
- Altmetric
Regulatory switches are wide spread in many biological systems. Uniquely among them, the switch of the bacterial flagellar motor is not an on/off switch but rather controls the motor’s direction of rotation in response to binding of the signaling protein CheY. Despite its extensive study, the molecular mechanism underlying this switch has remained largely unclear. Here, we resolved the functions of each of the three CheY‐binding sites at the switch in E. coli, as well as their different dependencies on phosphorylation and acetylation of CheY. Based on this, we propose that CheY motor switching activity is potentiated upon binding to the first site. Binding of potentiated CheY to the second site produces unstable switching and at the same time enables CheY binding to the third site, an event that stabilizes the switched state. Thereby, this mechanism exemplifies a unique combination of tight motor regulation with inherent switching flexibility.
A three‐step mechanism of CheY binding to the E. coli flagellar motor switches bacterial swimming behavior from swimming to tumbling to enable chemotaxis.
Introduction
Switches are vastly known throughout the field of biology, from transcription and expression of genes to controlling processes of signal transduction, cell fate and cell cycle, to mention a few (Cross et al, 2002; Laslo et al, 2006; Pomerening, 2008). Most of these switches turn processes on and off. An exception is the switch of the bacterial flagellar motor, which controls the motor’s direction of rotation rather than an on/off process (Eisenbach & Caplan, 1998). This dissimilarity combined with this switch’s unique properties—controlling a mechanical rather than a chemical process and being exceptionally ultrasensitive with respect to the switching signal (see below) (Cluzel et al, 2000), made it a challenging system of investigation. Indeed, in spite of decades of studies, the molecular mechanism underlying switching of the bacterial flagellar motor has remained obscure.
Switching of the motor enables bacterial cells to navigate. In bacteria like Escherichia coli, each cell contains multiple flagellar motors. When they rotate counterclockwise, the cell swims in a rather straight line, termed a “run”. When a considerable fraction of flagella switch from the default direction of rotation, counterclockwise, to clockwise, the cell preforms a chaotic‐like turning motion, termed a “tumble” (Berg & Brown, 1972; Turner et al, 2000) (Fig 1A), as a result of which the subsequent run (when the rotation switches back to counterclockwise) is in a randomly new direction. Conversely, very brief switching of some of the motors to clockwise generates slight changes in swimming direction without randomization rather than tumbles (Turner et al, 2000), thus maintaining directional swimming persistence (Vladimirov et al, 2010; Saragosti et al, 2011). This behavior was proposed to markedly improve the performance of collective migration (Saragosti et al, 2011), implying an evolutionary advantage. Maintenance of directional persistence requires extremely short intervals of clockwise rotation. However, it is unclear how the motor is regulated to produce both long clockwise intervals for tumbling and short intervals for directional persistence.

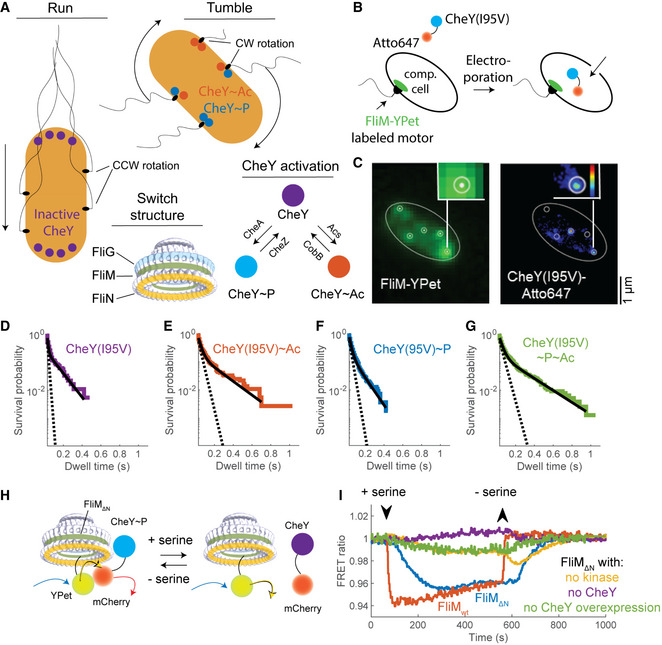
Two modes of CheY binding to the switch
ARun‐and‐tumble swimming modes in E. coli are regulated by CheY activation by phosphorylation or acetylation. The electron density map of the switch was produced by Thomas et al (2006) and downloaded from http://dx.doi.org/10.1093/nar/gkv1126.
BScheme demonstrating the internalization of Atto647‐labeled CheY(I95V) to single cells by electroporation.
CDemonstration of single‐molecule tracking in a single cell. Cell outline is shown as an ellipse. Left, Imaged FliM‐YPet fluorescence. White dots indicate the estimated motor locations. White circles illustrate a 75 nm radius around these locations. Right, Contour map of the normalized sum of probabilities of the localization events of CheY(I95V)‐Atto647 (from blue to red is low to high; black is zero probability). White circles show the motors’ locations from the left panel.
DSurvival probabilities of unmodified CheY(I95V)‐Atto647 at the switch (ΔcheA background; strain EW668). 63 cells were recorded with a total of 1,316 trajectories in which CheY was found to interact with FliM. Note the logarithmic scale of the ordinate. Black line, a bi‐exponential fit; dashed line, a fit of the fast decline process (see text for details).
EAs in (D) for acetylated CheY(I95V)‐Atto647 (presence of acetate, 50 mM, pH 7.0; strain EW668). 56 cells were recorded with a total of 1,904 trajectories.
FAs in (D) for phosphorylated CheY(I95V)‐Atto647 (ΔcheZ background; strain EW669). 82 cells were recorded with a total of 2,414 trajectories.
GAs in (D) for phosphorylated and acetylated CheY(I95V)‐Atto647 (presence of acetate, 50 mM, pH 7.0; ΔcheZ background, strain EW669). 40 cells were recorded with a total of 2660 trajectories.
HExperimental scheme of the FRET experiment. The attractant serine lowers the phosphorylation level of CheY. As a result, CheY dissociates from the switch and the energy transfer from YPet (conjugated to FliM) to mCherry (conjugated to CheY) is reduced.
ICheY interacts with FliM∆N and this interaction is sensitive to chemotactic stimuli. Each curve is the mean of two FRET measurements of cells in response to an attractant stimulus (0.1 mM serine; in the case of FliMwt, a single measurement was performed with serine; another measurement of FliMwt with 1mM aspartate produced similar results). See Appendix Fig S1 for the details of FRET analysis. FRET ratio is the ratio of mCherry to YPet fluorescence. CheY‐mCherry and mCherry concentrations were ~170 µM in the case of FliM∆N and ~15 µM in the case of FliMwt (Appendix Fig S2 for calibration of CheY concentration). Strains used: EW677 (FliMwt, red), EW659 (negative control of FliM∆N without CheY, purple), EW637 (FliM∆N, blue), and EW636 (FliM∆N ΔcheA, yellow).
While the mechanism of switching is not resolved, much is known about the components of the switching machinery and the interactions between them. The switch of the flagellar motor is a large complex at the motor’s base, consisting of multiple copies of the proteins FliM, FliN, and FliG (Fig 1A). Since chemotaxis of bacteria is achieved by modulating the direction of flagellar rotation, the main control target in chemotaxis is the switch. The switch shifts the direction of rotation from counterclockwise to clockwise in response to binding the signaling protein CheY, which shuttles back and forth between the chemotaxis receptor complex and the flagellar switch. Earlier studies of the corresponding author’s group revealed that phosphorylated CheY (CheY~P) mainly binds to the switch at the N terminus of FliM (FliMN) (Welch et al, 1993; Bren & Eisenbach, 1998). Subsequently, Blair’s group reported on two additional sites with weaker binding of CheY (termed hereafter “low‐affinity sites”), one at FliN, to which the binding is FliMN‐dependent and requires that CheY would be phosphorylated (Sarkar et al, 2010), and one at FliM at other location than FliMN (Mathews et al, 1998). On the basis of this evidence combined with mutational analysis and a structural model, this group further suggested that the interaction of CheY~P with FliMN serves to capture CheY~P and that switching to clockwise rotation involves the subsequent interaction of CheY~P with FliN (Sarkar et al, 2010). NMR analysis in Thermotoga maritima by Dahlquist’s group identified the middle domain of FliM (FliMM) as a low‐affinity binding site for CheY (Dyer et al, 2009). In view of this information, it is reasonable to assume that, also in E. coli, the other binding site is FliMM. (For simplicity, we will term hereafter this other site in E. coli FliMM even though its exact location is obscure.) It is not yet known how CheY binding to each of these sites affects the process of clockwise generation.
CheY is bound to receptor clusters at the cell’s poles. It is well established that it has to be activated by phosphorylation for switching the motor to clockwise. This activation results in CheY~P dissociation from the poles and, as mentioned above, in binding to FliMN. The level of CheY phosphorylation is regulated by CheA and CheZ as specific kinase and phosphatase, respectively (Fig 1A). A receptor‐mediated attractant response (or removal of a repellent) inhibits CheA activity; stimulation by repellents (or attractant removal) enhances its activity [for reviews—(Berg, 2003; Eisenbach, 2004; Terashima et al, 2008; Porter et al, 2011)]. Another covalent modification that activates CheY to generate clockwise rotation is lysine acetylation (Wolfe et al, 1988; Barak et al, 1992, 1998). The regulation of acetylation is known to involve Acs and CobB as acetyl‐transferase and deacetylase, respectively (Fig 1A) (Barak et al, 2004; Li et al, 2010). It has been shown that CheY acetylation is involved in bacterial chemotaxis (Barak & Eisenbach, 2001) and that it is inversely affected by CheA and CheZ (Barak & Eisenbach, 2004). Yet, the role that acetylated CheY (CheY~Ac) plays in chemotaxis is still obscure.
While the dependence of clockwise generation on the intracellular concentration of active CheY is highly cooperative, meaning that the motor is ultrasensitive (Cluzel et al, 2000), binding assays between CheY and the switch, carried out both in vivo and in vitro, found that the binding is non‐cooperative (Sourjik & Berg, 2002; Sagi et al, 2003). Subsequent studies, which employed a constitutively active CheY mutant protein, found that it binds better to clockwise‐rotating motors than to counterclockwise‐rotating motors (Fukuoka et al, 2014). This difference in binding may well result in cooperativity of binding (Duke et al, 2001).
Here, we addressed the question of the mechanism underlying the switch function. We demonstrate that delicate, hitherto unknown, steps of the switching mechanism are resolved when FliMN is truncated from the switch. We bring evidence for three sequential steps of CheY binding to distinct sites at the switch, each with a different outcome. Binding to the first site (FliMN) potentiates CheY at the switch. Binding of potentiated CheY to the second site (FliN) is short‐lived and generates transient motor switching. This seems to enable firm CheY binding to the third site (FliMM), with a resultant stabilization of the switched state.
