
The authors have declared that no competing interests exist.
- Altmetric
A diverse community of microorganisms inhabits various parts of a plant. Recent findings indicate that perturbations to the normal microbiota can be associated with positive and negative effects on plant health. In this review, we discuss these findings in the context of understanding how microbiota homeostasis is regulated in plants for promoting health and/or for preventing dysbiosis.
Defining the “core” microbiota in plants
In the past decade, many studies have surveyed microbiota composition in plants. Despite diverse microbes found in plants, overall bacterial composition seems to be conserved at the phylum level. In flowering plants that have been surveyed, the aboveground tissues (phyllosphere) are associated with bacterial assemblages dominated by Proteobacteria, Actinobacteria, Bacteroidetes, and Firmicutes. These bacterial phyla are also enriched in belowground tissues (rhizosphere) compared to bulk soil [1,2]. Nonvascular plants such as liverwort and moss are also dominated by these phyla [3–5], suggesting possible conservation of core microbiota across plant lineages. It should be pointed out that, so far, most microbiota surveys have been focused on bacterial components of plant-associated microbial communities. As fungi [6], viruses [7], and protozoa [8] are also common residents on or inside the plant, more efforts are needed in the future to systematically define fungal, viral, and protozoa microbiota members across plant taxa. Additionally, biogeography plays an important role in determining the reservoir of microbes from which a plant selects members of its microbiota [9]. Because different microbial taxa may provide functionally redundant traits to a plant host [10], detection of dissimilar taxa in different plants, especially at lower taxonomical levels, may not necessarily indicate functionally distinct microbiotas, an important topic that requires rigorous future investigations.
Regardless of whether there is a functionally conserved core microbiome in plants, increasing evidence suggests that microbiota homeostasis may be intimately linked to host processes and, in turn, plant health. Microbiota homeostasis in eukaryotic organisms is likely dynamic in both space and time. Here, we use eubiosis to describe the state of microbiota homeostasis that is necessary for maintaining host health under optimal, nonstressful conditions. The eubiotic state for an individual plant is not static, but instead dynamic over a plant’s lifetime. For example, microbiota of healthy plants can vary temporally based on the time of year [11] or developmental stage [12]. Stress or other perturbations may induce changes to the microbiota which could disrupt eubiosis. Disruption to microbiota eubiosis can be associated with negative impacts on host health and is often called dysbiosis in this context [13]. Deviation from eubiosis, however, is not always detrimental and may help plants cope with various forms of stress. Below, we focus our discussion on the interplay between microbiota homeostasis and plant health and resilience.
Microbiota homeostasis shifts toward dysbiosis
In humans, dysbiosis is associated with ailments such as inflammatory bowel disease, diabetes, allergies, and other health issues [14] and is often accompanied by a lower diversity microbial community with altered metabolic function [15]. However, broad use of the term “dysbiosis” in mammalian literature has come under scrutiny, particularly for its inconsistent definition and ambiguity as often no distinction can be made between it being a cause or effect of a specific disease [16]. A recent study showed an example of dysbiosis as the causal agent for tissue damages in plants. Several Arabidopsis immune-compromised mutants were found to harbor an increased amount and altered composition of phyllosphere microbiota and display leaf-tissue damage under high humidity [17]. The Shannon diversity index and the relative abundance of Firmicutes were markedly reduced, whereas Proteobacteria were enriched inside the leaves of these mutant plants, bearing cross-kingdom resemblance to some aspects of the dysbiosis that occurs in human inflammatory bowel disease. Importantly, bacterial community transplantation experiments showed that the application of the dysbiotic leaf bacterial community to otherwise healthy plants resulted in tissue damages, demonstrating that, in this case, dysbiosis is causative to negative impact on host health [17].
Tissue damage-associated microbiota shifts have also been observed during insect and pathogen attacks, which often compromise host immune responses. For example, herbivory of bittercress plants by the leaf-mining fly causes a significant shift in phyllosphere microbiota, resulting in an increased abundance of bacteria on damaged leaves. Growth of Pseudomonas spp. (belonging to Proteobacteria) was found to largely account for the increased abundance of microbiota [18]. Another study found that fungal pathogen Zymoseptoria tritici suppresses immune responses in susceptible wheat cultivars, resulting in an increase in bacteria members of the leaf microbiome near fungal infection sites [19]. Together, these examples imply that immune suppression during pathogen infections is associated with shifting the composition of microbiota, similar to what is observed in immune-compromised plant mutants [17]. This may be a broadly applicable principle. Indeed, microbiota changes have been described across many plant species upon biotic challenge, including citrus greening in citrus [20], parasitic nematode Meloidogyne graminicola infection in rice [21], Yellow Canopy Syndrome in sugarcane [21], and protist Plasmodiophora brassicae in Chinese cabbage [22]. However, in these cases, it is not yet known whether the observed changes in microbiota contribute causally to (or a consequence of) tissue damages in disease.
