
Competing Interests: The authors have declared that no competing interests exist.
Drosophila suzukii (spotted wing drosophila, SWD) is a pandemic quarantine pest that attacks mostly red fruits. The high number of life cycles per year, its ability to rapidly invade and spread across new habitats, and highly polyphagous nature, makes this a particularly aggressive invasive species, for which efficient control methods are currently lacking. The use of native natural predators is particularly promising to anchor sustainable and efficient measures to control SWD. While several field studies have suggested the presence of potential predatory species in infested orchards, only a few confirmed the presence of SWD DNA in predators’ gut content. Here, we use a DNA-based approach to identify SWD predators among the arthropod diversity in South Europe, by examining the gut content of potential predator specimens collected in SWD-infested berry fields in North Portugal. These specimens were morphologically identified to the family/order, and their gut content was screened for the presence of SWD DNA using PCR. New SWD predatory taxonomical groups were identified, as Opiliones and Hemerobiidae, in addition to known SWD predators, such as Hemerobiidae, Chrysopidae, Miridae, Carabidae, Formicidae and Araneae. Additionally, the presence of a spider family, Uloboridae, in the orchards was recorded for the first time, posing this family as another SWD-candidate predator. This study sets important bases to further investigate the potential large-scale use of some of these confirmed predator taxa for SWD control in South Europe.
The spotted wing drosophila (SWD), Drosophila suzukii (Matsumura), is a pandemic and highly invasive pest that recently arrived to Europe, with first reports in 2010 occurring in Italy and France (data from the European Plant Protection Organization (EPPO) [1]). SWD integrates since 2011 the A2 List of pests recommended for regulation as quarantine pests [2]. Being extremely polyphagous, this pest is able to develop in a wide range of cultivated and wild fruits [3], thus causing severe production losses in different fruit chains of value. Its most susceptible host fruits are thin-skinned berries, including several Vaccinum spp. such as cranberries and blueberries, strawberries, or table and wine grapes [4]. But SWD also attacks stone fruits, such as cherries or plums [5,6]. Each SWD female has the capacity to lay hundreds of eggs in its lifespan and a rapid life cycle that easily originates millions of descendants during the months of the fly’s reproductive season (up to 10 generations per year) [7]. In addition, as the larvae feed on the fruit flesh, infested fruits become unmarketable, which associated with loss of crops, pest management and fruit selection, leads to substantial economic losses [8].
Current SWD control methods rely on the use of broad-spectrum insecticides (spinosyns, organophosphates, pyrethroids and neonicotinoids), whose active ingredients work predominantly against adult flies, as the larvae are protected inside the fruits [9]. Other methods such as mass trapping, sanitary measures, netting or the use of kaolin/chalk have also been recommended for SWD control, but they are extremely time consuming and expensive, and end up not improving the control of the pest [10]. Moreover, insecticide resistance has been identified [11], making biological control strategies for SWD attractive approaches that promise to reduce long-term management costs, being also more sustainable and environmentally friendly [12,13].
The success of a biological invader such as SWD depends on reduced impacts of natural enemies in the invaded environments [5,14–19]. Predators and parasitoids are frequently used in biological control and may constitute a relevant strategy to reduce SWD populations, both in wild and crop host species [12,20,21]. A revision of 12 studies on potential predators showed 22 families of interest (excluding spider families, also SWD predators [10,22]), of which only eleven were confirmed as SWD predators, namely Forficulidae (earwigs) [10,23,24], Nabidae (damsel bugs) [10], Formicidae (ants) [25,26], Anthocoridae (pirate bugs) [20,24,25,27,28], Carabidae (ground beetles), Gryllidae (crickets) [29], Chrysopidae (green lacewing, larvae) [20,24] Staphylinidae (rove beetles) [10,30], Mantidae (praying mantis) [22], Miridae (mirid bugs) [20] and Labiduridae (striped earwigs) [28]. Only in three studies the confirmation of predatory activity by SWD DNA presence in the gut content of the predator was shown (S1 Table). Araneae families were addressed in five studies, and 20 families of potential spider predators were reported in literature, of which seven families were confirmed (S2 Table). Of these, only two studies used the presence of SWD DNA in predator’s gut content to confirm predation (S2 Table). Based on the same revision, most of the studies with SWD predators are focused on laboratory assays, while field predation remains relatively understudied. Wolf et al. [10] collected candidate SWD predators in Switzerland and detected feeding in earwigs, spiders and predatory bugs; in the United States of America (USA), Schmidt et al. [22] also collected field predators, and detected SWD DNA in spiders and in one individual from the Mantidae family. Ballman et al. [29], placed SWD pupae on a berry field (USA) in three methods of exposure to predators (fully-exposed, caged and buried) and determined that predation rates were higher in exposed pupae, and although potential predators were collected in the field, the identification of SWD predators was made as laboratory assays. Kamiyama et al. [31] detected natural field predation of SWD sentinel pupae in the USA, but specific predator groups were not identified.