Results
Two dwelling modes of CheY at the switch; phosphorylation affects one mode, acetylation affects both
Following the findings of Fukuoka et al (2014), mentioned just above, that constitutively active CheY binds differently to counterclockwise‐ and clockwise‐rotating motors, we investigated whether two modes of binding can also be observed with CheY activated by phosphorylation and acetylation. To this end, we measured in vivo the dwell time of single CheY molecules at motors whose FliM molecules were labeled with YPet. We compared between phosphorylating conditions, acetylating conditions, and conditions under which CheY was both phosphorylated and acetylated. We electroporated CheY(I95V) molecules labeled with a maleimide modification of the photo‐stable organic dye Atto647 into FliM‐YPet expressing cells (Fig 1B) (Di Paolo et al, 2016). The experiments were carried out in the following settings: in a ΔcheZ background to make CheY fully phosphorylated, in a ΔcheA background to make CheY non‐phosphorylated, and, in each of these settings, also in the presence of the acetyl donor acetate to make CheY acetylated (Barak et al, 1992, 2004). The cheY(I95V) mutation was designed to increase CheY affinity for FliMN (Schuster et al, 2000), thus enhancing sampling of otherwise rare binding events. Custom‐written software tracked CheY(I95V)‐Atto647 molecules and estimated switch locations in each cell using FliM‐YPet fluorescence images. We interpreted CheY(I95V)‐Atto647 molecules dwelling within 75 nm of a switch for >30 ms as binding to the motor (Fig 1C; see Materials and Methods). We observed CheY(I95V)‐Atto647 molecules dwelling at the motor both in the absence and presence of acetate. However, in the latter case, the dwell time was markedly longer (Fig 1D vs. E and Fig 1F vs. G for the survival probability, i.e., for the probability to remain bound to the switch; Movie EV1). Notably, the very long dwell events in Movie EV1 were only detected in the presence of acetate.
The survival distributions in all experiments appeared to be biphasic, i.e., each of them comprised two exponentially decaying distributions, fast and slow (note the logarithmic scale of the ordinates in Fig 1D–G). A biphasic distribution is indicative of two modes of CheY dissociation from the switch, fast and slow. These two modes can either reflect CheY binding to two different sites at the switch or to two different states of the same site.
To quantify the effect of phosphorylation and acetylation on each mode, we fitted each of the distributions in Fig 1D–G with a bi‐exponential expression [, with pre‐exponential factors A 1 > A 2, marking the fraction of each mode in the overall distribution, and k 1, k 2 being the rate constants of the modes; t is time]. From the fitted rate constants (and their inverse, the average dwell time of CheY at the switch), we could learn how phosphorylation, acetylation, or both modifications combined, affected each mode of CheY binding. Unmodified CheY exhibited the fastest decline rate in both modes (k 1 and k 2 being 99.5 and 9.7 s−1, and expected average dwell times being about 10 and 100 ms, respectively; Fig 1D). Phosphorylation markedly decreased the rate of the fast mode, but did not affect the slow mode (k 1 and k 2 being 45.3 and 11.1 s−1, and expected average dwell times being about 22 and 90 ms, respectively; Fig 1F). Acetylation alone decreased the decline rate of both modes (k 1 and k 2 being 32.8 and 5.6 s−1, and typical dwell times being about 30 and 180 ms, respectively; Fig 1E). Phosphorylation and acetylation combined yielded values comparable to acetylation alone (k 1 and k 2 being 28.7 and 4.9 s−1, and typical dwell times being about 35 and 200 ms, respectively; Fig 1G). Hence, it seems that phosphorylation mostly affects the fast mode, i.e., the mode with relatively short dwell times at the motor, whereas acetylation likely affects both modes.
CheY binds to the switch even in the absence of FliMN
To study the association of both binding modes with clockwise generation, we sought to read the direct output of these modes. We suspected that the high‐affinity binding of CheY to FliMN might mask fine outputs related to these binding modes. Therefore, we studied the binding and functional interaction of CheY with motors in which FliM had been truncated to remove FliMN (termed hereafter FliM∆N), i.e., with motors that only contained the low‐affinity binding sites of CheY. To examine whether CheY can at all bind to such motors, we employed in vivo Förster resonance energy transfer (FRET), measuring the interaction between overexpressed CheY‐mCherry and FliM∆N‐YPet‐labeled motors (Fig 1H for an explanatory scheme). First, to validate our FRET approach, we measured the response of non‐truncated FliM (termed hereafter FliMwt)‐YPet motors in cells that expressed CheY‐mCherry to approximately the endogenic CheY expression level (due to leaky promoter expression; no inducer added). With these cells, the addition and subsequent removal of the attractant serine caused reduction and enhancement of the FRET signal, respectively, thus validating the method (Fig 1I, red curve). To determine whether CheY binds to FliM∆N (within the switch or soluble in the cytoplasm), we repeated the experiment with FliM∆N‐YPet in cells expressing CheY‐mCherry, using ΔcheZ background to ensure that CheY‐mCherry was mostly phosphorylated. We observed a very weak, hardly detectable FRET response when the intracellular CheY‐mCherry concentration was comparable to the endogenic CheY expression level (Fig 1I, green). However, when we overexpressed CheY‐mCherry (inducer present), we observed a response similar in amplitude to that of FliMwt (Fig 1I, blue). This difference between cells containing FliMwt motors and cells containing FliM∆N motors was expected due to the absence of the high‐affinity binding site from FliM∆N motors. These results imply a low‐affinity interaction of CheY~P with FliM∆N. When, as a negative control, we overexpressed mCherry instead of CheY‐mCherry, we observed no response (Fig 1I, purple). This implies that the FRET responses, observed with CheY‐mCherry, did not emerge from an interaction between the fluorescent proteins themselves. The response of FliM∆N cells overexpressing CheY‐mCherry~P was slower than that of FliMwt cells (Fig 1I). This was likely because of the longer time required to phosphorylate and dephosphorylate such high CheY concentrations and because of the possible impairment of the on‐rate of CheY~P binding to the switch by the absence of FliMN. The response of CheY‐mCherry under conditions that do not allow its phosphorylation (ΔcheA background, i.e., the kinase is missing and the phosphatase is present) was minor (Fig 1I, yellow). This response possibly reflected the alternative pathway for chemotaxis, demonstrated in E. coli cells lacking most of the chemotaxis machinery but overexpressing CheY (Barak & Eisenbach, 1999).
To distinguish between CheY‐mCherry binding to the switch and binding to free FliM∆N‐YPet molecules within the cytoplasm, we attempted to measure CheY binding to individual motors, employing FRET photobleaching (Appendix Fig S3A and B). As detailed in Appendix 1, we indeed found an increased CheY~P binding at FliM∆N‐YPet spots (presumably spots of individual motors) when the CheY‐mCherry expression level was elevated (Appendix Fig S3C). However, as the CheY‐mCherry concentration increased, the fluorescence of the spots became diffusive. This avoided conclusive differentiation between switch‐originated and cytoplasm‐originated signals (Appendix 1).
To verify that CheY~P can, indeed, bind to FliM∆N motors, we examined whether it can generate clockwise rotation of such motors, relying on the fact that, for generating clockwise rotation, CheY~P must first bind to the motor. We tethered cells containing FliM∆N motors (FliM∆N‐YPet motors in some of the experiments) and~100 µM CheY in a ΔcheZ background (to ensure phosphorylation of CheY) to glass via their flagella (Fig 2A for experimental scheme and Fig 2B for a demonstration of a tethered cell and for a representative trace of the rotation rate) and analyzed their direction of rotation with an automated home‐made software. The mere overexpression of CheY was enough to produce clockwise rotation (Fig 2C prior to stimulation), indicating that CheY can bind to FliM∆N motors to generate clockwise rotation. The cells responded to positive stimuli (attractant addition or repellent removal) with reduced clockwise rotation (Fig 2C). Because positive stimuli work by lowering the phosphorylation level of CheY (Borkovich et al, 1989), the observed response implies that CheY had to be phosphorylated for binding to these FliM∆N motors. It also suggests that, as in cells containing FliMwt motors (Sourjik & Berg, 2002), lowering the level of CheY~P results in lowering the clockwise level. Repellent stimulation, known to work by elevating the phosphorylation level of CheY (Sourjik & Berg, 2002), had hardly any effect (Fig 2D). This is because the absence of CheZ caused CheY to be fully phosphorylated already prior to the repellent stimulation. Thus, CheY~P functionally binds to FliM∆N motors.

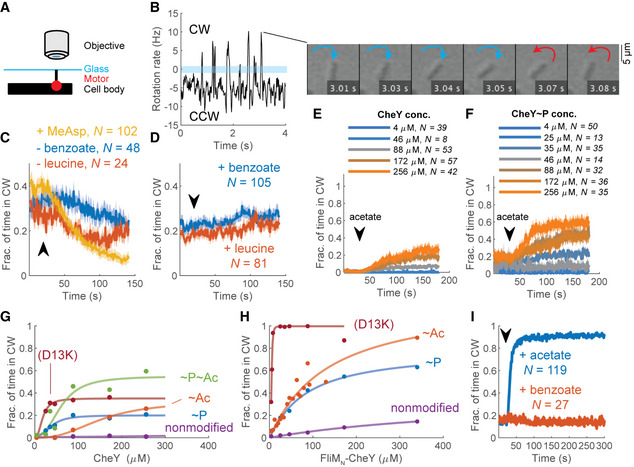
FliM∆N motors respond differently to various modifications of CheY and FliMN‐CheY
AExperimental scheme of flagellar motor tethering. The stub of sheared flagellum is tethered to glass by an anti‐flagellin antibody (Silverman & Simon, 1974) in a flow chamber (Berg & Block, 1984).
BDemonstration of rotation rate measured from a single cell; negative and positive values (below and above the blue bar) are counterclockwise and clockwise, respectively. The switch from negative to positive rotation rate at 3 s is shown in the montage to the right.
CThe response of tethered fliM ∆N ΔcheZ cells (strain EW635), induced for CheY expression from a plasmid (200 µM IPTG), to positive stimuli. Lines and shaded regions are the mean time spent in clockwise rotation ± SEM. The arrow marks the estimated time at which the stimulant arrived to, or left (as indicated), the flow chamber. N is the number of cells. α‐Methyl‐DL‐aspartate (MeAsp) and leucine were used at 1 mM, benzoate (pH 7.0) at 50 mM.
DAs in (C) for negative stimuli.
EResponse of tethered fliM ∆N ΔcheA cells (strain EW634) to acetate (50 mM, pH 7.0) at different CheY concentrations. Lines and shaded regions are mean ± SEM.
FResponse of tethered fliM ∆N ΔcheZ cells (strain EW635) to acetate. Details as in (E).
