
- Altmetric
There are glycopeptide structures missing in both Fig 2 and Fig 6. Please see the correct Fig 2 and Fig 6 here.
Also, in Fig 7 the m/z value 1255.8162 is associated with the wrong glycan composition and should be assigned to H5N6S1F2. Please see the correct Fig 7 here.

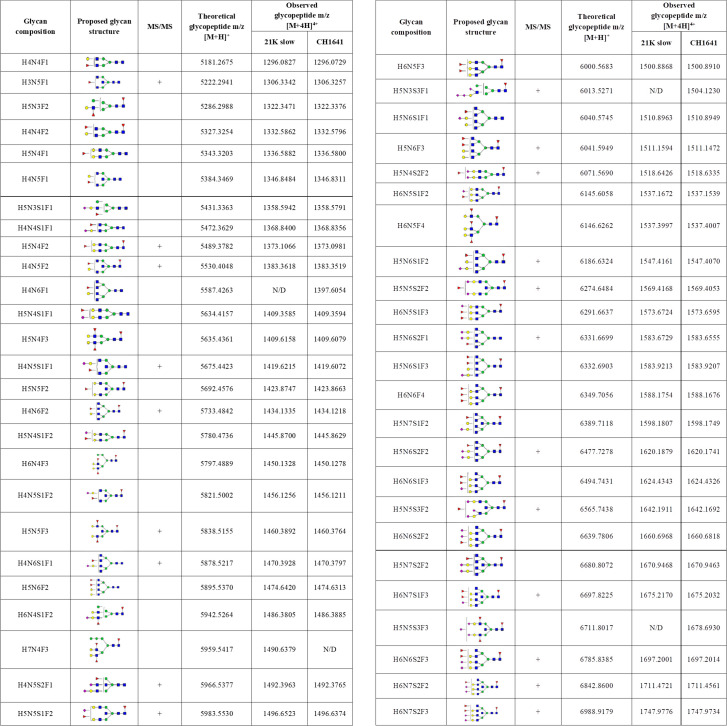
Detected N-184 glycopeptides on both prion strains.
Proposed glycan structures found on N-184 glycosylation site with the theoretical and observed m/z values of the detected glycopeptides. H–hexose, N–N-acetylhexosamine, F–fucose and S–N-acetylneuraminic acid (sialic acid). Blue square–N-acetylglucosamine (GlcNAc), green circle–mannose (Man), red triangle–fucose (Fuc), yellow circle–galactose (Gal), purple diamond–N-acetylneuraminic acid (Neu5Ac). The presence of MS/MS spectrum is indicated with +. N/D–not determined.

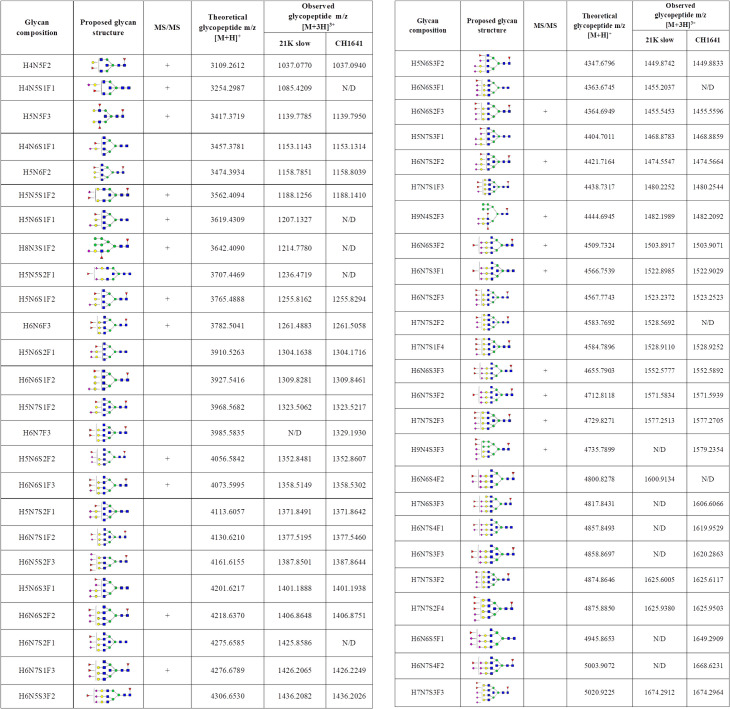
Identified N-200 glycopeptides on both prion strains.
Proposed glycan structures found on N-200 glycosylation site with the theoretical and observed m/z values of the detected glycopeptides. H–hexose, N–N-acetylhexosamine, F–fucose and S–N-acetylneuraminic acid (sialic acid). Blue square–N-acetylglucosamine (GlcNAc), green circle–mannose (Man), red triangle–fucose (Fuc), yellow circle–galactose (Gal), purple diamond–N-acetylneuraminic acid (Neu5Ac). The presence of MS/MS spectrum is indicated with +. N/D–not determined.

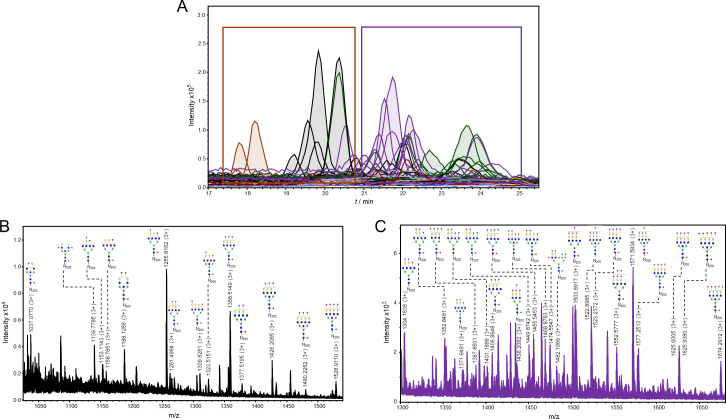
Representation of detected glycoforms on the N-200 glycosylation site of 21K slow prion strain.
A) Extracted ion chromatograms with mutual 35 glycoforms detected. B) Assigned glycoforms in MS spectrum with N-200 peptide backbone: neutral and monosialylated, C) disialylated and trisialylated glycoforms. Blue square–N-acetylglucosamine (GlcNAc), green circle–mannose (Man), red triangle–fucose (Fuc), yellow circle–galactose (Gal), purple diamond–N-acetylneuraminic acid (Neu5Ac).
Reference
1
 Correction: Site-specific analysis of N-glycans from different sheep prion strains
Correction: Site-specific analysis of N-glycans from different sheep prion strains