Even though previous information about SWD predators can be used to guide local studies, the identification of native field predators is essential to define adequate control measures, not only to avoid the introduction of non-native predators in a region, but also to improve and increase native populations of actual predators, as recommended under a conservation biological control approach. Despite still scarcely used, the confirmation of SWD-DNA in predators’ gut has emerged as the most reliable method to confirm predation [32].
The aim of this study was to identify potential SWD predators among the arthropod diversity in North Portugal, using as case study berry fields, and identify field SWD predation. The results obtained should represent an important first step to the management of the pest by supporting populations of natural enemies present in the region and further contribute to the general knowledge of SWD predators in other environments.
Arthropods were collected in the Northern region of Portugal, in August 2019, in four blueberry orchards and one blackberry orchard, in five distinct locations: Santiago de Piães (Location 1 (L1); 41.079528,-8.155139; blueberry), São Martinho de Mouros (L2; 41.119639,-7.890639; blueberry), Baião (L3; 41.143306,-8.066306; blueberry), Vieira do Minho (L4; 41.635250,-8.160306; blackberry) and Vale de Cambra (L5; 40.838972,-8.359917; blueberry) (temperature ranged from ≈20–30°C during collection time) (Fig 1). All orchards were developed and maintained under organic farming for 1–5 years upon arthropod collection time; time average size of the orchards was 2 ha, and an average of 6 years of age. All producers used homemade or commercial SWD traps to monitor the pest presence in the orchard; in L2, mass trapping was employed with a D. suzukii attractive bait. Additionally, the producers used mainly Spinosad as a SWD control agent, following all the legal procedures and recommendations. L1, L3 and L4 had wild vegetation between rows, and only L2 and L5 used mulching films—L2 along the blueberry bush rows, and L5 with the complete orchard’s soil covered. In L4 and L5 nets were used, but while in L4 the nets were only placed above the blackberry trees in order to provide protection against UV-rays, in L5 the net covered the entirety of the orchard as protection against birds. The orchards’ surrounding wild vegetation contained mainly oaks, pine trees, eucalyptus and olive trees; in some cases, there were also vineyards, cherry trees or other fruit trees (L2 and L4). Representative photos of each location are supplied in S1 Fig. The presence of D. suzukii in the orchard was confirmed by collecting the insect in arthropod samplings and/or in traps used by producers.

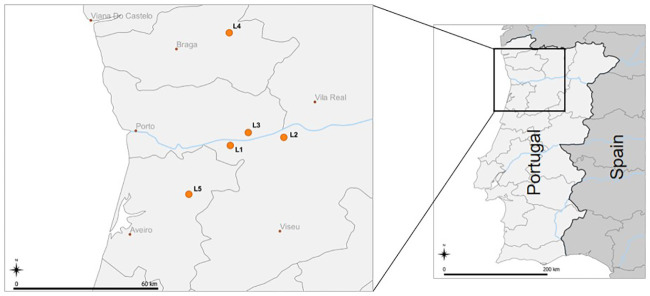
Arthropod sampling locations in Northern Portugal.
Scale bar: 60 km (left map), 200 km (right map) Map downloaded from Natural Earth (naturalearthdata.com) and edited in QGIS v3.16.0 (qgis.org).