GContribution of acetylation and phosphorylation of CheY as well as its D13K mutation to clockwise rotation. The points are the mean clockwise rotation calculated from panels E and F at time segments 0–20 s and 160–180 s. The variants shown are fliM∆N ΔcheZ cells expressing CheY in the absence and presence of acetate (10 mM, pH 7.0) (i.e., CheY~P and CheY~P~Ac; blue and green curves; strain EW635), fliM∆N ΔcheA cells expressing CheY, in the absence and presence of acetate (CheY and CheY~Ac; purple and red, respectively; EW634), and fliM∆N ΔcheA cells expressing CheY(D13K) (burgundy; EW737). Each data point is the average of all experiments at a given CheY concentration, weighted by the sample number of each experiment. For data, see Table EV1. The CheY concentrations shown are estimates based on the calibration curves in Appendix Fig S2, for which a similar CheY expression system was used.
HFliMN further activates CheY variants to produce clockwise rotation. The variants shown are fliM∆N ΔcheZ cells expressing FliMN‐CheY (i.e., FliMN‐CheY~P; blue curve; strain EW697), fliM∆N ΔcheA cells expressing FliMN‐CheY, in the absence and presence of acetate (10 mM, pH 7.0) (FliMN‐CheY and FliMN‐CheY~Ac; purple and red, respectively; EW696), and fliM∆N ΔcheA cells expressing FliMN‐CheY(D13K) (burgundy; EW739). See G for other details.
IResponse of fliM∆N ΔcheA cells expressing FliMN‐CheY (strain EW696) to acetate and benzoate (10 mM each; pH 7.0). FliMN‐CheY expression was induced with 800 μM IPTG. Lines and shaded regions are the mean time spent in clockwise rotation ± SEM. The black arrow indicates the estimated time point at which the stimulus entered the flow chamber.
FliM∆N motors respond differently to CheY~P and CheY~Ac
The observed different effects of phosphorylation and acetylation on the modes of CheY binding to FliMwt motors (Fig 1D–G) raised the possibility that CheY~P and CheY~Ac generate clockwise rotation by somewhat different mechanisms. To examine this possibility, we compared clockwise generation by CheY, CheY~P, CheY~Ac, and CheY~P~Ac in cells containing FliM∆N motors. Studying FliM∆N motors is advantageous in this case because it avoids complications caused by differences in binding—CheY~P binds well to FliMN (McEvoy et al, 1999) whereas CheY~Ac does not (Liarzi et al, 2010; Li et al, 2010). To assess the contribution of non‐phosphorylated CheY to clockwise generation, we measured the clockwise level of cells that lacked the kinase of CheY (ΔcheA cells); for CheY~P, we used cells that lacked the phosphatase of CheY (ΔcheZ cells); for CheY~Ac, we used ΔcheA cells supplied with the acetyl donor acetate (Appendix Fig S4 for a control of FliM∆N cells with intact chemotactic machinery, demonstrating that acetate acted as an acetyl donor rather than as a repellent); and for CheY~P~Ac, we employed ΔcheZ cells supplied with acetate. To ascertain that the effects are not limited by CheY availability, we measured each of these CheY forms at increasing CheY concentrations that were far beyond the endogenous concentration of chromosomally expressed CheY [estimated at ~10 μM (Li & Hazelbauer, 2004)]. While nonmodified CheY did not generate clockwise rotation (Fig 2E prior to acetate addition; Fig 2G, purple), CheY~P did (Fig 2F prior to acetate addition; Fig 2G, blue). Notably, the dependence of clockwise rotation on the CheY~P concentration had the shape of a saturation curve (Fig 2G, blue), unlike the ultrasensitive (sigmoidal, cooperative‐like) dependence observed in FliMwt motors (Cluzel et al, 2000). This suggests that the truncation of FliMN resulted in loss of ultrasensitivity. Also, the concentration‐dependence curve of clockwise rotation on CheY~P (Fig 2G, blue) further endorses the conclusion, made above, that the lack of a repellent response in Fig 2D was due to CheY being already fully phosphorylated rather than to a limiting CheY concentration.
The observation that CheY had to be activated by phosphorylation for generating clockwise rotation suggests that this clockwise response to CheY was due to specific interactions of CheY with the switch rather than to non‐specific interactions within the switch, promoted by overexpressed CheY. To validate this conclusion and to confirm that the observed clockwise response was neither limited by CheY availability nor by its activation level, we employed CheY(D13K). The latter is a constitutively active mutant of CheY, which can be hardly phosphorylated (Bourret et al, 1990). CheY(D13K), too, generated clockwise rotation (Fig 2G, burgundy), endorsing the conclusion that the generation of clockwise rotation by CheY was due to its specific interaction with the switch. Moreover, CheY(D13K) performed better than CheY~P in generating clockwise rotation, implying that the D13K mutation activates CheY better than does phosphorylation. The reason for this higher activity is unclear as, to the best of our knowledge, the mechanism of CheY(D13K) activation is not yet resolved. Nevertheless, as in the case of CheY~P, clockwise generation by CheY(D13K) saturated at intermediate clockwise levels, confirming that clockwise generation was not limited by CheY availability. Thus, at this point we conclude that FliMΔN motors differ from FliMwt motors in the sense that their response to CheY activated by phosphorylation or by D13K mutation is not ultrasensitive.
CheY~Ac also generated clockwise rotation (Fig 2E following acetate addition) but the dependence of clockwise generation on the CheY~Ac concentration was sigmoidal (cooperative‐like), reaching saturation at high concentrations (Fig 2G, red). Clockwise generation by CheY~P~Ac (Fig 2F) was roughly the sum of the individual contributions of CheY~P and CheY~Ac to clockwise generation (Fig 2G, green). Thus, consistent with the finding that phosphorylation and acetylation bring about different effects on CheY binding to FliMwt motors (Fig 1D–G), it seems that CheY~P and CheY~Ac differently generate clockwise rotation in FliM∆N motors (Fig 2G).
FliMN fusion to CheY compensates for FliMN truncation from the motor
The finding that FliMN is not essential for CheY binding to the switch and for clockwise generation raised the question of what its role is. FliMN promotes the interaction of CheY with the switch because, in its absence, CheY must be overexpressed for clockwise generation [Fig 2G; the chromosomally expressed CheY concentration is~10 μM (Li & Hazelbauer, 2004)]. Therefore, a reasonable possibility, suggested earlier (Dyer et al, 2009; Sarkar et al, 2010), is that FliMN tethers CheY to the switch, thereby elevating the local concentration of CheY at the low‐affinity sites. According to this possibility, FliMN acts as a “fishing line”, increasing the rate of CheY association with the clockwise‐generating low‐affinity sites at the switch. However, this might not be the only function of FliMN. The observation that CheY, activated by phosphorylation, acetylation, or D13K mutation, saturated much before reaching maximal clockwise rotation (Fig 2G), suggests that activated CheY is insufficiently potent to produce full clockwise rotation in FliMΔN motors and that FliMN is required for higher potency of CheY. It has been found that FliMN‐bound CheY adopts an intermediate conformation between the active and inactive states (Dyer & Dahlquist, 2006) and enhances the rate of CheY phosphorylation by small phosphodonors (Schuster et al, 2001). This begs the question of whether these observed features serve to increase CheY’s potency to generate clockwise rotation when it is tethered to FliMN. To examine this possibility, we studied the motor’s behavior in a FliM∆N strain that expresses CheY fused to FliMN (FliMN–CheY) from a plasmid prepared and employed earlier by Sarkar et al (2010). We compared it with the motor’s behavior of the same strain that expresses wild‐type CheY instead of FliMN–CheY. The expression of both proteins was induced in parallel and under identical conditions to different levels by an IPTG‐inducible promoter. We made this comparison in ΔcheZ and ΔcheA backgrounds, elevating and lowering CheY phosphorylation, respectively. In a ΔcheZ background, cells containing FliMN–CheY~P spent much more time in clockwise rotation than cells containing CheY~P (compare blue curves in Fig 2H and G). The observation that the half‐saturation concentrations (indicative of Kd) of these curves were similar, further suggests that the potentiation of CheY~P by FliMN to generate more clockwise rotation is not due to stronger binding of FliMN‐CheY~P to the switch. Nevertheless, the saturation point of FliMN‐CheY~P was about threefold higher than that of CheY~P. It, thus, appears that when FliMN is fused with CheY~P, the latter is more likely to be found in a conformation that is potent for clockwise generation. (We use here the term “potent” to distinguish from “active”, which denotes CheY activated by phosphorylation, acetylation, or mutation.) In a ΔcheA background, cells expressing FliMN–CheY could generate low levels of clockwise rotation whereas cells expressing wild‐type CheY could not produce clockwise rotation (purple curve in Fig 2H vs. G). This implies that CheY can acquire a potent clockwise‐generating conformation as a result of binding to FliMN.
To substantiate the conclusion that FliMN potentiates CheY independently of phosphorylation, we studied the dose‐dependent clockwise‐generating activity of FliMN‐CheY(D13K) under non‐phosphorylating conditions, i.e., in a ΔcheA background. The FliMN fusion extremely increased the clockwise generation potency of CheY(D13K) (burgundy curve in Fig 2H vs. G), so much so that clockwise rotation could be generated just by the leaky expression of the protein from the plasmid (levels that were roughly equivalent to endogenous CheY expression levels). This effect at so low concentrations suggests that CheY(D13K) binding to the switch was specific. Thus, FliMN fusion to CheY(D13K) substantially compensated for the FliMN deletion from the switch. This strongly supports the notion that FliMN functions to promote switching in a phosphorylation‐independent manner.
Switching of FliM∆N motors involves two phases of distinct interval lengths
Acetylation, mediated by saturating concentrations of acetate, greatly enhanced clockwise rotation of FliMN‐CheY in FliM∆N motors (red curve in Fig 2H vs. G). This clockwise‐generating effect of acetate was acetylation specific as benzoate, which acts as a repellent by the same mechanism as does acetate (Kihara & Macnab, 1981; Repaske & Adler, 1981) but does not serve as an acetyl donor (Barak et al, 1992, 2004), had no effect on the clockwise rotation (Fig 2I). In batches in which we measured the dependence of clockwise rotation on FliMN–CheY~Ac concentration at high resolution, we noticed that the dependence was biphasic, i.e., it comprised two consecutive saturation‐like curves (Fig 3A). To the best of our knowledge, two phases of motor response to CheY have not been hitherto described.