Microbiota homeostasis shifts benefiting plant stress resilience
Stress-induced deviation from eubiosis is not always associated with reduced plant performance. Microbiota changes (referred to hereafter as meliorbiosis; from the Latin root melior- meaning “to make better, improve”) that enable positive effects on plant performance under stressful conditions have been described. In the case of biotic stress, the fungal pathogen Fusarium graminearum was shown to induce shifts to rhizosphere microbiota of barley plants, including apparent recruitment of bacterial taxa that are enriched with antifungal traits [23]. In Arabidopsis, infection of leaves with oomycete Hyaloperonospora arabidopsidis, a causative agent of downy mildew, resulted in enrichment of specific rhizosphere bacteria that were able to induce systemic resistance against downy mildew [24]. Insect herbivory can also induce changes to the plant microbiota. For example, aphid [25] and whitefly [25,26] feeding of aboveground pepper plant tissues results in restructuring of rhizosphere microbiota and enhanced resilience to belowground bacterial pathogens.
Abiotic stress can also induce microbiota shifts. Drought stress, for instance, induces a large restructuring of belowground communities across diverse plant hosts [27–29]. This shift is generally associated with enrichment of Actinobacteria in the root endosphere relative to the rhizosphere or bulk soil [28]. The enrichment of specific strains under drought conditions, but not water replete conditions, is correlated with increased plant root biomass [29] which could contribute to improved drought resilience [30].
As is in the case of dysbiosis, the cause-and-effect relationship during meliorbiosis is still not so clear in many cases. While changes in microbiota composition are sometimes associated with positive effects on plant fitness, causality still needs to be demonstrated in most instances.
Plant factors regulating microbiota homeostasis
If proper microbiota homeostasis is critical for plant health, one would expect that plants would have evolved mechanisms to prevent health-damaging dysbiosis and allow health-promoting meliorbiosis under stressful conditions. Indeed, recent studies have begun to identify host factors that are involved in mediating microbiome homeostasis in plants (Fig 1). While it is well established that plant defense hormones salicylic acid (SA) and jasmonic acid (JA) play an important role in limiting the growth of virulent pathogens in plants, it is becoming increasingly evident that they also play a critical role in mediating the homeostasis of commensal microbiota members. Arabidopsis mutants with constitutively elevated levels of SA-mediated immune response harbor reduced bacterial diversity in the endophytic leaf microbiota, while mutants deficient in JA-mediated immune response harbor an increased bacterial diversity in epiphytic leaf microbiota [31]. Activation of JA pathways by application of methyl-JA also alters the composition of the rhizosphere microbiota [32], further implicating the role of JA pathways in the regulation of microbiome homeostasis in plants. A study involving multiple hormone mutants identified defense hormone SA is required to establish a normal rhizosphere microbiota and that SA-mediated modulation of the rhizosphere microbiota is likely to occur at the family level, instead of impacting only a select few largely abundant strains [33]. In addition to SA and JA, Arabidopsis ein2 mutants defective in ethylene signaling harbor distinct phyllosphere microbial communities compared to wild-type plants [34]. However, it remains to be determined whether the observed microbiota alterations in these defense hormone mutants causally impact plant fitness, either positively or negatively, an area of great interest for future research.

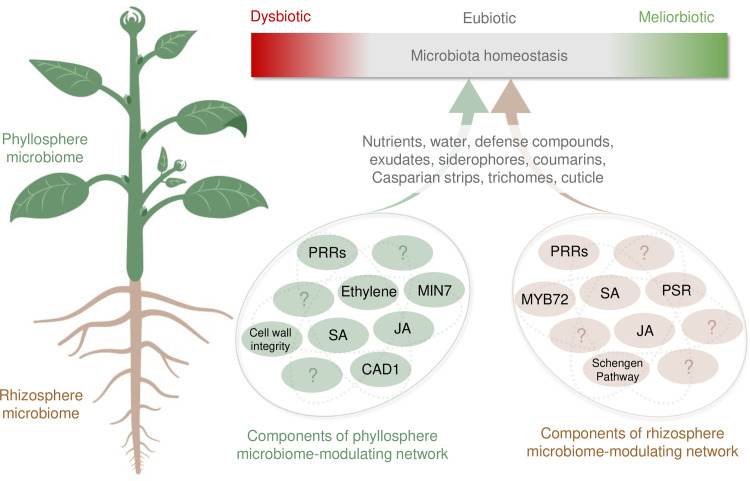
Host control of microbiota homeostasis in plants.
Microbiota eubiosis represents a normal range of microbiota abundance and composition in healthy plants grown under optimal conditions. If eubiosis is disrupted, either by host mutations, abiotic stress, or infections, or a combination thereof, homeostasis can shift toward a dysbiotic state associated with negative impacts on plant health or toward a meliorbiotic state associated with positive impacts on plant health. Examples of host factors that contribute to microbiota homeostasis in the phyllosphere (green) and rhizosphere (brown) are depicted in circles below. Not all known factors are depicted. CAD1, constitutively activated cell death 1; JA, jasmonic acid; MIN7, HOPM1-interactor 7; MYB72, MYB domain protein 72; PRRs, pattern recognition receptors; PSR, phosphate starvation response; SA, salicylic acid. Created with Biorender.