Arthropods were collected in three different rows of the orchard: in the first row of bushes in the orchards’ periphery; in the middle of the orchard; and in a row closer to wild vegetation, a water source and/or shade. The wild vegetation present in the last sampled row was characterized by large non-cultivated trees, usually surrounding a small water stream, which contributed to the presence of shade in the nearby rows. On the contrary, the first row in the orchards’ periphery did not have any shade or abundant wild vegetation in the surrounding area.
For the arthropod collection, two different methods were used: suction (with a Dietrick Vacuum insect net (D-Vac) machine (Rincon-Vitova Insectaries, Inc., Ventura, CA, US, model 122)), during ≈3min continuously walking along the row (orchards L1-L5), in which arthropods from the canopy were sucked into a meshed bag [17]; and pitfall traps, in which cups with 50% ethanol were inserted on the soil near the shrubs for 24h, for the collection of soil arthropods [33] (L2-L5) (Fig 2). Arthropods collected using D-Vac were etherized on the field by embedding a cotton ball in ether and placing it on a plastic bag with the meshed bag inside during the transportation to the laboratory; they were preserved at 4°C in 15 ml tubes with 96% ethanol until identification. The pitfall contents were transferred to 50 ml tubes and kept at 4°C until arthropod identification.

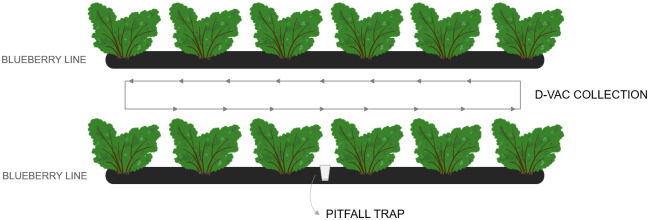
Visual representation of the arthropod sampling methods.
Collected arthropods were sorted into morphospecies and identified to the order or family level following Triplehorn et al. [34]. Moreover, in accordance with the same authors, arthropods were assigned to feeding guilds and grouped accordingly into four trophic categories: detritivores, phytophages, omnivores and predators. Spider family identification was based on Nentwig et al. [35] and specimens were further grouped following Cardoso et al. [36]. Thus, families of Araneae were grouped in 7 functional groups based on their foraging strategy (type of web and hunting method), prey range (stenophagous or euryphagous), vertical stratification (ground or foliage) and circadian activity (diurnal or nocturnal): (1) sheet web weavers (ShW), (2) space web weavers (SpW), (3) orb web weavers (OrW), (4) specialists (Sp), (5) ambush hunters (AH), (6) ground hunters (GH), and (7) other hunters (OH). After identification, arthropods belonging to predator and omnivores trophic groups were preserved in 96% ethanol, according to place of collection and family/order, and were kept at 4°C until DNA extraction.
Arthropods identified as potential predators were tested for field predation of D. suzukii. Potential predators were washed with a 1.5% bleach solution (v/v, in milliQ H2O) to remove any traces of D. suzukii DNA from their body surface. Screening of field predation was assessed by molecular gut-content analysis according to Wolf et al. [10]. DNA of potential predators was extracted with GeneMatrix Tissue Purification Kit (EURX Sp.zo.o., Poland) following the manufacturer’s protocol for insect DNA extraction. Arthropods (up to 50mg) were homogenized with PBS (pH = 7,6). The arthropod’s DNA was extracted as a pool within the same family, location and sampling method, except for Araneae families’ DNA, which was extracted according to functional group and sampling method. DNA quality and quantity were analyzed with μDrop™ (ThermoFischer Scientific, USA) on a microplate spectrophotometer Multiskan Go (ThermoFischer Scientific, USA). Predation was confirmed by PCR amplification with a primer specific for the identification of D. suzukii [10]. PCR amplification was employed twice in order to confirm predation vs. no predation results. The reaction was performed with the highest amount of DNA possible for the enzyme (DFS Taq MasterMix, Bioron Life Science, Germany) in order to avoid false negatives due to lack of DNA quantity in the reaction. The reaction was performed in a T100™ thermocycler (Bio-Rad Laboratories, USA) and the conditions were 94°C for 2 min, followed by 35 cycles of 94°C for 10 sec, 48°C for 20 sec, 72°C for 15 sec, and a final extension at 72°C for 3 min. A positive control consisting of DNA extracted from a single D. suzukii was used (previously phenotypically identified according to the EPPO identification key [37]). PCR results were visualized in an 1.4% agarose gel and results were considered positive if a band appeared in the 179bp region.