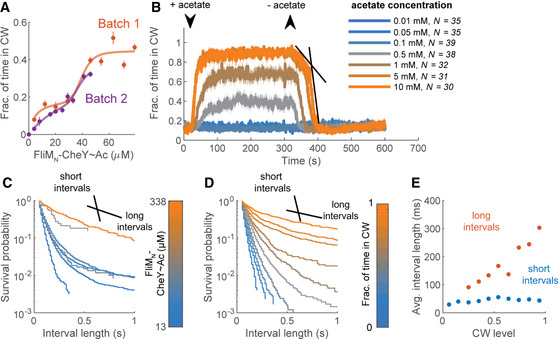
Biphasic switching of FliM∆N motors
ADependence of clockwise rotation on the level of FliMN‐CheY~Ac. The red and purple curves are two experimental days in which the clockwise production by fliM∆N ΔcheA cells expressing FliMN‐CheY in the presence of acetate (10 mM, pH 7.0) was measured as a function of FliMN‐CheY concentration (strain EW696; these curves are partial data of the red curve in Fig 2H). Concentrations are estimates based on the calibration curves in Appendix Fig S2, for which a similar CheY expression system was used. Each data point is a mean ± SEM of the data shown in Table EV1 for the second and third experimental days of this strain (in the presence of acetate).
BResponse of the strain shown in (A) to addition and removal of varying acetate concentrations (pH 7.0). FliMN‐CheY concentration was estimated to be~300 μM. Lines and shaded regions are the mean time spent in clockwise rotation ± SEM. The arrows indicate the estimated time points at which acetate entered or left the flow chamber. Black lines mark the linear part of the biphasic response to acetate removal. N is the number of cells.
CDistribution of clockwise interval lengths at different FliMN‐CheY~Ac concentrations in the strain shown in (A). FliMN‐CheY was acetylated by acetate (10 mM, pH 7.0). The black lines illustrate the slopes of the short and long clockwise interval distributions. The color‐bar indicates the estimated FliMN‐CheY~Ac concentration.
DAs in (C), but with distributions made with respect to the average clockwise levels of the cells.
EAverage clockwise interval length, calculated as the inverse of the fast (blue) and slow (red) rate constants from bi‐exponential fits of the distributions in (D).
When we measured, in the same strain, the motor’s response to acetate removal (a decay process that only depends on FliMN–CheY~Ac dissociation from the switch), we found that the decay was monophasic at low acetate concentrations and biphasic at high concentrations. Thus, at the two highest measured acetate concentrations (5 and 10 mM), a slower response to acetate removal preceded the fast one (Fig 3B, dark‐ and bright‐orange). When we fitted an exponential expression to the decay, we found that the fitted rate constants were similar at both concentrations (−0.095 and −0.006 s−1 for the fast and slow phases at 5 mM acetate—Fig 3B, black straight lines, and −0.091 and −0.005 s−1 for 10 mM acetate). However, only the fast phase was observed at lower concentrations (0.5 and 1 mM with fitted rate constants of −0.085 and −0.099 s−1, respectively). Thus, in FliM∆N motors, both the dependence of clockwise generation on the concentration of intracellular FliMN–CheY~Ac and the rate of the return to counterclockwise rotation appear to be biphasic.
A highly reasonable assumption is that switching of the motor is a random process, in which the life span of the clockwise state correlates with the lifetime of CheY binding to the motor. The apparent two phases, observed in Fig 3A, could potentially be a reflection of two concentration‐dependent binding modes of FliMN–CheY~Ac to the motor; one relatively weak binding and one relatively strong. If so, it might be expected that the life span of clockwise intervals would be distinct at low and high FliMN–CheY~Ac concentrations. Such short and long clockwise life spans should be seen as fast and slow exponentially decaying distributions of clockwise intervals, respectively (as in Fig 1D–G; for simplicity, we will refer hereafter to these distributions by their average interval length, i.e., fast and slow decay will be referred to as short and long clockwise intervals, correspondingly). To determine whether indeed the two phases in Fig 3A are associated with different distributions of clockwise intervals, we measured and constructed distributions from the clockwise interval lengths produced by FliM∆N motors, as a function of FliMN–CheY~Ac concentration (see Movie EV2 for a demonstration of rotation analysis). In Fig 3C, we present the interval lengths in survival distributions. In first approximation, at the lowest and highest FliMN‐CheY~Ac concentrations we observed roughly single‐exponential distributions, representing short and long clockwise intervals, respectively (Fig 3C blue and orange; note the logarithmic scale of the ordinate, where exponential curves are straight lines). At intermediate FliMN‐CheY~Ac concentrations, the distribution of clockwise intervals was biphasic (i.e., a mixture of short and long clockwise intervals; Fig 3C, gray). These observations are consistent with the possibility that low and high FliMN–CheY~Ac concentrations result in relatively weak and strong binding to the motor, respectively. Similar distributions of interval lengths were resolved when we clustered the interval lengths according to the average clockwise levels of the individual measured cells, which are likely a better proxy of CheY concentration at the switch (Fig 3D). To quantify the link between the phases of the clockwise intervals and the clockwise level, we fitted each of the distributions with a bi‐exponential expression (as done for the single‐molecule observations—Fig 1D–G), and plotted the average interval time of each phase, calculated from the fits, as a function of the average clockwise level of the cells (Fig 3E). We found that the fast‐decaying part of the distribution produced short intervals, ~40 ms long, independently of the clockwise level (Fig 3E, blue). This interval length is at the same order of magnitude as the short dwell time of CheY~Ac at wild‐type motors in the single‐molecule experiment (30 ms; Fig 1E). As of clockwise level of~0.25, the slowly decaying part of the distribution became apparent, and its clockwise intervals increased in length with the average clockwise level of the cells (Fig 3E, red). These intervals were relatively long, at the same order of magnitude as the long dwell time of CheY~Ac at wild‐type motors in the single‐molecule experiment (180 ms; Fig 1E). The different clockwise levels did not affect the rotation rate (Fig EV1A). The distribution of the counterclockwise intervals appeared to mirror the clockwise distributions (Fig EV1A). Similar results were obtained when CheY was activated by phosphorylation or by D13K substitution, with (Fig EV1, EV2) or without (Fig EV2A) FliMN fusion. This indicates that the mere appearance of short and long clockwise intervals is inherent to the switch’s response to CheY. It is independent of how CheY was activated.

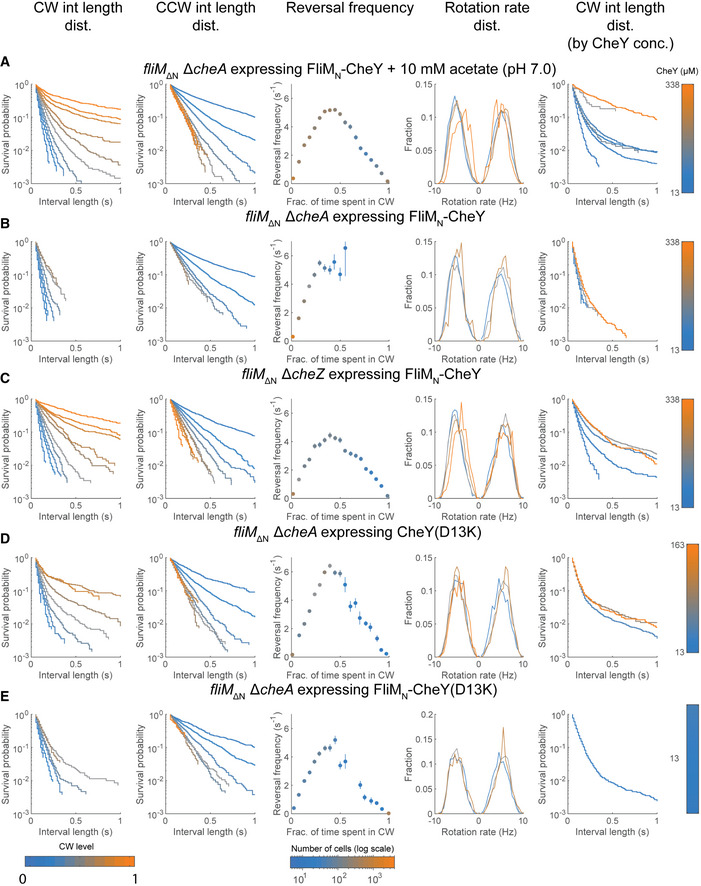
Interval analyses in FliM∆N strains expressing variants of FliMN‐CheY and CheY(D13K)
The results were pooled from all tethering experiments with the specified strains. First column, Distribution of clockwise interval lengths at 10 different clockwise levels (light blue is 0, orange is 1; see color‐bar legend at the bottom of this column). Second column, Distribution of counterclockwise interval lengths at 10 different clockwise levels (light blue is 0, orange is 1; see color‐bar legend at the bottom of the first column). Third column, Reversal frequency (mean ± SEM) as a function of clockwise levels. The color of each data point represents the number of cells averaged (see color‐bar at the bottom of this column). Fourth column, Distribution of rotation rates. Clockwise and counterclockwise rotations are shown as positive and negative values, respectively. Y‐axis stands for the fraction of intervals with the indicated rotation rate. The sum of fractions to the left of 0 as well as to the right of 0 is 1. Fifth column, As in the first column, only that the interval lengths were clustered according to the intracellular CheY concentration in the measured cells.
A fliM∆N ΔcheA cells expressing FliMN‐CheY in the presence of acetate (strain EW696). Note that the figure shown in the first column of this panel is Fig 3D.
B fliM∆N ΔcheA cells expressing FliMN‐CheY (strain EW696; no acetate present).
C fliM∆N ΔcheZ cells expressing FliMN‐CheY (strain EW697).
D fliM∆N ΔcheA cells expressing CheY(D13K) (strain EW737).
E fliM∆N ΔcheA cells expressing FliMN‐CheY(D13K) (strain EW738).

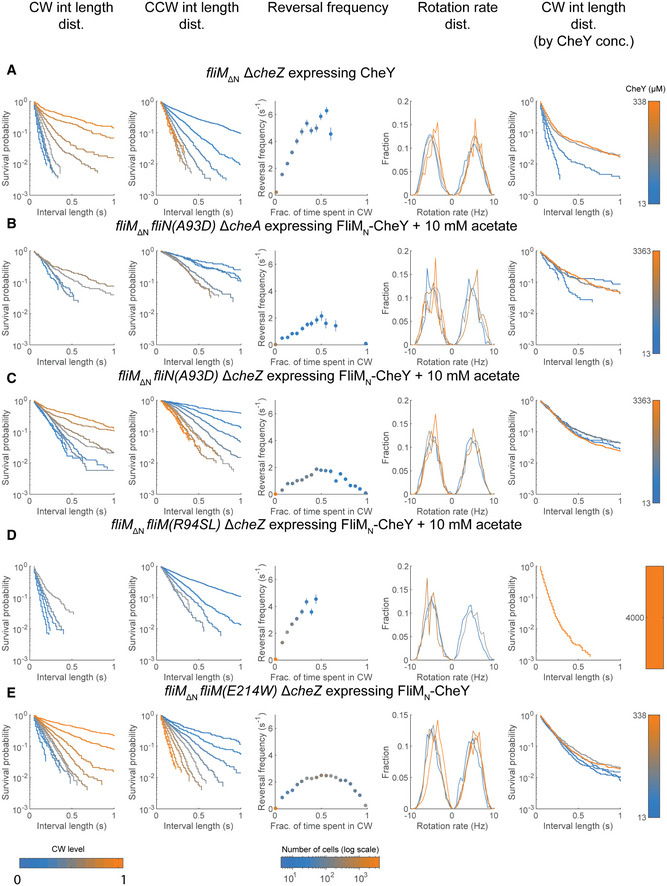
Interval analyses in FliM∆N strains expressing CheY~P and variants of FliMN‐CheY in cells containing motors with mutated FliN or FliM
See Fig EV1 for details.