In addition to defense hormones, recent studies have begun to show a critical role of pattern-triggered immunity (PTI) in regulating modulating microbiota homeostasis in plants [17,35,36]. PTI is a major form of plant immunity initiated by plant recognition of common microbial patterns by pattern-recognition receptors (PRRs) [37]. An Arabidopsis quadruple mutant lacking 3 PRRs/coreceptors (recognizing bacterial flagellin, elongation factor Tu, and peptidoglycan, respectively) and a vesicle traffic regulator, the MIN7 protein, displayed a dysbiotic shift in the quantity and composition of the endophytic leaf microbiota [38]. Similar alterations in endophytic leaf microbiota were found in an Arabidopsis mutant that carries a S205F mutation in a membrane attack complex/perforin domain protein, CAD1 [17]. The involvement of plant immunity in regulating some aspects of microbiota homeostasis illustrates a conceptual parallel to mammalian-microbiome interactions, as PRR gene mutations (e.g., NOD2) have been shown to be linked to dysbiosis in humans [39] and members of the MACPF protein family, such as human C9 and perforin in particular, have been shown to be involved in innate and adaptive immunity in mammals [40].
In addition to defense-related plant processes, physical barriers and leaf-surface structures such as the plant cell wall and trichomes may play a role in influencing microbiota homeostasis based on a genome-wide association study examining 196 accessions of Arabidopsis grown in the field and their associated bacterial and fungal communities [41]. Furthermore, Arabidopsis mutants with altered cuticle formation possess altered epiphytic phyllosphere bacterial communities [34,42]. A recent study further confirmed the role of physical structures in contributing to microbiota homeostasis [43]. Here, Arabidopsis mutants defective in genes controlling the function of endodermal root diffusion barriers, including those in the Schengen pathway required for Casparian strip formation and those involved with suberin deposition, possessed rhizosphere bacterial communities with altered composition [43].
Finally, pathways involved in plant nutrient response also play a role in microbiota homeostasis. For instance, under iron- and phosphate-limiting conditions, plants can induce the secretion of coumarins which, in addition to aiding in plant nutrient uptake, possess selective antimicrobial activity and can shape the root microbiota [44]. Arabidopsis, for instance, secretes iron-mobilizing coumarin scopoletin under iron-limiting conditions in response to beneficial bacteria in a manner dependent on the MYB72 transcription factor and a β-glucosidase, BGLU42. Scopoletin was found to have high antimicrobial activity against fungal pathogens Fusarium oxysporum and Verticillium dahlia, whereas many beneficial rhizobacteria are tolerant [45]. Additionally, several Arabidopsis mutants defective in components regulating phosphate starvation response (PSR) and inorganic phosphate availability in plant tissues were found to harbor endophytic root microbial communities that are significantly different compared to wild-type plants [46].
Future outlook
In this review, we have highlighted several examples of environmental and host influences on microbiota homeostasis in plants, a topic that is likely to become intensively studied in the coming decade. However, the current understanding is rather limited, and the cause–effect relationship has not been resolved in most cases. Further advances will likely hinge on progress in two critical areas. First, application of multiple methodologies will be needed to go beyond taxonomic knowledge of microbiota to gain insight into functional implications of microbiota shifts as traits rather than taxa may be selected during microbiome shifts. The application of methodologies such as metagenomics, metatranscriptomics, or metabolomics could inform on the functional implication of a particular microbiome. Further, incorporation of quantitative taxonomic analyses, either by direct quantification, DNA spike-in, or normalization to host DNA (i.e., chloroplasts and mitochondria as described in [18]), for instance, could help identify ecologically relevant fine-scale differences between microbial communities that may not be apparent in current studies.
Second, it is now time to more clearly define the taxonomical and functional features of eubiosis, dysbiosis, and meliorbiosis in plants and, perhaps more importantly, to resolve the cause–effect relationship between observed microbiota shifts and plant fitness. While methodologies, such as shotgun metagenomics, may enable researchers to generate hypotheses related to microbiome function, we anticipate increased use of rationally designed synthetic microbial communities [47] and innovative gnotobiotic plant growth systems [48] in future research in this area. Indeed, this is already happening [17,49–53], and we can expect significant progress in the next few years to reach a better understanding of how microbiota homeostasis is controlled in plants. Broad understanding of microbiota homeostasis should provide a critical knowledge base for the development of innovative plant- and/or microbiota-based solutions to globally improve pre- and post-harvest plant health and productivity.
Acknowledgements
We thank James Kremer, Timofy Arapov, Richard Hilleary, Jong Hum Kim, and Li Zhang for their critical comments to improve this manuscript.
References
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
 Toward understanding microbiota homeostasis in the plant kingdom
Toward understanding microbiota homeostasis in the plant kingdom