The number of arthropods identified as potential SWD predators was expressed as a percentage of the number of all arthropods collected. Differences in species richness and diversity were analysed with ANOVA. For the analysis of row diversity, the total number of potential natural enemies both from D-VAC and pitfall sampling locations was used based on each row location within the orchard. All graphs and statistical analysis were made using GraphPad Prism 8 (GraphPad Software, USA).
Different groups of arthropods were collected in four different blueberry orchards and one blackberry orchard (Table 1). In a total of 169 individuals identified as potential SWD predators collected with D-Vac (representing ≈25% of D-Vac collected arthropods), most belonged to the taxonomic group of Chrysopidae (larva and adults) followed by Araneae, Formicidae, Miridae, Coccinellidae (adults) and Hemerobiidae (adults) (Fig 3a). In the pitfall traps, from a total of 59 arthropods identified as potential predators (≈64% of pitfall collected arthropods), most belonged to Formicidae, followed by Araneae, Carabidae and Opiliones (Fig 3b).

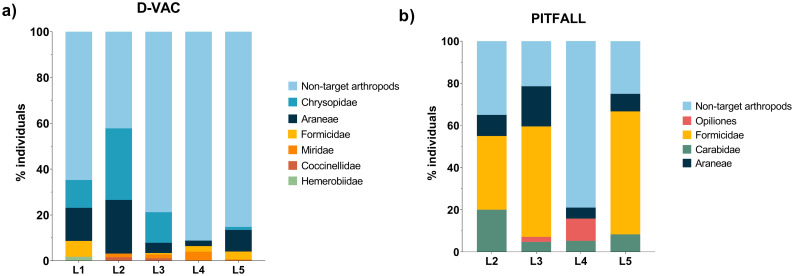
Arthropod diversity collected on berry fields.
a) D-Vac collected arthropods; b) Pitfall traps collected arthropods. Values are presented as percentage of individuals from each taxonomical group collected in each location.

| Trophic/Taxonomic group | D-Vac | Pitfall |
|---|---|---|
| Predators | ||
| Arachnida | ||
| Araneae | ||
| Agelenidae | 4 | 1 |
| Araneidae | 0 | 0 |
| Cheiracanthiidae | 1 | 0 |
| Dictynidae | 3 | 0 |
| Lycosidae | 0 | 8 |
| Oxyopidae | 35 | 0 |
| Philodromidae | 2 | 0 |
| Salticidae | 3 | 0 |
| Sparassidae | 1 | 0 |
| Tetragnathidae | 0 | 1 |
| Theridiidae | 4 | 0 |
| Thomisidae | 11 | 0 |
| Uloboridae | 1 | 0 |
| Zodariidae | 35 | 2 |
| Opiliones | 0 | 3 |
| Insecta | ||
| Coleoptera | ||
| Coccinellidae | 3 | 0 |
| Carabidae | 0 | 8 |
| Heteroptera | ||
| Miridae | 10 | 0 |
| Other | 11 | 0 |
| Neuroptera | ||
| Chrysopidae | 67 | 0 |
| Hemerobiidae | 3 | 0 |
| Omnivore | ||
| Insecta | ||
| Hymenoptera | ||
| Formicidae | 21 | 36 |
| Other | 28 | 1 |
| Phytophagous | ||
| Insecta | ||
| Coleoptera | ||
| Chrysomelidae | 11 | 2 |
| Mordellidae | 3 | 0 |
| Scolytidae | 0 | 1 |
| Tenebrionidae | 0 | 1 |
| Other | 0 | 1 |
| Hemiptera | ||
| Cicadomorpha | 4 | 0 |
| Cicadellidae | 307 | 9 |
| Cixiidae | 8 | 0 |
| Membracidae | 5 | 0 |
| Cydnidae | 0 | 2 |
| Indetermined | ||
| Insecta | ||
| Diptera | ||
| Brachycera | 115 | 17 |
| Nematocera | 23 | 0 |
| Lepidoptera | 5 | 0 |
Arthropods are grouped according to their feeding habits and taxonomical group.