A fliM∆N ΔcheZ cells expressing CheY (strain EW694).
B fliM∆N fliN(A93D) ΔcheA cells expressing FliMN‐CheY (strain EW714) in the presence of acetate. Note that the figure shown in the first column of this panel is Fig 4B.
C fliM∆N fliN(A93D) ΔcheZ cells expressing FliMN‐CheY (strain EW713) in the presence of acetate.
D fliM∆N fliM(R94SL) ΔcheZ cells expressing FliMN‐CheY (strains EW731 and EW733) in the presence of acetate. Note that the figure shown in the first column of this panel is Fig 5E.
E fliM∆N fliM(E214W) ΔcheZ cells expressing FliMN‐CheY (strain EW718). Note that the figure shown in the first column of this panel is Fig 5F.
To summarize our observations thus far, we detected multiple signatures of two switching processes in FliM∆N motors. These include the dependence of clockwise generation on the level of FliMN‐CheY~Ac (Fig 3A), the time‐dependent decrease in clockwise rotation upon CheY deacetylation (i.e., acetate removal; Fig 3B), and the sequential appearance of two types of clockwise interval lengths as a function of CheY levels (Fig 3C and D). Notably, a slow phase that precedes a fast phase (as in Fig 3B) may be indicative of a sequential response. Indeed, a biphasic response that mimics the observed response could be generated in a mathematical model by assuming two dependent CheY‐binding sites (Appendix Fig S5; see Discussion). Even though similar signatures of two switching processes were not noticed in earlier measurements (Scharf et al, 1998), probably due to the interfering effect of FliMN, we think that the observation of two CheY dwelling phases at FliMwt motors (Fig 1D–G) can be considered as such signatures in wild‐type motors.
The existence of two CheY‐binding sites other than FliMN at the switch (Mathews et al, 1998; Dyer et al, 2009; Sarkar et al, 2010), along with the observed two phases of motor response to CheY (Fig 3), raised the question of whether these two phases are associated with the two different CheY‐binding sites, FliN and FliMM. To investigate this possibility, we examined whether a perturbation of any of these sites affects a distinct phase of clockwise generation.
Impaired CheY binding to FliN eliminates short clockwise intervals
We examined the involvement of FliN by studying the generation of clockwise rotation in a fliN(A93D) mutant, in which the interaction of FliMN‐CheY~P with FliN is greatly impaired (Sarkar et al, 2010). Clearly, this mutant was unable to generate clockwise rotation by FliMN‐CheY~P (Fig 4A, blue). This supports the conclusion of Sarkar et al (2010) that FliN is involved in FliMN‐CheY~P binding to produce switching. However, FliMN‐CheY~Ac and, the more so, FliMN‐CheY~Ac~P, led to significant clockwise rotation (Fig 4A, red and green, respectively).

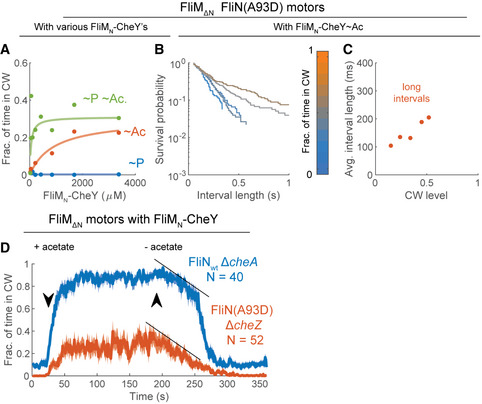
Impairment of FliMN‐CheY binding to FliN results in long clockwise intervals
AEffects of FliMN‐CheY acetylation and phosphorylation on clockwise rotation of tethered cells containing FliM∆N FliN(A93D) motors. Acetate concentration was 10 mM each (pH 7.0). FliMN‐CheY and FliMN‐CheY~P were produced by using ΔcheA and ΔcheZ backgrounds (strains EW713 and EW714), respectively. Each data point is the average of all measurements at a given FliMN‐CheY concentration, weighted by the sample number of each experiment. The concentrations shown are estimates based on the calibration curves in Appendix Fig S2, for which a similar CheY expression system was used. Concentrations larger than 500 μM were calculated by the linear extrapolation of the calibration curve. For data see Table EV1.
BDistribution of clockwise interval lengths at different average clockwise levels in ΔcheA cells containing FliM∆N FliN(A93D) motors and expressing FliMN‐CheY (strain EW713), in the presence of acetate (10 mM, pH 7.0).
CAverage clockwise interval length, calculated as the inverse of the rate constants from monophasic fits of the distributions in (B).
DResponse of tethered ΔcheZ cells having FliM∆N FliN(A93D) motors to acetate addition and removal (red; strain EW714; FliMN‐CheY concentration estimated at ~300 μM). Similar results were obtained with a ΔcheA strain (EW713). Tethered cells having FliM∆N FliNwt motors and containing FliMN‐CheY (strain EW696) are shown for reference (blue; this is the orange curve in Fig 3B after cutting some time segments before and after acetate removal to synchronize with the other response). Acetate concentration was 10 mM (pH 7.0). The data shown are the mean ± SEM. N is the number of cells. Black lines indicate the clockwise‐decay rates of FliNwt motors’ slow phase and of FliN(A93D) motors following acetate removal.
The impairment of CheY interaction with FliN in the mutant was also reflected in motor’s interval lengths. We calculated the distribution of the clockwise interval lengths of motors in this mutant under conditions like those in the experiment shown in Fig 3C and D (ΔcheA background in the presence of acetate). We found that the distribution of clockwise intervals slowly decayed (Fig 4B; Fig EV2B for additional parameters; Fig EV2C for cells containing FliMN‐CheY~Ac~P). The slope of the decay (Fig 4B) was similar to the decay slope of long clockwise intervals in FliNwt motors (Fig 3D). Excluding relatively rare events in cells with the highest measured clockwise level, none of the distributions could be fitted with a bi‐exponential expression. A mono‐exponential fit of the distributions resulted in average interval lengths similar to those observed for long intervals in FliNwt motors (Fig 4C; cf. Fig 3E, red; in the case of the highest clockwise distributions, only the first, faster decaying distribution was included in the fit). This observation is consistent with the much lower reversal frequency of FliN(A93D) motors than that of FliNwt motors (Fig EV2B and C vs. Fig EV1A and C, respectively). Thus, it appears that short clockwise intervals are relatively rare when CheY binding to FliN is impaired, suggesting that CheY binding to FliN generates short intervals of clockwise rotation. Since the dwell time of CheY at the switch is likely correlated with the length of the clockwise interval, this can also imply that CheY binding to FliN is short‐lived.
Consistently, the FliN(A93D) motors responded to acetate removal in a monophasic decay of clockwise rotation (Fig 4D, red; the decay's rate constant was 0.0135 ± 0.0027 s−1, mean ± SD of the three measurements), which was comparable in rate to that of the slow decay phase in FliM∆N FliNwt motors (Fig 4D, blue). The suppression of short clockwise intervals (Fig 4B and C) and of the fast decay rate (Fig 4D) when CheY interaction with FliN was impaired are in‐line with the proposition that CheY binding to FliN is short lived. They further suggest that long intervals are produced by CheY interaction with a switch site other than FliN.
Alteration of CheY binding to FliMM affects long clockwise intervals
As mentioned above, an obvious candidate for this other switch site is FliMM, shown to bind CheY in T. maritima (Dyer et al, 2009). To examine the plausibility of CheY binding to FliMM in E. coli, we employed in vivo crosslinking of cells expressing FliMΔN‐YPet, using a non‐specific crosslinker, glutaraldehyde. The advantage of using crosslinking is that, beyond being carried out in vivo, it can detect weak interactions. (We wish to point out that FRET experiments in vivo cannot distinguish between CheY binding to FliM and FliN due to being in close physical proximity to each other.) We tracked the crosslinking products by SDS–PAGE and scanning the gel for FliMΔN‐YPet fluorescence. This enabled us to quantify the extent of complex formation. Among many other crosslinking products, we obtained a product at the size of a complex between FliMΔN‐YPet and CheY (Fig 5A, lanes 1–3). When CheY was potentiated by FliMN fusion, the complex formed with FliM∆N was shifted by about the molecular mass of FliMN, and the amount of complex formation seemed higher (Fig 5A, lanes 4–6; the band of the shifted complex overlapped with an existing background band so the extent of excess complex formation due to CheY potentiation was hard to estimate). The formation of this complex was dependent on CheY overexpression (Appendix Fig S6), implying that the complex indeed contained CheY. These observations are consistent with the possibility of CheY binding to FliMM in vivo, but they do not rule out a possibility of CheY binding to another FliM domain (e.g., to the C terminus domain of FliM). Various direct in vitro binding assays between purified CheY and FliM∆N were not conclusive, probably due to the low affinity of FliM∆N for CheY.

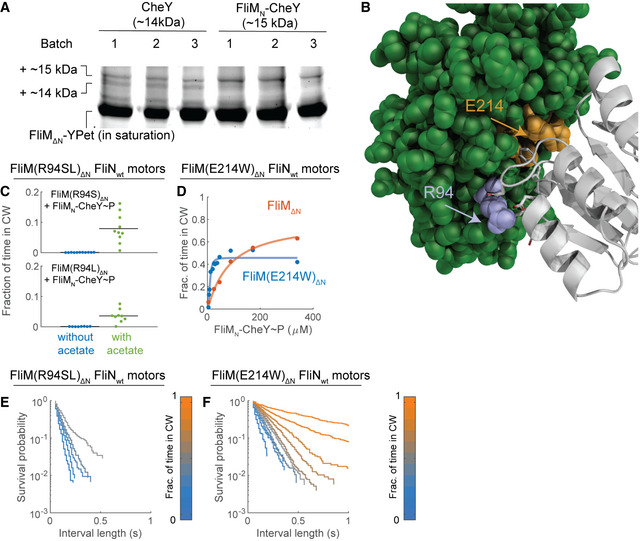
Mutations designed to impair and enhance CheY binding to FliMM produce short and long clockwise intervals, respectively
ACheY crosslinks with FliM∆N‐YPet in vivo. The cytoplasm of fliM ∆N‐YPet ΔcheZ cells overexpressing CheY (strain EW694) or FliMN‐CheY (strain EW697) from a plasmid was crosslinked by glutaraldehyde and resolved by SDS–PAGE. 1, 2, 3 stand for three different experiments that underwent this procedure. The plus sign before ~ 14 and ~ 15 stands for + 55 kDa of FliM∆N‐YPet. Note that gel running suffered from parabolic distortion in band positions. The annotations relate to the lowest positions of the bands. The gel was imaged for FliM∆N‐YPet fluorescence. To see the crosslinking products, the intense fluorescence of monomeric FliM∆N‐YPet is shown at saturation. See Appendix Fig S6 for additional details.
