise to notable changes and produced the starting coordinates for 10 ns MD runs performed every 100 degrees from 200 up to 900 K. Several parameters were systematically monitored over time: the hydrogen bond distances at each NO−TL contact, as well as the TS–TS, TL–TL, and piv–piv distances (Figure S29). Up to 400 K both C1 and C2 were found to be stable (Figure 4 a, Figures S30, S31a,b, movies S1, S2). Intermolecular hydrogen bonds did not disrupt and limited fluctuations allowed for the discrimination of two distinct C1 and C2 conformational ensembles. As can also be seen in the crystal structures (Figure 3), C2 is characterized by a TS–TS distance about half the piv–piv distance, whereas these two parameters are comparable in C1. These features are well reproduced by MD. Perhaps the most characteristic difference between the two is the relative positioning of the TL units: the one that is “above” the other in C1, is “below” in C2 (Figure 3 b vs. g). However, the TL–TL distance alone does not discriminate between these two states.

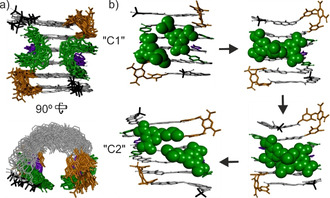
a) Overlay of ten snapshots of the 10 ns MD simulation of C1 at 400 K showing limited fluctuations at this temperature. b) Snapshots of a C1→C2 transition during the MD simulation of C1 at 500 K. Side chains and hydrogen atoms have been omitted except the isobutoxy side chains of TL in (b).
MD runs performed at higher temperatures (500–900 K) led to larger fluctuations. At the highest temperatures, disruption of the hydrogen bonds and local unfolding occurred frequently yet the structures remained stable. During these runs the fluctuations were large enough to create junctions between the C1 and C2 conformational spaces, and some C1→C2 or C2→C1 transitions occurred (Figure 4 b, Figures S31c,d, S32b, movie S3). All the parameters that discriminate C1 and C2 then changed in a concerted manner, in particular the piv–piv and TS–TS distances. Overall, the MD simulations are suggestive of sharp transition between C1 and C2, without dissociation, supporting the existence of (at least) two distinct ensembles of conformations. The C1↔C2 transition entails that the two TL units pass each other. During this process, the isobutoxy side chains that they carry (Figure 4 b) undergo extensive contacts which may then constitute a barrier. For future developments, one may take advantage of the proximity between these side chains to install functionalities conducive to attractive or repulsive forces, which would stabilize one or the other conformer. A step further would be to introduce functionalities that would allow for the controlled switching between the two. The concept of aromatic sheet self‐assembly may also be extended using turn units that do not allow for the intercalation of just one arene, such as TL, but of two or more. [15] Furthermore, the original half‐pipe architecture may be exploited for the purpose of molecular recognition, particularly with regards to the fact that functional groups in position 9 of the A monomers (here only methyl groups) all converge towards the interior of the cavity.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by the European Research Council under the European Union's Seventh Framework Programme (grant agreement no. ERC‐2012‐AdG‐320892). It benefited from the facilities and expertise of the Biophysical and Structural Chemistry platform at IECB, CNRS UMS3033, INSERM US001, Bordeaux University, France. We thank Dr. L. Sebaoun for preliminary experiments. Open access funding enabled and organized by Projekt DEAL.
References
2
4
4b
4d
4e
4g
4g
4h
4i
4j
4k
4m
4m
4n
4n
4o
5
6
6b
6b
6c
6c
6e
6e
7
7b
7b
7d
7f
7g
7i
7j
8
8a
9
9a
9c
9d
10
10a
10b
10c
10f
11
11a
11b
11c
12
14
15
15a
15a
 Large‐Amplitude Conformational Changes in Self‐Assembled Multi‐Stranded Aromatic Sheets
Large‐Amplitude Conformational Changes in Self‐Assembled Multi‐Stranded Aromatic Sheets