We were able to identify 14 different spider families, corresponding to 7 different functional groups (S3 Table), with families related to hunting functional groups being the most abundant in the collections (Fig 4). In all locations, adults of D. suzukii were collected with D-Vac, confirming the pest presence in the field. The highest number of D. suzukii was collected on the blackberry fields, with more than 100 individuals (results not shown). When considering the row of the orchard, most potential predators were collected in the middle rows of locations L1, L2 and L3, but in two locations the number was higher near the vegetation margin (L4) and in the periphery (L5) (Fig 5). Although no statistical differences were found between rows concerning species richness and diversity, the highest arthropod diversity was found in the middle row, with arthropods belonging to all of the taxonomic groups identified as potential SWD predators in this study.

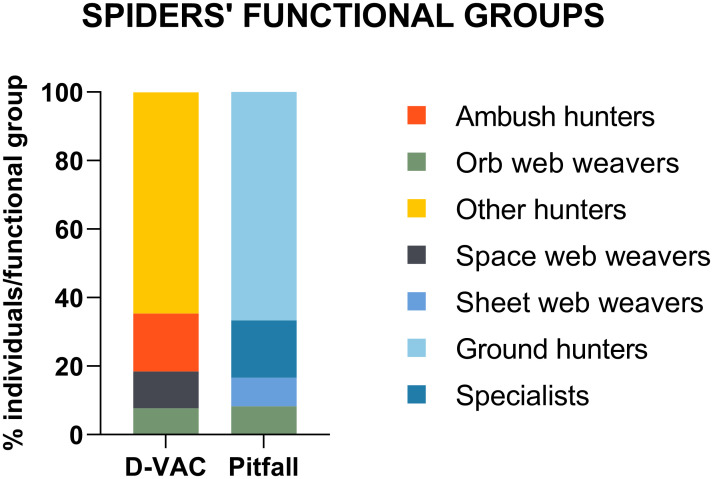
Spiders’ functional group diversity collected with D-Vac and pitfall traps in Portuguese berry fields.
Values are presented as percentage of individuals from each functional group collected with D-Vac and Pitfall traps.

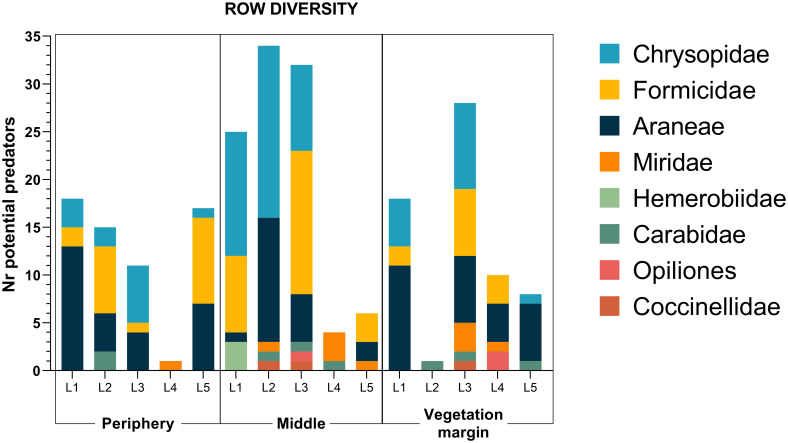
Arthropod row diversity (D-VAC+Pitfall traps).
Periphery—row in the periphery of the orchard; Middle—row in the middle of the orchard; Vegetation margin—row closer to wild vegetation, a water source and/or shade. Values are presented as the total number of individuals collected in each row of each location.