BPredicted binding interfaces of CheY (gray ribbon) and FliMM (green spheres) in E. coli. R94 of FliM is shown in blue. Part of the hydrophobic region of FliM (including E214) is shown in orange.
CClockwise rotation of tethered fliM(R94S)∆N ΔcheZ cells and fliM(R94L)∆N ΔcheZ cells expressing FliMN‐CheY (strains EW731 and EW733, respectively) in the absence or presence of acetate (10 mM, pH 7.0). FliMN‐CheY concentration was estimated to be ~ 4 mM by extrapolation of the calibration curves in Appendix Fig S2, for which a similar CheY expression system was used. No clockwise rotation was observed when benzoate substituted for acetate. Each data point is the mean of a separate experiment. Black line is the mean of all experiments. For sample number of each data point see Table EV1.
DClockwise rotation of tethered fliM(E214W)∆N ΔcheZ cells (strain EW718) at various concentrations of FliMN‐CheY. The concentrations shown are estimates based on the calibration curve in Appendix Fig S2, for which a similar CheY expression system was used. Each data point is the average of all measurements at similar FliMN‐CheY concentrations, weighted by the sample number of each experiment. The red points and curve are taken, as a reference, from Fig 2H. For data see Table EV1.
EDistribution of clockwise intervals of fliM(R94SL)∆N ΔcheZ cells expressing FliMN‐CheY (strains EW732 and EW734) in the presence of acetate (10 mM, pH 7).
FDistribution of clockwise intervals of fliM(E214W)∆N ΔcheZ cells expressing FliMN‐CheY (strain EW718) in the absence of acetate.
To examine the likelihood that CheY binding to FliMM is functional, we employed docking analysis between CheY and FliMM based on their binding interface in T. maritima (Appendix Supplementary Methods for detailed description). In the docking analysis, we produced a number of models of CheY binding to FliMM in T. maritima and translated those fitted well with the NMR results of Dyer et al (2009) to E. coli proteins by superposing the active and inactive forms of CheY upon the model of FliMM. We substantiated the superposition results by docking the E. coli proteins independently of the T. maritima model. The analysis predicted two prominent latching interfaces of CheY with FliMM: an electrostatic interface, mostly contributed by arginine at position 94 of FliM, and a hydrophobic pocket in FliM, predicted to face the phosphorylation site of CheY (Fig 5B, blue and orange for electrostatic and hydrophobic surfaces, respectively).
To determine the relevance of the binding interface predictions, we studied motors with substituted arginine at position 94 (Appendix Fig S7A) as well as motors with substituted glutamic acid at position 214 (Appendix Fig S7B). The first substitution was designed to diminish electrostatic interactions, whereas the substitution at position 214 was designed to enhance hydrophobic interactions by replacing a charged residue with a bulky hydrophobic one. Thus, to determine the relevance of the predicted electrostatic interaction, we examined the rotation of motors of cells expressing FliM∆N(R94S) or FliM∆N(R94L) in a ΔcheZ background and overexpressing FliMN‐CheY (i.e., FliMN‐CheY~P). The motors of these mutants did not rotate clockwise (Fig 5C, blue). Only when acetate was present, i.e., when CheY was both phosphorylated and acetylated, we observed low levels of clockwise rotation (Fig 5C, green). These results are in‐line with the docking model’s prediction that FliM(R94) is involved in CheY binding. To determine the functional relevance of the predicted hydrophobic interaction, we could not just diminish this interaction by a simple mutation, as we did for the electrostatic interaction, because the hydrophobic area is contributed by many residues. Therefore, we examined, instead, whether enhancement of hydrophobic interactions by replacement of a charged residue with a hydrophobic one in the hydrophobic CheY‐FliMM‐binding interface (E214W) would increase clockwise generation. Indeed, clockwise rotation in the fliM ∆N(E214W) mutant in a ΔcheZ background was observed at much lower FliMN‐CheY expression levels (Fig 5D). It appears that the mutation affected the motor’s sensitivity to CheY rather than affecting its intrinsic clockwise level. This is because the clockwise rotation levels of fliM ∆N(E214W) in a ΔcheA background, where FliMN‐CheY is mostly inactive and does not contribute much to clockwise generation, were low and comparable to cells containing nonmutated FliM∆N (Appendix Fig S8; blue). Yet, when FliMN‐CheY was activated by acetate, the response of cells with FliM∆N(E214W) motors exceeded that of cells with nonmutated FliM∆N (Appendix Fig S8; red).
To examine our prediction that CheY interaction with FliMM generates long clockwise intervals, i.e., stable clockwise rotation, we produced distributions of clockwise interval lengths in the fliM ∆N(R94SL) and fliM ∆N(E214W) mutants. Excluding cells with the highest measured clockwise level, fliM ∆N(R94SL) motors yielded a single, exponentially decaying distribution of short clockwise intervals (Fig 5E; Fig EV2D for additional parameters). This is in‐line with the expectation that attenuation of CheY interaction with FliMM would diminish long clockwise intervals. The slope of the distribution was like the slope of the short intervals’ phase in cells containing nonmutated FliM∆N motors (e.g., Fig 3D). Consistent with the anticipation that the elevation of CheY affinity for FliMM would generate longer clockwise intervals, distributions produced by the fliM ∆N(E214W) mutant were monophasic (Fig 5F; Fig EV2E for additional parameters). With the exception of the lowest clockwise level, the slopes of the distributions were similar to those of the long intervals’ phase in cells containing nonmutated FliM∆N motors (e.g., Fig 3D), meaning that short clockwise intervals were absent at most clockwise levels. Taken together, these results suggest that CheY binding to FliMM produces long clockwise intervals.
Discussion
Of the many known biological switches, the switch of the bacterial flagellar motor has been a focus of great interest due to its unique properties. It has also been a source of frustration due to lack of success in resolving its molecular mechanism. In the current study, we revealed the functions of each of the three CheY‐binding sites at the switch, FliMN, FliMM, and FliN. This led to uncovering processes at the switch that result in brief and long events of switching. Thus, we identified two modes of CheY binding to wild‐type motors as well as to FliM∆N motors, and a number of related processes that essentially consist of two phases. These included the dependence of clockwise generation on the level of activated CheY (FliMN‐CheY~Ac), the time‐dependent decrease in clockwise rotation upon CheY deactivation by acetate removal, and the motor’s switching kinetics. Studying motors carrying mutations in FliN and FliMM enabled us to detect the link between these sites and the biphasic processes, and to conclude that short clockwise intervals are mostly promoted by CheY binding to FliN, and long intervals—by CheY binding to FliMM. The observations that the average durations of the short and long clockwise intervals of FliM∆N motors are similar to the short and long dwell time of CheY at wild‐type motors, respectively, suggest that the conclusions drawn may be relevant to both FliM∆N motors and wild‐type motors. The finding that CheY has to be acetylated for producing long clockwise intervals in motors defective in CheY‐FliN binding further indicated that CheY~Ac is more effective than CheY~P in binding to FliMM. Below we discuss implications of these findings and we show how they are related to each other in a model of the switching mechanism.
Functions of FliMN
We found that the first site to which CheY binds, a high‐affinity site at FliMN (Welch et al, 1993; Bren & Eisenbach, 1998), is not essential for binding to the switch and for clockwise generation (Figs 1 and 2). Yet, the order‐of‐magnitude higher CheY concentration needed for binding and response in the absence of FliMN (Figs 1 and 2) indicated that FliMN is required for maximal sensitivity of the switch. FliMN may do it by tethering CheY to the switch, but it seems that its most pronounced effect is to potentiate CheY (Fig 2H). To the best of our knowledge, this is the first system in which a ligand (CheY) is potentiated by its receptor (FliM at the switch), rather than vice versa.
The conclusion that FliMN potentiates CheY is based on our studies with FliMN‐CheY fusion protein (Fig 2H). The published observation that CheY binds to FliMN [Kd = 27 and 680 µM for CheY~P and CheY, respectively (McEvoy et al, 1999)] strongly suggests that this potentiation also occurs in vivo with non‐fused proteins. CheY potentiation at the switch may be a preliminary step in the process of clockwise generation (Fig 6A), which may have evolved to filter out crosstalk with proteins having CheY‐like folds in other two‐component signaling pathways. (Binding of such proteins to FliMN is expected to be futile.)

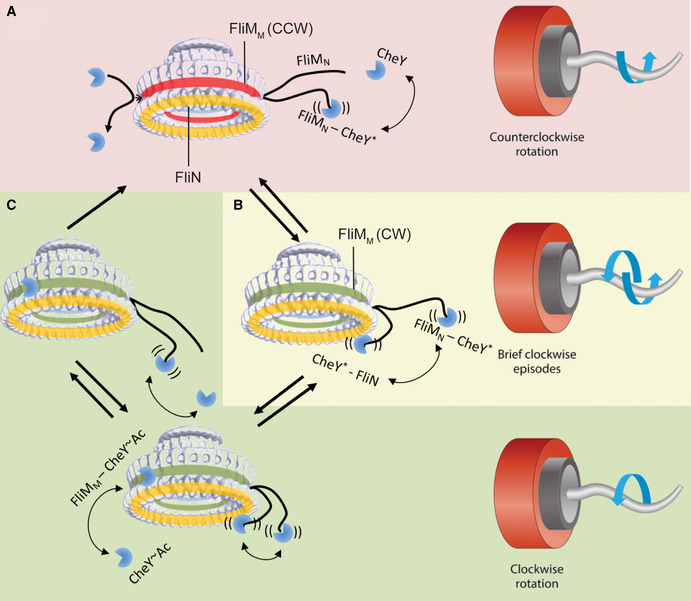
A model for clockwise generation by CheY~P and CheY~Ac
The model suggests the most likely interactions of CheY at the switch. Yellow ring, FliN. Green or red ring, FliMM in a conformation to which CheY can or cannot bind, respectively. Red background region is counterclockwise rotation. Yellow background region is frequent switching. Green‐shaded region is clockwise rotation.
AWhen the motor rotates counterclockwise, CheY most likely binds to FliMN because FliMN is a high‐affinity site and because it is the most accessible site. An interaction of CheY with FliN is not probable at physiological CheY concentrations, unless CheY first binds to FliMN. Binding to FliMN potentiates CheY, making it more likely to act on FliN. (Potentiated CheY is marked with an asterisk.) FliMM is probably sterically blocked for CheY binding or is in a conformation that does not favor CheY binding.