Potential D. suzukii predators’ gut content was analyzed by PCR amplification of D. suzukii cytochrome oxidase subunit I (COI) [10]. Of the 8 groups of potential D. suzukii predators, D. suzukii DNA was detected in 7, namely Chrysopidae, Formicidae, Miridae, Hemerobiidae, Carabidae, Opiliones and Araneae, with no detection of D. suzukii DNA in Coccinellidae individuals (Fig 6a). Regarding the Araneae functional groups, D. suzukii DNA was detected only on 2 of the 7 functional groups, namely orb web weavers (family Araneidae and Uloboridae) and space web weavers (family Dictynidae and Theridiidae), all collected with D-Vac (Fig 6b). The most abundant taxonomic groups with positive results were Chrysopidae, Formicidae and Araneae, however, the most abundant functional group of Araneae, Other hunters (Oxyopidae, Sparassidae, Cheiracanthiidae, Philodromidae and Salticidae), collected with D-Vac, did not predate on D. suzukii. If frequency of detection is considered, Formicidae was the only group collected in all locations, with positive results in individuals from 3 of the locations (Fig 7a), and both positive results in spiders were from groups collected with D-Vac (Fig 7b).

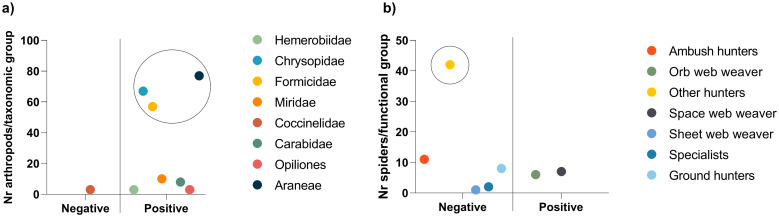
Molecular gut-content analysis of arthropods identified as potential SWD predators.
a) PCR amplification results for the 8 families/orders identified, with the representation of the number of identified individuals from each family/order; b) PCR amplification results of the 7 spider functional groups, with the representation of the number of identified individuals from each functional group. Circled are the most relevant taxonomic groups (a) and functional group (b), with the highest number of identified individuals.

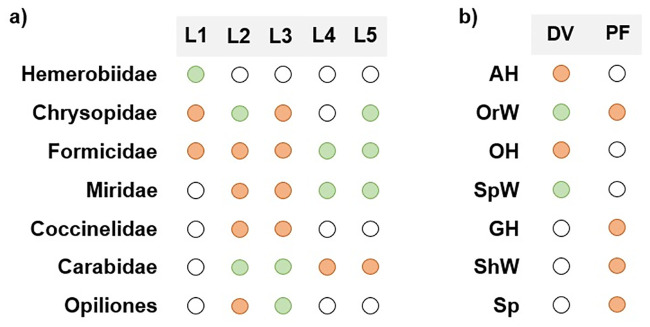
Frequency of D. suzukii DNA detection in potential predators’ gut-content.
a) Detection in 7 of the different familes/orders screened for D. suzukii DNA presence; b) detection in spiders’ functional groups screened for D. suzukii DNA presence. Blank dots—predator was not found in the location; orange dots—predator collected in the location but DNA detection was negative; green dots—predator collected in the location with positive DNA detection.
The need to develop new pest and disease management strategies is increasing. Biological control agents (BCAs) such as bacteria, fungi, parasitoids or predators [12] emerge as the most promising sustainable strategies. Here, we used a DNA-based strategy to produce a detailed catalogue of native predators of SWD in berry fields in Portugal, a critical first step to develop efficient and sustainable control measures.
D-Vac was used to collect predators in plants and surroundings, while pitfall traps collected the potential predators mostly on the ground. Hemerobiidae, Chrysopidae, Miridae and Coccinellidae families were collected in the vegetation, Opiliones and Carabidae on the ground, and Formicidae and Araneae in both cases. Comparing the three sampling rows, predator abundance was higher in the middle of the orchard. Schmidt et al. [22] found a higher abundance of natural enemies in a pine margin of a SWD infested orchard, which correlated with a higher activity of SWD in the same area. The fact that most natural enemies were collected in rows in the middle of the orchard suggests a higher activity of SWD in that area, probably due to higher amount of food.