BFliMN‐bound CheY has high probability of interacting with FliN. This interaction generates brief episodes of clockwise rotation, because the complex CheY‐FliN is unstable and is likely to quickly dissociate.
CDuring the short episodes of clockwise rotation shown in B, FliMM either becomes accessible to CheY or is changed to a conformation that favors CheY binding. As a result, CheY (or, most likely, CheY~Ac) binds to FliMM. This binding stabilizes the clockwise conformation of FliMM and prevents the latter from resuming its counterclockwise conformation or becoming again sterically blocked for CheY binding. As long as CheY is bound to FliMM, CheY dissociation from FliN no longer affects clockwise rotation. CheY dissociation from FliMM when CheY is already dissociated from FliN would cause an immediate return to counterclockwise rotation. See text for other details.
The meaning of CheY potentiation in molecular terms is not known. According to the view that confined segments of CheY switch asynchronously and locally between active and inactive conformations (McDonald et al, 2012), perhaps CheY binding to FliMN stabilizes a distinct conformation that is most potent for clockwise generation. Since FliMN‐bound CheY is more rapidly phosphorylated by small phosphodonors (Schuster et al, 2001), it is reasonable that in this clockwise‐promoting conformation, the phosphorylation site is more accessible to small phosphodonors. To test the possibility that CheY binding to FliMN indeed stabilizes a distinct conformation that is most potent for clockwise generation, it might be of interest to simulate or solve the conformation of an extremely potent CheY variant, such as FliMN‐CheY(D13K), and thereby derive the form of CheY that preforms the switching action at the motor.
Remarkably, the ultrasensitivity of the switch, observed in FliMwt motors as a sigmoidal (cooperative) clockwise dependence on the intracellular CheY~P level (Cluzel et al, 2000), is lost in FliM∆N motors (Fig 2G). This suggests that FliMN has a role here too. (Note, though, that CheY~Ac, which appears to bind to the switch more firmly than does CheY~P, exhibits sigmoidal dependence—Fig 2G; see below) The published finding that a constitutively active CheY mutant protein binds better to clockwise‐rotating motors than to counterclockwise‐rotating motors (Fukuoka et al, 2014) and the notion that this can generate cooperative binding (Duke et al, 2001), combined with the possibility, raised above, that CheY binding to FliMN may stabilize a distinct conformation that is most potent for clockwise generation, may explain why switch ultrasensitivity requires FliMN.
The switching mechanism: an apparent gating mechanism
This study suggests that CheY~P binds to FliN with resultant transient switches of the motor to clockwise rotation and that CheY~Ac preferentially binds to FliMM to produce stable clockwise rotation. [Note that an E. coli cell always contains some level of CheY~Ac, even in the absence of acetate (Yan et al, 2008; Fraiberg et al, 2015).] These are based on (i) the conclusions, made above, that short intervals are mostly promoted by CheY binding to FliN, and long intervals—by CheY binding to FliMM, (ii) the observation that the dwell time of CheY~P at the motor (Fig 1F) is shorter than that of CheY~Ac (Fig 1E), and (iii) the finding that CheY~Ac is more effective than CheY~P in clockwise generation (Fig 2G). We propose that, following CheY~P potentiation by FliMN, CheY~P mainly binds to FliN to make the first catalytic step of clockwise generation, producing brief episodes of clockwise rotation (Fig 6B), and that CheY~Ac mainly binds to FliMM and makes the second catalytic step of clockwise generation, stabilizing the rotation in the clockwise direction (Fig 6C). It is probable that CheY~P and CheY~Ac can also bind to FliMM and FliN, respectively, though at much lesser affinity. We assume that CheY, activated by phosphorylation or acetylation, can bind to FliN at any time, but it can only bind to FliMM when the motor already rotates clockwise to some extent. This assumption relies on the observation that the orientation of a FliM subunit within the switch is different in the counterclockwise and clockwise states (Paul et al, 2011), which may cause FliMM to be sterically blocked for CheY binding in the counterclockwise conformation. Thus, clockwise rotation, which results from CheY binding to FliN, exposes the CheY‐binding site at FliMM. We term such a mechanism, which involves a conditional binding, a “gating mechanism”. This mechanism is consistent with the kinetic model (Appendix Fig S5), proposed to explain the sequential appearance of two phases in the response of FliM∆N motors to acetate removal (Fig 3B). This kinetic model appears to be conceptually similar to the model of conformational spread (Duke et al, 2001; Bai et al, 2010).
The gating mechanism may provide an explanation of the different dependences of clockwise generation on the levels of CheY~P and CheY~Ac—saturation and sigmoidal curves, respectively (Fig 2G). Binding of CheY~P, according to this study, is mainly to FliN. The binding site at FliN is accessible to CheY~P, for which reason the dependence of clockwise generation (which reflects CheY~P binding to the switch) on the intracellular level of CheY~P would have the shape of a saturation curve. In contrast, CheY~Ac preferentially binds to FliMM, whose accessibility is, as explained above, conditional. The availability of more and more binding sites at FliMM as the level of intracellular CheY increases is likely to generate a sigmoidal, cooperative‐like dependence.
The gating mechanism may also support efficient swimming. As mentioned in the introduction, when some of the motors in a given bacterial cell exhibit extremely brief episodes of clockwise rotation, the swimming direction is maintained at large, and the outcome is directionally persistent migration of the cell population (Vladimirov et al, 2010; Saragosti et al, 2011). Such directional persistence may markedly improve collective migration (Saragosti et al, 2011). On the other hand, tumbling behavior, which reorients the swimming direction to support the classical run‐and‐tumble view of bacterial migration (Berg, 2003; Eisenbach, 2004), requires relatively long clockwise rotation intervals. The gating mechanism can generate both types of swimming behavior because it can produce both short and long clockwise intervals. Indeed, a small‐scale study, performed by us, suggested that the short intervals of clockwise rotation needed for directional persistence (Saragosti et al, 2011) could, indeed, be provided by CheY~P binding to FliN and that the long clockwise intervals for tumbling could be provided by CheY~Ac binding to FliMM (Appendix 2 and Appendix Fig S9). This means that the switching mechanism is wired to inputs from two different signaling pathways: One involves the chemotaxis machinery that regulates CheY phosphorylation, and the other involves the cellular metabolic pathway that regulates CheY acetylation. Hence, this switching mechanism apparently optimizes chemotactic performance according to the ambient conditions.
Unique features of the switching mechanism
The switching mechanism, revealed in this study, has a number of unique features. On the one hand, it is tightly regulated by three distinct binding sites and by two different covalent modifications. Binding to the second site, FliN, depends on phosphorylation of CheY, and binding to the third site, FliMM, mainly depends on acetylation. This binding can apparently occur only if it is preceded by CheY~P binding to FliN. On the other hand, this mechanism endows the motor with flexibility with respect to switching, as the intermediate stage at which CheY~P is bound to FliN provides a “go/no go” situation, in which the motor can either proceed to a stable clockwise rotation due to binding to FliMM or shift back to counterclockwise rotation. With this unique combination of seemingly conflicting, but complementing properties of the switching mechanism, it would not be surprising if similar mechanisms are found in the future in the output of other signaling systems.
Materials and Methods
Strains and plasmids
Strains and plasmids used in this study are listed in Appendix Tables S1 and S2, respectively. To produce fliMN truncation, fliM‐YPet was cloned from the strain JPA945 (Delalez et al, 2010) (kindly provided by J. Armitage) with genomic flanks of 500 base pairs to the Pst1 sites of pDS132 suicide plasmid (Philippe et al, 2004). FliM residues 1–16 were truncated from the resulting plasmid by RF cloning. The constructed plasmid was used to perform allelic exchange with strain JPA945 to produce strain EW566 bearing fliMΔ(1–16)‐YPet genomic mutation. The mutation was verified by PCR sequencing. Plasmidic mutations were produced by RF cloning. The genomic deletion mutations of cheA and cheZ were produced by subjecting cells to P1 transduction with phage containing the genetic background of strain JW1870 or JW1877, respectively.
Growth conditions
Strains, diluted 1:100 from overnight cultures, were cultured to mid‐late exponential phase at 30°C in tryptone broth with appropriate antibiotics to maintain the plasmids. For CheY expression, cells were grown to mid‐late exponential phase and induced by IPTG for 3–4 h. It appeared that overnight cultures which, prior to being diluted, had been left on the bench at room temperature for 1–2 days, gave rise to cultures that produced better responses to acetate.
Analysis of the direction of flagellar rotation
The direction of flagellar rotation was determined by the tethering assay (Silverman & Simon, 1974). Subsequent to flagellar partial truncation by passing the bacterial culture in a syringe several times, the resulting suspension was usually washed three times in motility buffer (10 mM KPi pH 7.0, 0.1 mM EDTA). The cells were tethered to a coverslip in a flow chamber as described (Berg & Block, 1984) and washed with motility buffer in the flow chamber for roughly 5 min prior to measurement. All chemicals used in behavioral assays were dissolved in the motility buffer. Documentation was done with a uEye digital camera on top of a Zeiss phase‐contrast microscope. Recording was typically done at 75 frames/s. The time of reagents’ entry to the flow chamber (~20 s from the time of introducing the reagent to the pump’s pipe) was estimated by the chemotactic response of wild‐type cells. The time needed for replacement of the total chamber’s volume was 6 s.
Automated analysis of flagellar motor direction of rotation
All the analyses were done with a pack of MATLAB scripts prepared for this study. These are freely available at https://github.com/OshriAfanzar/Afanzar‐et‐al‐2019. All samples of all experiments were analyzed in the exact same way and codes.
Frame and movie processing
Each frame was processed by the following scheme: Image binarization → Identification of connected pixels → Ellipse fitting to bodies of connected pixels. Image binarization process was written in MATLAB as the following code lines:
Pixels = vidFrame(:);
SortedPixels = sort(Pixels);
LinearBase = linspace(min(Pixels),max(Pixels),length(Pixels));
SubtractPixels = SortedPixels - LinearBase;
Tresh = mean(SortedPixels (find(SubtractPixels,min(SubtractPixels))));
vidFrame = (vidFrame – Tresh)>0;
Connected pixels were extracted by the “bwconncomp” function of MATLAB and ellipse fitting was done using the “regionprops” function of MATLAB. Trajectories of rotating cells were composed by identifying ellipses that shared common pixels area for at least 95% of the recording time. The value of 95% was chosen because during the recording there were sometimes events of missing acquisitions (e.g., out of focus events) and we reasoned that we can trust a cell recording only when it could be identified for at least 95% of the recording time. Trajectories in which the rotation was not smooth (e.g., in cases where the rotation frequently paused, or where the rotation was not in a 2D plain) and, therefore, contained over‐represented rotation angles, were spotted and discarded. The way to spot over‐represented angles was to calculate the extent by which the distribution of angles deviates from random distribution. Thus, we found empirically that when we employed bins of 10° for calculating the distribution of angles, trajectories that contained one or more bins with a number of counts exceeding the number [(total number of counts)*2/(number of bins)] were suspicious and discarded.