Of the collected arthropods, only those of the Coccinelidae family were negative for the presence of SWD-DNA specific sequence in the gut. Coccinellids are unspecific predators, and have been referenced as a potential BCA for other insect pests such as e.g., aphids, whiteflies, mealybugs or psyllids (reviewed by Kundoo and Khan [38]). Similar negative results for the presence of SWD in Coccinellids were previously reported in the USA and Switzerland [10,22], supporting coccinellids as unlikely SWD predators.
Families Chrysopidae, Miridae, Carabidae and Formicidae, which field individuals’ gut showed here to contain SWD DNA, had been proposed in laboratory assays as SWD predators. However, laboratorial experiments are often not supported by field data. For example, Chrysoperla carnea (Stephens) was in the laboratory a predator of SWD [20,24], but this predation was never confirmed in the field [22,31]. Also, the laboratory predatory activities of the Carabidae Pterostichus mutus (Say) and Bembidion quadrimaculatum (LeConte) and the Miridae Dicyphus hesperus Knight against SWD were not confirmed in the field [13,14]. Field predation of SWD by Formicidae was previously suggested, as ants were observed not only digging and carrying SWD pupae, but also preying on SWD pupae and larvae in dropped blueberries [25,26]. Our data confirm these taxonomic groups as field SWD predators, thus being an important clarification of those previous results.
Despite some inconsistent laboratory vs. field data for SWD predation [13,14,16,23], other studies have shown that Chrysopids are promising predators to be used in the control of other insect pests: Chrysoperla spp. larvae might be used to control Tuta absoluta (Meyrick) (tomato pinworm) [39], Frankliniella occidentalis Pergande (western flower thrips) [40], Melanaphis sacchari (Zehntner) (sugarcane aphid) [41] or Glycaspis brimblecombei Moore (eucalyptus aphid) [42]. However, the role of chrysopids remains unclear, as the collected individuals were adults, and not all adults of the family Chrysopidae are predators. For example, Chrysoperla spp. adults feed on nectar, pollen or honeydew, contrarily to adults of the genus Chrysopa that are predators [43].
Miridae, like chrysopsids, are potential BCAs of pests like T. absoluta [44], Bactericera cockerelli (Sulc), Spodoptera exigua (Hübner), Spodoptera frugiperda (JE Smith) [45,46], all tomato pests; or Cacopsylla pyri (Linnaeus), a pear psyllid [47]. In the only study of SWD predation by Miridae species [20], D. hesperus was confirmed as a SWD predator. Pérez-Hedo et al. [48] reviewed the use of mirid bugs as predators in horticultural crops, like tomato. Whilst commercially mirid bugs are already available, such as D. hesperus (in North America), Macrolophus pygmeus (Rambur) and Nesidiocoris tenuis (Reuter) (in Europe, Africa and Asia), their use as BCAs in pest management should be further explored [48].
Contrarily to chrysopids and mirid bugs, the use of carabid beetles as BCAs for insect pests has not been suggested. Carabids are generalist predators, feeding on insect pests but also weed seeds, which decreases consumption rates of one or the other [49]. Still, Carabidae species were already reported as predators of pests such as Ragholetis mendax Curran (blueberry fruit fly) [50], Itame argillacearia Packard (blueberry spanworm), Altica sylvia Malloch (blueberry flea beetle) [51] or Acalymma vittatum (Fabricius) (striped cucumber beetle) [52].
Among the taxa detected in our work, ants may be those with most potential as a BCA, considering its abundance and capacity to rapidly consume a large number of victims. The use of ant species as a BCA is not recent [53], and besides controlling pests in different agroecosystems [54], they also improve the soil quality [55]. Besides predating on D. suzukii, ants also predate other pests as C. pyri [56] and Hypothenemus hampei (Ferrari) [57].