Analysis of rotation
Angles extracted from the fitted ellipses were used to calculate rotation velocity as the difference in cell angle with respect to the frame rate. To ensure high‐quality data, events in which the fitted ellipse met the following stringent conditions were marked as erroneous: (i) Major axis/Minor Axis < 1.25. (ii) Major axis < 7 pixels. (iii) Major axis < mean(Major axis) – 2SD(Major axis). (iv) Major axis > mean(Major axis) + 2SD(Major axis). (v) Angular displacement was < 360°/frame rate (e.g., 360/75 for 75 frames/s) or > 10*360°/frame rate (e.g., 3600/75 for acquisition frequency of 75 frames/s). When a single erroneous event was flanked by intervals of a different type, the erroneous event was replaced by an event whose identity was determined by the calculated rate of rotation. For example, for clockwise = →, counterclockwise = ← and erroneous interval = E, for the sequence ←←←←E→→→→, which corresponds to the rotation rates (−5)(−5)(−5)(−5)(0.4)(5)(5)(5)(5) Hz, E would be replaced by → because the erroneous interval 0.4 is positive, i.e., with clockwise tendency.
Calculation of the fraction of time spent in clockwise rotation
The fraction of time that a motor spent in clockwise rotation was calculated as the sum of clockwise events in a unit time. For steady state rotation, the fraction of time spent in clockwise rotation was calculated for the whole time of acquisition (typically, 30 s) and averaged over all cells. For time‐resolved rotation (as in the response to stimuli), clockwise rotation of single cells was averaged in 1‐s intervals.
Analysis of interval length
We defined clockwise intervals as intervals that have at least four consecutive frames of clockwise rotation at a rate higher than 1 Hz. The rotation rate value was taken as two standard deviations away from the rotation rate control of non‐rotating cells [the rotation rate of these cells was 0 ± 0.5 Hz (±SD)]. This 4‐frame threshold excluded short events that might not be CheY‐mediated clockwise rotation. The choice of four frames (52 ms) for the threshold was according to another negative control, consisting of a ΔcheY strain, which cannot generate CheY‐mediated clockwise rotation. In this negative‐control strain, we studied 1‐min recordings of 344 cells, and found ~100 clockwise intervals, all shorter than 50 ms. To avoid artifacts that might have been caused by interactions between the cell body and the surface, we also excluded from the analysis intervals that were flanked by pause events. All excluded intervals were considered erroneous. We grouped clockwise intervals from cells that had similar average clockwise levels and produced from each group a distribution of interval lengths. Intervals were measured only when they were flanked by intervals of the opposite direction, and were not flanked by erroneous intervals. For example, for clockwise = →, counterclockwise = ←, and erroneous interval = E, the sequence of intervals ←←←←→→EE←←→→→→←← would resolve in only one interval of clockwise rotation at the length of four frames. When a single erroneous event was flanked by intervals of the same type, the erroneous event was replaced by an event of the same type as the flanks. For example, ←←←←E←←←← would be replaced by ←←←←←←←←←.
Analysis of reversal frequency
The reversal frequency was defined as the number of zero‐velocity crossing events per second (after filtering for noise).
In vivo FRET response to stimuli
FRET measurements in response to stimuli were carried out as described by Sourjik and Berg (2002).
Measurements of CheY expression levels
To assess CheY levels in the cytoplasm, cells expressing CheY‐mCherry or mCherry from a plasmid (strains EW575 and EW569, respectively) were grown to mid‐late exponential phase and induced by various IPTG concentrations for 4 or 6 h. The cells were washed three times in NaPi (10 mM, pH 7.6). A sample of the cells was plated in serial dilutions on LB‐agar plates to estimate, by colony forming units, the number of cells in the culture. The rest of the sample was sonicated and loaded in 200 μl aliquots to a 96‐well plate. In the same plate, a purified CheY‐mCherry protein at a known concentration was loaded in different dilutions. The plate was read by a Cytation‐5 plate reader (excitation and emission filters were 570 ± 20 nm and 610 ± 20 nm, respectively). The calculation of the cellular concentration of the expressed proteins assumed that the average volume of a single cell is 1 fl.
Single‐molecule observation and analysis
The protein 6xHis‐CheY(I95V) was purified by Protino Ni‐TED beads and labeled with a maleimide modification of the organic dye Atto647, previously shown to be bright, photo‐stable, not hydrophobic and, therefore, highly compatible with observation of single molecules in vivo (Plochowietz et al, 2014). The labeled protein was separated from the residual unreacted dye by size‐exclusion chromatography and was then electroporated into cells of strain EW669 in a low‐salinity buffer. The electroporation approach was carried out as described (Di Paolo et al, 2016). The electroporated cells were recovered for 5 min in Super Optimal broth with Catabolite repression, washed 4–5 times in motility buffer, and visualized on agar pads on a customized inverted Olympus IX‐71 microscope equipped with two lasers, a 637 nm diode laser (Vortran Stradus; Vortran Laser Technology, Sacramento, CA, USA) and a 532 nm DPSS laser (MGL‐III‐532 nm‐100 mW, CNI) (Appendix Fig S10A). Laser light was combined into a single‐mode optical fiber (Thorlabs, Newton, NJ, USA) and collimated before focusing on the objective. Highly inclined and laminated optical sheet (HILO) and total internal reflection fluorescence illuminations were achieved by adjusting the position of the focused excitation light on the back focal plane of the objective (UPLSAPO, 100×, NA 1.4, Olympus). Cellular fluorescence was collected through the same objective, filtered to remove excitation light through a long‐pass filter (HQ545LP; Chroma, Taoyuan Hsien, Taiwan) and a notch filter (NF02‐633S; Semrock, Rochester, NY, USA), and spectrally separated by a dichroic mirror (630DRLP, Omega, Brattleboro, VT, USA). Each channel was imaged onto separate halves of the chip of an EMCCD camera (iXon+, BI‐887, Andor, Belfast, UK). The illumination for bright‐field images comprised a white‐light lamp (IX2‐ILL100; Olympus, Shinjuku, Tokyo, Japan) and condenser (IX2‐LWUCD; Olympus) attached to the microscope. Movies and images were recorded at 100 frames/s using manufacturer’s software (Andor). All measurements were carried out in continuous wave mode for both green and red lasers. For all the experiments, the exposure time was 10 ms and the intensities used for YPet and Atto647 were 400 nW µm−2 and 1 µW µm−2, respectively.
The locations of CheY(I95V)‐Atto647 and FliM‐YPet were automatically estimated by a custom‐made MATLAB script. Switch locations were identified as peaks of fluorescence. The exact switch location was determined by identifying the location of the maximum of a 2D Gaussian, fitted to each switch spot. For the identification of switches, several images were acquired, normalized, and averaged before Atto647 imaging. The location of CheY(I95V)‐Atto647 was determined similarly to switch location in each frame. CheY(I95V)‐Atto647 was considered bound to the switch when it was within 75 nm of a switch location for at least 30 ms. The choice of 75 nm was based on the motor radius (25 nm) combined with the expected length of FliMN (assuming 15 nm FliMN length, considering about 3.5 Å per amino acid residue). The distance of 75 nm is more permissive than the expected binding radius (40 nm) because we expected some inaccuracy in the localization of the motor. Dwell times < 30 ms were excluded to avoid false‐positive measurements (e.g., molecules diffusing near the motor). The value of 30 ms (three frames) was chosen as the half of the expected mean dwell time of CheY(I95V)‐Atto647 with FliMN [~60 ms given a Kd = 3.9 µM for CheY(I95V)‐FliMN interaction (Schuster et al, 2000) and an estimated kon = 4 × 106 M−1 s−1 (Sourjik & Berg, 2002)]. The mean‐square displacement in two dimensions due to free diffusion is <r 2> = 4Dt. Setting <r 2> = 75 nm2 and t = 30 ms yields D = 0.047 µm2 s−1, which is much lower than the diffusion coefficient of CheY in the cell [estimated to be 50–100 µm2 s−1 (Segall et al, 1985)]. Thus, a false‐positive rate due to diffusing CheY molecules is expected to be negligible. Trajectories from the same type of experiments were joined for survival analysis (Fig S10B). The number of cells recorded was 82 (ΔcheZ no acetate), 40 (ΔcheZ with acetate), 63 (ΔcheA no acetate), and 56 (ΔcheA with acetate). The number of trajectories in which CheY was found to interact with FliM was 2414 (ΔcheZ no acetate), 2660 (ΔcheZ with acetate), 1316 (ΔcheA no acetate), and 1904 (ΔcheA with acetate).
Author contributions
Performing and analyzing the experiments: OA; Designing the experiments, result interpretation, and manuscript writing: OA, MEisenbach; Docking analysis: OA, MEisenstein; Some of the experiments and technical assistance: KL; Single‐molecule electroporation experiment analysis: DDP, OA; TIRF setup: ANK; Insight interpreting the single‐molecule results and providing important input for the data analysis of the tethering experiments and for writing the manuscript: RMB; Sharing a script that helped with image alignment: AP.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
We are indebted to Dr. David Blair and Paige Wheatley in his group for numerous intensive discussions, for critical reading of earlier versions of the manuscript, for disclosing their unpublished data, and for providing some of the mutants used in this study. We thank Dr. James E. Ferrell for his advice in the construction of the mathematical model, Dr. Ady Vaknin and Vered Frank in his group for allowing us to measure FRET in their laboratory and for their useful suggestions and technical assistance, Dr. S. Roy Caplan for helpful discussions, Vladimir Kiss for assistance with operating the confocal microscope, Dr. Judith Armitage for supplying bacterial strains, and Yana Gurevich for technical assistance. This study was supported by grant no. 2013197 from the US‐Israel Binational Science Foundation and by grant no. 66/14 from the Israel Science Foundation to ME, by a UK BBSRC grant no. BB/H01795X/1 and an ERC Starter grant no. ERC 261227 to ANK, and by a BBSRC grant to RMB. DDP was supported by a UK EPSRC DTC studentship. AP was supported by a UK EPSRC DTA studentship and the German National Academic Foundation (Studienstiftung) and the Phizackerley Senior Scholarship in Medical Sciences at Balliol College, Oxford. OA was supported by an EMBO short term travel fellowship to Oxford.
Data availability
All the source data from this publication have been deposited to the Biostudies database https://www.ebi.ac.uk/biostudies/ and assigned the identifier S‐BSST558 .
 The switching mechanism of the bacterial rotary motor combines tight regulation with inherent flexibility
The switching mechanism of the bacterial rotary motor combines tight regulation with inherent flexibility