This study identified for the first-time arthropods from the Opiliones and Hemerobiidae as SWD predators. Kamiyama et al. [31] had recovered Hemerobiidae individuals from traps with sentinel larvae, but did not confirm if the Hemerobiidae individuals were predators or predation was done by individuals from other taxa, such as Formicidae, Anthocoridae or Staphyilinidae [31]. Previous studies showed that Opiliones collected in orchards infested with SWD had negative results for the presence of SWD DNA [10,22]. Hemerobiids are predators of e.g., Pineus strobi (Hartig) (pine bark adelgid, a pine tree pest) [58], Aulacorthum solani (Kaltenbach) (foxglove aphid, in sweet peppers) [59] or Planococcus citri (Risso) (citrus mealybug) [60] but their predatory activity on SWD remains unknown. Similarly, harvestmen (Opiliones) were already identified as predators of Lobesia botrana (Denis & Schiffermüller) (grape berry moth) [61] or A. vittatum (striped cucumber beetle) [52]. Considering our results and that individuals belonging to this taxonomic group were already found in SWD infested orchards in North America and Europe, harvestmen should be considered for further predation studies on SWD, as they may be a suitable predator for the control of this pest.
Here, we also examined the presence of SWD DNA in spiders’ gut content. The use of spiders as BCAs has been explored for a long time now, as they are known as generalist predators, representing a good strategy for certain agroecosystems [62]. Updated insights highlight that the effect of spiders as BCAs depends on the type of pest or existence of alternative preys and environmental conditions (as with any other generalist predator), and on the phenotype of spiders, namely its hunting strategy or behavior (functional group) [63]. Spiders are commonly identified in the field as predators of various pest species, namely A. vittatum [52], Frankliniella spp. [64], or Phyllocnistis citrella (Stainton) (citrus leafminer) [65]. In this study, SWD DNA was found in the gut content of spider families belonging to two functional groups, orb web weavers (family Araneidae and Uloboridae) and space web weavers (family Dictynidae and Theridiidae), both web-building groups. Individuals belonging to the family Araneidae and Theridiidae had already been identified as predators of SWD, by testing positive for SWD DNA [10,22], while Dictynidae individuals, although collected in infested orchards, tested negative [22]. Thus, our work is the first report of Uloboridae spiders being collected in SWD infested orchards, although further studies are needed as our study, based on functional groups, did not identify which family specifically predated on SWD, which also applies to the Dictynidae individuals.
When considering the abundance of each predator group in the berry orchards of this study, the most abundant were green lacewings, ants and spiders, suggesting that these are the best candidates for SWD predator-based control, due to their natural abundance in these fields. However, considering the spiders’ functional groups, the most abundant individuals were identified as belonging to the group “other hunters”, which had no positive results for SWD predation. Nonetheless, in previous studies, individuals belonging to this functional group were found positive for SWD-predation [10,22], and it is important to take into consideration that negative results for D. suzukii DNA presence in predators’ gut may be influenced by a lack of predator-pest encounters at the time of sampling.
Our study did not identify anthocorids as SWD predators even though laboratory assays have focused on this group, namely Orius spp. [20,24,27,28]. Only in two instances anthocorids were considered potential SWD predators in field studies, but predation was not confirmed [26,31]. Woltz et al. [25] introduced a commercially available anthocorid species in an infested blueberry field with positive predation results, but there was no description if the species was already native to the orchard. Our results illustrated the need to conduct field studies, with the identification of native predators, in order to avoid the introduction of new species exclusively based in laboratory assays.
This study focused on the assessment of SWD field predation in infested Portuguese berry fields. The results confirmed the presence of SWD predators previously known or suspected, and importantly identified new predatory taxonomical groups, Opiliones and Hemerobiidae. This is the first study to confirm predation of SWD by individuals belonging to these families/orders and to identify the spider family Uloboridae in SWD infested orchards, also with the potential to be a SWD predator. Our results, together with previous studies, encourage additional field studies to further understand the role of the identified predator families/orders as Biological Control Agents, not only of SWD but also other insect pests. Only that will allow determining which native species would be more adequate for pest control, as field predation by these arthropods is still poorly understood.
The authors would like to thank the producers who kindly agreed to the sampling of arthropods in their fields.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65